Abstract
Mice lacking perilipin-2 (Plin2-null) are resistant to obesity, insulin resistance, and fatty liver induced by Western or high-fat diets. In the current study, we found that, compared with WT mice on Western diet, Plin2-null adipose tissue was more insulin sensitive and inguinal subcutaneous white adipose tissue (iWAT) exhibited profound browning and robust induction of thermogenic and carbohydrate-responsive genetic programs at room temperature. Surprisingly, these Plin2-null responses correlated with the content of simple carbohydrates, rather than fat, in the diet, and were independent of adipose Plin2 expression. To define Plin2 and sugar effects on adipose browning, WT and Plin2-null mice were placed on chow diets containing 20% sucrose in their drinking water for 6 weeks. Compared with WT mice, iWAT of Plin2-null mice exhibited pronounced browning and striking increases in the expression of thermogenic and insulin-responsive genes on this diet. Significantly, Plin2-null iWAT browning was associated with reduced sucrose intake and elevated serum fibroblast growth factor (FGF)21 levels, which correlated with greatly enhanced hepatic FGF21 production. These data identify Plin2 actions as novel mediators of sugar-induced adipose browning through indirect effects of hepatic FGF21 expression, and suggest that adipose browning mechanisms may contribute to Plin2-null resistance to obesity.
Keywords: lipid droplets, obesity, beige adipose tissue, adipocytes, nutrition/carbohydrate, insulin resistance, fatty acid, mass spectrometry, inguinal subcutaneous white adipose tissue
The obesity epidemic is recognized as one of the major public health challenges facing modern society, with 66% of adults in the United States being either overweight or obese (1). Health problems associated with obesity include insulin resistance and risk of diabetes development, fatty liver formation, alterations in circulating lipid profiles, cardiovascular disease, hypertension, and increased risk of certain cancers (2–5). As such, enormous effort has been put into understanding the molecular causes of obesity and identifying potential treatments (6). Traditional therapeutic approaches, such as dietary and lifestyle changes, have proven largely ineffective in the primary care setting, with the majority of patients unable to achieve statistically significant long-term weight loss (7). Additionally, many pharmacological agents developed to combat obesity have demonstrated unwanted side effects and toxicity (8, 9). Thus, the development of novel approaches to treat obesity is of critical importance.
In the last decade, the lipid droplet coat protein family of perilipins (PLINs) has been recognized as the physiological regulator of lipid accumulation in many tissues under both healthy and pathophysiological states (10). Five mammalian PLIN family members have been identified (PLIN1 through PLIN5), with differential tissue expression of the members under both healthy and disease states. PLIN2 is a constitutively associated intracellular lipid droplet coat protein expressed in many organs, including the liver, adipose tissue, and skeletal muscle (11). Our laboratory has previously generated mice with whole-body knockout of Plin2 (Plin2-null). These animals are resistant to obesity, adipose tissue inflammation, insulin resistance, and liver steatosis (nonalcoholic fatty liver disease) when chronically exposed to a Western diet (WD) or a high-fat (60%) diet (HFD) (12, 13). Additionally, others have shown that treatment of WT mice on a HFD with antisense oligonucleotides against Plin2 mRNA leads to a significant reduction in adiposity, improvement in liver steatosis, and attenuation of insulin resistance (14–16). Because loss of PLIN2 has beneficial effects on obesity development in both genetic and pharmacological intervention models, this protein appears to be a critical regulator of the physiological responses to diet that promote metabolic dysregulation, weight gain, and increased adiposity. Consequently, PLIN2 represents a potentially valuable therapeutic target for the treatment and prevention of metabolic disease. However, the mechanism by which loss of PLIN2 reduces adiposity is currently unknown.
Brown adipose tissue (BAT) has been recognized as an important modulator of metabolic parameters in both humans and rodents (17–19). Unlike white adipose tissue (WAT), BAT is composed of specialized heat-generating adipocytes that express a program of genes that serve to increase energy expenditure by fluxing substrates through uncoupled mitochondrial oxidation. This is accomplished primarily through increased expression of PPARγ coactivator (PGC)1α (involved in mitochondrial biogenesis) and uncoupling protein 1 (UCP1) (an uncoupling protein that allows for mitochondrial heat output instead of ATP production) (20). Increased BAT activity has been shown to lower circulating triglycerides, improve glucose tolerance, and decrease adiposity (21, 22). During the last few decades, it has been shown that WAT can undergo a white-to-brown conversion under various conditions in rodents (23). During this process, known as browning (or beiging), unilocular white adipocytes convert to multilocular cells that express the same thermogenic gene program as brown adipocytes. The net effect of this is an increase in energy expenditure and heat production in white adipose depots (24). Browning of WAT has been shown to have beneficial effects on obesity and other metabolic parameters similar to those observed with BAT activation (17).
To date, a variety of mechanisms have been shown to induce white-to-brown conversion of WAT. The first fully defined mechanism involved browning of WAT in response to cold exposure (25). Similar to activation of BAT, cold exposure leads to a sympathetic nervous system release of norepinephrine into WAT depots, which causes a white-to-brown phenotypic switch. Mammals are able to use this mechanism to increase nonshivering thermogenesis and heat output during cold stress. Additionally, other mechanisms for browning of WAT have since been described, including a variety of endogenous hormones. Irisin, a hormone primarily secreted by skeletal muscle, has powerful effects on WAT browning and is currently the subject of therapeutic interest (26). Fibroblast growth factor (FGF)19 (and its rodent homolog FGF15) are hormones secreted from the intestine and have also been shown to promote browning of subcutaneous WAT (27). Next among the most potent hormones regulating WAT browning is FGF21 (28–31). FGF21 is secreted at high levels by the liver during fasting and carbohydrate feeding, although it has also been shown that FGF21 produced within adipose tissue itself is able to induce WAT browning by autocrine/paracrine mechanisms (32–34). Finally, carbohydrate response element binding protein (ChREBP) has recently been shown to exert powerful effects on white-to-brown adipose conversion (35, 36). ChREBP is a protein that is responsive to carbohydrates and controls transcription of a suite of lipogenic genes, including FASN and acetyl-CoA carboxylase (ACC). Transgenic increase of ChREBP in adipose tissue greatly enhances insulin sensitivity, PPAR activity, and transcription of the thermogenic program.
Here, we identify PLIN2 as a novel determinant of inguinal subcutaneous WAT (iWAT) browning at ambient temperature. Mice lacking whole-body Plin2 exhibit drastic differences in subcutaneous iWAT histology and thermogenic program gene/protein expression compared with WT animals. Remarkably, the degree of browning in subcutaneous iWAT in Plin2-null mice is strongly influenced by the diet these animals are fed, with diets high in simple carbohydrates evoking the strongest iWAT browning responses. Significantly, our data implicate liver-derived FGF21 in the mechanism of Plin2 regulation of these diet-induced browning effects and possibly effects of Plin2 on caloric intake. Taken together, our data provide critical insights into the molecular mechanisms of obesity resistance in Plin2-null animals.
MATERIALS AND METHODS
Animal housing and feeding
All animal protocols were approved by the IACUC of the University of Colorado Denver. Plin2-null mice were generated as previously described (12). Adipose-specific Plin2 knockout mice were generated by crossing the Plin2-floxed mouse with one expressing Cre recombinase driven by the FABP4 promoter [B6.Cg-Tg(Fabp4-cre)1Rev/J]. Mice were housed in the University of Colorado Anschutz Medical Campus animal facility at ambient temperatures (22–24°C) on a 10:14 h light-dark cycle. Feeding was ad libitum with free access to food and water. For the 30 week feeding studies, mice were fed standard chow from weaning until 8 weeks, after which they were fed either control diet (CD) (Teklad TD.08485), WD (Teklad TD.88137), HFD (Research Diets D12492), low-fat diet (LFD) (Research Diets D12450B), or standard chow (Teklad 2920X) for 30 weeks. For the sucrose supplementation study, animals were raised on standard chow until 8 weeks of age. Mice were then split into two groups and given either water or 20% sucrose in water bottles for 6 weeks with free access to chow. Liquid intake, food intake, and body weights (BWs) were monitored weekly. Upon completion of the feeding experiments, mice were fasted for 4 h and euthanized by CO2 exposure and cardiac puncture.
Histological analysis
Samples of freshly excised adipose tissue were fixed for 24–36 h in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with H&E as described previously (12). Processing of histological samples was performed by the Pathology Core at the University of Colorado Denver Anschutz Medical Campus.
Gene expression analysis
RNA was extracted from snap-frozen tissue using a modified phenol-chloroform method. Approximately 50–75 mg of tissue were homogenized in 1 ml of ice-cold QIAzol® (Qiagen) using bead homogenization with two rounds of shaking at 30 Hz for 2 min each. To maximize RNA yield, homogenates were quantitatively transferred to Phase Lock Gel Heavy tubes (Quantabio Inc.), 400 μl of chloroform were added, and samples were shaken for 30 s. Samples were then centrifuged at 21,000 g for 15 min at 4°C. The RNA-containing aqueous layer above the gel was transferred to a new tube, mixed 1:1 with Buffer RLT Plus (Qiagen), mixed with 1 ml 70% ethanol, and subjected to RNA cleanup using the RNeasy Plus Mini kit (Qiagen). Genomic DNA was removed by on-column DNase digestion. RNA quality was assessed by the Genomics and Microarray Core Facility at the University of Colorado Denver Anschutz Medical Campus. cDNA was synthesized from 900 ng RNA using the iScript cDNA synthesis kit (Bio-Rad). Quantitative (q)RT-PCR was performed on a Bio-Rad CFX96 instrument using SYBR green probes and melting curve analysis to ensure single-product amplification. The qRT-PCR master mix was 2× SYBR Green qPCR Mastermix (BioTool). All gene expression was normalized to 18S rRNA, and relative expression levels were calculated using the ΔΔCt method. All primer sequences used for qRT-PCR can be found in the supplemental materials (supplemental Table S1).
Western blot analysis
Protein extraction was performed by homogenizing ∼100 mg of frozen tissue directly into 2:1 chloroform:methanol in a bead homogenizer for two cycles at 30 Hz for 2 min each. The insoluble protein pellet was spun down, washed with an additional 2 ml of 2:1 chloroform:methanol to remove residual lipids, washed twice with ice-cold methanol, briefly dried, and resuspended in 10% SDS containing protease and phosphatase inhibitors (Thermo Pierce). Protein was quantified with the BCA assay (Thermo Pierce) with standards prepared in 10% SDS. For Western blotting, 37.5 μg of protein were separated on 4–20% Tris-glycine gels (Bio-Rad) and transferred to 0.2 μM nitrocellulose membranes (Bio-Rad) at 100 V for 1 h and 10 min. All membranes were blocked with 5% BSA in Tris-buffered saline containing 0.1% Tween-20, pH 7.3. All antibodies and dilutions used for Western blotting can be found in the supplemental materials (supplemental Table S2). Immunoreactive bands were detected with corresponding labeled secondary antibodies (Li-COR) and quantified by imaging on a Li-COR CLx instrument. Protein intensities were normalized to α-tubulin for quantitative analysis.
Adipose tissue lipid analysis
Approximately 10 mg of frozen adipose tissue were homogenized in 1 ml 2:1 chloroform:methanol containing 300 μg of tritridecanoin reference standard (Nu-Check Prep Inc.) using bead homogenization with two cycles at 30 Hz for 2 min each cycle. Homogenates were diluted into a total of 4 ml 2:1 chloroform:methanol in glass tubes, mixed with 800 μl of 0.9% sodium chloride, vortexed, and centrifuged at 3,250 g for 5 min. The lower phase was removed and dried under N2 gas. Total lipids were resuspended in 330 ul 100% chloroform and applied to HyperSep SI SPE columns (Thermo Scientific) preequilibrated with 15 column volumes of chloroform. Neutral lipid (NL) was eluted with 3 ml of chloroform followed by 5 ml of acetone:methanol (9:1) to elute glycolipids. Phospholipids (PLs) were then eluted with 3 ml of 100% methanol. All lipid fractions were dried under N2 gas and resuspended in 1 ml of methanol containing 2.5% H2SO4. Fatty acid methyl ester (FAME) production was initiated by heating at 80°C for 1.5 h with occasional vortexing. One milliliter of HPLC-grade water was added to quench the reactions, and FAMEs were extracted in 400 μl of hexane by vortexing. A Trace 1310 gas chromatograph with a TG-5MS column (Thermo Scientific) was used to separate FAMEs chromatographically, and lipids were analyzed with an ISQ single quadrupole mass spectrometer (Thermo Scientific). For each lipid species analyzed, serial dilutions were performed to ensure that peak areas were within linear range. Xcalibur software (Thermo Scientific) was used to calculate peak areas. Areas were normalized to the reference standard and then to total protein.
Serum FFA analysis
Mice were fasted for 4 h starting at 8:00 AM and then split into two groups, with one receiving saline and the other receiving insulin (0.5 U/kg BW) via intraperitoneal injection. The insulin used was Humulin N® (Eli Lilly). Blood was taken 20 min postinjection, and FFA was measured calorimetrically in serum using the Serum/Plasma FFA Detection kit (ZenBio Inc., product number SFA-1).
Serum FGF21 analysis
Serum FGF21 was assessed with the mouse FGF21 ELISA kit (Abcam, product number ab212160). Pilot dilutions were performed to ensure that samples were within the range of standards, and development of TMB was monitored kinetically at 600 nM in 20 s intervals for 20 min on a spectrophotometer equipped with shaking capabilities (Molecular Devices). Data were analyzed using a four-parameter curve fit with SoftMax Pro software (Molecular Devices).
Statistics
All data are presented as mean ± SEM and were analyzed by two-way ANOVA unless otherwise indicated. Two-way ANOVA with post hoc Tukey’s multiple comparison testing was performed with GraphPad Prism software. Three-way ANOVA was performed with STATA v15 software. For lipidomic analysis under individual dietary conditions, Student’s t-test was performed with Bonferroni correction using GraphPad Prism. P < 0.05 was considered to be statistically significant.
RESULTS
PLIN2 deletion induces profound histological browning of iWAT at ambient temperature
WT and Plin2-null mice were placed on either a WD containing 42% kcal from fat, 42.7% kcal from carbohydrate, 15.2% kcal from protein, and 0.2% cholesterol or a matched CD containing 13% kcal from fat, 67.9% kcal from carbohydrate, 19.1% kcal from protein, and no added cholesterol for 30 weeks at ambient temperature. We previously reported that WD-fed Plin2-null mice are completely protected from the obesity, glucose intolerance, and nonalcoholic fatty liver formation found in WT mice on this diet (13). In this study, we investigated the molecular differences that underlie the resistance of Plin2-null animals to obesity. We first performed histological examination of various adipose tissues of WT and Plin2-null mice, including interscapular BAT (iBAT), iWAT, and epididymal (perigonadal) WAT (eWAT). As expected, iBAT from both WT and Plin2-null mice exhibited classic widespread and homogenous cytoplasmic lipid droplet multilocularity on both diets (Fig. 1A), although WT mice appeared to have larger lipid droplets in their iBAT on WD compared with CD. No major differences were observed in the iBAT of Plin2-null mice on either diet. Next, we examined the iWAT of WT and Plin2-null mice on these diets (Fig. 1B). WT mice demonstrated classic iWAT, with unilocular adipocytes that were slightly larger on WD compared with CD. Remarkably, iWAT from Plin2-null mice fed either WD or CD very closely resembled the characteristics of iBAT, with primarily small multilocular adipocytes dominating the specimens. These multilocular droplets were homogenous throughout the bulk of the tissue, suggesting that the response was widespread. Interestingly, lipid droplets in iWAT from WD-fed Plin2-null mice appeared to be slightly smaller than those from CD-fed animals. Overall, the histological appearance and multilocularity of iWAT from Plin2-null mice at ambient temperatures was reminiscent of the browning response in iWAT normally observed in mice exposed to cold temperatures (37). Finally, we examined eWAT from WT and Plin2-null mice on WD and CD (Fig. 1C). Compared with CD-fed mice, eWAT from WT mice fed the WD exhibited larger adipocytes and the presence of crown-like structures indicative of inflammation (38). Conversely, Plin2-null mice showed much smaller adipocytes on both diets compared with WT, and no crown-like structures were observed in these mice. Importantly, we did not observe multilocular adipocytes or browning in eWAT from either genotype.
Fig. 1.
Plin2-null mice exhibit differences in histological beiging and PLIN family member gene expression in their iWAT compared with WT mice. A–C: H&E staining of adipose tissues from WT and Plin2-null mice on 30 weeks of CD and WD. The depots shown include iBAT, iWAT, and eWAT. iBAT and iWAT are shown at 400× in order to clearly see the multilocularity of cytoplasmic lipid droplets, while eWAT is shown at 200×. Plin2-null mice exhibit beige adipose tissue on both CD and WD compared with WT mice. The scale bar indicates 100 microns. D–F: PLIN family member gene expression in iBAT, iWAT, and eWAT from WT and Plin2-null mice on CD and WD. Plin2 was not detected in Plin2-null adipose tissue. iWAT from Plin2-null mice exhibits statistically significant reductions in Plin4 on both diets, while Plin5 is increased on both diets. Plin5 expression is significantly higher on WD compared with CD in Plin2-null mice. Data are expressed as mean ± SEM; statistically significant comparisons are indicated by a solid bold line (P < 0.05, two-way ANOVA with post hoc Tukey’s multiple comparisons test); n = 4 for WT WD, WT CD, and Plin2-null WD; n = 3 for Plin2-null CD.
We also performed mass analysis of iBAT, iWAT, and eWAT fat pads from Plin2-null and WT mice on long-term WD and CD. We previously reported quantitative MRI data from these animals (13). Consistent with previous data that showed that the reduction in BW in Plin2-null mice is due to reduced fat mass rather than lean mass, we found statistically significant reductions in all fat pad masses in Plin2-null mice on WD compared with WT counterparts (supplemental Fig. S1A). For Plin2-null mice on WD, iBAT was reduced 85% (P < 0.0001) and iWAT was reduced 78% (P < 0.0001). Additionally, we found significant mass reductions in iWAT and eWAT in Plin2-null mice on CD compared with WT mice on that diet, with a 45% reduction in iWAT mass (P = 0.0453) and a 61% reduction in eWAT mass (P = 0.0275). Fat pad masses normalized to BW are also provided in supplemental Fig. S1B.
Effects of diet and Plin2 deletion on PLIN family member gene expression in iWAT
Next, we were interested in whether loss of whole-body PLIN2 imparted changes in PLIN family member expression in adipose tissue. As expected, we did not detect Plin2 transcripts by qRT-PCR in any adipose tissues from Plin2-null mice. In iBAT, we observed significant dietary modulation of Plin3, but not in other PLIN family members, in WT and Plin2-null mice (Fig. 1D). Compared with WD fed animals, Plin3 transcript levels were increased ∼5-fold (P < 0.05) in both WT and Plin2-null mice fed the CD. However, loss of Plin2 did not significantly affect iBAT expression of other PLIN family members on either diet (Fig. 1D). In iWAT (Fig. 1E), we did not detect genotype or diet effects on Plin1 or Plin3 expression. However, in WD-fed WT mice, mRNA for Plin2 and Plin4 was significantly increased 1.5-fold (P = 0.0038) and ∼2-fold (P = 0.0447), respectively, over their values in CD-fed mice. In addition, we found that loss of Plin2 significantly reduced Plin4 expression by 75% (P = 0.0012) in WD-fed mice, and resulted in a 50% decrease in Plin4 in CD-fed animals that did not reach significance (P = 0.18, two-way ANOVA). Interestingly, we found that Plin2 deletion led to a profound statistically significant 15-fold increase (P < 0.0001) in Plin5 mRNA in WD-fed mice, and a 5-fold increase in Plin5 in CD-fed mice that did not reach significance by two-way ANOVA (P = 0.134). Importantly, we also noticed significant dietary modulation of Plin5 expression in Plin2-null mice, with Plin5 mRNA expression being 3-fold (P = 0.0011) higher in the iWAT of Plin2-null mice on WD compared with CD. In eWAT (Fig. 1F), we observed dietary modulation of Plin2 expression in WT mice, with levels of this gene experiencing a 2-fold (P = 0.0051) increase on WD compared with CD. However, the expression of other PLIN family members was not affected by diet or Plin2 deletion.
Plin2 deletion promotes expression of thermogenic genes and markers of beige and brown adipocytes in iWAT
Because we observed histological differences that suggested profound iWAT browning in Plin2-null mice, we investigated PLIN2- and diet-mediated effects on thermogenic program gene expression in iBAT, iWAT, and eWAT. We first examined Ucp1, elongase of very long-chain fatty acid protein 3 (Elovl3), cell death-inducing DNA fragmentation factor-α-like effector α (CideA), type II iodothyronine deiodinase (Dio2), carnitine palmitoyltransferase (Cpt)1a, Cpt1b, and Pgc1α mRNA expression in iBAT. These genes are commonly associated with the thermogenic program and are activated during cold exposure, β3-adrenergic agonism, and other reported browning mechanisms (39–41). In iBAT, we noticed trending increases in Ucp1, CideA, Dio2, and Cpt1b in Plin2-null mice on both diets. However, these responses did not reach statistical significance (Fig. 2A). The use of a log-scale should be noted for the results in Fig. 2. Compared with WT mice, loss of Plin2 was associated with significant increases in transcript levels for Elovl3 of ∼5-fold (P = 0.0144) and 5.7-fold (P = 0.0309) for WD- and CD-fed animals, respectively, and ∼3-fold for Pgc1α for animals on WD (P = 0.0048) and CD (P = 0.0112). We did not observe statistically significant dietary modulation of these genes in iBAT from WT or Plin2-null animals. We next analyzed the expression of Fgf21, Pparα, Pparγ2, and Lpl, which are implicated in the modulation of adipose thermogenic responses (42, 43). Intriguingly, Fgf21 mRNA expression in iBAT, which has been shown to be upregulated by cold exposure and adrenergic stimulation to induce PGC1α (29, 44), increased 27-fold (P = 0.0035) and 32-fold (P = 0.0237), respectively, with loss of Plin2 in WD- and CD-fed mice. However, we did not observe any differences across diet or genotype in mRNA expression of Pparα, Pparγ2, and Lpl.
Fig. 2.
iWAT from Plin2-null mice exhibits greatly enhanced thermogenic program gene expression and markers of beige and brown adipocytes. A: Thermogenic program mRNA expression in iBAT from WT and Plin2-null mice on 30 week CD and WD. All genes were normalized to 18S expression. Ucp1, Elovl3, CideA, Dio2, Cpt1b, and Pgc1α are classic thermogenic program members. Also shown are mRNA expression levels for Pgc1β, Fgf21, Pparα, Pparγ2, and Lpl. B: Thermogenic gene expression for iWAT from WT and Plin2-null mice on CD and WD. C: Thermogenic program expression in eWAT from WT and Plin2-null mice on both diets. Note the use of log scales in panels A–C. D–F: Gene expression for markers of white (Asc1), beige (Pat2), and brown (P2rx5) adipocytes in iBAT, iWAT, and eWAT from WT and Plin2-null mice on CD and WD. Data are expressed as mean ± SEM; statistically significant comparisons are indicated by a solid bold line (P < 0.05, two-way ANOVA with post hoc Tukey’s multiple comparisons test); n = 4 for WT WD, WT CD, and Plin2-null WD; n = 3 for Plin2-null CD.
In contrast to iBAT, we observed profound increases in almost all thermogenic program genes in the iWAT of Plin2-null animals on WD (Fig. 2B). Again, the use of log-scale should be noted for this figure. For mice on the WD, PLIN2 deletion resulted in profound increases in mRNA expression of 12,000-fold (P = 0.003) for Ucp1, 564-fold (P < 0.0001) for Elovl3, 116-fold (P = 0.0002) for CideA, 157-fold (P = 0.0003) for Dio2, 60-fold (P = 0.0018) for Cpt1b, and 6.8-fold (P < 0.0001) for Pgc1α. In addition, loss of PLIN2 increased mRNA expression 10-fold (P = 0.0001) for Pgc1β, 20-fold (P = 0.0035) for Fgf21, and 22-fold (P < 0.0001) for Pparα for WD-fed animals. However, contrary to expectations, PLIN2 deletion was associated with a 50% (P = 0.0074) decrease in Cpt1a transcript levels for WD-fed animals. We observed no statistical differences in Pparγ2 or Lpl expression across diet or genotype. For CD-fed mice, Plin2 deletion was associated with large trending increases in thermogenic program genes that did not reach significance by two-way ANOVA. Unexpectedly, we observed powerful dietary modulation of iWAT thermogenic program genes in Plin2-null mice (Fig. 2B). Compared with Plin2-null mice on CD, Plin2-null animals on WD experienced statistically significant increases in mRNA expression for Ucp1 (8-fold, P = 0.0018), Elovl3 (8-fold, P = 0.0007), CideA (4.4-fold, P = 0.0025), Dio2 (5.6-fold, P = 0.0022), Cpt1b (6.7-fold, P = 0.0089), Pgc1α (2.2-fold, P = 0.0037), Fgf21 (8.4-fold, P = 0.0101), and Pparα (3.3-fold, P = 0.0008). Finally, in eWAT, many thermogenic program genes, including Ucp1, Elvol3, CideA, and Dio2, trended higher in Plin2-null animals on both diets, but none reached significance (Fig. 2C).
Consistent with histological and thermogenic gene expression evidence of iWAT browning under ambient temperature conditions in Plin2-null mice, we found that loss of PLIN2 significantly increased transcript levels of proton/amino acid cotransporter 2 (Pat2) (2-fold, P = 0.0026) and purinergic receptor P2X ligand-gated ion channel 5 (P2rx5) (2.5-fold, P = 0.0032), which are well-validated markers of beige and brown adipose, respectively (45–49), in WD-fed animals (Fig. 2E). In addition, we found that PLIN2 deletion produced significant corresponding decreases (50%, P = 0.0477) in mRNA levels of the white adipose marker, Asc-type amino acid transporter 1 (Asc1) (48), in WD-fed mice (Fig. 2E). In CD-fed mice, Plin2 deletion also significantly decreased Asc1 mRNA (50%, P = 0.0477) and produced a modest nonsignificant 1.5-fold increase in the levels of P2rx5 mRNA. However, under these conditions, iWAT mRNA levels of Pat2 were not affected by Plin2 loss (Fig. 2E). Importantly, we also observed dietary modulation of iWAT Pat2 mRNA levels in Plin2-null mice, with Pat2 expression being ∼2-fold (P = 0.0079) higher on WD compared with CD. P2rx5 experienced a trending dietary modulation in Plin2-null mice, with a 1.6-fold increase on WD compared with CD, although this difference did not reach significance (P = 0.07). iWAT levels of Asc1 mRNA were not affected by diet in Plin2-null mice (Fig. 2C). We did not observe differences in mRNA expression for these markers in iBAT or eWAT across diets or genotypes (Fig. 2D, F). Collectively, these data identify a role for PLIN2 in regulating aspects of the thermogenic program in BAT, as well as activation of the thermogenic program and white-to-beige conversion of mouse iWAT at ambient temperatures.
Plin2-null mice demonstrate evidence of iWAT browning at the protein and lipid metabolism levels
To further confirm histological and gene expression evidence of iWAT browning in Plin2-null mice and establish functional changes associated with this response, we quantified the effects of PLIN2 deletion on protein levels of several thermogenic program-related genes in CD- and WD-fed mice at ambient temperatures by Western blotting (Fig. 3A, B). UCP1 protein was found to be significantly upregulated in Plin2-null compared with WT mice on both diets. On WD, iWAT UCP1 levels increased 81-fold (P < 0.0001) in Plin2-null compared with WT, and 9.3-fold in Plin2-null compared with WT animals on CD (P = 0.0003) (Fig. 3C). However, we did not observe dietary modulation of UCP1 in Plin2-null mice at the protein level. For iWAT CIDEA, we observed a 32-fold (P < 0.0001) increase in Plin2-null compared with WT animals on WD and an ∼4-fold (P = 0.0168) increase in iWAT CIDEA in Plin2-null compared with WT mice on CD. Unlike UCP1, we found significant dietary modulation of CIDEA protein in the iWAT of Plin2-null mice, with a 2-fold (P = 0.002) increase for Plin2-null mice on WD compared with CD. We only detected miniscule levels of CPT1B protein in WT iWAT on both diets. For animals on WD, loss of Plin2 was associated with a 235-fold increase in CPT1B (P = 0.0007). Although Plin2 deletion was also associated with increased iWAT CPT1B in CD-fed animals, the effect did not reach significance by two-way ANOVA due to variability in CPT1B protein in Plin2-null tissues. However, we did observe significant dietary modulation of CPT1B protein in Plin2-null mice, with a 6-fold (P = 0.0221) increase in Plin2-null animals on WD. Similarly, iWAT from Plin2-null mice on WD exhibited a 100-fold increase in DIO2 protein compared with WT mice on that diet (P = 0.0003), and we observed a dietary modulation of this protein in Plin2-null animals, with levels being 2.9-fold higher in Plin2-null mice on WD compared with CD (P = 0.0131). Next, because we observed differences in Plin5 mRNA expression across diets and genotypes, and because PLIN5 is highly expressed in BAT and beige adipocytes, we also checked levels of this protein (50). For animals on WD, PLIN5 was increased 33-fold (P = 0.0014) in Plin2-null iWAT. PLIN5 trended slightly higher in Plin2-null animals on CD compared with WT, but this difference was not significant. Like CIDEA, CPT1B, and DIO2, there was strong dietary modulation of PLIN5 in Plin2-null animals, with expression being 7-fold higher on WD (P = 0.0064). Importantly, we also measured significant changes in PGC1α protein in Plin2-null iWAT, as well. iWAT from Plin2-null mice on WD exhibited a 6.5-fold increase in PGC1α protein compared with WT mice on WD (P < 0.0001), and PGC1α was 2.3-fold higher in Plin2-null animals on WD compared with Plin2-null animals on CD. Given the observed changes in iWAT Pgc1α mRNA and protein expression, and that PGC1α is a key regulator of mitochondrial biogenesis, we hypothesized that we would see increased markers of mitochondrial number. As such, we examined voltage-dependent ion channel (VDAC) protein levels in WT and Plin2-null iWAT. Compared with WT mice, VDAC protein was 11-fold (P < 0.0001) higher in Plin2-null iWAT for WD-fed animals, and 3-fold (P = 0.0462) higher in Plin2-null iWAT for CD-fed animals. VDAC protein also experienced dietary modulation in Plin2-null iWAT, with levels being 2-fold (P = 0.0040) higher for animals on WD. Finally, as predicted by gene expression data, we did not observe differences in total PPARγ protein across diet or genotype (Fig. 3C).
Fig. 3.
Plin2-null mice demonstrate iWAT beiging at the protein and lipid levels. A, B: Western blot analysis of UCP1, CIDEA, CPT1b, DIO2, PLIN5, PGC1α, VDAC, PPARγ, and α-tubulin in iWAT from WT and Plin2-null mice on 30 week WD and CD. C: Quantification of Western blots from panels A and B. Protein intensities were normalized to α-tubulin. Graphs are shown with WT WD arbitrarily set to 1. Statistically significant comparisons are indicated by a solid bold line (P < 0.05, two-way ANOVA with post hoc Tukey’s multiple comparisons test). D, E: GC-MS analysis of long-chain saturated FAMEs, C20:0, C22:0, and C24:0, derived from NLs extracted from the iWAT of WT and Plin2-null mice on long-term CD/WD. These long-chain species are the products resulting from ELOVL3 elongation and are expressed as percent change compared with WT on each diet. *Significant differences compared with WT (P < 0.05, Student’s t-test). F: PL to NL ratios in iWAT of WT and Plin2-null mice on long-term CD/WD. Higher ratios suggest increased cytoplasmic lipid droplet multilocularity. G: Quantification of NL normalized to total protein in the iWAT of WT and Plin2-null mice on WD and CD. Data are expressed as mean ± SEM; statistically significant comparisons are indicated by a solid bold line (P < 0.05, two-way ANOVA with post hoc Tukey’s multiple comparisons test); n = 4 for WT WD, WT CD, and Plin2-null WD; n = 3 for Plin2-null CD.
Because Elovl3 mRNA levels were significantly higher in Plin2-null mice on WD compared with WT, we hypothesized that iWAT from Plin2-null mice would contain increased products of ELOVL3 enzyme activity. ELOVL3 is highly expressed in BAT and activated beige iWAT, and it catalyzes the elongation of primarily saturated fatty acids to very long chain products that include C20:0, C22:0, and C24:0 (51–53). Thus, we expected to see enrichment of these products in iWAT from Plin2-null mice. Accordingly, we performed GC-MS lipidomic analysis of iWAT NLs from WT and Plin2-null mice on WD and CD. For Plin2-null mice on WD, as a percentage of total NL-derived FAMEs, we observed a 115% increase in C20:0 (P = 0.0177), a 78% increase in C22:0 (P = 0.0028), and a 131% increase in C24:0 (P = 0.0003) compared with WT (Fig. 3D). In Plin2-null mice on CD, we observed a 230% increase in C20:0 (P = 0.0057), a 250% increase in C22:0 (P = 0.0214), and a 227% increase in C24:0 (P = 0.0361) compared with WT (Fig. 3E). Due to the multilocularity of cytoplasmic lipid droplets that we observed in Plin2-null iWAT with histological methods, we also hypothesized that iWAT from Plin2-null mice would exhibit significantly higher PL to NL ratios because the tissues appeared to have smaller lipid droplets with more surface area. For Plin2-null mice on WD, we observed a PL:NL ratio 2.5-fold higher than that seen in WT mice (P = 0.0139), while Plin2-null mice on CD exhibited a ratio 2.1-fold higher (P = 0.0201) compared with WT on that diet (Fig. 3F). We did not observe PL:NL ratio differences between WT animals on CD or WD, or between Plin2-null mice on these diets. It should be noted that PL:NL ratio calculations were based on total PL quantities present in iWAT extracts, which include PLs from the plasma membrane and organelle membranes, as well as free PLs, rather than those solely derived from lipid droplets. Thus, it is likely that actual differences in PL:NL ratios of lipid droplets from WT and Plin2-null mice are larger. Finally, we used GC-MS to measure iWAT NL levels normalized to total protein in Plin2-null and WT mice on WD and CD (Fig. 3G). For animals on WD, Plin2-null iWAT contained 67% less NL per milligram protein compared with WT (P = 0.0027), and Plin2-null iWAT from mice on CD contained 84% less NL per milligram protein compared with WT on that diet (P = 0.0035). We did not observe statistically significant dietary modulation of NL per milligram protein across diets for either genotype. Together with the histological, gene expression, and protein evidence, the increased very long chain fatty acids, increased PL:NL ratio, and the massively decreased NL per milligram protein measurements together suggest the presence of profoundly beige iWAT in Plin2-null mice on long-term diets compared with WT counterparts.
Plin2-null animals exhibit markers of enhanced iWAT insulin sensitivity and de novo lipogenesis
Because we previously showed improved glucose tolerance and insulin sensitivity in Plin2-null mice on long-term WD compared with WT animals, we hypothesized that we would be able to see biochemical markers of enhanced insulin sensitivity in adipose tissues from these mice (12, 13). ChREBP is a glucose-responsive gene in adipose tissue, and its other isoform transcribed from an alternate promoter, ChREBPβ, is a well-validated marker of adipose tissue insulin sensitivity in both humans and mice (54–56). As such, we first looked at levels of ChREBP (pan) and ChREBPβ in iBAT, iWAT, and eWAT from WT and Plin2-null WD- and CD-fed mice (Fig. 4A–C). In the iBAT of animals on WD, we did not observe significant differences by two-way ANOVA in ChREBP or ChREBPβ mRNA expression between WT and Plin2-null mice. However, for Plin2-null mice on CD, we observed a trending 1.5-fold increase in ChREBP expression and a significant ∼3.5-fold (P = 0.0002) increase in ChREBPβ expression compared with WT mice on that diet (Fig. 4A). We also observed dietary modulation of ChREBP in Plin2-null mouse iBAT, with transcript levels of this gene being 2.1-fold (P = 0.029) higher in Plin2-null mice on CD compared with WD. Because ChREBP is a major determinant of lipogenic gene expression in adipose tissues, we also examined expression levels of the ChREBP target genes, Fasn and Acc1, in iBAT. In iBAT from mice on CD, Fasn experienced a 2-fold trending increase in Plin2-null mice that did not quite reach significance (P = 0.0562), while Acc1 exhibited a significant 2.6-fold (P = 0.0143) increase compared with WT mice on CD. Like ChREBPβ, we saw dietary modulation of this mRNA in Plin2-null animals, with levels being 2.6-fold (P = 0.0152) higher in Plin2-null mice on CD compared with WD (Fig. 4A). We also checked mRNA expression levels of glucose transporter (Glut)1 and Glut4. GLUT1 has been shown to increase during cold-mediated browning of adipose tissue and has also been shown to be essential for increased β3-adrenergic agonist-stimulated glucose uptake in BAT (57, 58). However, we did not find differences in iBAT Glut1 expression across diets or genotypes. Next, we examined Glut4 expression. Modulation of GLUT4 has been shown to regulate ChREBP mRNA expression in adipose tissue (54). Similar to iBAT ChREBP and ChREBPβ expression, we observed a significant 2-fold (P = 0.0295) increase in Plin2-null iBAT Glut4, but only on CD. Because FASN and ACC1 are also targets of SREBP1C, we checked mRNA levels of this transcription factor and its well-known target gene, stearoyl-CoA desaturase 1 (Scd1), but did not observe differences in expression across diets or genotypes. Therefore, we suspect that the observed effects on lipogenic genes are primarily ChREBP-driven.
Fig. 4.
Plin2-null animals exhibit markers of enhanced adipose tissue insulin sensitivity and de novo lipogenesis. A–C: mRNA expression of ChREBP, ChREBPβ, Fasn, Acc1, Glut1, Glut4, Srebp1c, and Scd1 in iBAT, iWAT, and eWAT of WT and Plin2-null mice on 30 week WD and CD. D: Serum FFAs in WT and Plin2-null mice on long-term WD and CD given either saline or insulin. Each genotype was fasted 4 h and split into two groups, with one receiving saline and the other receiving insulin (0.5 U/kg BW) via intraperitoneal injection. Blood was taken 20 min postinjection and FFAs were measured calorimetrically in serum. Three-way ANOVA was used to look for interaction between diet, genotype, and insulin injection status. E, F: Western blot analysis of ChREBPα, FASN, phospho-ACC (Ser79), total-ACC, phospho:total ACC, and α-tubulin in iWAT of WT and Plin2-null mice on 30 week WD and CD. Mice were fasted for 4 h prior to harvest. G: Quantification of Western blots from panels E and F. Proteins were normalized to α-tubulin. H: Adipose-specific Plin2 knockout mice (FatKO) were generated by crossing the Plin2-floxed mouse with one expressing Cre recombinase driven by the FABP4 promoter. Floxed and FatKO mice were placed on long-term WD or CD, and gene expression was performed in iWAT for Plin2, Ucp1, Elovl3, CideA, Dio2, Cpt1b, Pgc1α, Pparα, Fgf21, and Acc1. Except for Dio2 for CD mice, no differences were detected between genotypes. Most genes are modulated by diet, with higher expression levels observed in mice on CD. Data are expressed as mean ± SEM; statistically significant comparisons are indicated by a solid bold line (P < 0.05, two-way ANOVA with post hoc Tukey’s multiple comparisons test). Data are expressed as mean ± SEM; statistically significant comparisons are indicated by a solid bold line (P < 0.05, two-way ANOVA with post hoc Tukey’s multiple comparisons test); n = 4 for WT WD, WT CD, and Plin2-null WD; n = 3 for Plin2-null CD; n = 4 for all floxed and FatKO groups.
In contrast to iBAT results, we found that Plin2 deletion resulted in profound statistically significant increases in ChREBP, ChREBPβ, Fasn, and Acc1 mRNA expression in iWAT on both diets (Fig. 4B). For mice on WD, Plin2 deletion resulted in a 3-fold (P = 0.0103) increase in ChREBP, a striking 46,000-fold (P = 0.0477) increase in ChREBPβ, a 3.6-fold (P = 0.0327) increase in Fasn, and a 5-fold (P = 0.0019) increase in Acc1. While loss of Plin2 did not affect iWAT Glut1 mRNA expression in WD-fed mice, it increased Glut4 by 1.75-fold (P = 0.0066) on this diet. Plin2 deletion also did not significantly alter Srebp1c or Scd1 expression in WD-fed mice. For mice on the CD, Plin2 deletion produced a trending 2-fold (P = 0.221) increase in iWAT ChREBP and significant increases in ChREBPβ (37-fold, P = 0.022), Fasn (4.4-fold, P = 0.0021), and Acc1 (4-fold, P < 0.0001). Additionally, we observed significant dietary modulation of Acc1 in Plin2-null mice, with levels of this gene being 1.6-fold (P = 0.0235) higher in Plin2-null mice on WD compared with CD. Plin2 deletion did not affect iWAT Glut1 expression for mice on CD, but it increased Glut4 mRNA by 1.5-fold (P = 0.029). Like WD-fed mice, the expression of Srebp1c or Scd1 was not affected by Plin2 deletion in CD-fed mice. In eWAT, the only significant difference in the expression of carbohydrate-responsive genes was a dietary modulation of ChREBP in Plin2-null mice, with levels being 2-fold (P = 0.029) higher in Plin2-null mice on CD compared with WD (Fig. 4C). However, we did observe large trending increases in eWAT ChREBPβ in Plin2-null animals on both WD and CD that did not reach significance.
The ability of insulin to shut down adipose tissue lipolysis is a hallmark of adipose insulin sensitivity (59, 60). Because we observed biochemical markers of enhanced insulin sensitivity in adipose tissue from Plin2-null mice on long-term diets, we hypothesized that these animals should exhibit direct improvements in lipolysis suppression after acute insulin treatment. Accordingly, we treated 30 week WD and CD mice with insulin and then measured serum FFAs 20 min afterwards. We were unable to observe significant reductions in serum FFAs in WT mice on WD or CD after this brief duration in insulin action (Fig. 4D). However, insulin treatment significantly (three-way ANOVA) reduced serum FFA levels in WD- and CD-fed Plin2-null mice by 50% (P = 0.0237) and 56% (P = 0.0016), respectively. Combined with gene expression data (Fig. 4A–C), the reductions in serum FFAs after insulin treatment provide direct evidence that Plin2-null mice experience enhanced adipose tissue insulin sensitivity.
Finally, we examined lipogenic protein levels in the iWAT of Plin2-null and WT mice on long-term WD and CD. To confirm increases in lipogenic gene expression at the protein level, we examined ChREBPα, FASN, phosphorylated-ACC Ser79 (P-ACC), and total-ACC (T-ACC) in iWAT after a 4 h fast (Fig. 4E, F). For mice on WD, Plin2 deletion increased ChREBPα by 4.7-fold (P = 0.0003), whereas FASN was increased 2.8-fold (P = 0.0007) (Fig. 4G). Similarly, Plin2 deletion significantly increased P-ACC and T-ACC by 4-fold (P = 0.0242 and P = 0.0013, respectively). The P-ACC:T-ACC ratio was not affected by Plin2 loss for mice on WD. For mice on CD, Plin2 deletion increased ChREBPα by 6.7-fold (P < 0.0001), FASN by 2.7-fold (P = 0.0003), P-ACC by 4.4-fold (P < 0.0001), and T-ACC by 3-fold (P = 0.0012). Unlike the WD, we did observe a slight trending increase in the P/T-ACC ratio in Plin2-null mice on CD compared with WT on CD, but this did not reach significance by two-way ANOVA (P = 0.189). We also observed dietary modulation of P-ACC levels in Plin2-null mice, with levels of P-ACC being 2.5-fold (P = 0.0005) higher in mice on CD compared with WD (Fig. 4G). Notably, the effects of Plin2 deletion on ChREBPα, FASN, and T-ACC protein levels in iWAT from WD and CD animals correlate with Plin2-null effects on iWAT transcripts of these genes, which suggest that the actions of Plin2 on iWAT lipogenesis are regulated at the transcriptional level.
iWAT browning observed in Plin2-null mice is not due to loss of Plin2 in adipocytes
In light of evidence that Plin2 has cell autonomous roles in adipose function, we hypothesized that the iWAT browning phenotype observed in Plin2-null mice might be due to loss of Plin2 specifically within adipocytes (61). Accordingly, we generated adipose-specific PLIN2 knockout mice (FatKO) by crossing a PLIN2-floxed mouse with one expressing Cre recombinase driven by the FABP4 promoter. Both floxed and FatKO mice were placed on WD and CD and then iWAT was assessed for Plin2, thermogenic program, and lipogenic genes (Fig. 4H). Unlike the whole-body knockout, we did detect minimal Plin2 expression in the iWAT of FatKO mice on both diets. We attribute this to the fact that other cell types in adipose tissue, such as macrophages, still express Plin2 in the FatKO mouse. Nevertheless, we observed a 93% (P = 0.0022) decrease in Plin2 mRNA in FatKO mice on WD compared with floxed mice and a 93% (P = 0.0355) decrease in Plin2 in FatKO animals on CD. Next, we examined thermogenic genes in floxed and FatKO mice on WD and CD. For animals on WD, we observed no statistical differences (two-way ANOVA) in Ucp1, Elovl3, CideA, Dio2, Cpt1b, Pgc1α, or Pparα expression between the two genotypes. In fact, thermogenic genes in FatKO iWAT trended lower than those for floxed mice on the WD. Additionally, levels of all thermogenic genes in all mice were orders of magnitude lower than those observed in Plin2-null mice on WD. Next, we examined thermogenic gene expression for mice on CD. Again, we did not find statistically significant differences between genotypes in transcript levels of Ucp1, Elovl3, CideA, Cpt1b, Pgc1α, or Pparα by two-way ANOVA. We did observe a two-fold (P = 0.0336) increase in Dio2 mRNA in FatKO animals on CD. However, this difference in Dio2 expression was not seen for animals on WD, so further investigation is warranted to examine this phenomenon.
Thermogenic genes were lower in the iWAT from both FatKO and floxed mice on WD compared with CD, although not all of them reached significance by two-way ANOVA. For floxed mice, iWAT transcript levels of Elovl3 were 11.5-fold (P = 0.042) and Pparα were 3.5-fold (P = 0.0016) higher on CD compared with WD. All other thermogenic genes trended substantially lower in iWAT from floxed mice on WD compared with CD, but differences were not significant. For FatKO mice on the CD, mRNA levels were significantly increased over WD levels for Ucp1 (26.5-fold, P = 0.0097), Elovl3 (73-fold, P = 0.0108), CideA (4-fold, P = 0.0376), Dio2 (30-fold, P = 0.0009), Cpt1b (7.8-fold, P = 0.0009), and Pparα (6.6-fold, P < 0.0001). No significant differences were observed for Fgf21 expression across diets or genotypes. Finally, we examined Acc1 expression in the iWAT of floxed and FatKO animals. We did not observe differences in Acc1 expression between genotypes on CD or WD, although we did observe dietary modulation of this gene within genotypes, with Acc1 transcript levels being 5.2-fold (P < 00001) higher in floxed animals and 5.1-fold (P < 0.0001) higher in FatKO animals on CD compared with WD. Because we did not see genotypic differences in thermogenic or lipogenic gene expression among floxed and FatKO animals, we conclude that the browning phenotype observed in Plin2-null mice is not due to specific loss of PLIN2 in adipocytes.
Dietary sucrose is a powerful modulator of iWAT browning in Plin2-null mice
Because we observed strong dietary modulation of browning in Plin2-null mice on WD and CD, we were interested in determining which dietary component(s) were responsible for this phenomenon. Increased caloric intake and certain fatty acids, such as DHA and EPA, have been shown to activate traditional BAT thermogenesis (62, 63). Additionally, levels of circulating hormones that have been shown to mediate WAT browning, such as FGF21 or FGF19, can be modulated by dietary fat or carbohydrates (29, 33, 34, 64, 65). To attempt to parse out which dietary component mediates browning in Plin2-null mice, we placed both WT and Plin2-null mice on three additional diets: a 60% HFD containing 8% sucrose; a 10% LFD containing 35% sucrose; and standard chow lacking simple carbohydrates (Fig. 5) for 30 weeks, after which we assessed Ucp1 and Elovl3 expression in iWAT and compared their relative responses to those of animals fed WD and CD, as previously shown in Fig. 2. Both genes were higher in Plin2-null mice compared with WT on all diets (Fig. 5). Interestingly, the greatest increase in iWAT Ucp1 and Elovl3 expression occurred in Plin2-null mice on WD and LFD. Plin2-null mice placed on CD, HFD, and chow had comparable levels of Ucp1 and Elovl3 expression, which were significantly lower than those found in WD- and LFD-fed animals. Because Plin2-null mice on HFD had comparable levels of expression to those on CD and chow, it is unlikely that dietary fat mediates iWAT browning in Plin2-null animals. Based on these data, we hypothesized that dietary sucrose modulates browning in Plin2-null mice. We designed a 6 week study in which 8-week-old chow-fed WT and Plin2-null mice were given either water or 20% sucrose ad libitum. Because our chow had no sucrose, this strategy allowed us to easily manipulate dietary sucrose composition. At the end of the feeding study, mice were euthanized and examined for iWAT browning.
Fig. 5.
Dietary sucrose is a powerful modulator of iWAT beiging in Plin2-null mice. A: Ucp1 and Elovl3 mRNA expression in WT and Plin2-null mice on 30 week WD, CD, HFD, LFD, and chow. All data are relative, with WT on WD arbitrarily set to 1. Data for WD and CD are the same as shown in Fig. 3, and are presented again for purposes of comparison to the other diets. Genes were normalized to 18S expression. B: Table showing the company, product number, and sucrose content of WD, CD, HFD, LFD, and chow. C: Experimental design for the sucrose-supplementation study. Chow-fed WT and Plin2-null mice were divided into two groups, with one receiving 20% sucrose in water and the other receiving 0% sucrose in water for 6 weeks. All groups had free access to chow and their respective liquids during the course of the study. D: Total caloric intake for WT and Plin2-null animals on either water or 20% sucrose. Calories derived from both chow and sucrose are shown. E: Liquid intake (milliliters per day) for WT and Plin2-null mice during the course of the 6 week sucrose study. F: Feeding efficiency (total daily calories normalized to BW) for WT and Plin2-null mice receiving water or 20% sucrose. G: H&E staining of iWAT from WT and Plin2-null mice on either water or 20% sucrose for 6 weeks. The scale bar represents 100 microns. H: Ucp1, Elovl3, CideA, Dio2, Cpt1a, Cpt1b, Pgc1a, Pgc1b, Pparα, Plin5, and Pparγ2 mRNA expression in iWAT of WT and Plin2-null mice on 20% sucrose or water for 6 weeks. I: mRNA expression of markers for white (Asc1), beige (Pat2), and brown (P2rx5) adipocytes in iWAT of WT and Plin2-null mice from the 6 week sucrose study. Quantification of Western blots from panels E and F. Proteins were normalized to α-tubulin. Data are expressed as mean ± SEM; statistically significant comparisons are indicated by a solid bold line (P < 0.05, two-way ANOVA with post hoc Tukey’s multiple comparisons test); n = 5 for all groups.
Figure 5D shows total caloric intake for this 6 week study. Consistent with other studies (66), we found that mice that were provided 20% sucrose water ad libitum showed decreased chow intake with a concomitant increase in calories derived from sucrose (Fig. 5D). WT animals obtained ∼57% of their calories from sucrose, while Plin2-null mice on sucrose derived ∼48% of their daily calories from sugar. Both genotypes experienced increased daily caloric intake compared with their water-only counterparts. WT animals on sucrose consumed 30% more kilocalories per day compared with WT on water (P < 0.0001), whereas Plin2-null animals on sucrose consumed 21% more kilocalories per day compared with Plin2-null on water (P = 0.002). Interestingly, WT animals on sucrose consumed an extra 2.21 kcal/day compared with Plin2-null mice on sucrose (P = 0.001), with the extra calories coming from sucrose rather than chow (Fig. 5D). There was no statistical difference in chow intake for animals on water, nor was there a difference in chow intake for animals on sucrose. When daily liquid intake was examined, it was found that WT mice consumed an extra 2.9 ml of sucrose water per day compared with Plin2-null animals (P < 0.0001) (Fig. 5E). WT and Plin2-null mice on water both consumed approximately 4.6 ml/day, with no statistical difference between genotypes. When total caloric intake per day was normalized to BW, there was no statistical difference between genotypes on sucrose, nor was there a difference between animals on water (Fig. 5F). However, WT animals on sucrose consumed an extra 0.12 kcal/day/g compared with WT animals on water (P < 0.0001), and Plin2-null animals on sucrose consumed an extra 0.14 kcal/day/g compared with Plin2-null mice on water (P < 0.0001). Statistically significant reductions in iBAT, iWAT, and eWAT masses were also observed in Plin2-null animals on sucrose compared with WT mice on that diet (supplemental Fig. S2A). For Plin2-null animals on sucrose, iBAT mass was reduced 41% (P = 0.0476), iWAT was reduced by 50% (P = 0.0440), and eWAT was reduced 62% (P = 0.0108). Finally, we observed a significant 45% reduction in eWAT mass in Plin2-null mice fed water compared with WT mice on that diet (P = 0.0472). Fat pad masses normalized to BW are also provided in supplemental Fig. S2B.
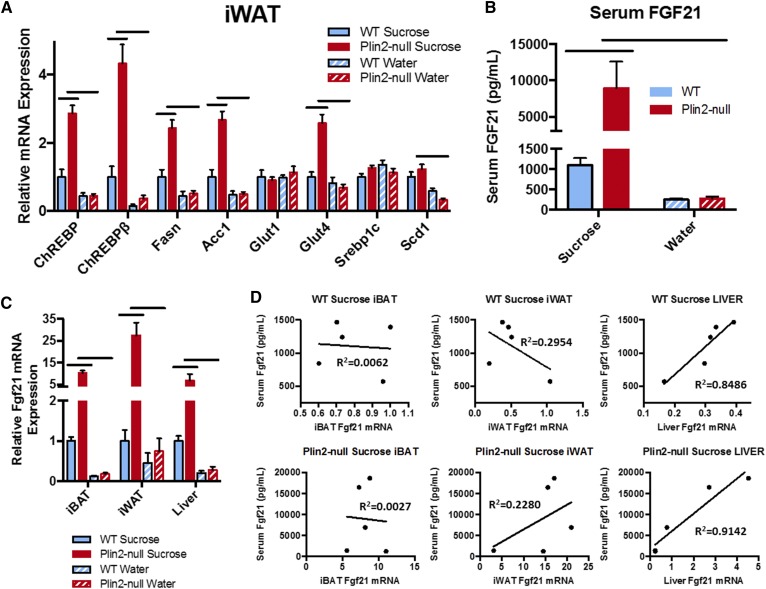
Next, we examined iWAT from animals on sucrose for signs of browning both histologically and by gene expression. Histologically, iWAT from WT and Plin2-null animals on water appeared to be largely unilocular (Fig. 5G), although Plin2-null mice tended to have slightly more pockets of multilocular beige adipocytes. Additionally, unilocular adipocytes from Plin2-null mice on water tended to be slightly smaller than those from WT animals on water. By contrast, we found large increases in multilocular beige adipocytes in the iWAT from both genotypes on 20% sucrose. Nevertheless, WT animals on sucrose retained significant numbers of unilocular adipocytes, suggesting partial beige transformation. In contrast, beige transformation of iWAT in Plin2-null mice on sucrose appeared to be nearly complete and closely resembled that found in Plin2-null mice on WD (see Fig. 1). To confirm the browning observed histologically, we next checked thermogenic program-related genes in the iWAT of animals placed on either water or sucrose (Fig. 5H). We did not observe differences in thermogenic gene expression for animals on water. For WT mice, almost all thermogenic genes trended higher on sucrose compared with water, but these did not reach significance by two-way ANOVA. On the other hand, Plin2-null mice on sucrose experienced large and significant upregulation of iWAT thermogenic program genes compared with Plin2-null mice on water or WT mice on sucrose (Fig. 5H). For mice on sucrose, Plin2 deletion increased Ucp1 6.5-fold (P < 0.0001), Elovl3 13.2-fold (P = <0.0001), CideA 4.5-fold (P < 0.0001), Dio2 23-fold (P < 0.0001), Cpt1b 4-fold (P < 0.0001), Pgc1α 3.3-fold (P = 0.0002), Pgc1β 2.3-fold (P = 0.0002), Pparα 2.4-fold (P = 0.0004), and Plin5 3.5-fold (P < 0.0001). No differences were detected for Cpt1a or Pparγ between WT and Plin2-null mice on sucrose. Compared with animals on water, sucrose feeding of Plin2-null mice significantly increased iWAT Ucp1 10-fold (P < 0.0001), Elovl3 25-fold (P < 0.0001), CideA 6-fold (P < 0.0001), Dio2 23-fold (P < 0.0001), Cpt1b 5-fold (P < 0.0001), Pgc1α 4.3-fold (P < 0.0001), Pgc1β 3.4-fold (P < 0.0001), Pparα 5-fold (P < 0.0001), and Plin5 3.9-fold (P < 0.0001).
Finally, we checked iWAT for markers of white (Asc1), beige (Pat2), and brown (P2rx5) adipocytes in WT and Plin2-null mice on sucrose and water (Fig. 5I). For Plin2-null mice, sucrose consumption did not affect Asc1 mRNA. However, it significantly enhanced expression of Pat2 (2.7-fold, P < 0.0001) and P2rx5 (2.4-fold, P < 0.0001) compared with mice on water. Conversely, for WT mice, sucrose consumption significantly decreased Asc1 expression by 40% (P = 0.0049), but did not alter expression of Pat2 or P2rx5 compared with mice consuming water only. In addition, we found that loss of PLIN2 significantly decreased Asc1 expression 50% (P = 0.0271), and increased expression of Pat2 (1.9-fold, P = 0.0014) and P2rx5 (2.4-fold, P < 0.0001) for mice on sucrose. For mice on water, loss of Plin2 decreased Asc1 expression by 60% (P = 0.0001), but did not affect expression of Pat2 or P2rx5. Given these histological and gene expression data, it was evident that sucrose is a powerful modulator of iWAT browning in Plin2-null mice.
Enhanced markers of browning and insulin sensitivity in iWAT of Plin2-null mice given dietary sucrose are likely driven by increased circulating FGF21 derived from the liver
Next, as with the 30 week WD and CD study, we checked markers of adipose insulin sensitivity and de novo lipogenesis in the iWAT from mice on water or 20% sucrose for 6 weeks (Fig. 6A) and found significant effects of genotype and diet. For sucrose-supplemented mice, Plin2 deletion significantly (two-way ANOVA) increased ChREBP 3-fold (P < 0.0001), ChREBPβ 4.2-fold (P < 0.0001), Fasn 2.4-fold (P = 0.0002), Acc1 2.7-fold (P < 0.0001), and Glut4 2.6-fold (P < 0.0001). PLIN2 loss did not affect transcript levels of Glut1, Srebp1c, or Scd1 on this diet. Plin2 deletion did not significantly affect transcript levels of any of these genes in mice consuming water without sucrose. When we compared the effects of sucrose supplementation on the expression of these genes within WT and Plin2-null mice, we found that sucrose supplementation modestly increased expression of ChREBP, ChREBPβ, Fasn, Acc1, and Scd1 in WT mice, but the effects were not significant (two-way ANOVA). Sucrose supplementation did not affect iWAT expression of Glut4, Glut1, or Srebp1c in WT mice compared with those that consumed water without sucrose. For Plin2-null mice, sucrose supplementation produced large statistically significant inductions of ChREBP (6.3-fold, P < 0.0001), ChREBPβ (12-fold, P < 0.0001), Fasn (4.8-fold, P < 0.0001), Acc1 (5.4-fold, P < 0.0001), Glut4 (3.8-fold, P < 0.0001), and Scd1 (3.7-fold, P = 0.0001). Overall, sucrose supplementation exerted the same changes in markers for insulin sensitivity and lipogenic gene expression that were observed for Plin2-null animals on WD and CD. Although it should be noted that Scd1 expression was not affected by diet or genotype in WD- and CD-fed mice.
Fig. 6.
Enhanced markers of beiging and insulin sensitivity in iWAT of Plin2-null mice given dietary sucrose are likely driven by increased circulating FGF21 derived from the liver. A: mRNA expression of ChREBP, ChREBPβ, Fasn, Acc1, Glut1, Glut4, Srebp1c, and Scd1 in iWAT of WT and Plin2-null mice on 6 weeks of water or 20% sucrose. All gene expression is normalized to 18S. B: Serum FGF21 levels in WT and Plin2-null mice given either water or 20% sucrose for 6 weeks as assessed by kinetic ELISA. C: Fgf21 mRNA expression in iBAT, iWAT, and liver of WT and Plin2-null mice on water or 20% sucrose for 6 weeks. D: Linear regression analysis of tissue-specific Fgf21 mRNA expression with serum FGF21 levels. With the best R2 coefficients, liver is the likely driver of the observed FGF21 serum levels. Data are expressed as mean ± SEM; statistically significant comparisons are indicated by a solid bold line (P < 0.05, two-way ANOVA with post hoc Tukey’s multiple comparisons test); n = 5 for all groups.
Because increased circulating FGF21 has been shown to induce iWAT browning, enhanced carbohydrate uptake, and markers of insulin sensitivity in WAT/BAT, and decreased ad libitum sucrose intake in mice, we hypothesized that increased circulating levels of this hormone could be driving our iWAT phenotype in Plin2-null mice (29, 64, 67–71). Accordingly, we measured circulating levels of this hormone in our mice on sucrose or water (Fig. 6B). For animals given sucrose, serum FGF21 levels in Plin2-null mice were massively increased over those of WT animals, and serum levels of FGF21 in Plin2-null mice were more than 10 times greater than those reported previously for WT C57Bl/6 mice supplemented with sucrose (33). Despite significantly decreased sucrose intake, Plin2-null animals on sucrose exhibited serum FGF21 levels of ∼10,000 pg/ml compared with 1,000 pg/ml observed in WT mice on this diet (P = 0.0085). Compared with animals given water, we found that sucrose supplementation resulted in a significant 32-fold (P = 0.0044) increase in serum FGF21 levels in Plin2-null mice, and an ∼4-fold increase in serum FGF21 in WT mice, which did not reach significance by two-way ANOVA due to variance in FGF21 values observed for Plin2-null animals on sucrose. No difference was observed in serum FGF21 for animals given only water.
Given the drastically increased serum FGF21 levels in Plin2-null mice provided sucrose, we then sought to determine the origin of these elevated hormone levels (Fig. 6C). Liver, BAT, and WAT have all been shown to express FGF21, and adipose-derived FGF21 has been demonstrated to be a potent autocrine/paracrine factor exerting important effects in those tissues (29, 71, 72). Even so, the liver has consistently been shown to be the predominant source of circulating FGF21 in mice and humans. Indeed, iBAT, iWAT, and livers from mice exposed to 20% sucrose over the course of 6 weeks all showed increases in Fgf21 mRNA expression (Fig. 6C). For animals given sucrose, PLIN2 deletion increased Fgf21 expression 10.2-fold (P < 0.0001) in iBAT, 28-fold (P < 0.0001) in iWAT, and 7-fold (P = 0.0408) in liver. There was also significant dietary modulation of Fgf21 mRNA in these tissues in Plin2-null mice. Compared with Plin2-null mice given water, Fgf21 mRNA increased 56-fold (P < 0.0001) in iBAT, 37-fold (P < 0.0001) in iWAT, and 25-fold (P = 0.0201) in the liver of Plin2-null mice given sucrose. By two-way ANOVA, no statistically significant differences were observed for Fgf21 expression in iBAT, iWAT, or liver of WT mice given either water or sucrose. Given the fact that hepatic Fgf21 expression has been shown to be the major driver of serum FGF21 levels, we performed linear regression analysis of tissue-specific Fgf21 mRNA expression compared with FGF21 serum levels (Fig. 6D). We did not find significant correlations between iBAT or iWAT Fgf21 mRNA levels and serum levels of the hormone itself in either genotype. However, liver Fgf21 mRNA expression closely correlated with serum levels of the hormone for both genotypes given 20% sucrose for 6 weeks. In WT mice, liver Fgf21 mRNA, when correlated to serum FGF21 levels, exhibited an R2 of 0.85. Similarly, Plin2-null liver Fgf21 mRNA correlated with serum levels of the hormone with an R2 of 0.91. Therefore, we concluded that hepatic Fgf21 mRNA expression is the most likely driver of FGF21 serum levels.
DISCUSSION
Despite extensive study, the mechanisms by which loss of PLIN2 imparts resistance to obesity have remained elusive (12, 13, 73). Here, we show that Plin2-null mice exhibit profound browning of iWAT even at ambient temperatures. Importantly, we find this browning to be heavily controlled by levels of simple carbohydrates in the diet. Browning of WAT has been shown to have beneficial effects on adipocity, energy expenditure, serum lipid profiles, and glucose tolerance (17, 31). Additionally, dozens of mouse models that are resistant to diet-induced obesity display beige WAT, suggesting that browning of WAT may very well play a role in resistance to this disorder (74). Of key relevance, Plin2-null animals appear to be highly sensitive to sucrose-induced FGF21 expression, with serum levels of this protein achieving levels ∼10-fold higher than WT counterparts.
Originally, we believed another PLIN family member may have been compensating for loss of Plin2 in adipocytes in order to cause the observed browning phenotype. Although we observed large increases in Plin5 mRNA and protein in iWAT from Plin2-null mice on WD, CD, and 20% sucrose, it is important to note that we did not observe this change in iBAT or eWAT. Furthermore, we did not see a difference in iWAT Plin5 in young animals given water instead of sucrose. Plin5 appears to be a gene that is regulated along with other members of the thermogenic program, and indeed, others have shown this to be the case in iWAT of mice exposed to cold temperatures (75). This makes mechanistic sense from two standpoints. First, PLIN5 is highly expressed in BAT, so if white adipocytes become more phenotypically “brown,” then the increase in PLIN5 would be expected. Second, PLIN5 has been shown to promote interaction of lipid droplets with mitochondria, a key process necessary for increased energy expenditure during the browning process (50). Given our observations, we conclude that loss of PLIN2 within adipocytes does not result in an inherent compensation by another PLIN family member to cause our observed browning phenotype. Interestingly, it has previously been shown that PLIN2 protein is upregulated in BAT upon cold exposure; although it is unknown whether the presence of PLIN2 is actually necessary for stimulated thermogenesis in that tissue (76). Although browning of iWAT is also associated with cold exposure, it remains to be seen whether the diet-mediated browning mechanism reported in our study is similar to the type of browning that occurs with cold treatment. Thus, further investigation is warranted to study the response of Plin2-null mice to cold temperatures and whether the presence of PLIN2 is necessary for physiological homeostasis under that specific type of treatment.
Although loss of PLIN3 has been shown to result in WAT browning, this phenotype was only significant when mice were exposed to cold rather than ambient temperatures, and no dietary modulation of iWAT beiging was noted in that study (77). For such a system to be effective in combating diet-induced obesity, subjects lacking PLIN3 would need to be cold-exposed. In Plin2-null mice, profound browning occurs at room temperature, meaning that no such exposure is necessary. Second, in vitro studies suggest that adipocyte-specific loss of PLIN3 is responsible for the browning phenotype observed in PLIN3 knockout mice, whereas we have shown, with an adipose-specific PLIN2 knockout model, that inherent loss of PLIN2 in adipocytes is not responsible for our observed browning phenotype. This is important for possible future therapeutic development, as targeting adipose tissue is more difficult than other approaches that genetically target other tissues (78).
Interestingly, we did not detect dietary modulation of markers of iWAT insulin sensitivity (ChREBP, ChREBPβ, Acc1, and Fasn) in Plin2-null mice during our long-term 30 week study despite significant differences in the sucrose contents of the CD (12%) and WD (34%). Moreover, when we compared iWAT transcript levels of these genes normalized to 18S RNA from Plin2-null mice fed CD, WD, or 20% sucrose, in which they were deriving ∼50% of their calories from sucrose, we found that these treatments resulted in similar levels of expression (data not shown). These observations raise the possibility that there is a maximum threshold that these markers might reach in iWAT, and even 12% dietary sucrose could be enough to reach this threshold in Plin2-null mice. Nevertheless, it is also possible that the increased dietary carbohydrates in the WD could be fluxing through this iWAT at a higher rate compared with CD, and this will be a focus of future studies in our laboratory.
It is also worth noting that the relative effects of Plin2 deletion on mRNA levels of ChREBP, ChREBPβ, Acc1, and Fasn were vastly different between long-term WD-fed mice and mice fed sucrose for 6 weeks. In the case of ChREBPβ, for instance, Plin2-null mice exhibited an ∼50,000-fold increase in iWAT ChREBPβ compared with WT in the 30 week WD/CD study (Fig. 4B), whereas knockout animals demonstrated only a 4.5-fold increase in this gene in the 6 week sucrose feeding study (Fig. 6A). While multiple mechanisms may contribute to these differences, a primary cause appears to be large decreases in the expression of these genes in WT relative to Plin2-null animals as they aged during the long-term WD feeding studies. In the long-term study, levels of markers of adipose insulin sensitivity were very low in WT animals, who were 38 weeks old at the time of tissue harvest; whereas mice used in the sucrose study were 14–16 weeks old at harvest. Natural age-associated loss of peripheral insulin sensitivity has been documented in WT mice, and we conclude that we were able to observe these effects in our long-term study (79).
Next, the trending increases in lipolysis observed in Plin2-null mice without insulin treatment (Fig. 4D) appear to contradict our previous study (13), in which we found significant decreases in Plin2-null serum FFA after prolonged overnight fasting; whereas our current study utilized a short-term (4 h) fasting model prior to saline or insulin treatment. Additionally, the trending increases in basal lipolysis observed in this study are in line with what has been seen in cell culture models of differentiated adipocytes lacking PLIN2 (80, 81). In Plin2-null mice, it is possible that, during the early stages of fasting, there is more surface area for intracellular lipases to act on due to the multilocularity of the lipid droplets. This could explain the trending increase in serum FFA that was observed in the absence of insulin administration. It is also possible that Plin2-null animals may exhibit differences in prolonged fasting autophagy, peripheral FFA uptake, or regulation of hormones (e.g., cortisol, glucagon, etc.) that can mediate rates of adipose tissue lipolysis under prolonged fasting conditions. Future studies will be directed at elucidating the basis for differences between the effects of short- versus long-term fasting on serum FFA in Plin2-null mice.
Of key significance to our study was the observation of increased serum FGF21 and enhanced biochemical markers of adipose insulin sensitivity when Plin2-null mice were supplemented with sucrose in their diet. In addition to being a powerful WAT browning agent, FGF21 is able to induce beneficial effects, including resistance to weight gain and improved glucose tolerance, even in mice lacking traditional thermogenic capabilities (82). Indeed, increased FGF21 or enhanced peripheral sensitivity to FGF21 signaling may partially explain the improved glucose tolerance observed in Plin2-null mice observed previously (12, 13, 16). Others have attributed this improved glucose tolerance to reductions in hepatic diacylglycerol (DAG), although our laboratory has observed that this reduction comes predominantly from decreases in 1,3-DAG rather than the 1,2-DAG that has been implicated in hepatic insulin resistance (unpublished observations) (16). Accordingly, increased FGF21 may be a previously overlooked factor in the improved glucose tolerance in these mice.
The fact that loss of a lipid droplet coat protein could have such profound impacts on dietary carbohydrate metabolism was certainly unexpected. We had originally anticipated that lipid would be the most likely dietary factor mediating WAT browning responses in Plin2-null mice. However, simple dietary carbohydrates, rather than dietary fat, proved to be very potent browning agents in these animals. Furthermore, the cause of the enormous increase in hepatic FGF21 expression in Plin2-null mice in response to dietary sucrose deserves further exploration. FGF21 is controlled by a variety of transcription factors in the liver, including ChREBP, PPARα, FXR, RORα, RXR, ATF4, thyroid receptor β, and CREBH (83–88). Preliminary studies in our laboratory using transcription factor and target gene expression have ruled out hepatic ChREBP, PPARα, FXR, ATF4, and thyroid receptor β as likely candidates for the increased Fgf21 expression in our sucrose-fed mice. We did, however, find large significant increases in the endoplasmic reticulum-associated transcription factor, CREBH, in livers of Plin2-null mice fed sucrose. Nevertheless, this has yet to be confirmed at the protein level, and future work will involve ChIP analysis to confirm increased CREBH protein at the FGF21 promoter in knockout mice. Elucidating the mechanism for the increased FGF21 expression may provide novel therapeutic pathways for future manipulation.
Finally, an important function of FGF21 is to decrease caloric intake of foods rich in carbohydrates, such as sucrose, by acting on neurons in the periventricular nucleus of the hypothalamus, and we were able to observe large reductions in sucrose intake in Plin2-null mice concomitant with massively increased serum FGF21 levels (33, 34, 89). Additionally, we have reported significantly decreased caloric intake of sucrose-containing diets in Plin2-null mice previously (12, 13). Thus, increased circulating FGF21 might possibly explain decreased caloric intake that would be paramount to combating obesity development in Plin2-null animals. Finally, the fact that our studies suggest that the increased FGF21 is derived from the liver should not be overlooked. While we did observe large increases in iBAT and iWAT Fgf21 expression, correlation analysis suggests that the circulating levels of this hormone in our mice are liver-derived. This is especially important because it suggests that existing commercial antisense oligonucleotides could potentially be used to target hepatic PLIN2 in combination with a low-fat high-carbohydrate diet in a therapeutic setting because those drugs accumulate in and are most effective against targets in the liver (78). Nevertheless, adipose-derived FGF21 is still able to act in an autocrine/paracrine manner to improve metabolic homeostasis in that tissue (32). Future work in our laboratory will focus on testing the increased FGF21 production in liver-specific PLIN2 knockout mice as well as confirming increased FGF21 signaling in the hypothalamus of Plin2-null animals.
Supplementary Material
Acknowledgments
The authors would like to thank Andy Bradford for helpful comments during manuscript preparation. The authors would also like to thank Tânia Reis for use of mass spectrometry instrumentation. The authors appreciate the contribution to this research made by E. Erin Smith, Jenna Van Der Volgen, Allison Quador, and Jessica Arnold of the University of Colorado Denver Histology Shared Resource, which is supported in part by the University of Colorado Cancer Center Support Grant P30CA046934.
Footnotes
Abbreviations:
- ACC
- acetyl-CoA carboxylase
- ASC1
- Asc-type amino acid transporter 1
- BAT
- brown adipose tissue
- BW
- body weight
- CD
- control diet
- ChREBP
- carbohydrate response element binding protein
- CIDEA
- cell death-inducing DNA fragmentation factor-α-like effector α
- CPT
- carnitine palmitoyltransferase
- DAG
- diacylglycerol
- DIO2
- type II iodothyronine deiodinase
- ELOVL3
- elongase of very long-chain fatty acid protein 3
- eWAT
- epididymal white adipose tissue
- FAME
- fatty acid methyl ester
- FGF
- fibroblast growth factor
- GLUT
- glucose transporter
- HFD
- high-fat diet
- iBAT
- interscapular brown adipose tissue
- iWAT
- inguinal subcutaneous white adipose tissue
- LFD
- low-fat diet
- NL
- neutral lipid
- P-ACC
- phosphorylated-acetyl-CoA carboxylase Ser79
- PAT2
- proton/amino acid cotransporter 2
- PGC
- PPARγ coactivator
- PL
- phospholipid
- PLIN
- perilipin
- Plin2-null
- mice lacking perilipin-2
- P2RX5
- purinergic receptor P2X ligand-gated ion channel 5
- SCD1
- stearoyl-CoA desaturase 1
- T-ACC
- total-acetyl-CoA carboxylase
- UCP1
- uncoupling protein 1
- VDAC
- voltage-dependent ion channel
- WAT
- white adipose tissue
- WD
- Western diet
This work was supported by National Institutes of Health Grants 2RO1- HD045965 and RO1-HD075285 (to J.L.M.) and TL1-TR001081 (University of Colorado CCTSI, to A.E.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Nguyen D. M., and El-Serag H. B.. 2010. The epidemiology of obesity. Gastroenterol. Clin. North Am. 39: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catenacci V. A., Hill J. O., and Wyatt H. R.. 2009. The obesity epidemic. Clin. Chest Med. 30: 415–444. [DOI] [PubMed] [Google Scholar]
- 3.Polsky S., and Ellis S. L.. 2015. Obesity, insulin resistance, and type 1 diabetes mellitus. Curr. Opin. Endocrinol. Diabetes Obes. 22: 277–282. [DOI] [PubMed] [Google Scholar]
- 4.Hubert H. B., Feinleib M., McNamara P. M., and Castelli W. P.. 1983. Obesity as an independent risk factor for cardiovascular disease: a 26-year follow-up of participants in the Framingham Heart Study. Circulation. 67: 968–977. [DOI] [PubMed] [Google Scholar]
- 5.Vucenik I., and Stains J. P.. 2012. Obesity and cancer risk: evidence, mechanisms, and recommendations. Ann. N. Y. Acad. Sci. 1271: 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanovski S. Z., and Yanovski J. A.. 2014. Long-term drug treatment for obesity: a systematic and clinical review. JAMA. 311: 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadden T. A., Volger S., Tsai A. G., Sarwer D. B., Berkowitz R. I., Diewald L. K., Carvajal R., Moran C. H., and Vetter M.; POWER-UP Research Group. 2013. Managing obesity in primary care practice: an overview with perspective from the POWER-UP study. Int. J. Obes. (Lond.). 37 (Suppl. 1): S3–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iqbal M. B., Fisher N. G., Lyne J. C., and McDonagh T. A.. 2008. Thiazolinediones and heart failure: the potential to precipitate irreversible cardiac dysfunction. Int. J. Cardiol. 123: e35–e37. [DOI] [PubMed] [Google Scholar]
- 9.Kang J. G., and Park C. Y.. 2012. Anti-obesity drugs: a review about their effects and Safety. Diabetes Metab. J. 36: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducharme N. A., and Bickel P. E.. 2008. Lipid droplets in lipogenesis and lipolysis. Endocrinology. 149: 942–949. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg A. S., Coleman R. A., Kraemer F. B., McManaman J. L., Obin M. S., Puri V., Yan Q. W., Miyoshi H., and Mashek D. G.. 2011. The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Invest. 121: 2102–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McManaman J. L., Bales E. S., Orlicky D. J., Jackman M., MacLean P. S., Cain S., Crunk A. E., Mansur A., Graham C. E., Bowman T. A., et al. 2013. Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J. Lipid Res. 54: 1346–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libby A. E., Bales E., Orlicky D. J., and McManaman J. L.. 2016. Perilipin-2 deletion impairs hepatic lipid accumulation by Interfering with sterol regulatory element-binding protein (SREBP) activation and altering the hepatic lipidome. J. Biol. Chem. 291: 24231–24246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai Y., Boyle S., Varela G. M., Caron E., Yin X., Dhir R., Dhir R., Graham M. J., and Ahima R. S.. 2012. Effects of perilipin 2 antisense oligonucleotide treatment on hepatic lipid metabolism and gene expression. Physiol. Genomics. 44: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai Y., Varela G. M., Jackson M. B., Graham M. J., Crooke R. M., and Ahima R. S.. 2007. Reduction of hepatosteatosis and lipid levels by an adipose differentiation-related protein antisense oligonucleotide. Gastroenterology. 132: 1947–1954. [DOI] [PubMed] [Google Scholar]
- 16.Varela G. M., Antwi D. A., Dhir R., Yin X., Singhal N. S., Graham M. J., Crooke R. M., and Ahima R. S.. 2008. Inhibition of ADRP prevents diet-induced insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G621–G628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harms M., and Seale P.. 2013. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19: 1252–1263. [DOI] [PubMed] [Google Scholar]
- 18.Virtanen K. A., Lidell M. E., Orava J., Heglind M., Westergren R., Niemi T., Taittonen M., Laine J., Savisto N. J., Enerback S., et al. 2009. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360: 1518–1525. [DOI] [PubMed] [Google Scholar]
- 19.Saito M., Okamatsu-Ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-Kobayashi J., Iwanaga T., Miyagawa M., Kameya T., Nakada K., et al. 2009. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 58: 1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajimura S., and Saito M.. 2014. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu. Rev. Physiol. 76: 225–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannon B., and Nedergaard J.. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84: 277–359. [DOI] [PubMed] [Google Scholar]
- 22.Bartelt A., Bruns O. T., Reimer R., Hohenberg H., Ittrich H., Peldschus K., Kaul M. G., Tromsdorf U. I., Weller H., Waurisch C., et al. 2011. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17: 200–205. [DOI] [PubMed] [Google Scholar]
- 23.Bartelt A., and Heeren J.. 2014. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 10: 24–36. [DOI] [PubMed] [Google Scholar]
- 24.Cohen P., Levy J. D., Zhang Y., Frontini A., Kolodin D. P., Svensson K. J., Lo J. C., Zeng X., Ye L., Khandekar M. J., et al. 2014. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 156: 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J., Cohen P., and Spiegelman B. M.. 2013. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev. 27: 234–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boström P., Wu J., Jedrychowski M. P., Korde A., Ye L., Lo J. C., Rasbach K. A., Boström E. A., Choi J. H., Long J. Z., et al. 2012. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 481: 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang S., Suh J. M., Reilly S. M., Yu E., Osborn O., Lackey D., Yoshihara E., Perino A., Jacinto S., Lukasheva Y., et al. 2015. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat. Med. 21: 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chau M. D., Gao J., Yang Q., Wu Z., and Gromada J.. 2010. Fibroblast growth factor 21 regulates energy metabolism by activating the AMPK-SIRT1-PGC-1alpha pathway. Proc. Natl. Acad. Sci. USA. 107: 12553–12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher F. M., Kleiner S., Douris N., Fox E. C., Mepani R. J., Verdeguer F., Wu J., Kharitonenkov A., Flier J. S., Maratos-Flier E., et al. 2012. FGF21 regulates PGC-1alpha and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 26: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potthoff M. J., Inagaki T., Satapati S., Ding X., He T., Goetz R., Mohammadi M., Finck B. N., Mangelsdorf D. J., Kliewer S. A., et al. 2009. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc. Natl. Acad. Sci. USA. 106: 10853–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlein C., Talukdar S., Heine M., Fischer A. W., Krott L. M., Nilsson S. K., Brenner M. B., Heeren J., and Scheja L.. 2016. FGF21 lowers plasma triglycerides by accelerating lipoprotein catabolism in white and brown adipose tissues. Cell Metab. 23: 441–453. [DOI] [PubMed] [Google Scholar]
- 32.Itoh N. 2014. FGF21 as a hepatokine, adipokine, and myokine in metabolism and diseases. Front. Endocrinol. (Lausanne). 5: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Holstein-Rathlou S., BonDurant L. D., Peltekian L., Naber M. C., Yin T. C., Claflin K. E., Urizar A. I., Madsen A. N., Ratner C., Holst B., et al. 2016. FGF21 mediates endocrine control of simple sugar intake and sweet taste preference by the liver. Cell Metab. 23: 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soberg S., Sandholt C. H., Jespersen N. Z., Toft U., Madsen A. L., von Holstein-Rathlou S., Grevengoed T. J., Christensen K. B., Bredie W. L. P., Potthoff M. J., et al. 2017. FGF21 is a sugar-induced hormone associated with sweet intake and preference in humans. Cell Metab. 25: 1045–1053.e6. [DOI] [PubMed] [Google Scholar]
- 35.Witte N., Muenzner M., Rietscher J., Knauer M., Heidenreich S., Nuotio-Antar A. M., Graef F. A., Fedders R., Tolkachov A., Goehring I., et al. 2015. The glucose sensor ChREBP links de novo lipogenesis to PPARgamma activity and adipocyte differentiation. Endocrinology. 156: 4008–4019. [DOI] [PubMed] [Google Scholar]
- 36.Nuotio-Antar A. M., Poungvarin N., Li M., Schupp M., Mohammad M., Gerard S., Zou F., and Chan L.. 2015. FABP4-Cre mediated expression of constitutively active ChREBP protects against obesity, fatty liver, and insulin resistance. Endocrinology. 156: 4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbatelli G., Murano I., Madsen L., Hao Q., Jimenez M., Kristiansen K., Giacobino J. P., De Matteis R., and Cinti S.. 2010. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am. J. Physiol. Endocrinol. Metab. 298: E1244–E1253. [DOI] [PubMed] [Google Scholar]
- 38.Lumeng C. N., Bodzin J. L., and Saltiel A. R.. 2007. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117: 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villanueva C. J., Vergnes L., Wang J., Drew B. G., Hong C., Tu Y., Hu Y., Peng X., Xu F., Saez E., et al. 2013. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell Metab. 17: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye L., Wu J., Cohen P., Kazak L., Khandekar M. J., Jedrychowski M. P., Zeng X., Gygi S. P., and Spiegelman B. M.. 2013. Fat cells directly sense temperature to activate thermogenesis. Proc. Natl. Acad. Sci. USA. 110: 12480–12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mössenböck K., Vegiopoulos A., Rose A. J., Sijmonsma T. P., Herzig S., and Schafmeier T.. 2014. Browning of white adipose tissue uncouples glucose uptake from insulin signaling. PLoS One. 9: e110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seale P. 2015. Transcriptional regulatory circuits controlling brown fat development and activation. Diabetes. 64: 2369–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dijk W., Heine M., Vergnes L., Boon M. R., Schaart G., Hesselink M. K., Reue K., van Marken Lichtenbelt W. D., Olivecrona G., Rensen P. C., et al. 2015. ANGPTL4 mediates shuttling of lipid fuel to brown adipose tissue during sustained cold exposure. eLife. 4: e08428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chartoumpekis D. V., Habeos I. G., Ziros P. G., Psyrogiannis A. I., Kyriazopoulou V. E., and Papavassiliou A. G.. 2011. Brown adipose tissue responds to cold and adrenergic stimulation by induction of FGF21. Mol. Med. 17: 736–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Jong J. M., Larsson O., Cannon B., and Nedergaard J.. 2015. A stringent validation of mouse adipose tissue identity markers. Am. J. Physiol. Endocrinol. Metab. 308: E1085–E1105. [DOI] [PubMed] [Google Scholar]
- 46.Garcia R. A., Roemmich J. N., and Claycombe K. J.. 2016. Evaluation of markers of beige adipocytes in white adipose tissue of the mouse. Nutr. Metab. (Lond.). 13: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rockstroh D., Landgraf K., Wagner I. V., Gesing J., Tauscher R., Lakowa N., Kiess W., Buhligen U., Wojan M., Till H., et al. 2015. Direct evidence of brown adipocytes in different fat depots in children. PLoS One. 10: e0117841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ussar S., Lee K. Y., Dankel S. N., Boucher J., Haering M. F., Kleinridders A., Thomou T., Xue R., Macotela Y., Cypess A. M., et al. 2014. ASC-1, PAT2, and P2RX5 are cell surface markers for white, beige, and brown adipocytes. Sci. Transl. Med. 6: 247ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakhuda A., Josse A. R., Gburcik V., Crossland H., Raymond F., Metairon S., Good L., Atherton P. J., Phillips S. M., and Timmons J. A.. 2016. Biomarkers of browning of white adipose tissue and their regulation during exercise- and diet-induced weight loss. Am. J. Clin. Nutr. 104: 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimmel A. R., and Sztalryd C.. 2014. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr. Opin. Lipidol. 25: 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitazawa H., Miyamoto Y., Shimamura K., Nagumo A., and Tokita S.. 2009. Development of a high-density assay for long-chain fatty acyl-CoA elongases. Lipids. 44: 765–773. [DOI] [PubMed] [Google Scholar]
- 52.Westerberg R., Mansson J. E., Golozoubova V., Shabalina I. G., Backlund E. C., Tvrdik P., Retterstol K., Capecchi M. R., and Jacobsson A.. 2006. ELOVL3 is an important component for early onset of lipid recruitment in brown adipose tissue. J. Biol. Chem. 281: 4958–4968. [DOI] [PubMed] [Google Scholar]
- 53.Tucci S., Behringer S., and Spiekerkoetter U.. 2015. De novo fatty acid biosynthesis and elongation in very long-chain acyl-CoA dehydrogenase-deficient mice supplemented with odd or even medium-chain fatty acids. FEBS J. 282: 4242–4253. [DOI] [PubMed] [Google Scholar]
- 54.Herman M. A., Peroni O. D., Villoria J., Schon M. R., Abumrad N. A., Bluher M., Klein S., and Kahn B. B.. 2012. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 484: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Y., Wallace M., Sanchez-Gurmaches J., Hsiao W. Y., Li H., Lee P. L., Vernia S., Metallo C. M., and Guertin D. A.. 2016. Adipose tissue mTORC2 regulates ChREBP-driven de novo lipogenesis and hepatic glucose metabolism. Nat. Commun. 7: 11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iizuka K. 2013. Recent progress on the role of ChREBP in glucose and lipid metabolism. Endocr. J. 60: 543–555. [DOI] [PubMed] [Google Scholar]
- 57.Jia R., Luo X. Q., Wang G., Lin C. X., Qiao H., Wang N., Yao T., Barclay J. L., Whitehead J. P., Luo X., et al. 2016. Characterization of cold-induced remodelling reveals depot-specific differences across and within brown and white adipose tissues in mice. Acta Physiol. (Oxf.). 217: 311–324. [DOI] [PubMed] [Google Scholar]
- 58.Olsen J. M., Sato M., Dallner O. S., Sandstrom A. L., Pisani D. F., Chambard J. C., Amri E. Z., Hutchinson D. S., and Bengtsson T.. 2014. Glucose uptake in brown fat cells is dependent on mTOR complex 2-promoted GLUT1 translocation. J. Cell Biol. 207: 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruan H., and Lodish H. F.. 2003. Insulin resistance in adipose tissue: direct and indirect effects of tumor necrosis factor-α. Cytokine Growth Factor Rev. 14: 447–455. [DOI] [PubMed] [Google Scholar]
- 60.Nilsson N. Ö., Strålfors P., Fredrikson G., and Belfrage P.. 1980. Regulation of adipose tissue lipolysis: effects of noradrenaline and insulin on phosphorylation of hormone-sensitive lipase and on lipolysis in intact rat adipocytes. FEBS Lett. 111: 125–130. [DOI] [PubMed] [Google Scholar]
- 61.Xu G., Sztalryd C., Lu X., Tansey J. T., Gan J., Dorward H., Kimmel A. R., and Londos C.. 2005. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J. Biol. Chem. 280: 42841–42847. [DOI] [PubMed] [Google Scholar]
- 62.Okla M., Kim J., Koehler K., and Chung S.. 2017. Dietary factors promoting brown and beige fat development and thermogenesis. Adv. Nutr. 8: 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sidossis L., and Kajimura S.. 2015. Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Invest. 125: 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang Z., Zhong L., Lee J. T. H., Zhang J., Wu D., Geng L., Wang Y., Wong C. M., and Xu A.. 2017. The FGF21-CCL11 axis mediates beiging of white adipose tissues by coupling sympathetic nervous system to type 2 immunity. Cell Metab. 26: 493–508.e4. [DOI] [PubMed] [Google Scholar]
- 65.Fu L., John L. M., Adams S. H., Yu X. X., Tomlinson E., Renz M., Williams P. M., Soriano R., Corpuz R., Moffat B., et al. 2004. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology. 145: 2594–2603. [DOI] [PubMed] [Google Scholar]
- 66.Ishimoto T., Lanaspa M. A., Le M. T., Garcia G. E., Diggle C. P., Maclean P. S., Jackman M. R., Asipu A., Roncal-Jimenez C. A., Kosugi T., et al. 2012. Opposing effects of fructokinase C and A isoforms on fructose-induced metabolic syndrome in mice. Proc. Natl. Acad. Sci. USA. 109: 4320–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kharitonenkov A., Shiyanova T. L., Koester A., Ford A. M., Micanovic R., Galbreath E. J., Sandusky G. E., Hammond L. J., Moyers J. S., Owens R. A., et al. 2005. FGF-21 as a novel metabolic regulator. J. Clin. Invest. 115: 1627–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Wu G., Fang Q., Zhang M., Hui X., Sheng B., Wu L., Bao Y., Li P., Xu A., et al. 2018. Fibroblast growth factor 21 increases insulin sensitivity through specific expansion of subcutaneous fat. Nat. Commun. 9: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurosu H., Choi M., Ogawa Y., Dickson A. S., Goetz R., Eliseenkova A. V., Mohammadi M., Rosenblatt K. P., Kliewer S. A., and Kuro-o M.. 2007. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 282: 26687–26695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coskun T., Bina H. A., Schneider M. A., Dunbar J. D., Hu C. C., Chen Y., Moller D. E., and Kharitonenkov A.. 2008. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology. 149: 6018–6027. [DOI] [PubMed] [Google Scholar]
- 71.Markan K. R., Naber M. C., Ameka M. K., Anderegg M. D., Mangelsdorf D. J., Kliewer S. A., Mohammadi M., and Potthoff M. J.. 2014. Circulating FGF21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes. 63: 4057–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hondares E., Iglesias R., Giralt A., Gonzalez F. J., Giralt M., Mampel T., and Villarroya F.. 2011. Thermogenic activation induces FGF21 expression and release in brown adipose tissue. J. Biol. Chem. 286: 12983–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Najt C. P., Senthivinayagam S., Aljazi M. B., Fader K. A., Olenic S. D., Brock J. R., Lydic T. A., Jones A. D., and Atshaves B. P.. 2016. Liver-specific loss of Perilipin 2 alleviates diet-induced hepatic steatosis, inflammation, and fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 310: G726–G738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kozak L. P. 2011. The genetics of brown adipocyte induction in white fat depots. Front. Endocrinol. (Lausanne). 2: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barneda D., Frontini A., Cinti S., and Christian M.. 2013. Dynamic changes in lipid droplet-associated proteins in the “browning” of white adipose tissues. Biochim. Biophys. Acta. 1831: 924–933. [DOI] [PubMed] [Google Scholar]
- 76.Yu J., Zhang S., Cui L., Wang W., Na H., Zhu X., Li L., Xu G., Yang F., Christian M., et al. 2015. Lipid droplet remodeling and interaction with mitochondria in mouse brown adipose tissue during cold treatment. Biochim. Biophys. Acta. 1853: 918–928. [DOI] [PubMed] [Google Scholar]
- 77.Lee Y. K., Sohn J. H., Han J. S., Park Y. J., Jeon Y. G., Ji Y., Dalen K. T., Sztalryd C., Kimmel A. R., and Kim J. B.. 2018. Perilipin 3 deficiency stimulates thermogenic beige adipocytes through PPARα activation. Diabetes. 67: 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Geary R. S., Norris D., Yu R., and Bennett C. F.. 2015. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 87: 46–51. [DOI] [PubMed] [Google Scholar]
- 79.Rogers N. H., Landa A., Park S., and Smith R. G.. 2012. Aging leads to a programmed loss of brown adipocytes in murine subcutaneous white adipose tissue. Aging Cell. 11: 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang B. H., Li L., Paul A., Taniguchi S., Nannegari V., Heird W. C., and Chan L.. 2006. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 26: 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi Y., Shinoda A., Kamada H., Shimizu M., Inoue J., and Sato R.. 2016. Perilipin2 plays a positive role in adipocytes during lipolysis by escaping proteasomal degradation. Sci. Rep. 6: 20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Véniant M. M., Sivits G., Helmering J., Komorowski R., Lee J., Fan W., Moyer C., and Lloyd D. J.. 2015. Pharmacologic effects of FGF21 are independent of the “browning” of white adipose tissue. Cell Metab. 21: 731–738. [DOI] [PubMed] [Google Scholar]
- 83.Vernia S., Cavanagh-Kyros J., Garcia-Haro L., Sabio G., Barrett T., Jung D. Y., Kim J. K., Xu J., Shulha H. P., Garber M., et al. 2014. The PPARalpha-FGF21 hormone axis contributes to metabolic regulation by the hepatic JNK signaling pathway. Cell Metab. 20: 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fisher F. M., Kim M., Doridot L., Cunniff J. C., Parker T. S., Levine D. M., Hellerstein M. K., Hudgins L. C., Maratos-Flier E., and Herman M. A.. 2016. A critical role for ChREBP-mediated FGF21 secretion in hepatic fructose metabolism. Mol. Metab. 6: 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y., Solt L. A., and Burris T. P.. 2010. Regulation of FGF21 expression and secretion by retinoic acid receptor-related orphan receptor alpha. J. Biol. Chem. 285: 15668–15673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park J. G., Xu X., Cho S., Hur K. Y., Lee M. S., Kersten S., and Lee A. H.. 2016. CREBH-FGF21 axis improves hepatic steatosis by suppressing adipose tissue lipolysis. Sci. Rep. 6: 27938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adams A. C., Astapova I., Fisher F. M., Badman M. K., Kurgansky K. E., Flier J. S., Hollenberg A. N., and Maratos-Flier E.. 2010. Thyroid hormone regulates hepatic expression of fibroblast growth factor 21 in a PPARalpha-dependent manner. J. Biol. Chem. 285: 14078–14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cyphert H. A., Ge X., Kohan A. B., Salati L. M., Zhang Y., and Hillgartner F. B.. 2012. Activation of the farnesoid X receptor induces hepatic expression and secretion of fibroblast growth factor 21. J. Biol. Chem. 287: 25123–25138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Talukdar S., Owen B. M., Song P., Hernandez G., Zhang Y., Zhou Y., Scott W. T., Paratala B., Turner T., Smith A., et al. 2016. FGF21 regulates sweet and alcohol preference. Cell Metab. 23: 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.