 The ‘superbug’ infection caused by metallo-β-lactamases (MβLs) including L1 has grown into an emerging threat.
The ‘superbug’ infection caused by metallo-β-lactamases (MβLs) including L1 has grown into an emerging threat.
Abstract
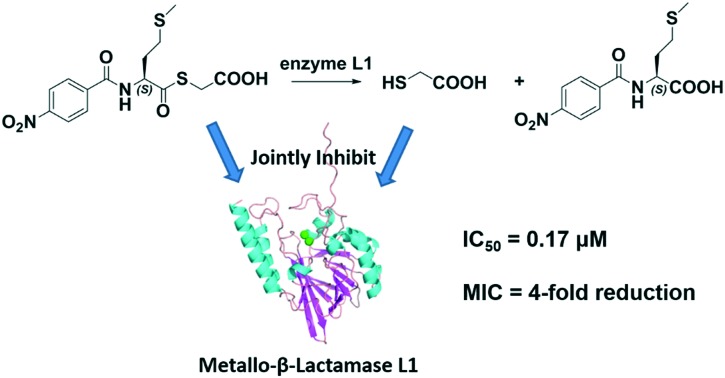
The ‘superbug’ infection caused by metallo-β-lactamases (MβLs) including L1 has grown into an emerging threat. To probe whether mercaptoacetate thioesters inhibiting L1 is a contribution of the thioester itself or its hydrolysate, ten mercaptoacetate thioesters 1–10 were synthesized, which specifically inhibited L1, exhibiting IC50 values ranging from 0.17 to 1.2 μM, and 8 was found to be the best inhibitor (IC50 = 0.17 μM). These thioesters restored the antimicrobial activity of cefazolin against E. coli expressing L1 by 2–4-fold. UV-vis monitoring showed that 1, 8 and 9 were unhydrolyzed in Tris buffer (pH 6.0–8.5), but hydrolyzed by L1; further HPLC monitoring indicated that 1/3 of the thioester 9 was converted to mercaptoacetic acid. STD-NMR monitoring suggested that both the thioester and its hydrolysate mercaptoacetic acid jointly inhibited L1.
Introduction
The effectiveness of β-lactam antibiotics has been threatened by the emergence of pathogenic bacteria producing β-lactamases in clinical settings.1,2 These enzymes are divided into four classes (A–D) based upon DNA sequence similarity.3 The class A, C and D enzymes are called serine β-lactamases (SβLs), which utilize an active-site serine as a nucleophile to attack the β-lactam carbonyl. The class B enzymes whose mechanism relies upon the presence of one or two active site zinc ions to hydrolyze β-lactams are thus called metallo-β-lactamases (MβLs). MβLs have been further subdivided into three subclasses (B1–B3), based on their amino acid sequence and metal occupancies.4 The strategy of co-administration of antibiotic adjuvants capable of maintaining the activity of existing antibiotics is widely effective in treating infections due to bacteria expressing SβLs. Clinically relevant SβL inhibitors include clavulanic acid, sulbactam, tazobactam and avibactam which effectively protect β-lactam antibiotics from inactivation when administered as combination therapies. By comparison, there are no inhibitors of MβLs available in clinics to date.5
At present, MβLs are of increasing clinical concern, because they hydrolyze almost all β-lactam antibiotics.6 Ongoing attempts have been made to prevent the hydrolysis of antibiotics by MβLs; various small molecule inhibitors have been reported,7 including thiols,8 sulfates,9 dicarboxylic acids,10 hydroxamates,11 tetrazoles,12 ebselen13 and AMA.14 Mercaptoacetate thioesters have been reported to inhibit MβLs through mercaptoacetic acid, the hydrolysate of thioester, which binds to the metal coordinating cysteine of the enzyme. However, the presence of the phenyl substituent attenuated the cleavage of the thioester bond and inhibition of MβLs was not accomplished through chelation of the Zn(ii) ions.15 In addition, MALDI-TOF showed that two molecules of mercaptoacetate bind to L1 when the enzyme was incubated with the thioester.16 Recently, our studies indicated that mercaptoacetate thioesters are highly promising scaffolds for the development of L1 inhibitors, exhibiting IC50 values in the submicromolar range. These scaffolds were further optimized to yield more potent L1 inhibitors.17,18 Also, the carbamylmethyl mercaptoacetate thioether has been reported to be a potential inhibitor of L1.19
There have been many reports on mercaptoacetate thioesters as inhibitors of L1 to date. However, almost all these reports indicated that the essence of thioesters inhibiting L1 is the contribution of either the thioester itself, or its hydrolysate mercaptoacetic acid. To probe the truth, in this work, we constructed a series of phenyl substituted mercaptoacetate thioesters and used them in combination with β-lactams to combat antibiotic resistant bacteria that produce MβLs. These thioesters were tested as inhibitors against the purified L1; STD-NMR was employed to monitor the interaction of L1 with the thioester and its hydrolysate, and their binding affinity was evaluated by isothermal titration calorimetry (ITC). Also, the capacity of these inhibitors to restore the antibacterial activity of cefazolin against E. coli expressing L1 was evaluated, and their cytotoxicity against L-929 mouse fibroblastic cells was tested.
Results and discussion
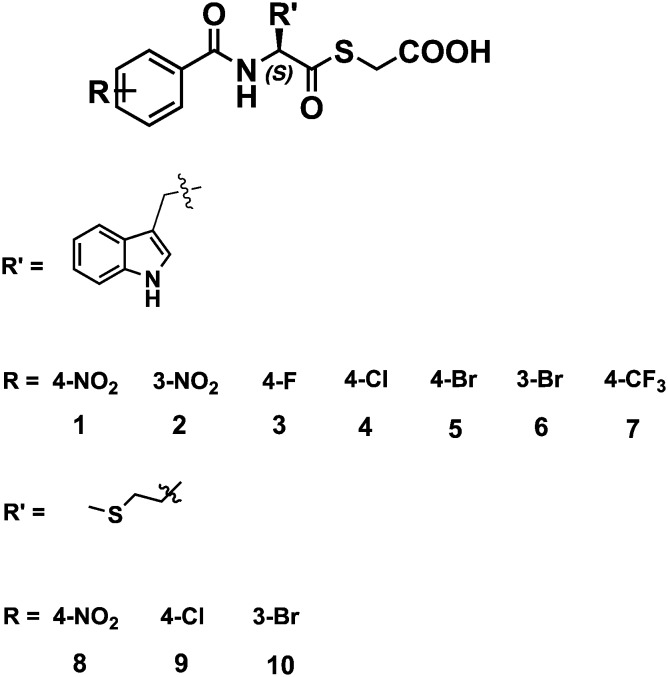
Towards the above goal, ten mercaptoacetate thioesters were synthesized through the synthetic pathway shown in Scheme S1.† The structures of the synthesized thioesters are shown in Fig. 1. These thioesters were all characterized by 1H and 13C NMR and confirmed by MS (see the ESI†).
Fig. 1. Structures of the synthesized mercaptoacetate thioesters.
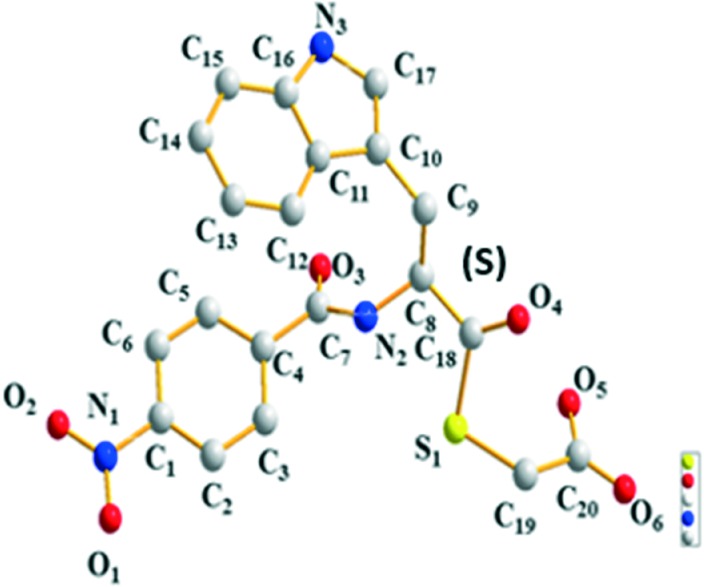
To confirm the molecular structures of the mercaptoacetate thioesters, yellow crystals suitable for X-ray analysis were obtained through slow evaporation of a solution of 1 in methanol–ethyl acetate. The crystal structures are given in Fig. 2, and the resulting structures based on X-ray diffraction confirmed the S-configuration.
Fig. 2. Crystal structure of compound 1.
To test whether these thioesters were MβL inhibitors, MβLs from subclasses B1 (NDM-1), B2 (ImiS) and B3 (L1) were over-expressed and purified by previously described methods.20 Steady-state kinetic studies of thioesters as inhibitors against MβL were conducted on an Agilent UV8453 spectrometer. Hydrolysis of the substrate (cefazolin for NDM-1 and L1; imipenem for ImiS) was monitored at 262 and 300 nm. The initial reaction rates were determined in the absence and presence of inhibitors in triplicate, and the average values were recorded. Percent inhibition, defined as the enzyme activity without the inhibitor (100%) minus the residual activity with 20 μM inhibitor, is shown for all the enzymes tested in Fig. S1.†
The percent inhibition data indicated that all these thioesters exhibited more than 85% inhibition against L1, but no obvious inhibition on NDM-1 or ImiS was observed at an inhibitor concentration of up to 20 μM (Fig. S1†). Therefore, for the remainder of this report, we focused on inhibition studies of L1. The inhibitor concentration causing a 50% decrease of enzyme activity (IC50) of compounds 1–10 against L1 was determined in 30 mM Tris, pH 7.0, using cefazolin as the substrate. The collected IC50 data (Table 1) indicated that all the inhibitors have excellent inhibitory activity against L1, exhibiting IC50 values ranging from 0.17 to 1.2 μM, and 8 showed the lowest IC50 value of 0.17 μM, revealing that mercaptoacetate thioesters potentially inhibit L1, which is consistent with their high %inhibition (90–95%) at an inhibitor dose of 20 μM. These results are comparable to what we have previously reported. Also, we performed assays of time- and concentration-dependent inhibition of L1 by thioester 9; the results indicated that the thioester exhibited maximum inhibition after incubation with the enzyme for 1.5 h and at a concentration of around 1.0 μM (Fig. S2†).
Table 1. Inhibitory activities of mercaptoacetate thioesters 1–10 against MβL L1.
| Inhibitor | IC50 (μM) | Inhibitor | IC50 (μM) |
| 1 | 0.37 ± 0.04 | 6 | 0.62 ± 0.07 |
| 2 | 0.80 ± 0.12 | 7 | 1.03 ± 0.14 |
| 3 | 0.75 ± 0.06 | 8 | 0.17 ± 0.03 |
| 4 | 0.19 ± 0.02 | 9 | 0.94 ± 0.09 |
| 5 | 0.88 ± 0.08 | 10 | 1.20 ± 0.15 |
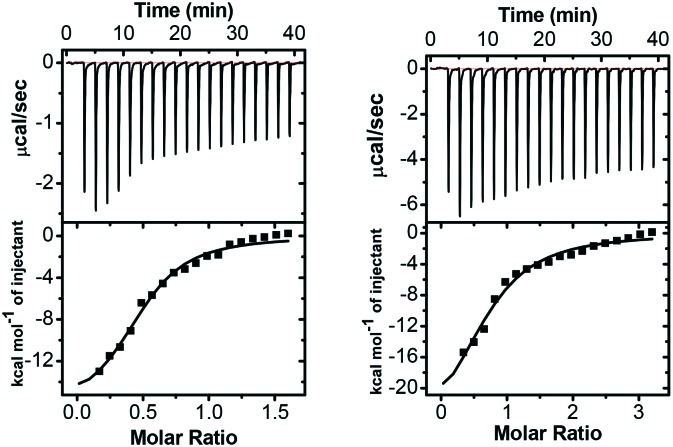
Isothermal titration calorimetry (ITC) is an essential technique to probe protein–ligand interactions by directly titrating one solution into another, and the key is to identify the source of the heat absorbed or released. MicroCal-ITC200 was employed to investigate the interaction of mercaptoacetate thioesters with L1 in multiple injection mode at 25 °C.21 The calorimetry profiles for the binding of two inhibitors 8 and 9 with L1 are shown in Fig. 3. In the top panels, each peak represents a single injection of the inhibitor into the protein solution, while the bottom panels show integrated plots of the amount of heat liberated per injection as a function of the molar ratio of the inhibitor to the protein. As shown in Fig. 3, negative heat deflection was observed, indicating that the binding is an exothermic process. The enthalpy changes (ΔH), binding affinities (Kb), entropy changes (ΔS) and binding stoichiometries (N) shown in Table 2 were obtained by fitting the data for single sites, using the sequential binding model, and the dissociation constants (Kd) and the Gibbs energy changes (ΔG) were calculated using the equations Kd = 1/Kb and ΔG = ΔH – TΔS. The Kd values of 8–L1 and 9–L1 bindings are 5.5 and 19.7 μM, respectively, which are fully consistent with the IC50 data (0.17 and 0.94 μM) of 8 and 9 against L1.
Fig. 3. ITC profiles of titration of a solution of inhibitor 8 (left) or 9 (right) into an L1 protein solution at 25 °C. The concentration of 8 or 9 was 0.5 mM, and the concentration of the protein was 60 μM.
Table 2. ITC derived thermodynamic parameters for the binding of mercaptoacetate thioesters 8 and 9 with L1 at 25 °C, pH 7.0.
| Compd | K d (μM) | N (site) | ΔH (kcal mol–1) | TΔS (kcal mol–1) | ΔG (kcal mol–1) |
| 8 | 5.5 | 0.49 | –16.78 | –9.60 | –7.18 |
| 9 | 19.7 | 0.71 | –28.24 | –21.81 | –6.43 |
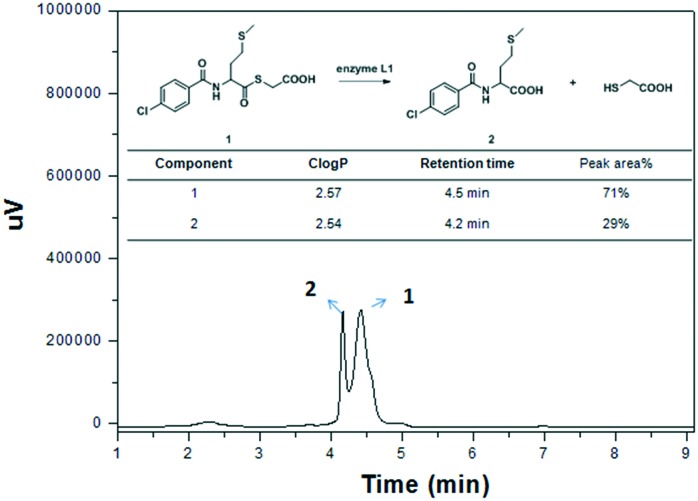
To probe the mechanism of the mercaptoacetate thioester interaction with L1, we firstly evaluated the effect of pH and L1 enzyme on thioester stability (Fig. S3†), and found that thioesters 1, 8 and 9 were unhydrolyzed when incubated in Tris buffer with pH ranging from 6.0 to 8.5 over 24 h. However, they were hydrolyzed when incubated with L1 as monitored by UV-vis spectroscopy (Fig. S3†). Further, the hydrolysis of the thioesters in the presence of enzyme L1 was monitored by HPLC (Fig. 4), and it was found that 1/3 of the thioesters were converted to mercaptoacetic acids under the same conditions as the above IC50 assay. This implied that the thioesters as well as their hydrolysate mercaptoacetic acids may jointly inhibit L1.
Fig. 4. The hydrolysis of mercaptoacetate thioester 9 in the presence of enzyme L1 as monitored by HPLC under the conditions of the IC50 assay.
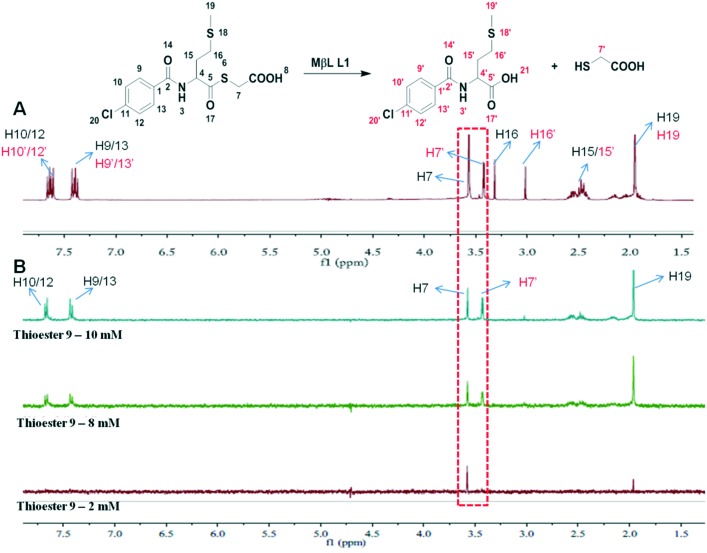
To verify this hypothesis, STD-NMR was employed to monitor the binding of thioester and/or mercaptoacetate to L1. STD experiments were performed on a Bruker Avance III 400 MHz spectrometer at 25 °C.22 The selective saturation for STD was achieved by a train of Gauss-shaped pulses with a length of 50 ms, alternating between on and off resonance with water suppression using excitation sculpting with gradients and spin-lock pulses to suppress protein signals (STDDIFFESGP.3). A sinc1.1000 shaped pulse was applied to suppress the solvent signal. The subtraction of on- and off-resonances is performed after every scan using phase cycling which gives the STD signal. Fig. 5A and B present the reference 1H NMR spectrum and the STD-NMR spectra of L1 mixed with thioester 9. It is clearly observed that there are three components, thioester 9, its hydrolysate N-substituted methionine, and mercaptoacetic acid, in the reference 1H NMR spectrum (Fig. 5A). On the other hand, as shown in Fig. 5B, the resonances of proton-7, 9/13, 10/12, and 19 of 9 and proton-7′ of the mercaptoacetic acid were enhanced in the STD spectrum with the increase of inhibitor concentration from 2 to 10 mM, indicating that they bind to L1 specifically with a defined binding conformation, whereas another hydrolysate N-substituted methionine does not present STD-NMR signals, indicating that it does not bind with L1, which is consistent with its inhibitory activities (Table S1†). These results suggested that thioester 9 and its hydrolysate mercaptoacetate jointly targeted the active sites of L1.
Fig. 5. Reference 1H NMR (A) and STD-NMR (B) spectra for the variation of the STD amplification factor with the increase of inhibitor 9 concentration. The L1 stock solution was prepared by dissolving 10 mg of protein in 3 mL of PBS buffer in D2O (50 μM) and was stored at 4 °C. Thioester 9 was dissolved in 5 mL of DMSO-d6 (final concentration = 2–10 mM). STD experiments were performed on a 400 MHz spectrometer in DMSO-d6/D2O PBS, pH 7.5, at 298 K.
The ability of the mercaptoacetate thioesters to restore the antibacterial activity of cefazolin against E. coli expressing L1 was investigated by determining the minimum inhibitory concentrations (MICs) of the antibiotic in the presence and absence of 1–10. A bacterial strain of E. coli BL21(DE3) containing plasmids pET26b-L1 and E. coli BL21 control without plasmids were used to assess these inhibitors. The concentration of the inhibitor was 16 μg mL–1. The collected MIC data are listed in Table 3. It is indicated that inhibitors 1–10 resulted in a 2–4-fold reduction in the MIC of cefazolin on E. coli expressing L1, indicating successful inhibition of L1 in vivo and effectively restoring the antibacterial activity of the antibiotic.
Table 3. MICs of cefazolin (μg mL–1) against E. coli cells expressing L1 in the presence and absence of mercaptoacetate thioesters at a concentration of 16 μg mL–1.
| Inhibitor | E. coli | E. coli-L1 | Inhibitor | E. coli | E. coli-L1 |
| Blank | 4 | 32 | 6 | 4 | 16 |
| 1 | 4 | 8 | 7 | 4 | 16 |
| 2 | 4 | 16 | 8 | 4 | 8 |
| 3 | 4 | 16 | 9 | 4 | 16 |
| 4 | 4 | 8 | 10 | 4 | 16 |
| 5 | 4 | 16 |
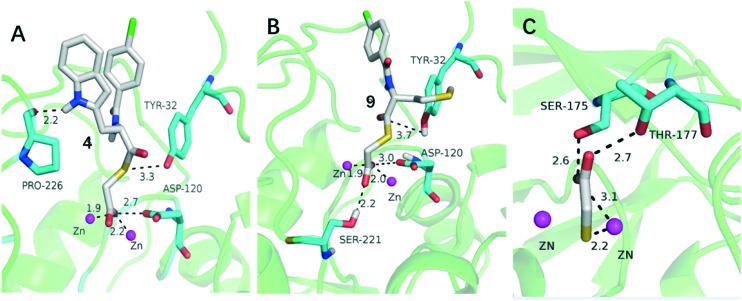
To explore the potential binding mode of mercaptoacetate thioesters alone as potent L1 inhibitors, two typical representatives, thioesters 4 and 9, were docked into the active sites of L1.23,24 The conformations shown in Fig. 6 are the lowest-energy conformations of those clusters, with binding energies of –12.1 and –10.8 kcal mol–1 for the L1/4 and L1/9 complexes, respectively.
Fig. 6. Lowest-energy docking conformations of compounds 4 and 9 (A and B) and the binding mode of mercaptoacetic acid targeting the subclass B3 metallo-β-lactamase SMB-1(PDB: ; 3VQZ) (C). The enzyme backbone is shown as a cartoon in green, and selected residues are shown as sticks colored by element (H, white; C, cyan; N, blue; O, red; S, yellow). Zn(ii) ions are shown as magenta spheres; the lower (front) one is Zn2 and the upper (back) one is Zn1. Compounds 4 and 9 are also shown as sticks with the same color code as the amino acid residues except that C is in white and Cl is in green. Characteristic short distances between the inhibitors and the protein are indicated by dashed lines. These figures were generated with PyMOL.
For complexes L1/4 and L1/9 shown in Fig. 6A and B, the carboxylate oxygen of the two compounds coordinated with the two Zn(ii) ions (1.9 and 2.2 Å for L1/4, 1.9 and 2.0 Å for L1/9, respectively) and formed a H-bond with Asp-120 (2.7 in L1/4 and 3.0 Å in L1/9), tightly anchoring these inhibitors in the active site, as seen previously with amino acid thioesters.17,18 Also, the sulfur atom and ketonic oxygen of 4 and 9 interacted with Tyr 32 in L1 (3.3 and 3.7 Å, respectively) via a H-bond. For compound 4, the tyrosine side chain forms an extra H-bond with Pro-226 (2.2 Å), compared with the methionine side chain in 9. The hydrolysate mercaptoacetic acid may coordinate with the Zn(ii) ions of L1 similar to its binding mode when targeting B3 subclass MβL SMB-1 (Fig. 6C).25
The potential toxicity of enzyme inhibitors is a major biomedical concern. A selection of mercaptoacetate thioesters 8 and 9 was subjected to a cytotoxicity assay with mouse fibroblast cells (L929) with different working concentrations (12.5, 25, 50, 100, 200, and 400 μM). As shown in Fig. S5,† none of them affected the viability of the L-929 mouse fibroblastic cells at a concentration of up to 200 μM, indicating that these compounds have low cytotoxicity.
Conclusions
In summary, to probe whether the essence of mercaptoacetate thioesters inhibiting L1 is the contribution of the thioester itself or its hydrolysate, ten mercaptoacetate thioesters 1–10 were synthesized and characterized. The obtained molecules specifically inhibited MβL L1, exhibiting IC50 values ranging from 0.17 to 1.2 μM, and 8 was found to be the best inhibitor, with an IC50 value of 0.17 μM using cefazolin as the substrate. MIC assays demonstrated that all the compounds restored the antimicrobial activity of cefazolin against E. coli expressing L1 by 2–4-fold. UV-vis monitoring showed that thioesters 1, 8 and 9 were unhydrolyzed in Tris buffer with pH ranging from 6.0 to 8.5, but hydrolyzed with L1, and further HPLC monitoring indicated that 1/3 of the thioester 9 was hydrolyzed by enzyme L1 under the conditions of the IC50 assay. STD-NMR monitoring suggested that the thioester and its hydrolysate mercaptoacetic acid jointly inhibited L1. Docking studies suggested that the carboxyl oxygen of 4 and 9 interacts with the metal at the Zn2 site of L1; the sulfur atom and ketonic oxygen of 4 and 9 interact with Tyr 32 of the enzyme via a H-bond. Cytotoxicity tests show that 8 and 9 did not affect the viability of mammalian cells at a dose up of to 200 μM.
E. coli BL21(DE3) cells and mouse fibro-blast cells (L929) were purchased from the Cell Bank, Chinese Academy of Sciences (Shanghai). Plasmids pET26b-NDM-1, pET26b-ImiS and pET26b (+)-L1 were obtained from professor Michael Crowder at Miami University, USA.
Conflicts of interest
The authors declare no competing interest.
Supplementary Material
Acknowledgments
This work was supported by grants 81361138018 and 21572179 (to K. W. Y.) from the National Natural Science Foundation of China.
Footnotes
†Electronic supplementary information (ESI) available: 1H, 13CNMR and MS characterization, stability assays of thioesters, and cytotoxicity assay. CCDC 1544802. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8md00091c
References
- King D. T., Strynadka N. C. Future Med. Chem. 2013;5:1243. doi: 10.4155/fmc.13.55. [DOI] [PubMed] [Google Scholar]
- Bush K. Ann. N. Y. Acad. Sci. 2013;1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- Ambler R. P. Philos. Trans. R. Soc., B. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Drawz S. M., Bonomo R. A. Clin. Microbiol. Rev. 2010;23:160–172. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornaglia G., Giamarellou H., Rossolini G. M. Lancet Infect. Dis. 2011;11:381. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- Lassaux P., Hamel M., Gulea M., Delbrück H., Mercuri P. S., Horsfall L., Dehareng D., Kupper M., Frère J. M., Hoffmann K. J. Med. Chem. 2010;53:4862–4872. doi: 10.1021/jm100213c. [DOI] [PubMed] [Google Scholar]
- Mcgeary R. P., Schenk G., Guddat L. W. Eur. J. Med. Chem. 2014;45:132–140. doi: 10.1016/j.ejmech.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Mohamed M. S., Hussein W. M., Mcgeary R. P., Vella P., Schenk G., El-Hameed R. H. A. Eur. J. Med. Chem. 2011;46:6075–6082. doi: 10.1016/j.ejmech.2011.10.030. [DOI] [PubMed] [Google Scholar]
- Simm A. M., Loveridge E. J., Crosby J., Avison M. B., Walsh T. R., Bennett P. M. Biochem. J. 2005;387:585–590. doi: 10.1042/BJ20041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toney J. H., Hammond G. G., Fitzgerald P. M. D., Sharma N., Balkovec J. M., Rouen G. P., Olson S. H., Hammond M. L., Greenlee M. L., Gao Y. J. Biol. Chem. 2001;276:31913–31918. doi: 10.1074/jbc.M104742200. [DOI] [PubMed] [Google Scholar]
- Walter M. W., Valladares M. H., Adlington R. M., Amicosante G., Baldwin J. E., Frère J. M., Galleni M., Rossolini G. M., Schofield C. J. Bioorg. Chem. 1999;27:35–40. [Google Scholar]
- Toney J. H., Fitzgerald P., Groversharma N., Olson S. H., May W. J., Sundelof J. G., Vanderwall D. E., Cleary K. A., Grant S. K., Wu J. K. Chem. Biol. 1998;5:185–196. doi: 10.1016/s1074-5521(98)90632-9. [DOI] [PubMed] [Google Scholar]
- Chiou J., Wan S., Chan K. F., So P. K., He D., Chan E. W., Chan T. H., Wong K. Y., Tao J., Chen S. Chem. Commun. 2015;51:9543–9546. doi: 10.1039/c5cc02594j. [DOI] [PubMed] [Google Scholar]
- King A. M., Reidyu S. A., Wang W., King D. T., Pascale G. D., Strynadka N. C., Walsh T. R., Coombes B. K., Wright G. D. Nature. 2014;510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. J., Bateson J. H., Gasson B. C., Khushi T., Proctor D., Pearson S. C., Reid R. FEMS Microbiol. Lett. 1997;157:171–175. doi: 10.1111/j.1574-6968.1997.tb12769.x. [DOI] [PubMed] [Google Scholar]
- Yang K. W., Crowder M. W. Arch. Biochem. Biophys. 1999;368:1–5. doi: 10.1006/abbi.1999.1293. [DOI] [PubMed] [Google Scholar]
- Liu X. L., Shi Y., Kang J. S., Oelschlaeger P., Yang K. W. ACS Med. Chem. Lett. 2015;6:660–665. doi: 10.1021/acsmedchemlett.5b00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. L., Yang K. W., Zhang Y. J., Ge Y., Xiang Y., Chang Y. N., Oelschlaeger P. Bioorg. Med. Chem. Lett. 2016;26:4698–4701. doi: 10.1016/j.bmcl.2016.08.048. [DOI] [PubMed] [Google Scholar]
- Chang Y. N., Xiang Y., Zhang Y. J., Wang W. M., Chen C., Oelschlaeger P., Yang K. W. ACS Med. Chem. Lett. 2017;8:527–532. doi: 10.1021/acsmedchemlett.7b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder M. W., Walsh T. R., Banovic L., Pettit M., Spencer J. Antimicrob. Agents Chemother. 1998;42:921–925. doi: 10.1128/aac.42.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Ali A., Danishuddin, Srivastava G., Sharma A. Sci. Rep. 2017;7:9207–9220. doi: 10.1038/s41598-017-09588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna A. M., Rodrigues T., Schmidt T. P., Böhm M., Stutz K., Reker D., Pfeiffer B., Altmann K. H., Backert S., Wessler S. Angew. Chem., Int. Ed. 2015;54:10244–10248. doi: 10.1002/anie.201504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer J., Read J., Sessions R. B., Howell S., Blackburn G. M., Gamblin S. J. J. Am. Chem. Soc. 2005;127:14439–14444. doi: 10.1021/ja0536062. [DOI] [PubMed] [Google Scholar]
- Zhang Y. L., Yang K. W., Zhou Y. J., Lacuran A. E., Oelschlaeger P., Crowder M. W. ChemMedChem. 2014;9:2445–2448. doi: 10.1002/cmdc.201402249. [DOI] [PubMed] [Google Scholar]
- Wachino J., Yamaguchi Y., Mori S., Kurosaki H., Arakawa Y., Shibayama K. Antimicrob. Agents Chemother. 2013;57:101–109. doi: 10.1128/AAC.01264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.