 Concerns of emerging resistance and the search for molecules with potential for single exposure radical cure and prophylaxis have spurred research into compounds with target profiles for clinical development into antimalarial drugs.
Concerns of emerging resistance and the search for molecules with potential for single exposure radical cure and prophylaxis have spurred research into compounds with target profiles for clinical development into antimalarial drugs.
Abstract
Though morbidity and mortality due to malaria have declined in the last 15 years, emerging resistance to first-line artemisinin-based antimalarials, absence of efficacious vaccines and limited chemotherapeutic alternatives imperil the consolidation of these gains. As a blueprint to steer future designs of new medicines, malaria drug discovery recently adopted a descriptive proposal for the ideal candidate molecules and drugs likely to successfully progress into the final stages of clinical development. As an audit of recent developments in the chemotherapy of malaria in the last five years, this review captures a landscape of diverse molecules at various stages of drug development and discusses their progress. In brief, we also discuss how omics data on Plasmodium has been extensively leveraged to identify potential vaccine candidates and putative targets of molecules in development and clinical use as well as map loci implicit in their modes of resistance. Future perspective on malaria drug development should involve a reconciliation of some of the challenges of the target candidate profiles (TCPs), specifically TCP3, with the promise of effective anti-hypnozoite medicines. Similarly, with the recent development of a humanized mouse model that can evaluate the prophylactic potential of candidate drugs, we argue for increased effort at identifying more liver-stage molecules, which are often only secondarily prioritized in conventional screening programs.
Introduction
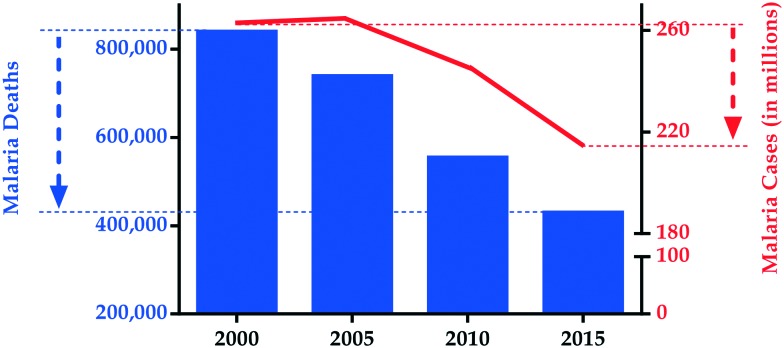
Malaria constitutes one of the leading causes of death worldwide, despite decades of strategic interventions aimed at reducing incidence and related mortality. This parasitic disease continues to heavily impact on public health and economies, with most severe consequences in countries with high poverty indices.1 Of the five Plasmodium species that cause malaria, Plasmodium falciparum is the most virulent with severe manifestation of clinical symptoms. The most recent empirical estimates from the World Health Organization (WHO) indicate that there were ∼216 million malaria cases and ∼445 000 related deaths globally in 2016.2 Although unprecedented progress has been made in reducing malaria morbidity and mortality in the past decade3 (Fig. 1), the latest statistics represent a worrying increase of ∼4 million cases relative to 2015 and call attention to the multiple challenges that increasingly complicate control strategies. For instance, the emergence of P. falciparum strains with reduced sensitivity to the WHO-recommended artemisinin combination therapy (ACT)4–6 and suboptimal responses to insecticides used for Anopheles vector management threaten control efforts. In addition, malaria is often co-endemic with other infectious tropical diseases like schistosomiasis in most resource-strained settings, hence co-infections are not uncommon. Novel, safe and effective therapeutic options to tackle this disease are therefore urgently needed.
Fig. 1. The evolution of malaria mortality and incidence between 2000 and 2015.
A significant proportion of the aforementioned progress in reducing malaria burden in the last 15 years is attributable to investments in vector control, diagnostic-led case management and chemoprevention. Specifically, the technological revolution in molecular platforms in the last decade has stimulated tremendous advances in research and development of novel drug candidates that would serve as viable alternatives to the current clinically-approved medicines. In this regard, the recent publication of a proposal by Burrows and colleagues at Medicines for Malaria Venture (MMV) in 2013 profiling the type of molecules (target candidate) and medicines (target product) ideal for new malaria therapy,7 helped establish a crucial blueprint for malaria drug design by defining the minimum acceptable profiles of ideal candidate molecules and drugs. Target candidate profile (TCP) 1 covers molecules that would clear asexual plasmodial blood stages in patients, while TCP3 profiles molecules that can prevent relapses due to P. vivax or P. ovale parasite infections by clearing or reactivating (and then killing) the dormant hypnozoites associated with disease latency. TCP4 anticipates chemoprotective molecules that can clear liver stage infection, an obligatory but transient stage preceding the symptomatic blood stage, thus potentially preventing infection in migrants moving into regions of high endemicity. TCP5 and TCP6 cover the profile of transmission blockers that either render the gametocytes non-functional (TCP5) or kill the mosquito vector following a blood meal (TCP6). Undoubtedly, these criteria have raised the bar for developing a new antimalarial drug and MMV envisages that only ∼2% of new antimalarial projects will progress into late-stage clinical development. These put into sharper focus the need for a more intensive search for new effective treatment options against malaria.
This review provides an abridged update on some of the important developments in research published within the past lustrum, aimed at identifying new treatments for malaria. On the basis of selected literature examples of molecules in advanced stages of clinical development and drug leads in pre-clinical evaluation, we catalogue some recent progress in drug development against this disease. Additionally, we discuss recent milestones in the exploitation of omics data in malaria for vaccine and drug development, and conclude with a perspective on the future direction in the development of new treatments against malaria.
Current status of vaccine research and chemotherapy in malaria
Vaccine development
As previously mentioned, the reduction in malaria burden in the recent past has largely been consequent to a scale-up of combinations of control strategies. In the main, these comprise long-lasting insecticide-treated nets, indoor residual spraying and intermittent preventive therapy for pregnant women (IPTp) for prevention and effective treatments using ACTs. Significant strides have also been made in malaria vaccine development, with the imminent large-scale roll-out of the pre-erythrocytic vaccine RTS,S/AS01 following a large phase III clinical trial in seven African countries highlighting this progress.8 However, the efficacy of RTS,S/AS01 against severe malaria is reportedly limited (18–36%) and immunity appears to quickly wane,8,9 thus restricting long-term protection. Combined with existing malaria interventions, this vaccine – albeit with its short-term and partial efficacy, still has potential to disrupt transmission in areas of low endemicity.
In appreciation of the shortcomings of RTS,S/AS01, more vaccine candidates are presently under evaluation. These include irradiated sporozoite vaccines like PfSPZ, which is closer to pivotal phase III trials,10 and subunit pre-erythrocytic and blood stage vaccines, which could be developed into multistage and multicomponent formulations.11 Such limitations in efficacy, absence of standard predictive biomarkers of protective efficacy and the need to constantly update vaccine formulations due to antigenic polymorphism further underscore the current reliance on chemotherapy.
Chemotherapy
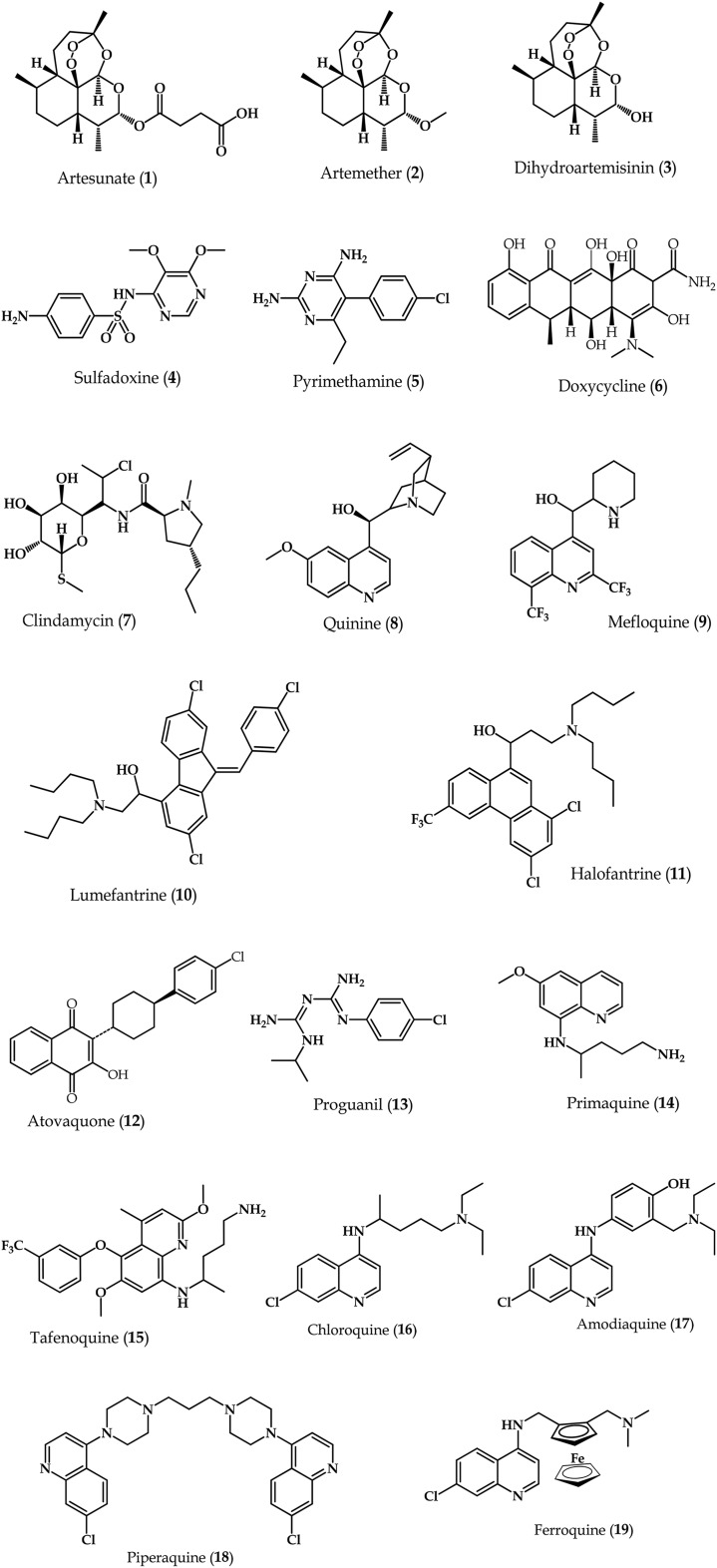
The sophistication in the lifecycle of P. falciparum presents different stages of the parasite's development that can be targeted using different types of drugs or drug combinations to treat malaria. Current antimalarials (Fig. 2) can be grouped into multiple classes namely: 4-aminoquinolines, 8-aminoquinolines, aryl-amino alcohols, antifolates, artemisinins, antibiotics and inhibitors of the cytochrome bc1 complex in the parasitic electron transport chain. ACTs constitute the current WHO-recommended treatment option for uncomplicated malaria.12 Artemisinins, exemplified by synthetic derivatives such as artesunate (1), artemether (2) and dihydroartemisinin (3), act on both early and late stages of the malaria parasite's lifecycle in the human host.13 They are fast-acting, efficacious and safe, and commonly combined with other longer-acting antimalarials in a strategy aimed at delaying the onset of resistance. The antifolates sulfadoxine (4) and pyrimethamine (5) inhibit enzymes within the folate pathway,14 and their combination (which is synergistic) constitutes a WHO-recommended IPTp course. Antibiotic antimalarials, like doxycycline (6) or clindamycin (7), inhibit protein synthesis and due to their slow onset of action, are often partnered with faster-acting antimalarials such as the artemisinins. Quinine (8), mefloquine (9), lumefantrine (10) and halofantrine (11) are examples of aryl-amino alcohols. Compounds 9 and 10 constitute ACT partner drugs in combination with 1 and 2, respectively, while 11 is not widely used due to concerns of severe cardiotoxicity. Atovaquone (12), a respiratory chain inhibitor,15 is mostly used for chemoprevention in combination with the antifolate proguanil (13), and for treatment of uncomplicated P. falciparum malaria. Primaquine (14) is currently the only 8-aminoquinoline drug available and is the drug of choice against P. vivax and relapsing forms of malaria.16 Tafenoquine (15) is a more potent and less toxic analogue of 14 that has since undergone phase II and III clinical evaluation.17–19 These 8-aminoquinolines form part of the broader quinoline-based drugs comprising the 4-aminoquinolines, the most-widely known of which is chloroquine (16) – currently used to treat uncomplicated vivax malaria. Following widespread clinical resistance to 16, amodiaquine (17) was designed as an alternative, although its prolonged use on its own has since been shown to result in an unacceptably high risk of agranulocytosis and hepatitis.20,21 Currently, this drug is used as part of ACTs where it is combined with 1 for treatment of uncomplicated malaria. Piperaquine (18), another 4-aminoquinoline, is currently part of an ACT treatment regimen with 3 in a fixed-dose combination, which has shown excellent tolerability and high malaria cure rates.22–24 Discovered more recently is ferroquine (19), which has completed phase IIa clinical trials,25 and has recently shown efficacy in combination with 1.26
Fig. 2. Chemical structures of antimalarial drugs in current clinical use or late stage of development.
Recent developments in antimalarial drug discovery
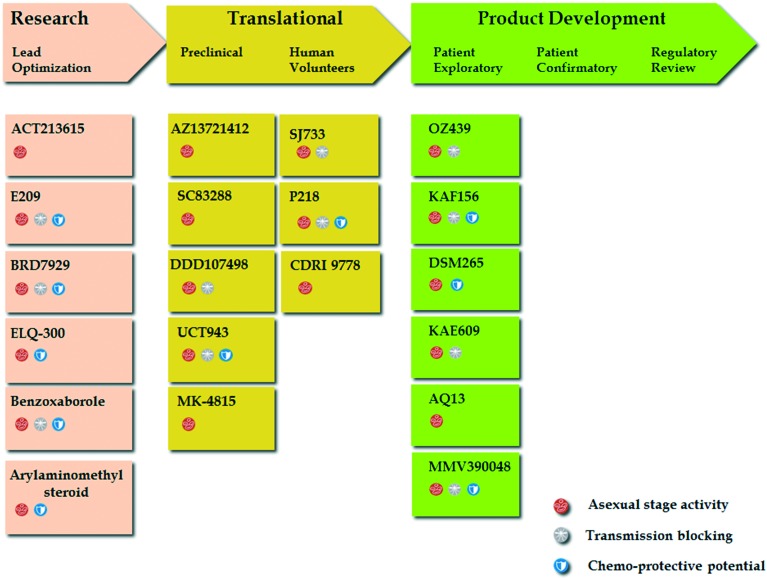
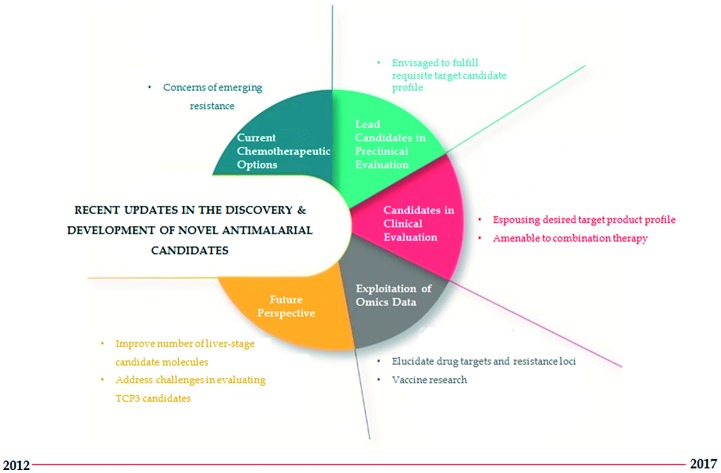
Increasing evidence of tolerance to ACTs and the mainstay antimalarials, and a dearth in treatment options against non-falciparum malaria have heightened the urgency for new compounds that are impervious to resistance and endowed with supplementary properties necessary to prevent transmission and provide radical cure against infections. In this section, we review recent progress in drug candidates described in the MMV-supported Global Portfolio of Antimalarial Medicine 2Q/2017 and the NIH-hosted repository for clinical trials, ClinicalTrials.gov, for which there exists recently updated information. Using the MeSH term “Antimalarial Drug Development AND Preclinical AND Clinical Candidate” we additionally searched literature for research articles published in the last 5–6 years in the biomedical and life sciences journal archive, PubMed, describing antimalarial candidate molecules currently under various stages of consideration for development as summarised in Fig. 3.
Fig. 3. Drug development stages and MMV-defined target candidate profiles of the antimalarial compounds discussed in this review.
Novel antimalarial drug candidates in lead optimization and preclinical stages
MK-4815
A recent screening of the Merck proprietary compound library for chemical entities with antimalarial activity resulted in the discovery of a Mannich base compound, 2-aminomethyl-3,5-di-tert-butylphenol, coded MK-4815 (20) (Fig. 4).27 This compound showed potent in vitro activity against a panel of geographically diverse, drug-resistant P. falciparum clinical isolates and 100% survival was observed in mice orally treated with 25, 12.5, or 6.25 mg kg–1 of the drug once on the day of infection then twice daily for an additional 4 days. Though comparable volume of distribution of the drug at steady state was observed in mice and rhesus monkey, the compound exhibited lower clearance and extended half-life in the monkeys, indicating superior pharmacokinetic parameters in the higher species. Even though its precise mode of action (MoA) still remains unclear, it appears to involve the mitochondrial electron transport chain of the parasite. Due to its structural simplicity, efficacy against multidrug-resistant P. falciparum strains (fulfils TCP1 characterization), good pharmacokinetic profile and ability to cure acute P. berghei infection at a single 50 mg kg–1 dose, compound 20 has potential as an antimalarial drug and in fact, is presently under further evaluation as a preclinical candidate by MMV.
Fig. 4. Chemical structures of the antimalarial leads and related compounds 20–32.
ACT-213615
In the quest for new antimalarial compounds with a MoA distinct from that of the current registered ones, Brunner et al., recently described the identification of the piperazine-containing ACT-213615 (21) (Fig. 4) from a phenotypic screen of aspartic protease inhibitors and compounds with undefined targets.28 The compound was fast-acting against the asexual erythrocytic stages of the parasite in vitro and equipotent to 1 (artesunate) against a panel of drug-resistant strains. This compound also exhibited oral efficacy against murine malaria and its activity was comparable to 16 (chloroquine) in the P. falciparum severe combined immunodeficiency (PfSCID) mouse model, underscoring the potential of this class of compounds as new antimalarial agents. To establish its putative MoA, microarray analysis was used to assess genome-wide transcriptional responses induced by 21 in comparison to 20 established profiles. The chemical perturbation signature of this compound did not cluster as artemisinin and was dissimilar to that of 8 (quinine) and 16 (chloroquine), thus hinting at a different putative target. As follow-up on the likely target of this compound, a functional interaction between 21 and P. falciparum multidrug resistance protein 1 was recently established,29,30 and postulated a potential role of this transporter protein on the activity of 21. These data warrant further research on this compound and/or its chemical class.
AZ13721412
Using a high-content image-based approach, a high-throughput screen (HTS) of 500 000 compounds from the AstraZeneca corporate library against asexual blood stage P. falciparum parasites identified a potent triaminopyrimidine series.31 In 2015, Hameed et al., reported subsequent structure activity relationship (SAR) exploration of the initial hit in this series that led to identification of the lead compound, AZ13721412, (22) (Fig. 4) bearing a N-methylpiperazine moiety at the C-5 position of the pyrimidine core and possessing a fast-killing and long-acting profile.32 This compound displayed the best potency in the series combined with >1000-fold selectivity against human Ether-a-go-go-Related Gene (hERG) protein and acetylcholinesterase as well as a significantly improved bioavailability (80%), which translated into improved in vivo efficacy. Studies on its putative MoA suggested it could be an inhibitor of the essential proton pump, V-type H+-ATPase, which mediates ATP-dependent H+ efflux and pH regulation. These data, coupled with the novelty of its MoA and low frequency of spontaneous resistance selection, supported its progression into further preclinical evaluation. Due to its potential for single dose cure in combination with suitable partner drugs, MMV and Zydus Cadila collaboratively announced plans in 2016 to progress this compound into active preclinical development.
SC83288
In the absence of parenteral artesunate (1) or artemether (2), current treatment for severe malaria comprises intravenous or intramuscular administration of 1 for at least 24 h or with 8 (quinine). However, this treatment strategy is blighted by safety reports associating intravenous administration of 1 with delayed haemolysis in 7–21% of the treated patients,33,34 while 8, aside from its unpleasant taste, has long been implicated in tinnitus and other audio-visual disorders.35 In this light, there is an equally pressing need for a new generation of drugs for the treatment of severe malaria, in addition to novel drugs for uncomplicated malaria. A recent report on medicinal chemistry optimization of amicarbalides led to the identification of a clinical development candidate molecule, SC83288 (23) (Fig. 4) with enhanced pharmacological and antiparasitic properties.36 Compound 23 was shown to be fast-acting and cured P. falciparum infection in a humanized NOD/SCID mouse model, with preclinical pharmacokinetic and toxicological studies revealing no apparent shortcomings. Sarco/endoplasmic reticulum Ca2+ transporting PfATP6 was identified as a putative determinant of resistance to 23. Its distinct chemotype, ability to rapidly kill parasites, potentially novel MoA and good safety indices support the clinical development of 23 as an intravenous application for the treatment of severe malaria. Presently, Heidelberg University Hospital and the German Centre for Infection Research are collaboratively in the process of conducting the regulatory preclinical procedures with the hope of initiating clinical trials in due course.
E209
Polymorphisms in the P. falciparum gene encoding the kelch-13 (PfK13) propeller domain are primary markers of clinical resistance to artemisinin.37 Recently, O'Neill and colleagues published the identification of a tetraoxane-based antimalarial drug candidate, E209 (24) (Fig. 4) that can overcome PfK13 Cys-580-Tyr dependent artemisinin resistance.38 To circumvent the liabilities that riddle the artemisinin class, single-dose survival time, intrinsic clearance and stability in blood were employed as the fundamental guides to steer SAR studies towards lead series progression from which 24 emerged as one of the front-runners. Further evaluation revealed retention of in vitro potency against a panel of geographically diverse sensitive and multidrug-resistant P. falciparum parasite isolates, with no observable cross-resistance. Compound 24 also exhibited equipotent ex vivo activity against P. vivax and P. falciparum Indonesian clinical isolates while screening for gametocytocidality showed a transmission reducing profile consistent with the endoperoxides. Critical to its development, evaluation of its activity against artemisinin-resistant parasites showed that the highly prevalent Cys-580-Tyr PfK13 mutation did not confer cross-resistance to 24. Equally important, in vivo experiments in P. berghei-infected mice demonstrated complete parasite clearance with an estimated oral ED50 of 4 mg kg–1 after three doses and a 66% cure rate after a single oral dose of 30 mg kg–1. This compound therefore has the potential for deployment in a superior combination treatment with a partner drug devoid of existing in vivo resistance liabilities hence offers a significant improvement on current ACTs and provides an urgently required alternative TCP1 drug for malaria treatment and elimination. Furthermore, its effectiveness against P. vivax and gametocytocidal activity fulfils TCP3 and TCP5 criteria for potential to prevent relapse and block transmission, respectively.
DDD107498
Chemical optimization of a compound series based on a 2,6-disubstituted quinoline-4-carboxamide scaffold identified through phenotypic screening of the Dundee protein kinase scaffold library against the 3D7 strain of P. falciparum recently led to the identification of DDD107498 (25) (Fig. 4).39 Compound 25 showed excellent activity against 3D7 and several drug-resistant strains, and exhibited greater potency than 1 (artesunate) in ex vivo assays against P. falciparum and P. vivax clinical isolates from Indonesia. The compound also displayed excellent pharmacokinetic properties in preclinical species, including good oral bioavailability and long plasma half-life (critical for chemoprophylaxis and single-dose treatment). Compound 25 was active in several mouse models of malaria, with comparable or greater efficacy than current antimalarials. In addition, 25 showed sub-micromolar activity against P. berghei and P. yoelii liver schizonts, potently inhibited gametocyte formation and showed potential to act as a transmission-blocker within a clinical context. Mechanistic studies found that 25 targets translation elongation factor 2 which is essential for protein synthesis, specifically the GTP-dependent translocation of the ribosome along messenger RNA. Due to its excellent drug-like features, potent activity profile against multiple P. falciparum lifecycle stages and novel MoA, compound 25 has potential for single-dose treatment where it would constitute the longer-acting partner to complete parasite clearance in a combination. Presently, this compound has been progressed into preclinical development, with the aim of entering into human clinical trials.
BRD7929
In an attempt to identify antimalarial compounds with novel MoAs, Kato and colleagues assessed ∼100 000 compounds synthesized at the Broad Institute using the build/couple/pair strategy of diversity-oriented synthesis,40 against the Dd2 strain using a phenotypic blood-stage growth-inhibition assay, which models a human blood-stage infection.41 This screening identified a series of hits acting on known antimalarial-target proteins as well as bicyclic azetidines which, through drug pressure studies and whole genome sequencing (WGS), were shown to target the cytosolic P. falciparum phenylalanyl-tRNA synthetase. This enzyme serves to enable transfer-RNAs deliver the amino acid phenylalanine to nascent proteins during RNA translation and protein synthesis. The lead bicyclic azetidine, BRD7929 (26) (Fig. 4), exhibited in vivo efficacy in P. falciparum and P. berghei-infected mice at single, low doses. The compound was also highly potent against the liver and asexual stages of P. falciparum and showed transmission-blocking ability at concentrations that achieved single-dose cures of asexual blood-stage infections. Though 26 displayed good oral bioavailability (80%), improved aqueous solubility and a long half-life in mice (∼32 hours), moderate cytotoxicity was observed in the HepG2 cell line and hERG channel thus presenting possible setbacks, which would have to be addressed during further optimization. Nevertheless, the ability of 26 to eliminate blood-stage (asexual and sexual) and liver-stage parasites suggests that this compound (and by extension the bicyclic azetidines) has the potential to cure the disease, provide prophylaxis and block transmission.
UCT943
Research at the University of Cape Town (UCT) has also recently led to the identification of a piperazine amide derivative, UCT943 (27) (Fig. 4), with potent in vitro activity against the erythrocytic stages of P. falciparum in both CQ-sensitive (CQS) and multidrug resistant strains.42 An extended evaluation of the activity of 27 in the P. berghei liver and P. falciparum gametocytic stages revealed that the compound was highly potent against the liver stage (IC50: 0.92 nM) and displayed good activity against the early and late stage gametocytes (IC50 = 134/66 nM, respectively). In vivo efficacy evaluation in P. berghei-infected and P. falciparum-infected NOD-scid IL-2Rγnull mouse models of malaria showed that 27 afforded complete cures (3/3 mice cured and mean survival days >30 days) with ≥99.9% activity at 4 × 10 mg kg–1. In fact, this efficacy was achievable at a very low ED90 (dose that reduces parasitemia at day 7 after infection by 90% with respect to vehicle-treated mice) of 1 mg kg–1. This potent in vivo efficacy is partly due to the compound's good pharmacokinetics properties, including a long half-life and bioavailability of 98%. This compound also showed a good preclinical safety profile, which collectively demonstrates its potential to become a drug candidate for the treatment of malaria. Of note, this molecule is a potential back-up to a previously-published 2-aminopyridine inhibitor of the Plasmodium phosphatidylinositol 4-kinase (PfPI4K), MMV390048 (covered later in this review) which is currently in clinical trials for the treatment of malaria. Compound 27 was selected by MMV for preclinical development in 2016.
ELQ-300
In 2008, Yeates and colleagues published a report describing a novel series of diphenylether-substituted pyridones that, like 12 (atovaquone), are thought to target the Qo site of the Plasmodium cytochrome bc1 complex in the mitochondrial electron transport chain, with the lead compound in this series, GW844520 (28), showing in vivo potency in P. yoelli-infected mice.43 However, around the same time, a structurally related pyridone analogue 29, which was in phase 1 clinical evaluation, was withdrawn due to toxicity concerns attributed to inhibition of mammalian mitochondrial cytochrome bc1 complex. Structural re-engineering of 29 by researchers from Oregon Health and Science University to rid it of this safety liability afforded the 4(1H)-quinolone derivative, ELQ-300 (30) (Fig. 4). This compound potently inhibited erythrocytic stages of P. falciparum and P. vivax in clinical field isolates as well as liver stages and transmissible stages of the parasite.44 Crucially, 30 proved highly selective against plasmodial cytochrome bc1 complexes (S. index: ≥10 000), suggesting diminished potential for side effects due to inhibition of the host enzyme. Compound 30 was also established as a slow-acting drug with a delayed parasite reduction ratio similar to 12 (atovaquone), and exhibited strong synergy with 13 (proguanil). Mutant selection experiments failed to achieve variants, suggesting a significantly low propensity for resistance. The compound was highly potent in the Thompson test in vivo model of P. berghei-infected mice with an ED50 of 0.016 mg kg–1 per day and cures produced with doses as low as 0.1 mg kg–1 per day, thus offering the potential as a combination partner aimed at a single-dose cure. Additional safety evaluation indicated no significant off-target pharmacological activities. A major obstacle to the clinical development of 30 relates to its relatively poor aqueous solubility which limits absorption to the extent that only low blood concentrations can be achieved with oral doses. Although these low blood concentrations suffice for therapy, the levels remain too low to establish an acceptable safety margin required for clinical development. An approach aimed at designing bioreversible alkoxycarbonate ester prodrugs has recently been successfully explored to overcome the physicochemical hurdles of 30 and attain bloodstream concentrations sufficient for safety and toxicology studies, as well as achieving single-dose cures.45,46
Benzoxaboroles
Benzoxaboroles are boron-containing compounds that have demonstrated potent activity against a number of pathogens, including bacteria,47 fungi,48 and trypanosomes.49 Recently, Anacor Pharmaceuticals and Shanghai ChemPartner reported on the identification of a set of carboxamide pyrazinyloxy benzoxaboroles with a satisfactory blend of antimalarial activity, physicochemical properties, pharmacokinetic profile, in vivo efficacy, and safety profile.50 Optimization efforts on this series yielded the lead compound 31 (Fig. 4), which fulfilled the target candidate profile of excellent in vitro activity against eleven cultured strains and in vivo efficacy in P. falciparum (ED90 = 0.85 mg kg–1) and P. berghei (ED90 = 1.92 mg kg–1) infected mice. The compound exhibited rapid in vitro parasite reduction and an in vivo parasite clearance profile comparable to artemisinin and chloroquine. Further, compound 31 showed good pharmacokinetic properties in mice, rats, and dogs as well as no evidence of genotoxicity in an in vivo rat micronucleus assay when dosed orally up to 2000 mg kg–1. Recent genetic and biochemical evidence using a benzoxaborole derivative suggests that the antiplasmodial activity of this class of compounds involves inhibition of a homologue of mammalian cleavage and polyadenylation specific factor subunit 3 in P. falciparum.51 On the basis of the in vitro and in vivo data, this novel benzoxaborole has been selected as a preclinical development candidate.
Arylmethylamino steroids
One approach to discover new biologically active compounds is to combine a steroid skeleton with structural elements endowed with appropriate biological activities. In a previous screening for potential antiprotozoal drugs, the naturally occurring steroid 3β-amino-22,26-epiminocholest-5-ene (sarachine) from the leaves of Saraca punctate, was identified as an antiplasmodial hit,52 and subsequent amino steroids bearing side chains similar to that of sarachine have been explored. Recently, Krieg and co-workers reported on low molecular weight arylmethylamino steroids with varying constitutions of the basic gonane core and exhibiting excellent antimalarial activity.53 The most promising derivative, 1o (32) (Fig. 4), showed in vitro activity against human and murine Plasmodium asexual blood stages as well as against P. falciparum gametocytes. Speed of growth inhibition analyses revealed that 32 is fast-acting, with MoA studies hinting at potential influence of 32 on glutathione redox homeostasis, though this would need further corroboration. Furthermore, 32 exhibited in vivo activity in P. berghei-infected mice as well as reduction of parasite transmission to mosquitoes, which could be as a result of the compound's ability to impair gametocyte maturation or commitment. Though low cytotoxicity and plasma stability further support the potential of this compound as a preclinical candidate, next steps for optimization aim at further improvement of lipophilicity and ADME properties including oral bioavailability (which was <5%) and enhancement of in vivo potency since only 2/3 of the treated mice were cured on oral administration of 4 × 100 mg kg–1 of compound 32 despite 99.8% suppression of parasitemia and a mean survival period of 27 days.
Antimalarial drug candidates in clinical evaluation
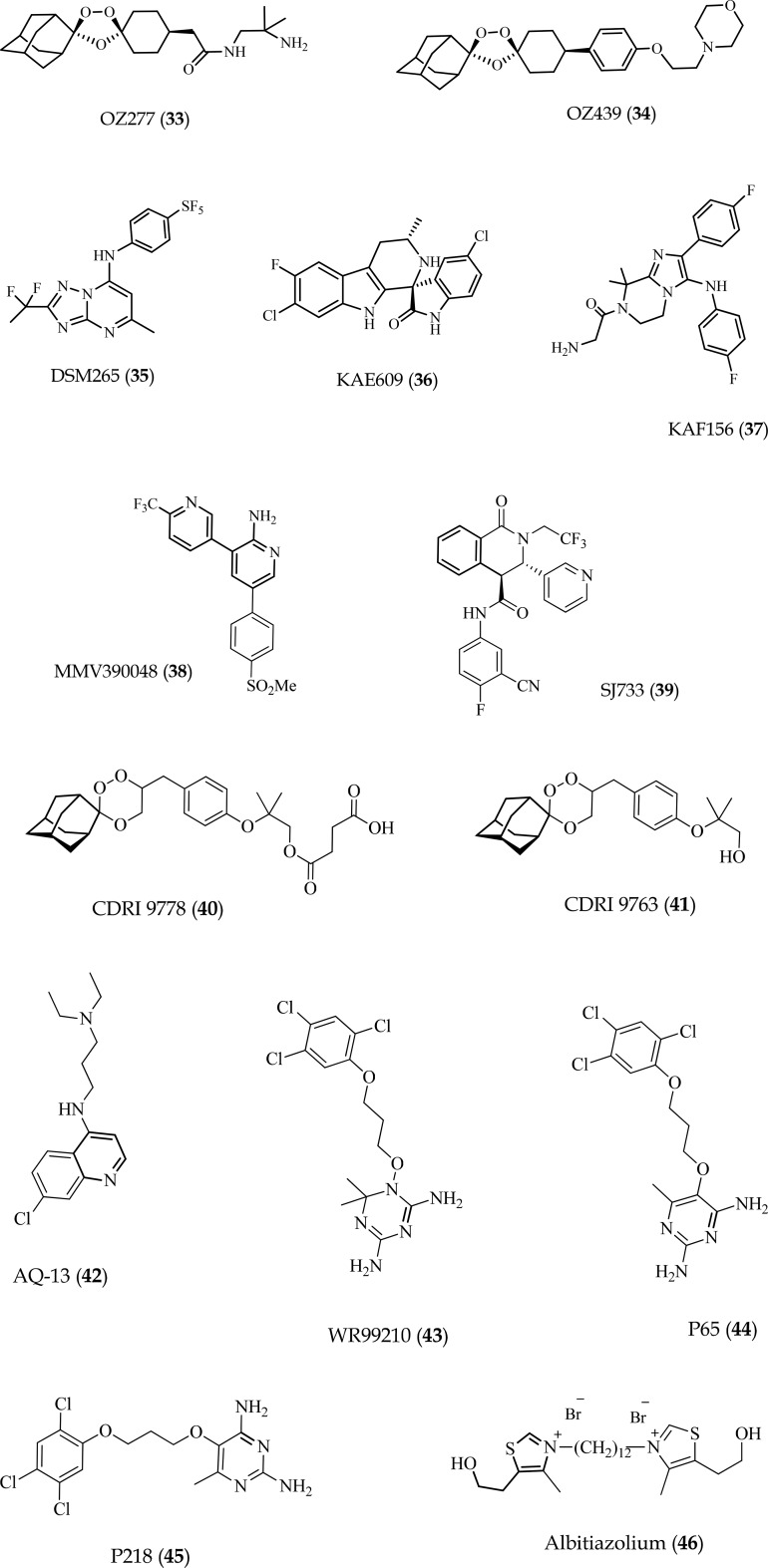
OZ439
A collaborative drug discovery project involving University of Nebraska, Monash University and MMV led to the development of OZ277 (33),54 a safe and well-tolerated synthetic ozonide. In phase II trials, monotherapy using 33 showed insufficient activity in patients due to low exposure relative to healthy volunteers. Nevertheless, further clinical development in combination with 18 (piperaquine) showed higher blood levels and clinical activity, with no recrudescence observed after therapy.55 However, to bypass the initial liability of low exposure related with 33, a programme to identify and develop new ozonides led to the identification of OZ439 (34) (Fig. 5), an orally active synthetic peroxide drug candidate with exceptional antimalarial and pharmacokinetic properties in vitro and in animal models.55 Unlike all other synthetic peroxides and artemisinin derivatives, 34 completely cured P. berghei-infected mice at a single oral dose of 20 mg kg–1 and showed superior prophylactic activity compared to most antimalarials.56 In 2015, following reports on the safety and pharmacokinetic profiles of 34, a combination with 19 (ferroquine) was progressed into a phase IIb trial, to evaluate the efficacy of a single oral dose regimen in adults and children aimed at replacing the current three doses prescribed for artemisinin derivatives (; clinicaltrials.gov/ct2/show/NCT02497612).
Fig. 5. Chemical structures of the clinical candidates and related compounds (33–46).
DSM265
In 2015, Phillips and co-workers reported on a long-duration triazolopyrimidine-based inhibitor of the P. falciparum pyrimidine biosynthetic enzyme, dihydroorotate dehydrogenase (PfDHODH), for prevention and treatment of malaria.57 This compound, DSM265 (35) (Fig. 5), was identified from optimization of the series using an X-ray structure-guided medicinal chemistry approach and showed high selectivity for PfDHODH, potency against drug-resistant parasite isolates as well as efficacy against both blood and liver stages of P. falciparum. Pharmacokinetic predictions further showed single oral doses in the range of 200–400 mg kg–1 would provide therapeutic concentrations for more than 8 days. The preclinical safety profiles, blood and liver-stage activity as well as predicted favorable pharmacokinetics, position 35 as a new potential drug combination partner for single-dose treatment as it espouses clear advantages over current treatment options that involve daily dosing or possess no liver-stage activity. Phase IIa evaluation of 35 to examine its efficacy in uncomplicated P. vivax and P. falciparum blood-stage malaria in adults was completed in 2016 (; clinicaltrials.gov/ct2/show/NCT02123290) while trials on its prophylactic potential were concluded in 2017 (; clinicaltrials.gov/ct2/show/NCT02562872). It is remains to be seen, from this clinical efficacy data, whether the reported half-life and dose predictions accurately reflect what is observed in humans.
Cipargamin (KAE609)
The synthetic spiroindolone analogue, cipargamin (36) (Fig. 5) represents a new class of potent, fast-acting, schizonticidal agents that has been shown to be safe and well-tolerated in healthy subjects up to as high a single dose as 300 mg and multiple doses of 150 mg daily for 3 days.58 In fact, evaluation in adults with uncomplicated P. vivax or P. falciparum malaria showed that once-daily dosing at 30 mg for 3 days was well-tolerated and resulted in a rapid median parasite clearance time of approximately 12 hours for both,59 placing this compound in the same range as ACTs such as Coartem™. Mechanistically, 36 inhibits P. falciparum P-type cation-translocating ATPase ATP4 (PfATP4) thus disturbing parasite sodium homeostasis leading to osmotic dysregulation,3,60 and this potentially explains the rapid parasite clearance following treatment with this compound.1,59 While the results of the preclinical and first clinical evaluations of 36 show promise (; clinicaltrials.gov/ct2/show/NCT03334747), its actual potency will be examined in subsequent trials with plans underway to advance it into phase IIb clinical development.
KAF156
KAF156 (37) (Fig. 5) is an imidazolopiperazine molecule previously identified by HTS and shown to have potent in vitro activity against both asexual and sexual blood stages and the pre-erythrocytic liver stages of the malarial parasite.61 Though its MoA is still unclear, in vitro resistance has been selected and is associated with mutations in the Golgi-associated P. falciparum cyclic amine resistance locus – encoding a conserved protein of as yet unknown function.62 A recent phase II study at five centres in Thailand and Vietnam to assess the efficacy, safety, and pharmacokinetic profile of this molecule in adults with acute P. vivax or P. falciparum malaria showed that 37 cleared parasites more rapidly than sulfadoxine–pyrimethamine or atovaquone–proguanil combinations, though this rate was slightly slower than artemisinin and compound 35 (DSM265).63 Further, therapeutic responses to treatment with 37 suggested effectiveness against P. falciparum and P. vivax infections resistant to all currently available antimalarial drugs without evident safety concerns. Novartis and MMV recently launched a patient trial to test the efficacy of 37 in combination with a new, improved formulation with 10 (lumefantrine), with the first trial centre already operational in Mali and to be followed by another sixteen across nine countries in Africa and Asia (; clinicaltrials.gov/ct2/show/NCT03167242).
MMV390048
A set of 3,5-diaryl-2-aminopyridines were identified from an image-based HTS of a commercial SoftFocus64 kinase library as a promising starting point to develop new antimalarial drugs. Stemming from this series, UCT identified the derivative, MMV390048 (38) (Fig. 5),65 which lacked cross-resistance with current antimalarial drugs and was active against all Plasmodium lifecycle stages, apart from late hypnozoites in the liver where it nevertheless delayed relapse. Efficacy was shown in the humanized P. falciparum mouse model, and modest reductions in mouse-to-mouse transmission observed in the P. berghei mouse model. Experiments in P. cynomolgi-infected rhesus macaques revealed the ability of 38 to be used for full chemoprotection. Similar to its potential back-up molecule, 27 (UCT943), this compound was recently shown to target PfPI4K.66 Due to its ability to block all lifecycle stages of the malaria parasite, 38 is presently in further clinical development and, in combination with an appropriate partner drug, is envisaged to be part of a single-dose regime for treatment of malaria in one instead of the current three days. Phase I study of ascending doses in healthy adult volunteers has been completed (; clinicaltrials.gov/ct2/show/NCT02230579) and recruitment for ‘first patient first visit’ phase IIa for patients with P. vivax and uncomplicated P. falciparum infection is presently underway (; clinicaltrials.gov/ct2/show/NCT02880241). The results of this trial will identify active, well-tolerated doses for further investigation in combination with a partner drug within a phase IIb clinical trial.
SJ733
Pharmacological and chemical optimization of a series of dihydroisoquinolones, identified in a HTS by researchers from St Jude's Children Hospital led to the selection of the clinical candidate, (+)-SJ733 (39) (Fig. 5), for development as a fast clearance component (TCP1) of a single-exposure radical cure and prophylaxis drug. Compound 39 was well-tolerated, orally bioavailable in multiple species (%F > 30%) and potently suppressed parasitemia in both a P. berghei model (ED50's < 50 mg kg–1) and P. falciparum SCID mouse model (ED50's < 5 mg kg–1), with 90% effective dose and exposure superior to 1 (artesunate), 5 (pyrimethamine) and 16 (chloroquine) in the SCID mouse.67 Subsequent mechanistic analyses showed that this molecule targets the Plasmodium cation-transporting ATPase, PfATP4, consequently affecting Na+ and H+ flux between the erythrocytic and the parasitic cytosols thus causing significant increase in cytosolic Na+ within the parasite. This process results in the immediate arrest of parasite motility and blockade of intracellular parasite replication. This elucidation of its MoA together with the fact that the compound meets all other criteria for a clinical candidate, including high oral bioavailability, high safety margins, and ability to block transmission support the continued development of this molecule as a potential new therapy for malaria. MMV, in partnership with Eisai Pharmaceuticals, are presently recruiting volunteers for First in Human (FIH) studies with 3 day multiple, rising dose to assess its safety, tolerability and blood levels in healthy adult volunteers (; clinicaltrials.gov/ct2/show/NCT02661373).
CDRI 9778
CDRI 9778 (40) (Fig. 5) is a trioxane peroxide originally developed by the Central Drug Research Institute in India as a potential substitute for the currently used artemisinin derivatives. Though this compound, like the artemisinins, contains the 1,2,4-trioxane nucleus responsible for pharmacological activity, it clusters among inhibitors of plasmodial phospholipid metabolism, most of whose members have shown high efficacy against multidrug resistant P. falciparum strains.68 Compound 40 successfully completed preclinical regulatory pharmacology and toxicity studies which established its safety. The first in-human phase I single-dose study was recently completed, with the data indicating the compound is safe and well-tolerated in normal healthy subjects albeit with a few adverse effects that were neither dose-dependent nor required intervention.69 Instructively, due to its poor bioavailability, 40 was synthesized as a hemisuccinate salt which, upon administration, is converted into its active in vivo metabolite CDRI 9763 (41). Pharmacokinetic data revealed rapid absorption and conversion to 41, with a long half-life that may help maintain the therapeutic efficacy and reduce recrudescence during therapy and follow-up. If these results can be replicated in multiple dose studies and benefit shown in confirmatory trials, 40 is likely to be an asset in malarial therapy.
AQ-13
Studies of 4-aminoquinoline activity against P. falciparum have shown that aminoquinolines with side chains shorter (2–3 carbons) or longer (10–12 carbons) than the 5-carbon isopentyl side chain of 16 (chloroquine) retain activity against CQ-resistant (CQR) P. falciparum strains.70 Indeed, AQ-13 [N′-(7-chloro-4-quinolinyl)-N,N-diethyl-1,3-propanediamine dihydrochloride trihydrate] (42) (Fig. 5), which has a linear three carbon n-propyl side chain linker has confirmed activity in CQS and CQR parasite strains,71 and has proven to be safe in human participants.72 Recently, this antimalarial candidate was advanced into a randomised, phase II, non-inferiority clinical trial to compare its efficacy to the combination of 2 (artemether) and 10 (lumefantrine) in semi-immune Malian men with uncomplicated malaria (; clinicaltrials.gov/ct2/show/NCT01614964).73 The data showed no serious adverse events and asexual parasites were cleared by day 7 in both groups. On the basis of the per-protocol analysis, compound 42 and the combination of 2 and 10 groups had similar proportions cured (28/28 [100%] for 42vs. 31/33 [93.9%] for the combination), and 42 was not inferior to the combination of 2 and 10. However, since semi-immune participants, such as those included in the mentioned study, can competently clear parasites resistant to the test drugs, additional studies in non-immune populations, such as children, are needed before recommendations for further advancement of 42.
P218
Various diaminoheterocycles, including pteridines, quinazolines, pyridopyrimidines, pyrimidines and triazines have served as scaffolds for good inhibitors of P. falciparum dihydrofolate reductase (PfDHFR). In an attempt to circumvent bioavailability challenges associated with the 4,6-diamino-1,2-dihydro-1,3,5-triazine antifolate, WR99210 (43), a BIOTEC research team in collaboration with MMV designed P65 (44), a 2,4-diaminopyrimidine analogue of 43.74 Indeed, the oral bioavailability of 44 in rats was found to be 83%, compared to <1% for 43. However, the X-ray co-crystal structure of 44 with mutant PfDHFR suggested that replacement of the trichlorophenyl group by an alkyl or alkyloxy group carrying a terminal carboxylate could favour better interaction with Arg122, a conserved region in the active site, which forms hydrogen bonds with the α-carboxylate of the dihydrofolate substrate. This led to the development of P218 (45) (Fig. 5), bearing the envisaged 2′-carboxyethylphenyl replacement. This compound demonstrated potent in vitro antiplasmodial and in vivo antimalarial activity as well as suitable pharmacological, metabolic, and safety profiles to warrant further consideration for clinical development. Preclinical assessment of 45 was made in 2014–2015 under the collaboration between the National Science and Technology Development Agency and MMV and the results displayed satisfactory safety profiles across a panel of required toxicology package in both in vitro and animal models (rodent and dog models) to demonstrate feasibility for FIH study which is currently recruiting (; clinicaltrials.gov/ct2/show/NCT02885506).
Albitiazolium
As a new class of antimalarial drugs, choline analogues have previously been documented to mechanistically inhibit parasite phospholipid metabolism.75,76 Among these analogues, the bisthiazolium series have been optimized into salts which inhibit P. falciparum asexual blood stages at concentrations in the low-nanomolar range and cure in vivo malaria infections at very low doses.77,78 The choline derivative, albitiazolium (46) (Fig. 5), was developed in 2004,78 and has been evaluated in preclinical studies and human phase I and II clinical trials. Owing to the absence of an oral formulation to treat uncomplicated malaria, Sanofi developed 46 for the treatment of severe malaria by intramuscular administration. However, following successful phase II trials in adults, trials in a paediatric population revealed higher drug clearance without a significant change in the highest (maximum) observed concentration (Cmax). The clinical development of 46 has since been halted due to this constraint since this high Cmax could not permit increase in dose without risking potential toxic effects.
Recent developments in the exploration of the P. falciparum omics landscape for drug discovery and vaccine research
The radical evolution of omics-based technologies has meant that the subsequent wealth of data could significantly accelerate antimalarial drug discovery and development if systematically integrated to address some of the often-cited challenges inherent in this process as well as vaccine development. The completion and subsequent publication of the P. falciparum genome79 has been a milestone and revolutionised malaria research in a manner that has contributed to better understanding of this pathogen. Advances in next generation sequencing and cost reliefs in WGS techniques have further facilitated elucidation of the genomes of other infectious human Plasmodium species,80,81 subsequently providing the molecular fabric from which to decipher the genetic basis of parasite resistance to antimalarial drugs, identify enzymatic targets and/or processes that would modulate the activity of novel lead compounds, and explore vaccine development.
Elucidation of drug targets and resistance loci
Recently, using whole genome data on individual sequenced samples and their clinical outcomes after artemisinin treatment, an association was drawn between a particular locus in chromosome 13 and delayed parasite clearance.82,83 The exact gene, kelch13 – encoding the kelch (K13) propeller domains, was identified through mutant selection and WGS of artemisinin-tolerant clinical isolates.84 More recently, a genome-wide association study using field isolates from the China–Myanmar border identified polymorphic loci that correlated with resistance to 10 commonly-used antimalarials.85 Particularly interesting was the elucidation of a locus on chromosome 10 containing the autophagy-related protein 18 (PfATG18), a phosphatidylinositol-3-phosphate binding protein involved in apicoplast biogenesis and associated with decreased sensitivities to the ACT component drugs 2 (artemether), 3 (dihydroartemisinin) and 18 (piperaquine) in this study. The identification of these correlates of artemisinin tolerance has provided a potent tool for monitoring the emergence of resistance in populations under artemisinin selective pressure and outlining methods for future surveillance as was recently applied in a worldwide survey to assess the global distribution of K13 polymorphisms in samples from malaria-endemic regions of the world.86 Also, using the dual-RNA-guided clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 recombinases, mutations introduced into the sodium efflux channel PfATP4 by transfection of a purified CRISPR/Cas9-guide RNA ribonucleoprotein complex have been shown to confer resistance to compound 39.87
The P. falciparum kinome contains ∼65 eukaryotic-like protein kinase sequences belonging to different established families as well as a unique family of atypical kinases to which the ribosome biogenesis regulators, RIO kinases, cluster. Transcriptome studies of intra-erythrocytic stages indicate that PFD0975w, a RIO-like kinase, is present in P. falciparum and is constitutively expressed throughout the life-cycle stages in red blood cells.88 Exploiting this information, Chakraborty and colleagues recently identified the localization patterns of this kinase in response to stress conditions such as starvation and antimalarial drug pressure.89 The recombinant PFD0975w exhibited a punctuate pattern in drug-treated parasite during trophozoite and schizont stages compared to untreated parasites, indicating a putative role of PFD0975w in cellular stress handling, thus flagging it as a potential drug target in the malaria parasite. Active recombinant PFD0975w can therefore be exploited to identify, validate or design novel inhibitors.
Vaccine research
Exploration of genomic data has also aided in the identification of potential malaria vaccine candidates as WGS data have, for instance, recently shed more light on the sequence structure and binding interactions between the malaria vaccine candidate P. falciparum reticulocyte binding protein homologue 5 and its human receptor, basigin.90 Even more recently, a genome-wide study using integrative bioinformatics approaches on the P. falciparum genome screened 5370 proteins to identify the presence of signal peptide/anchor and GPI anchor motifs favourable in vaccine design.91 From the 22 proteins consensually predicted in this analysis as antigens, 4944 peptides were predicted as CD8+ T cell epitopes and subsequently advanced for optimization studies in malaria-endemic regions. The predicted epitope 13ILFYFFLWV21 of antigen 6-cysteine was binding to the HLA-A*0201 allele with the highest fraction (26%) of immunogenicity in the target populations thereby highlighting the potential of these epitopes in vaccine development.
Conclusion and future perspectives
Though the landscape of malaria burden is changing due to the numerous control interventions presently underway in endemic countries, increasingly-compelling evidence of drug resistance and the paucity of clinical alternatives still remain a concern. For chemotherapy to still hold relevant beyond malaria control, it is essential that drugs and drug combinations comprise molecules showing activity beyond the asexual blood stage and as such, current focus of malaria drug design aims at conformity with the MMV-defined TCPs and TPPs to which a number of drugs and drug combinations fitting these ‘ideal medicine’ attributes are in clinical evaluation. However, this has not been without challenges as, for instance, design of TCP3 molecules is still constrained by the absence of established protocols for continuous in vitro culturing of P. vivax parasites or P. vivax animal models. Also, the diversity of P. vivax strains confounds standardization as different strains differ in hypnozoite to schizont ratios and relapse times. Lastly, current testing of P. vivax liver stage activity of candidate molecules is conducted against hypnozoites growing in primary hepatocytes, which are metabolically active and thus can breakdown test compounds (into inactive metabolites) leading to false negative signals.
A related concern involves the limited progress thus far in the development of liver stage drugs to address TCP4 objectives. This lack of success presumably mirrors the systemic design of most current drug discovery programs which involve initial HTS analyses of molecules and only selecting active hits for progression into anti-liver stage screening.92 Molecules targeting the liver stage have the dual advantage of first, acting at a stage with lower parasite biomass (compared to the blood stage) hence reduced likelihood for development of resistance and secondly, having the potential to inhibit new targets that are absent or non-essential during the blood stage phase. As further support for the development of more liver stage-acting antimalarials, Kappe and colleagues developed a human liver-chimeric mouse model that supports complete P. falciparum liver stage development with kinetics similar to those in humans.93 This humanized model has since been used by the same group to describe the efficacy of clinically-used and candidate antimalarials with prophylactic activity as well as measure liver stage-to-blood stage transition of the parasite.94 This model therefore effectively provides a platform for preclinical evaluation of drug candidates for liver-stage causal prophylactic activity, pharmacokinetic/pharmacodynamics studies, and MoA in vivo.
Overall, structural modifications of known heterocyclic scaffolds, exploitation of natural products and deeper excavation of genomic data all have potential to provide and reify advancements in leads and drug candidates for radical cure of malaria. The last five years have witnessed even more encouraging strides in the development of chemotherapeutic molecules for the treatment of malaria since the turn of the millennium. However, these recent breakthroughs will still need the insulation of long-term financial investments, political will and scientific endeavor to ensure sustainability and translate to further reduction in global disease burden.
Abbreviations
- ACT
Artemisinin combination therapy
- CRISPR
Clustered regularly interspaced short palindromic repeats
- ED50
Median effective dose
- FIH
First in human
- hERG
Human Ether-a-go-go-Related Gene
- HTS
High throughput screening
- IC50
Half-maximal inhibitory concentration
- IPTp
Intermittent preventive treatment (in pregnancy)
- MMV
Medicines for Malaria Venture
- MoA
Mode of action
- PfDHODH
P. falciparum dihydroorotate dehydrogenase
- PfMDR1
P. falciparum multidrug resistance protein 1
- PfSCID
P. falciparum severe combined immunodeficiency
- SAR
Structure activity relationship
- TCAMS
Tres Cantos Antimalarial Set
- TCP
Target candidate profile
- TPP
Target product profile
- WGS
Whole-genome sequencing
- WHO
World Health Organization
Conflicts of interest
There are no conflicts of interest to declare.
Acknowledgments
The Novartis Research Foundation, Bill and Melinda Gates Foundation, MMV, University of Cape Town, South African Medical Research Council, and South African Department of Science and Technology/National Research Foundation Research Chairs Initiative are gratefully acknowledged for support.
References
- Ridley R. G. Nature. 2002;415:686–693. doi: 10.1038/415686a. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO), World Malaria Report 2017, http://www.who.int/malaria/publications/world-malaria-report-2017/en/, Geneva, Switzerland (accessed December 2017).
- Bhatt S., Weiss D. J., Cameron E., Bisanzio D., Mappin B., Dalrymple U., Battle K., Moyes C. L., Henry A., Eckhoff P. A., Wenger E. A., Briet O., Penny M. A., Smith T. A., Bennett A., Yukich J., Eisele T. P., Griffin J. T., Fergus C. A., Lynch M., Lindgren F., Cohen J. M., Murray C. L. J., Smith D. L., Hay S. I., Cibulskis R. E., Gething P. W. Nature. 2015;526:207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A. M., Nosten F., Yi P., Das D., Phyo A. P., Tarning J., Lwin K. M., Ariey F., Hanpithakpong W., Lee S. J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S. S., Yeung S., Singhasivanon P., Day N. P., Lindegardh N., Socheat D., White N. J. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Culleton R., Zhang M., Ramaprasad A., von Seidlein L., Zhou H., Zhu G., Tang J., Liu Y., Wang W., Cao Y., Xu S., Gu Y., Li J., Zhang C., Gao Q., Menard D., Pain A., Yang H., Zhang Q., Cao J. N. Engl. J. Med. 2017;376:991–993. doi: 10.1056/NEJMc1612765. [DOI] [PubMed] [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B. L., Socheat D., Fukuda M. M. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Burrows J. N., van Huijsduijnen R. H., Möhrle J. J., Oeuvray C., Wells T. N. Malar. J. 2013;12:187. doi: 10.1186/1475-2875-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RTS,S Clinical Trials Partnership Lancet. 2015;386:31–45. [Google Scholar]
- Neafsey D. E., Juraska M., Bedford T., Benkeser D., Valim C., Griggs A., Lievens M., Abdulla S., Adjei S., Agbenyega T., Agnandji S. T., Aide P., Anderson S., Ansong D., Aponte J. J., Asante K. P., Bejon P., Birkett A. J., Bruls M., Connolly K. M., D'Alessandro U., Dobano C., Gesase S., Greenwood B., Grimsby J., Tinto H., Hamel M. J., Hoffman I., Kamthunzi P., Kariuki S., Kremsner P. G., Leach A., Lell B., Lennon N. J., Lusingu J., Marsh K., Martinson F., Molel J. T., Moss E. L., Njuguna P., Ockenhouse C. F., Ogutu B. R., Otieno W., Otieno L., Otieno K., Owusu-Agyei S., Park D. J., Pelle K., Robbins D., Russ C., Ryan E. M., Sacarlal J., Sogoloff B., Sorgho H., Tanner M., Theander T., Valea I., Volkman S. K., Yu Q., Lapierre D., Birren B. W., Gilbert P. B., Wirth D. F. N. Engl. J. Med. 2015;373:2025–2037. doi: 10.1056/NEJMoa1505819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie T. L., Billingsley P. F., Sim B. K., James E. R., Chakravarty S., Epstein J. E., Lyke K. E., Mordmuller B., Alonso P., Duffy P. E., Doumbo O. K., Sauerwein R. W., Tanner M., Abdulla S., Kremsner P. G., Seder R. A., Hoffman S. L. Vaccine. 2015;33:7452–7461. doi: 10.1016/j.vaccine.2015.09.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper S. J., Angov E., Horii T., Miller L. H., Srinivasan P., Theisen M., Biswas S. Vaccine. 2015;33:7433–7443. doi: 10.1016/j.vaccine.2015.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO), Antimalarial Drug Combination Therapy: Report of a Technical Consultation, World Health Organization, Geneva, Switzerland, 2001.
- Wilson D. W., Langer C., Goodman C. D., McFadden G. I., Beeson J. G. Antimicrob. Agents Chemother. 2013;57:1455–1467. doi: 10.1128/AAC.01881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzila A. J. Antimicrob. Chemother. 2006;57:1043–1054. doi: 10.1093/jac/dkl104. [DOI] [PubMed] [Google Scholar]
- Winter R. W., Kelly J. X., Smilkstein M. J., Dodean R., Hinrichs D., Riscoe M. K. Exp. Parasitol. 2008;118:487–497. doi: 10.1016/j.exppara.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird J. K., Hoffman S. L. Clin. Infect. Dis. 2004;39:1336–1345. doi: 10.1086/424663. [DOI] [PubMed] [Google Scholar]
- Llanos-Cuentas A., Lacerda M. V., Rueangweerayut R., Krudsood S., Gupta S. K., Kochar S. K., Arthur P., Chuenchom N., Mohrle J. J., Duparc S., Ugwuegbulam C., Kleim J. P., Carter N., Green J. A., Kellam L. Lancet. 2014;383:1049–1058. doi: 10.1016/S0140-6736(13)62568-4. [DOI] [PubMed] [Google Scholar]
- Walsh D. S., Looareesuwan S., Wilairatana P., Heppner Jr. D. G., Tang D. B., Brewer T. G., Chokejindachai W., Viriyavejakul P., Kyle D. E., Milhous W. K., Schuster B. G., Horton J., Braitman D. J., Brueckner R. P. J. Infect. Dis. 1999;180:1282–1287. doi: 10.1086/315034. [DOI] [PubMed] [Google Scholar]
- Walsh D. S., Wilairatana P., Tang D. B., Heppner Jr. D. G., Brewer T. G., Krudsood S., Silachamroon U., Phumratanaprapin W., Siriyanonda D., Looareesuwan S. Clin. Infect. Dis. 2004;39:1095–1103. doi: 10.1086/424508. [DOI] [PubMed] [Google Scholar]
- Lind D. E., Levi J. A., Vincent P. C. Br. Med. J. 1973;1:458–460. doi: 10.1136/bmj.1.5851.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel K. A., Woodtly W., Schmid M., Frick P. G., Fehr J. Br. Med. J. 1986;292:721–723. doi: 10.1136/bmj.292.6522.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arinaitwe E., Sandison T. G., Wanzira H., Kakuru A., Homsy J., Kalamya J., Kamya M. R., Vora N., Greenhouse B., Rosenthal P. J., Tappero J., Dorsey G. Clin. Infect. Dis. 2009;49:1629–1637. doi: 10.1086/647946. [DOI] [PubMed] [Google Scholar]
- Grande T., Bernasconi A., Erhart A., Gamboa D., Casapia M., Delgado C., Torres K., Fanello C., Llanos-Cuentas A., D'Alessandro U. PLoS One. 2007;2:e1101. doi: 10.1371/journal.pone.0001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T. H., Dolecek C., Pham P. M., Nguyen T. D., Nguyen T. T., Le H. T., Dong T. H., Tran T. T., Stepniewska K., White N. J., Farrar J. Lancet. 2004;363:18–22. doi: 10.1016/s0140-6736(03)15163-x. [DOI] [PubMed] [Google Scholar]
- Supan C., Mombo-Ngoma G., Kombila M., Ospina Salazar C. L., Held J., Lell B., Cantalloube C., Djeriou E., Ogutu B., Waitumbi J., Otsula N., Apollo D., Polhemus M. E., Kremsner P. G., Walsh D. S. Am. J. Trop. Med. Hyg. 2017;97:514–525. doi: 10.4269/ajtmh.16-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held J., Supan C., Salazar C. L., Tinto H., Bonkian L. N., Nahum A., Moulero B., Sie A., Coulibaly B., Sirima S. B., Siribie M., Otsyula N., Otieno L., Abdallah A. M., Kimutai R., Bouyou-Akotet M., Kombila M., Koiwai K., Cantalloube C., Din-Bell C., Djeriou E., Waitumbi J., Mordmuller B., Ter-Minassian D., Lell B., Kremsner P. G. Lancet Infect. Dis. 2015;15:1409–1419. doi: 10.1016/S1473-3099(15)00079-1. [DOI] [PubMed] [Google Scholar]
- Powles M. A., Allocco J., Yeung L., Nare B., Liberator P., Schmatz D. Antimicrob. Agents Chemother. 2012;56:2414–2419. doi: 10.1128/AAC.05326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner R., Aissaoui H., Boss C., Bozdech Z., Brun R., Corminboeuf O., Delahaye S., Fischli C., Heidmann B., Kaiser M., Kamber J., Meyer S., Papastogiannidis P., Siegrist R., Voss T., Welford R., Wittlin S., Binkert C. J. Infect. Dis. 2012;206:735–743. doi: 10.1093/infdis/jis418. [DOI] [PubMed] [Google Scholar]
- Brunner R., Ng C. L., Aissaoui H., Akabas M. H., Boss C., Brun R., Callaghan P. S., Corminboeuf O., Fidock D. A., Frame I. J., Heidmann B., Le Bihan A., Jeno P., Mattheis C., Moes S., Muller I. B., Paguio M., Roepe P. D., Siegrist R., Voss T., Welford R. W., Wittlin S., Binkert C. J. Biol. Chem. 2013;288:22576–22583. doi: 10.1074/jbc.M113.453159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C. L., Siciliano G., Lee M. C., de Almeida M. J., Corey V. C., Bopp S. E., Bertuccini L., Wittlin S., Kasdin R. G., Le Bihan A., Clozel M., Winzeler E. A., Alano P., Fidock D. A. Mol. Microbiol. 2016;101:381–393. doi: 10.1111/mmi.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Hameed P. S., Srivastava A., Shanbhag G., Morayya S., Rautela N., Awasthy D., Kavanagh S., Bharath S., Reddy J., Panduga V., Prabhakar K. R., Saralaya R., Nanduri R., Raichurkar A., Menasinakai S., Achar V., Jimenez-Diaz M. B., Martinez M. S., Angulo-Barturen I., Ferrer S., Sanz L. M., Gamo F. J., Duffy S., Avery V. M., Waterson D., Lee M. C., Coburn-Flynn O., Fidock D. A., Iyer P. S., Narayanan S., Hosagrahara V., Sambandamurthy V. K. J. Med. Chem. 2014;57:6642–6652. doi: 10.1021/jm500715u. [DOI] [PubMed] [Google Scholar]
- Hameed P. S., Solapure S., Patil V., Henrich P. P., Magistrado P. A., Bharath S., Murugan K., Viswanath P., Puttur J., Srivastava A., Bellale E., Panduga V., Shanbag G., Awasthy D., Landge S., Morayya S., Koushik K., Saralaya R., Raichurkar A., Rautela N., Roy Choudhury N., Ambady A., Nandishaiah R., Reddy J., Prabhakar K. R., Menasinakai S., Rudrapatna S., Chatterji M., Jimenez-Diaz M. B., Martinez M. S., Sanz L. M., Coburn-Flynn O., Fidock D. A., Lukens A. K., Wirth D. F., Bandodkar B., Mukherjee K., McLaughlin R. E., Waterson D., Rosenbrier-Ribeiro L., Hickling K., Balasubramanian V., Warner P., Hosagrahara V., Dudley A., Iyer P. S., Narayanan S., Kavanagh S., Sambandamurthy V. K. Nat. Commun. 2015;6:6715. doi: 10.1038/ncomms7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman K., Lotsch F., Kremsner P. G., Ramharter M. Int. J. Infect. Dis. 2014;29:268–273. doi: 10.1016/j.ijid.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Rolling T., Agbenyega T., Krishna S., Kremsner P. G., Cramer J. P. Travel. Med. Infect. Dis. 2015;13:143–149. doi: 10.1016/j.tmaid.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Achan J., Talisuna A. O., Erhart A., Yeka A., Tibenderana J. K., Baliraine F. N., Rosenthal P. J., D'Alessandro U. Malar. J. 2011;10:144. doi: 10.1186/1475-2875-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro S., Duffey M., Otto T. D., Wang Y., Rosemann R., Baumgartner R., Fehler S. K., Lucantoni L., Avery V. M., Moreno-Sabater A., Mazier D., Vial H. J., Strobl S., Sanchez C. P., Lanzer M. Nat. Commun. 2017;8:14193. doi: 10.1038/ncomms14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straimer J., Gnadig N. F., Witkowski B., Amaratunga C., Duru V., Ramadani A. P., Dacheux M., Khim N., Zhang L., Lam S., Gregory P. D., Urnov F. D., Mercereau-Puijalon O., Benoit-Vical F., Fairhurst R. M., Menard D., Fidock D. A. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill P. M., Amewu R. K., Charman S. A., Sabbani S., Gnadig N. F., Straimer J., Fidock D. A., Shore E. R., Roberts N. L., Wong M. H., Hong W. D., Pidathala C., Riley C., Murphy B., Aljayyoussi G., Gamo F. J., Sanz L., Rodrigues J., Cortes C. G., Herreros E., Angulo-Barturen I., Jimenez-Diaz M. B., Bazaga S. F., Martinez-Martinez M. S., Campo B., Sharma R., Ryan E., Shackleford D. M., Campbell S., Smith D. A., Wirjanata G., Noviyanti R., Price R. N., Marfurt J., Palmer M. J., Copple I. M., Mercer A. E., Ruecker A., Delves M. J., Sinden R. E., Siegl P., Davies J., Rochford R., Kocken C. H. M., Zeeman A. M., Nixon G. L., Biagini G. A., Ward S. A. Nat. Commun. 2017;8:15159. doi: 10.1038/ncomms15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baragana B., Hallyburton I., Lee M. C., Norcross N. R., Grimaldi R., Otto T. D., Proto W. R., Blagborough A. M., Meister S., Wirjanata G., Ruecker A., Upton L. M., Abraham T. S., Almeida M. J., Pradhan A., Porzelle A., Luksch T., Martinez M. S., Bolscher J. M., Woodland A., Norval S., Zuccotto F., Thomas J., Simeons F., Stojanovski L., Osuna-Cabello M., Brock P. M., Churcher T. S., Sala K. A., Zakutansky S. E., Jimenez-Diaz M. B., Sanz L. M., Riley J., Basak R., Campbell M., Avery V. M., Sauerwein R. W., Dechering K. J., Noviyanti R., Campo B., Frearson J. A., Angulo-Barturen I., Ferrer-Bazaga S., Gamo F. J., Wyatt P. G., Leroy D., Siegl P., Delves M. J., Kyle D. E., Wittlin S., Marfurt J., Price R. N., Sinden R. E., Winzeler E. A., Charman S. A., Bebrevska L., Gray D. W., Campbell S., Fairlamb A. H., Willis P. A., Rayner J. C., Fidock D. A., Read K. D., Gilbert I. H. Nature. 2015;522:315–320. doi: 10.1038/nature14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J. T., Lee M. D. T., Akella L. B., Davoine E., Donckele E. J., Durak L., Duvall J. R., Gerard B., Holson E. B., Joliton A., Kesavan S., Lemercier B. C., Liu H., Marie J. C., Mulrooney C. A., Muncipinto G., Welzel-O'Shea M., Panko L. M., Rowley A., Suh B. C., Thomas M., Wagner F. F., Wei J., Foley M. A., Marcaurelle L. A. J. Org. Chem. 2012;77:7187–7211. doi: 10.1021/jo300974j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Comer E., Sakata-Kato T., Sharma A., Sharma M., Maetani M., Bastien J., Brancucci N. M., Bittker J. A., Corey V., Clarke D., Derbyshire E. R., Dornan G. L., Duffy S., Eckley S., Itoe M. A., Koolen K. M., Lewis T. A., Lui P. S., Lukens A. K., Lund E., March S., Meibalan E., Meier B. C., McPhail J. A., Mitasev B., Moss E. L., Sayes M., Van Gessel Y., Wawer M. J., Yoshinaga T., Zeeman A. M., Avery V. M., Bhatia S. N., Burke J. E., Catteruccia F., Clardy J. C., Clemons P. A., Dechering K. J., Duvall J. R., Foley M. A., Gusovsky F., Kocken C. H., Marti M., Morningstar M. L., Munoz B., Neafsey D. E., Winzeler E. A., Wirth D. F., Scherer C. A., Schreiber S. L. Nature. 2016;538:344–349. doi: 10.1038/nature19804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Manach C., Nchinda A. T., Paquet T., Gonzalez Cabrera D., Younis Y., Han Z., Bashyam S., Zabiulla M., Taylor D., Lawrence N., White K. L., Charman S. A., Waterson D., Witty M. J., Wittlin S., Botha M. E., Nondaba S. H., Reader J., Birkholtz L. M., Jimenez-Diaz M. B., Martinez M. S., Ferrer S., Angulo-Barturen I., Meister S., Antonova-Koch Y., Winzeler E. A., Street L. J., Chibale K. J. Med. Chem. 2016;59:9890–9905. doi: 10.1021/acs.jmedchem.6b01265. [DOI] [PubMed] [Google Scholar]
- Yeates C. L., Batchelor J. F., Capon E. C., Cheesman N. J., Fry M., Hudson A. T., Pudney M., Trimming H., Woolven J., Bueno J. M., Chicharro J., Fernandez E., Fiandor J. M., Gargallo-Viola D., Gomez de las Heras F., Herreros E., Leon M. L. J. Med. Chem. 2008;51:2845–2852. doi: 10.1021/jm0705760. [DOI] [PubMed] [Google Scholar]
- Nilsen A., LaCrue A. N., White K. L., Forquer I. P., Cross R. M., Marfurt J., Mather M. W., Delves M. J., Shackleford D. M., Saenz F. E., Morrisey J. M., Steuten J., Mutka T., Li Y., Wirjanata G., Ryan E., Duffy S., Kelly J. X., Sebayang B. F., Zeeman A. M., Noviyanti R., Sinden R. E., Kocken C. H. M., Price R. N., Avery V. M., Angulo-Barturen I., Jimenez-Diaz M. B., Ferrer S., Herreros E., Sanz L. M., Gamo F. J., Bathurst I., Burrows J. N., Siegl P., Guy R. K., Winter R. W., Vaidya A. B., Charman S. A., Kyle D. E., Manetsch R., Riscoe M. K. Sci. Transl. Med. 2013;5:177ra137. doi: 10.1126/scitranslmed.3005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frueh L., Li Y., Mather M. W., Li Q., Pou S., Nilsen A., Winter R. W., Forquer I. P., Pershing A. M., Xie L. H., Smilkstein M. J., Caridha D., Koop D. R., Campbell R. F., Sciotti R. J., Kreishman-Deitrick M., Kelly J. X., Vesely B., Vaidya A. B., Riscoe M. K. ACS Infect. Dis. 2017;3:728–735. doi: 10.1021/acsinfecdis.7b00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miley G. P., Pou S., Winter R., Nilsen A., Li Y., Kelly J. X., Stickles A. M., Mather M. W., Forquer I. P., Pershing A. M., White K., Shackleford D., Saunders J., Chen G., Ting L. M., Kim K., Zakharov L. N., Donini C., Burrows J. N., Vaidya A. B., Charman S. A., Riscoe M. K. Antimicrob. Agents Chemother. 2015;59:5555–5560. doi: 10.1128/AAC.01183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. H., Liu R. J., Fang Z. P., Zhang J., Ding Y. Y., Tan M., Wang M., Pan W., Zhou H. C., Wang E. D. Sci. Rep. 2013;3:2475. doi: 10.1038/srep02475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock F. L., Mao W., Yaremchuk A., Tukalo M., Crepin T., Zhou H., Zhang Y. K., Hernandez V., Akama T., Baker S. J., Plattner J. J., Shapiro L., Martinis S. A., Benkovic S. J., Cusack S., Alley M. R. Science. 2007;316:1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- Jacobs R. T., Plattner J. J., Nare B., Wring S. A., Chen D., Freund Y., Gaukel E. G., Orr M. D., Perales J. B., Jenks M., Noe R. A., Sligar J. M., Zhang Y. K., Bacchi C. J., Yarlett N., Don R. Future Med. Chem. 2011;3:1259–1278. doi: 10.4155/fmc.11.80. [DOI] [PubMed] [Google Scholar]
- Zhang Y. K., Plattner J. J., Easom E. E., Jacobs R. T., Guo D., Freund Y. R., Berry P., Ciaravino V., Erve J. C. L., Rosenthal P. J., Campo B., Gamo F. J., Sanz L. M., Cao J. J. Med. Chem. 2017;60:5889–5908. doi: 10.1021/acs.jmedchem.7b00621. [DOI] [PubMed] [Google Scholar]
- Sonoiki E., Ng C. L., Lee M. C., Guo D., Zhang Y. K., Zhou Y., Alley M. R., Ahyong V., Sanz L. M., Lafuente-Monasterio M. J., Dong C., Schupp P. G., Gut J., Legac J., Cooper R. A., Gamo F. J., DeRisi J., Freund Y. R., Fidock D. A., Rosenthal P. J. Nat. Commun. 2017;8:14574. doi: 10.1038/ncomms14574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti C., Sauvain M., Lavaud C., Massiot G., Bravo J., Munoz V. J. Nat. Prod. 1998;61:1390–1393. doi: 10.1021/np9800654. [DOI] [PubMed] [Google Scholar]
- Krieg R., Jortzik E., Goetz A. A., Blandin S., Wittlin S., Elhabiri M., Rahbari M., Nuryyeva S., Voigt K., Dahse H. M., Brakhage A., Beckmann S., Quack T., Grevelding C. G., Pinkerton A. B., Schonecker B., Burrows J., Davioud-Charvet E., Rahlfs S., Becker K. Nat. Commun. 2017;8:14478. doi: 10.1038/ncomms14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennerstrom J. L., Arbe-Barnes S., Brun R., Charman S. A., Chiu F. C., Chollet J., Dong Y., Dorn A., Hunziker D., Matile H., McIntosh K., Padmanilayam M., Santo Tomas J., Scheurer C., Scorneaux B., Tang Y., Urwyler H., Wittlin S., Charman W. N. Nature. 2004;430:900–904. doi: 10.1038/nature02779. [DOI] [PubMed] [Google Scholar]
- Charman S. A., Arbe-Barnes S., Bathurst I. C., Brun R., Campbell M., Charman W. N., Chiu F. C., Chollet J., Craft J. C., Creek D. J., Dong Y., Matile H., Maurer M., Morizzi J., Nguyen T., Papastogiannidis P., Scheurer C., Shackleford D. M., Sriraghavan K., Stingelin L., Tang Y., Urwyler H., Wang X., White K. L., Wittlin S., Zhou L., Vennerstrom J. L. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4400–4405. doi: 10.1073/pnas.1015762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phyo A. P., Jittamala P., Nosten F. H., Pukrittayakamee S., Imwong M., White N. J., Duparc S., Macintyre F., Baker M., Mohrle J. J. Lancet Infect. Dis. 2016;16:61–69. doi: 10.1016/S1473-3099(15)00320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. A., Lotharius J., Marsh K., White J., Dayan A., White K. L., Njoroge J. W., El Mazouni F., Lao Y., Kokkonda S., Tomchick D. R., Deng X., Laird T., Bhatia S. N., March S., Ng C. L., Fidock D. A., Wittlin S., Lafuente-Monasterio M., Benito F. J., Alonso L. M., Martinez M. S., Jimenez-Diaz M. B., Bazaga S. F., Angulo-Barturen I., Haselden J. N., Louttit J., Cui Y., Sridhar A., Zeeman A. M., Kocken C., Sauerwein R., Dechering K., Avery V. M., Duffy S., Delves M., Sinden R., Ruecker A., Wickham K. S., Rochford R., Gahagen J., Iyer L., Riccio E., Mirsalis J., Bathhurst I., Rueckle T., Ding X., Campo B., Leroy D., Rogers M. J., Rathod P. K., Burrows J. N., Charman S. A. Sci. Transl. Med. 2015;7:296ra111. doi: 10.1126/scitranslmed.aaa6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong F. J., Li R., Jain J. P., Lefèvre G., Magnusson B., Diagana T. T., Pertel P. Antimicrob. Agents Chemother. 2014;58(10):6209–6214. doi: 10.1128/AAC.03393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N. J., Pukrittayakamee S., Phyo A. P., Rueangweerayut R., Nosten F., Jittamala P., Jeeyapant A., Jain J. P., Lefevre G., Li R., Magnusson B., Diagana T. T., Leong F. J. N. Engl. J. Med. 2014;371:403–410. doi: 10.1056/NEJMoa1315860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillman N. J., Allen R. J., McNamara C. W., Yeung B. K., Winzeler E. A., Diagana T. T., Kirk K. Cell Host Microbe. 2013;13:227–237. doi: 10.1016/j.chom.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhen K. L., Chatterjee A. K., Rottmann M., Gagaring K., Borboa R., Buenviaje J., Chen Z., Francek C., Wu T., Nagle A., Barnes S. W., Plouffe D., Lee M. C., Fidock D. A., Graumans W., van de Vegte-Bolmer M., van Gemert G. J., Wirjanata G., Sebayang B., Marfurt J., Russell B., Suwanarusk R., Price R. N., Nosten F., Tungtaeng A., Gettayacamin M., Sattabongkot J., Taylor J., Walker J. R., Tully D., Patra K. P., Flannery E. L., Vinetz J. M., Renia L., Sauerwein R. W., Winzeler E. A., Glynne R. J., Diagana T. T. Antimicrob. Agents Chemother. 2014;58:5060–5067. doi: 10.1128/AAC.02727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonte G., Lim M. Y., Wree M., Reimer C., Nachon M., Corey V., Gedeck P., Plouffe D., Du A., Figueroa N., Yeung B., Bifani P., Winzeler E. A. mBio. 2016;7(4):e00696-16. doi: 10.1128/mBio.00696-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N. J., Duong T. T., Uthaisin C., Nosten F., Phyo A. P., Hanboonkunupakarn B., Pukrittayakamee S., Jittamala P., Chuthasmit K., Cheung M. S., Feng Y., Li R., Magnusson B., Sultan M., Wieser D., Xun X., Zhao R., Diagana T. T., Pertel P., Leong F. J. N. Engl. J. Med. 2016;375:1152–1160. doi: 10.1056/NEJMoa1602250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C. J., Hill R. D., Sheppard D. W., Slater M. J., Stouten P. F. Comb. Chem. High Throughput Screening. 2011;14:521–531. doi: 10.2174/138620711795767802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis Y., Douelle F., Feng T. S., Gonzalez Cabrera D., Le Manach C., Nchinda A. T., Duffy S., White K. L., Shackleford D. M., Morizzi J., Mannila J., Katneni K., Bhamidipati R., Zabiulla K. M., Joseph J. T., Bashyam S., Waterson D., Witty M. J., Hardick D., Wittlin S., Avery V., Charman S. A., Chibale K. J. Med. Chem. 2012;55:3479–3487. doi: 10.1021/jm3001373. [DOI] [PubMed] [Google Scholar]
- Paquet T., Le Manach C., Cabrera D. G., Younis Y., Henrich P. P., Abraham T. S., Lee M. C. S., Basak R., Ghidelli-Disse S., Lafuente-Monasterio M. J., Bantscheff M., Ruecker A., Blagborough A. M., Zakutansky S. E., Zeeman A. M., White K. L., Shackleford D. M., Mannila J., Morizzi J., Scheurer C., Angulo-Barturen I., Martinez M. S., Ferrer S., Sanz L. M., Gamo F. J., Reader J., Botha M., Dechering K. J., Sauerwein R. W., Tungtaeng A., Vanachayangkul P., Lim C. S., Burrows J., Witty M. J., Marsh K. C., Bodenreider C., Rochford R., Solapure S. M., Jimenez-Diaz M. B., Wittlin S., Charman S. A., Donini C., Campo B., Birkholtz L. M., Hanson K. K., Drewes G., Kocken C. H. M., Delves M. J., Leroy D., Fidock D. A., Waterson D., Street L. J., Chibale K. Sci. Transl. Med. 2017;9:eaad9735. doi: 10.1126/scitranslmed.aad9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Diaz M. B., Ebert D., Salinas Y., Pradhan A., Lehane A. M., Myrand-Lapierre M. E., O'Loughlin K. G., Shackleford D. M., Justino de Almeida M., Carrillo A. K., Clark J. A., Dennis A. S., Diep J., Deng X., Duffy S., Endsley A. N., Fedewa G., Guiguemde W. A., Gomez M. G., Holbrook G., Horst J., Kim C. C., Liu J., Lee M. C., Matheny A., Martinez M. S., Miller G., Rodriguez-Alejandre A., Sanz L., Sigal M., Spillman N. J., Stein P. D., Wang Z., Zhu F., Waterson D., Knapp S., Shelat A., Avery V. M., Fidock D. A., Gamo F. J., Charman S. A., Mirsalis J. C., Ma H., Ferrer S., Kirk K., Angulo-Barturen I., Kyle D. E., DeRisi J. L., Floyd D. M., Guy R. K. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E5455–5462. doi: 10.1073/pnas.1414221111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C., Verma V. P., Naikade N. K., Singh A. S., Hassam M., Puri S. K. J. Med. Chem. 2008;51:7581–7592. doi: 10.1021/jm801006v. [DOI] [PubMed] [Google Scholar]
- Shafiq N., Rajagopalan S., Kushwaha H. N., Mittal N., Chandurkar N., Bhalla A., Kaur S., Pandhi P., Puri G. D., Achuthan S., Pareek A., Singh S. K., Srivastava J. S., Gaur S. P., Malhotra S. Malar. Res. Treat. 2014;2014:372521. doi: 10.1155/2014/372521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De D., Krogstad F. M., Cogswell F. B., Krogstad D. J. Am. J. Trop. Med. Hyg. 1996;55:579–583. doi: 10.4269/ajtmh.1996.55.579. [DOI] [PubMed] [Google Scholar]
- De D., Krogstad F. M., Byers L. D., Krogstad D. J. J. Med. Chem. 1998;41:4918–4926. doi: 10.1021/jm980146x. [DOI] [PubMed] [Google Scholar]
- Mzayek F., Deng H., Mather F. J., Wasilevich E. C., Liu H., Hadi C. M., Chansolme D. H., Murphy H. A., Melek B. H., Tenaglia A. N., Mushatt D. M., Dreisbach A. W., Lertora J. J., Krogstad D. J. PLoS Clin. Trials. 2007;2:e6. doi: 10.1371/journal.pctr.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koita O. A., Sangare L., Miller H. D., Sissako A., Coulibaly M., Thompson T. A., Fongoro S., Diarra Y., Ba M., Maiga A., Diallo B., Mushatt D. M., Mather F. J., Shaffer J. G., Anwar A. H., Krogstad D. J. Lancet Infect. Dis. 2017;17:1266–1275. doi: 10.1016/S1473-3099(17)30365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuthavong Y., Tarnchompoo B., Vilaivan T., Chitnumsub P., Kamchonwongpaisan S., Charman S. A., McLennan D. N., White K. L., Vivas L., Bongard E., Thongphanchang C., Taweechai S., Vanichtanankul J., Rattanajak R., Arwon U., Fantauzzi P., Yuvaniyama J., Charman W. N., Matthews D. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16823–16828. doi: 10.1073/pnas.1204556109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin M. L., Calas M., Bompart J., Cordina G., Martin D., Ben Bari M., Jei T., Druilhe P., Vial H. J. Blood. 1998;91:1426–1437. [PubMed] [Google Scholar]
- Calas M., Cordina G., Bompart J., Ben Bari M., Jei T., Ancelin M. L., Vial H. J. Med. Chem. 1997;40:3557–3566. doi: 10.1021/jm9701886. [DOI] [PubMed] [Google Scholar]
- Hamze A., Rubi E., Arnal P., Boisbrun M., Carcel C., Salom-Roig X., Maynadier M., Wein S., Vial H., Calas M. J. Med. Chem. 2005;48:3639–3643. doi: 10.1021/jm0492608. [DOI] [PubMed] [Google Scholar]
- Vial H. J., Wein S., Farenc C., Kocken C., Nicolas O., Ancelin M. L., Bressolle F., Thomas A., Calas M. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15458–15463. doi: 10.1073/pnas.0404037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., Paulsen I. T., James K., Eisen J. A., Rutherford K., Salzberg S. L., Craig A., Kyes S., Chan M. S., Nene V., Shallom S. J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M. W., Vaidya A. B., Martin D. M., Fairlamb A. H., Fraunholz M. J., Roos D. S., Ralph S. A., McFadden G. I., Cummings L. M., Subramanian G. M., Mungall C., Venter J. C., Carucci D. J., Hoffman S. L., Newbold C., Davis R. W., Fraser C. M., Barrell B. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J. M., Adams J. H., Silva J. C., Bidwell S. L., Lorenzi H., Caler E., Crabtree J., Angiuoli S. V., Merino E. F., Amedeo P., Cheng Q., Coulson R. M., Crabb B. S., Del Portillo H. A., Essien K., Feldblyum T. V., Fernandez-Becerra C., Gilson P. R., Gueye A. H., Guo X., Kang'a S., Kooij T. W., Korsinczky M., Meyer E. V., Nene V., Paulsen I., White O., Ralph S. A., Ren Q., Sargeant T. J., Salzberg S. L., Stoeckert C. J., Sullivan S. A., Yamamoto M. M., Hoffman S. L., Wortman J. R., Gardner M. J., Galinski M. R., Barnwell J. W., Fraser-Liggett C. M. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge G. G., Bohme U., Sanders M., Reid A. J., Cotton J. A., Maiga-Ascofare O., Djimde A. A., Apinjoh T. O., Amenga-Etego L., Manske M., Barnwell J. W., Renaud F., Ollomo B., Prugnolle F., Anstey N. M., Auburn S., Price R. N., McCarthy J. S., Kwiatkowski D. P., Newbold C. I., Berriman M., Otto T. D. Nature. 2017;542:101–104. doi: 10.1038/nature21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. H., Miller B. A., Nair S., Nkhoma S., Tan A., Tan J. C., Al Saai S., Phyo A. P., Moo C. L., Lwin K. M., McGready R., Ashley E., Imwong M., Stepniewska K., Yi P., Dondorp A. M., Mayxay M., Newton P. N., White N. J., Nosten F., Ferdig M. T., Anderson T. J. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S., Clark T. G., Jacob C. G., Cummings M. P., Miotto O., Dondorp A. M., Fukuda M. M., Nosten F., Noedl H., Imwong M., Bethell D., Se Y., Lon C., Tyner S. D., Saunders D. L., Socheat D., Ariey F., Phyo A. P., Starzengruber P., Fuehrer H. P., Swoboda P., Stepniewska K., Flegg J., Arze C., Cerqueira G. C., Silva J. C., Ricklefs S. M., Porcella S. F., Stephens R. M., Adams M., Kenefic L. J., Campino S., Auburn S., MacInnis B., Kwiatkowski D. P., Su X. Z., White N. J., Ringwald P., Plowe C. V. Proc. Natl. Acad. Sci. U. S. A. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A. C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C. M., Bout D. M., Menard S., Rogers W. O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J. C., Fairhurst R. M., Benoit-Vical F., Mercereau-Puijalon O., Menard D. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cabrera M., Yang J., Yuan L., Gupta B., Liang X., Kemirembe K., Shrestha S., Brashear A., Li X., Porcella S. F., Miao J., Yang Z., Su X. Z., Cui L. Sci. Rep. 2016;6:33891. doi: 10.1038/srep33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D., Khim N., Beghain J., Adegnika A. A., Shafiul-Alam M., Amodu O., Rahim-Awab G., Barnadas C., Berry A., Boum Y., Bustos M. D., Cao J., Chen J. H., Collet L., Cui L., Thakur G. D., Dieye A., Djalle D., Dorkenoo M. A., Eboumbou-Moukoko C. E., Espino F. E., Fandeur T., Ferreira-da-Cruz M. F., Fola A. A., Fuehrer H. P., Hassan A. M., Herrera S., Hongvanthong B., Houze S., Ibrahim M. L., Jahirul-Karim M., Jiang L., Kano S., Ali-Khan W., Khanthavong M., Kremsner P. G., Lacerda M., Leang R., Leelawong M., Li M., Lin K., Mazarati J. B., Menard S., Morlais I., Muhindo-Mavoko H., Musset L., Na-Bangchang K., Nambozi M., Niare K., Noedl H., Ouedraogo J. B., Pillai D. R., Pradines B., Quang-Phuc B., Ramharter M., Randrianarivelojosia M., Sattabongkot J., Sheikh-Omar A., Silue K. D., Sirima S. B., Sutherland C., Syafruddin D., Tahar R., Tang L. H., Toure O. A., Tshibangu-wa-Tshibangu P., Vigan-Womas I., Warsame M., Wini L., Zakeri S., Kim S., Eam R., Berne L., Khean C., Chy S., Ken M., Loch K., Canier L., Duru V., Legrand E., Barale J. C., Stokes B., Straimer J., Witkowski B., Fidock D. A., Rogier C., Ringwald P., Ariey F., Mercereau-Puijalon O. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford E. D., Quan J., Horst J. A., Ebert D., Wu W., DeRisi J. L. PLoS One. 2017;12:e0178163. doi: 10.1371/journal.pone.0178163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P., Equinet L., Packer J., Doerig C. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A., Balaji S. N., Sahasrabuddhe A. A., Trivedi V. Infect. Disord.: Drug Targets. 2016;16:121–134. doi: 10.2174/1871526516666160506150651. [DOI] [PubMed] [Google Scholar]
- Wright K. E., Hjerrild K. A., Bartlett J., Douglas A. D., Jin J., Brown R. E., Illingworth J. J., Ashfield R., Clemmensen S. B., de Jongh W. A., Draper S. J., Higgins M. K. Nature. 2014;515:427–430. doi: 10.1038/nature13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. P., Srivastava D., Mishra B. N. 3 Biotech. 2017;7:318. doi: 10.1007/s13205-017-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazier D., Renia L., Snounou G. Nat. Rev. Drug Discovery. 2009;8:854–864. doi: 10.1038/nrd2960. [DOI] [PubMed] [Google Scholar]
- Vaughan A. M., Mikolajczak S. A., Wilson E. M., Grompe M., Kaushansky A., Camargo N., Bial J., Ploss A., Kappe S. H. J. Clin. Invest. 2012;122:3618–3628. doi: 10.1172/JCI62684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery E. L., Foquet L., Chuenchob V., Fishbaugher M., Billman Z., Navarro M. J., Betz W., Olsen T. M., Lee J., Camargo N., Nguyen T., Schafer C., Sack B. K., Wilson E. M., Saunders J., Bial J., Campo B., Charman S. A., Murphy S. C., Phillips M. A., Kappe S. H., Mikolajczak S. A. JCI Insight. 2018;3(1):e92587. doi: 10.1172/jci.insight.92587. [DOI] [PMC free article] [PubMed] [Google Scholar]