Abstract
Background:
The epigenome may be an important interface between environmental chemical exposures and human health. However, the links between epigenetic modifications and health outcomes are often correlative and do not distinguish between cause and effect or common-cause relationships. The Adverse Outcome Pathway (AOP) framework has the potential to demonstrate, by way of an inference- and science-based analysis, the causal relationship between chemical exposures, epigenome, and adverse health outcomes.
Objective:
The objective of this work is to discuss the epigenome as a modifier of exposure effects and risk, perspectives for integrating toxicoepigenetic data into an AOP framework, tools for the exploration of epigenetic toxicity, and integration of AOP-guided epigenetic information into science and risk-assessment processes.
Discussion:
Organizing epigenetic information into the topology of a qualitative AOP network may help describe how a system will respond to epigenetic modifications caused by environmental chemical exposures. However, understanding the biological plausibility, linking epigenetic effects to short- and long-term health outcomes, and including epigenetic studies in the risk assessment process is met by substantive challenges. These obstacles include understanding the complex range of epigenetic modifications and their combinatorial effects, the large number of environmental chemicals to be tested, and the lack of data that quantitatively evaluate the epigenetic effects of environmental exposure.
Conclusion:
We anticipate that epigenetic information organized into AOP frameworks can be consistently used to support biological plausibility and to identify data gaps that will accelerate the pace at which epigenetic information is applied in chemical evaluation and risk-assessment paradigms. https://doi.org/10.1289/EHP2322
Introduction
The epigenome is a factor that can be modified by environmental chemical exposures. Epigenetic alterations (or the “epigenome”, when considering epigenetics on a genome-wide scale) include modifications to DNA, histone proteins that serve as a genome scaffold, and other factors including noncoding RNAs that collectively regulate the expression of genes without changing DNA sequence. In short, modification to the epigenome can lead to a change in phenotype without a change in genotype. Epigenetic states are malleable and are influenced by both intrinsic (e.g., age, sex, genetic polymorphisms) and extrinsic (e.g., environmental exposures, stress, diet) factors (Bowers and McCullough 2017). These “environmental factors” have quantitative effects on the epigenome that influence health outcomes across life stages and possibly across generations (Chamorro-García et al. 2013; Manikkam et al. 2012; Skinner et al. 2013; Tracey et al. 2013). The study of the epigenome causing adverse health effects of environmental exposures, or “toxicoepigenetics,” is a rapidly expanding field that has made marked progress in the past decade (Hansen et al. 2011; Roadmap Epigenomics Consortium et al. 2015). Yet, the possible biological pathways linking environmental exposures, their impacts on the epigenome, and the health effects from exposure are not entirely clear.
To help simplify the roles that epigenetics may play in mediating biological effects after exposure, epigenetic modifications can be reduced into three effect groups. The first, an “adverse effect,” can be defined as an exposure-induced epigenetic change that results in either an adverse outcome or a new epigenetic threshold vulnerable to a subsequent exposure. The second, an “adaptive effect,” can be defined as an exposure-induced change that primes the epigenome into a state protected from a subsequent exposure. The third, a “null effect,” can be defined as an exposure-induced epigenetic change with no adverse or adaptive outcome. Although null effects may pose no or low immediate risk to the affected individual, it is possible that null effects may emerge during sensitive life stages and/or propagate across generations. Further, with respect to all three effect types, the contribution of an exposure-induced epigenetic change to an adverse outcome can vary depending on the biological context (e.g., developmental windows of susceptibility and modulating factors such as disease). Nevertheless, the fundamental challenge to including epigenetic information in a risk assessment is identifying measurable causal relationships between epigenetic modifications and health outcomes.
A step toward discovering these causal relationships could include collecting and organizing existing scientific evidence into a framework that establishes evidence-based biological plausibility. The Adverse Outcome Pathway (AOP) paradigm (Ankley et al. 2010) may provide the framework needed to do this, that is, i.e., to link epigenetic study evidence (that may not inform mechanisms) with apical toxicological outcomes. AOPs are meant to describe how perturbation of a biological system leads to a particular adverse health outcome using components called molecular initiating events (MIEs), Key Events (KEs), Key Event Relationships (KERs), and Adverse Outcomes (AOs) that are supported by both biological plausibility and scientific evidence (Table 1) (Villeneuve et al. 2014a). Progression along the AOP may occur as compensatory mechanisms or feedback loops are overcome. AOPs originated from ecotoxicology as a means to extend toxicity pathways and modes of action into more holistic, yet systematic, descriptions of systems biology and ecology interactions that inform pathology (Ankley et al. 2010; Vinken et al. 2017). AOP development has been described previously (Villeneuve et al. 2014a, b), and specific guidance on developing AOPs is available (OECD 2013). AOPs are generally accepted by regulatory agencies, and they are being extended as a systematic framework for incorporating new and alternative methods into toxicity testing and risk assessment (OECD 2013, 2016; Tollefsen et al. 2014). Conceptually, individual AOPs are a network of components consisting of overlapping modules that can be experimentally measured (i.e., end points or biomarkers) and used to draw inferences about how a system will respond to a perturbation. AOP components are often depicted as linear paths, but in reality, the components are networked objects within a dimensional biological space. This concept is critical when considering that AOPs can be thought of as layered pages describing different pathways (e.g., DNA methylation, histone modification, gene expression) that collectively tell a story (supported by scientific evidence) relating exposure, complex biology, and outcome. A more detailed description of AOP networks and their applications can be found in two related papers (Knapen et al. 2018; Villeneuve et al. 2018). Collective AOP knowledge is integrated and disseminated to make it tractable for domestic and international stakeholders to map measured end-point or biomarker evidence to identify AOP networks and infer policy-relevant outcomes (see http://aopkb.org). AOPs, however, lack dosimetry and are qualitative; therefore, they must be transformed into a quantitative framework for practical application toward risk assessment.
Table 1.
Description of adverse outcome pathway (AOP) components (OECD 2016; Villeneuve et al. 2014a).
| Component | Description |
|---|---|
| Molecular initiating event (MIE) | The initial chemical–molecular interaction that is the starting point of an AOP. |
| Key event (KE) | A measurable change in biological state that is essential for progression along an AOP. |
| Key event relationship (KER) | A scientifically based connection that describes a directional relationship from one key event to another. |
| Adverse outcome (AO) | An apical end point that is generally viewed as having regulatory significance. |
Nonetheless, the AOP paradigm may yield a clear, transparent, scientific, and evidence-based approach to causally linking epigenome toxicity to otherwise correlative human health effects. This approach is generally applicable to exposure-related disease but requires expert curation. Therefore, this work extends the field of toxicoepigenetics by not only describing the biology of epigenetic modifications but also organizing those epigenome toxicity pathways into an AOP-like framework. Using this structured approach helps to clarify challenges to practical application of this knowledge as well as to experimental detection methods that might inform epigenetic mechanisms and other data gaps. The goal is to integrate existing science using an approach that provides evidence-based biological plausibility that is needed to apply observational and experimental information into the U.S. Environmental Protection Agency (EPA) risk assessment paradigm (NRC 2009).
Epigenetic Modifications
Epigenetic effects are often evaluated by examining modification of DNA, histones, noncoding RNA, and changes in chromatin structure. These modifications shape the genomic information that is used to control DNA accessibility and to regulate cell- and tissue-specific transcription, replication, and DNA repair. The hypothesis is that exposure to environmental factors leads to epigenetic (local) or epigenomic (global) effects that alter regulation or expression (or both) of exposure-responsive genes. Changing spatial and temporal patterns of gene regulation can affect organismal development and physiologic (e.g., metabolic, endocrine, cardiovascular, neurological, respiratory) function. However, although some may be causative of disease, many are merely transient and may have no phenotypic impact on health. Therefore, before organizing epigenetic information into a biologically plausible framework, it is important to understand what these modifications are and their impacts on biology.
DNA methylation.
To date, DNA methylation (5-methylcytosine; “5-mC”) is the most-studied epigenetic modification associated with exposure. Genomic regions enriched in 5-mC are typically transcriptionally repressed, compacted regions of the genome known as heterochromatin. Conversely, genomic regions where 5-mC is sparse or absent are typically associated with genes that are either active or poised to be induced. DNA methylation is dynamic and is mediated by DNA methyltransferases (DNMTs) and DNA demethylases (Ludwig et al. 2016). In mammals, DNMT3a and DNMT3b establish patterns of DNA methylation de novo during early embryo and germ cell development, and DNMT1 maintains DNA methylation during replication. DNA demethylation is mediated by passive dilution during DNA replication or by the Ten-eleven translocation (TET) family of DNA demethylases, which actively oxidize 5-mC to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine (Bochtler et al. 2017).
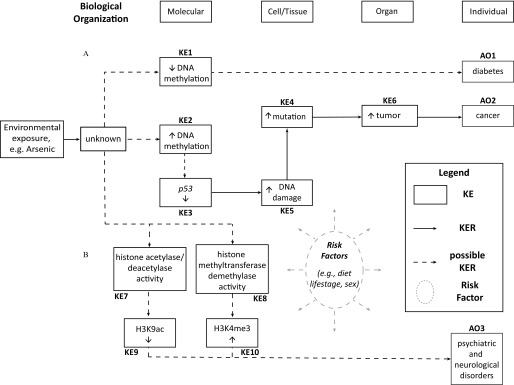
These mechanisms are by no means all-inclusive, but they are important when considering studies examining how extrinsic factors such as diet, early-life exposures, socioeconomic status (SES), and aging have affected DNA methylation patterns. For example, S-adenosyl methionine (SAM) is a methyl group source used during DNA methylation and is also consumed during the detoxification of inorganic arsenic (iAs) to mono- and dimethylated metabolites (MMA and DMA, respectively) (Bailey et al. 2013). In a cohort from Zamapan, Mexico, exposed to iAs [average ] in drinking water, (Bailey et al. 2013) reported a positive association between urinary MMA and DMA levels and peripheral blood leukocyte DNA hypomethylation in the promoters of type 1 and type 2 diabetes mellitus target genes. The same study participants were also evaluated for glycated hemoglobin (Hba1c), and 15 out of the 16 subjects had levels () indicative of prediabetes or diabetes status. The evidence can be mapped to an AOP-like framework with the caveat that AOPs are intended to be chemically agnostic. For example, people were exposed to arsenic in drinking water (Figure 1, left-most box), leading to an unknown molecular initiating effect. The ingested arsenic was metabolized and eliminated as urinary MMA and DMA that were positively associated with decreased DNA methylation in people with diabetes (Figure 1, KE1 and AO1). Although this example AOP is supported by limited and correlative evidence, it is an example of how one may begin organizing study data in a biologically plausible framework that is clearly and transparently supported by accompanying evidence tables (Tables 2 and 3).
Figure 1.
Putative Adverse Outcome Pathway (AOP) network for arsenic exposure. (A) Changes in DNA methylation leading to cancer and (B) changes in histone modifications leading to psychiatric and neurological disorders. The gray oval and gray dashed lines indicate modifying factors or “risk” factors, all dashed lines for weak weight of evidence or areas where there are data gaps. The up and down arrows within a key event (KE) box indicate the direction of change of a KE. Note: AO, adverse outcome; KER, key event relationship.
Table 2.
Evidence supporting adverse outcome pathway (AOP) network for DNA methylation (Figure 1A).
| Key event | Event description | Key event relationship | Reference | |||
|---|---|---|---|---|---|---|
| Exposure | ||||||
| IE | Arsenic Exposure | Sources of human exposure to inorganic arsenic (iAs) include drinking water, diet, air, and soils (which can contain naturally occurring arsenic or contamination from anthropogenic sources). | (ATSDR 2007) | |||
| Molecular | ||||||
| KE1 | Decreased DNA methylation | S-adenosyl methionine (SAM) is a methyl group source used during DNA methylation and is also consumed during the detoxification of iAs to mono- and dimethylated metabolites (MMA and DMA, respectively). The consumption of SAM during As metabolism may limit the availability of methyl donors. | (Bailey et al. 2013; Vahter and Marafante 1988). | |||
| KE2 | Increased DNA methylation of TP53 promoter | In a cohort from Zamapan, Mexico, exposed to iAs (average ) in drinking water, a positive association between hypermethylation of a TP53 network and arsenic-induced skin lesions was identified. | (Chanda et al. 2006; Smeester et al. 2011) | |||
| KE3 | Decreased TP53 gene expression | TP53 is a tumor suppressor and its loss of activity propagates DNA damage, the accumulation of mutations, and cancer. | (Levine and Oren 2009) | |||
| Cellular/Tissue | ||||||
| KE4 | Increased DNA damage | Loss of TP53 activity propagates DNA damage. | (Levine and Oren 2009) | |||
| KE5 | Increased DNA mutation | DNA damage may lead to the accumulation of mutations. | (Levine and Oren 2009). | |||
| Organ | ||||||
| KE6 | Increased tumors | Alterations in the activity of tumor suppressor genes is positively correlated with tumor progression. | (Yokota 2000) | |||
| Individual | ||||||
| AO1 | Diabetes | In a cohort from Zamapan, Mexico, exposed to iAs (average ) in drinking water, urinary MMA and DMA levels were positively associated with peripheral blood leukocyte DNA hypomethylation in the promoters of type 1 and type 2 diabetes mellitus target genes. | (Bailey et al. 2013). | |||
| AO2 | Cancer | Increased incidence of skin cancer was observed in populations drinking water with high iAs concentration. | (Tseng et al. 1968; Tseng 1977) | |||
Note: AO, adverse outcome; DMA, dimethylarsinic acid; IE, initiating event; KE, key event; MMA, monomethylarsonic acid.
Table 3.
Evidence supporting adverse outcome pathway (AOP) network for histone methylation (Figure 1B).
| Key event | Event description | Key event relationship | Reference |
|---|---|---|---|
| Exposure | |||
| IE | Arsenic exposure | Sources of human exposure to inorganic arsenic include drinking water, diet, air, and soils (which can contain naturally occurring arsenic or contamination from anthropogenic sources). | (ATSDR 2007) |
| Molecular | |||
| KE7 | Decreased histone acetylase or increased histone deacetylase activity | Arsenic exposure increased the histone acetyltransferase GCN5 in the DG, whereas GCN5 was decreased in the FC. | (Chervona et al. 2012) |
| KE8 | Increased histone methyltransferase or decreased histone demethylase activity | Developmental arsenic exposure ( decreased histone demethylase KDM5B and increased expression of the histone methyltransferase MLL in the DG and FC of male mice only. | (Tyler et al. 2015a, 2015b) |
| KE9 | Decreased H3K9ac | In vivo arsenic exposure was negatively correlated with global H3K9ac levels in peripheral blood mononuclear cells in a sex-specific manner. Histone acetyltransferase GCN5 was increased in the DG, whereas H3K9ac and GCN5 were decreased in the FC. | (Chervona et al. 2012) |
| KE10 | Increased H3K4me3 | Developmental arsenic exposure () increased global H3K4me3 levels. | (Tyler et al. 2015a, 2015b) |
| Individual | |||
| AO3 | Psychiatric and neurological disorders | Arsenic exposure was positively correlated with developing psychiatric disorders and cognitive dysfunction. | (Brinkel et al. 2009; Zierold et al. 2004) |
Note: AO, adverse outcome; DG, dentate gyrus; FC, frontal cortex; IE, initiating event; KE, key event.
Interestingly, the same cohort was used to identify a positive association between hypermethylation (Figure 1, KE2) of a TP53 network and arsenic-induced skin lesions (Smeester et al. 2011). This result was consistent with the findings of Chanda et al. (2006), who also identified an association between arsenic-induced skin lesions and hypermethylation of the TP53 promoter leading to decreased TP53 expression (KE3). TP53 is a tumor suppressor, and its loss of activity propagates DNA damage (KE4), the accumulation of mutations (KE4), and cancer (KE5) (Levine and Oren 2009) (Figure 1A). It may be that arsenic limits methyl donors, yet in mice prenatal folate supplementation worsened health outcomes in male offspring exposed to iAs in utero (Tsang et al. 2012). The lack of coherence between evidence streams is clarified by evidence mapping and may provide a platform for streamlining resources to address data gaps that might have otherwise been missed.
Histone modifications and chromatin accessibility.
DNA strands are more than 2 in length, yet they are condensed into a nucleus with a diameter of by packaging DNA onto nucleosomes that contain two molecules of the histone proteins H2A, H2B, H3, and H4. Nucleosomal packaging of DNA allows for high-order compaction but also creates the need for controlled decompaction so that cellular functions such as transcription, DNA replication, and DNA repair can occur. Histone proteins have N- and C-terminal tails that extend outward from the nucleosome core and are posttranslationally modified (Kouzarides 2007). Histone tail modifications act as the letters in an alphabet that direct the use of information encoded in the genome; this is mediated by alterations to histone-DNA interactions and the binding of specialized domains in chromatin-interacting proteins (so-called “readers”) (Strahl and Allis 2000). These modifications are added or removed by classes of enzymes (so-called “writers” and “erasers”) that are regulated by cellular signaling networks and thus are responsive to the effects of environmental exposures. Generally speaking, individual histone modifications are described as “activating” (facilitating open chromatin structure and accessibility of DNA to RNA polymerase) or “repressing” (facilitating a closed or condensed chromatin structure that is not accessible to RNA polymerase); however, studies have demonstrated a role for chromatin regions containing both activating and repressing modifications (“bivalent” domains) that regulate gene expression (Bernstein et al. 2006). Chromatin states can also vary depending on histone variants in nucleosomes. These variants are targeted by variant-specific modifications and chromatin-remodeling enzymes that regulate the density and distribution of nucleosomes within gene regulatory regions (Clapier and Cairns 2009; Venkatesh and Workman 2015). The collective effects of histone modifications, DNA methylation, histone variants, and nucleosome remodeling at a given locus (or globally) leads to regulation of both DNA accessibility and transcriptional initiation, elongation, and termination.
Histone acetylation.
Histone acetylation plays a key role in transcriptional regulation by modulating chromatin accessibility as well as transcriptional activator recruitment to gene regulatory regions. The acetylation of histone lysine residues is a reversible modification that is added by histone acetyltransferases (HATs) and removed by histone deacetylases (HDACs). Both HAT and HDAC enzymes, such as GCN5/KAT2A and HDAC1, respectively, are subunits in multiprotein complexes (Grant et al. 1997; Lee and Workman 2007; Yang and Seto 2008) that target and coordinate multiple histone reader, writer, and eraser functions. The acetylation of lysine residues on histone tails aids chromatin accessibility through two primary mechanisms. The first neutralizes positively charged lysine residues, leading to weakened histone-DNA interactions (Li and Reinberg 2011). This effect is most pronounced in H3 and H4, and the acetylation of a single residue on H4 (H4K16) is sufficient to disrupt proper chromatin folding (Kan et al. 2009). The second mechanism occurs when acetylated lysine residues are bound by bromodomains, a motif common in histone modifying and remodeling complexes. Bromodomain binding is directed by acetyl lysine and is further affected by other adjacent histone modifications (Filippakopoulos and Knapp 2012).
Histone methylation.
Histone tails are methylated and demethylated by histone methyltransferases (HMTs) and histone demethylases (HDMs), respectively, similar to histone acetylation. However, unlike the single acetylation of individual lysine residues, histone tails can be modified by mono- (me1), di- (me2), or trimethylation (me3) of the terminal amine group of lysine residues and monomethylation, asymmetric dimethylation (dimethylation of one of the two terminal amide groups), or symmetric dimethylation (monomethylation of both the terminal amide groups) of arginine residues (Kouzarides 2007). Further, whereas histone acetylation is generally associated with open chromatin, histone methylation is found in both open and condensed chromatin. The distribution of specific methylated lysine residues is often associated with distinct gene regulatory regions and function. Enhancers and active promoters are typically enriched with H3K4me1 and H3K4me3, respectively, whereas H3K36me2 and me3 are most abundant within the bodies of actively transcribed genes (Guenther et al. 2007; Heintzman et al. 2007). In contrast, H3K9me3, H3K27me3, and H4K20me3 are commonly associated with chromatin condensation (Kouzarides 2007). The location of these modifications is important for transcriptional regulation and is determined by the recruitment and binding of a broad range of histone methyl-lysine binding domains (Berger 2007; Martin and Zhang 2005). These interactions are the foundation of downstream biological implications of histone methylation.
A broad range of toxic exposures, from arsenic to air pollution, have been shown to alter histone acetylation and methylation patterns through the dysregulation of either HAT/HDAC and HMT/HDM expression or enzymatic activity. These effects can be similarly applied to the AOP framework and are outlined in Figure 1B and Table 3. Specifically using in vivo arsenic exposures as an example, global H3K9me2 and H3K9ac (KE9) levels in peripheral blood mononuclear cells were positively and negatively correlated with arsenic and supported that arsenic exposure led to the repression of gene expression in a sex-specific manner (Chervona et al. 2012). These arsenic-mediated sex-specific epigenetic differences were also observed during murine development in the dentate gyrus (DG) and the frontal cortex (FC) (Tyler et al. 2015a). Specifically, a developmental arsenic exposure increased global H3K4me3 (KE10) levels, decreased histone demethylase KDM5B, and increased the histone methyltransferase MLL expression in the DG and FC of male mice only (KE8) (Figure 1B). The H3K4me3 enrichment mapped to the regulatory regions of genes involved in cell development and growth, cell death and survival, and neurological disorders (AO3) (Tyler et al. 2015b). In a similar sex-specific manner, arsenic exposure increased H3K9ac and the histone acetyltransferase GCN5 in the DG, whereas H3K9ac (KE7) and GCN5 (KE8) were decreased in the FC of male mice. Evidence suggests that arsenic exposure during early development is associated with psychiatric and cognitive dysfunction, yet the biological pathway linking epigenetic mechanism to outcome is unclear.
Noncoding RNAs.
Noncoding RNAs include microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), PIWI-interacting RNAs (piRNAs) and long noncoding RNAs ( nucleotides, e.g., large intergenic noncoding RNAs (lincRNAs), circular and noncircular competitive endogenous RNAs (ceRNAs), and enhancer RNAs (eRNAs). Noncoding RNAs are derived from various parts of the genome (exonic, intronic, intergenic, etc.) and are processed posttranscriptionally in different ways. Long noncoding RNA can be processed into its mature form by RNase P, which cleaves transfer RNA (tRNA)-like subunits from termini of the mature lncRNA and is then stabilized by formation of a triple RNA helix (Wilusz et al. 2012). miRNA biogenesis has been shown to occur through Microprocessor complex processing of lncRNAs (Dhir et al. 2015), but miRNAs are primarily generated from intronic and exonic regions of protein-coding genes. Commonly, a long primary miRNA (pri-miRNA) is cleaved by the Microprocessor complex, resulting in a precursor miRNA (pre-miRNA), and exported from the nucleus by Exportin-5 and RanGTP (Slezak-Prochazka et al. 2010). Pre-miRNA is then further processed into a mature miRNA duplex by Dicer cleavage (Hutvágner et al. 2001). The sense or antisense strand is then loaded into the RNA-induced silencing complex (RISC) and is then used to target messenger RNA for degradation or to inhibit translation of the gene product.
MicroRNAs are altered by environmental chemical exposures, and these changes have been linked to disease and susceptibility. A panel of miRNAs were associated with short-term exposure of foundry workers to mixed particulate matter (PM) components: miR-122 was associated with lead exposure, miR-21 with 8-OH-dG, and miR-146 with lead and cadmium exposures (Bollati et al. 2010). In another observational study, miR-21 and miR-221 urine levels were positively associated with arsenic and lead levels in school-age children with microalbuminuria (Kong et al. 2012). Further, a panel of miRNAs found in cord blood were correlated with in utero arsenic exposure—including let-7a, miR-107, and miR-26b—that were also associated with cancer, diabetes, and immune response networks (Rager et al. 2014). Related to the AOP example in Figure 1, in vitro experimental studies do provide mechanistic evidence in support of both miR-200b– (Wang et al. 2011, 2014) and miR-190– (Beezhold et al. 2011) mediated cancer pathogenesis in response to arsenic dosing. Yet, collectively, the evidence streams lack coherence for anchoring exposure to possible pathways leading to adverse outcomes.
Despite the evidence that miRNA expression profiles were altered by environmental exposure, evidence that these alterations directly affected molecular progression to an adverse health outcome was unclear. However, these measurements may be valuable biomarkers of exposure. For example, miRNAs are detected in a number of different biofluids (e.g., blood, urine, sputum) and are stable in these matrices (Arroyo et al. 2011; Wang et al. 2010). These biofluid miRNA measurements are particularly valuable because they can indicate tissue-specific toxicity sooner and more robustly than traditional clinical protein biomarkers (Harrill et al. 2016). Other noncoding RNAs might also be important rapid or stable biomarkers of cellular response to exposures. For example, eRNAs are a type of lncRNA that promote gene enhancer and promoter interaction and transcriptional machinery recruitment through chromatin remodeling (Li et al. 2016). After cellular perturbation, eRNA response is rapid (), as with peroxisome proliferator activated receptor gamma ()-mediated up-regulation with rosiglitazone exposure in mature adipocytes, and is strongly associated with steady-state mRNA levels that mediate cellular response (Step et al. 2014). Similarly, circular competitive endogenous RNA (or circRNA) is a looped lncRNA derived from various regions of the genome. circRNAs may regulate miRNA (and other transcripts) by acting as a molecular “sponge” through multiple binding sites for specific miRNA, thereby limiting miRNA seed regions to interact and subsequently suppress messenger RNA targets (Hansen et al. 2013; Memczak et al. 2013). circRNAs have been linked to diseases such as cancer and, similar to miRNA, are stable, are present in circulating whole blood, and have tissue-specific origins (Hou and Zhang 2017).
Notably, there is debate whether noncoding RNAs should be considered an epigenetic mechanism because miRNA can be regulated by epigenetic mechanisms, and the epigenetic machinery itself can be regulated by miRNA expression (Saito et al. 2006; Tuddenham et al. 2006). For example, the miRNA (miR)-29 family can target the de novo methyltransferases DNMT3A and DNMT3B (Fabbri et al. 2007). In non-small cell lung cancer, this targeting leads to an aberrant pattern of DNA methylation in cancer cells. Further, DNA hypermethylation and increased lysine methylation of histones H2K9 and H3K27 can affect the persistent and decreased expression of miR-375 in mouse liver after withdrawal of the nongenotoxic carcinogen furan (de Conti et al. 2016). However, the interdependency of miRNA and other mechanisms that direct epigenetic phenotype is well documented and, therefore, is considered in the context of epigenetic events leading to adverse health outcomes.
Gap-Filling with Epigenetic and Epigenomic Methods
Early toxicoepigenetic studies have demonstrated that environmental exposures can alter the epigenome, yet the mechanistic linkage between exposure and epigenetic modification is not clear (iAs example, Figure 1). Chemicals could have a direct impact on the activity of epigenetic modification enzymes (HDACS, DNMTs, etc.), could block enzymes (such as DNMT) from accessing DNA by activating transcription factors that bind DNA, or both. These potential mechanisms are further complicated by the functional consequence of biallelic epigenetic-directed gene regulation (Cheung et al. 2017) and extrinsic factors, such as diet, that might limit the availability of methyl donors (Tsang et al. 2012). Furthermore, the premise that all epigenetic modifications lead to changes in gene expression questions the practical limitations and scope of some epigenetic assays that are expensive and do not readily allow for evaluation of environmental effects on multiple epigenetic modifications from a single sample. However, changes in gene expression may not always correlate with epigenetic modification. Further, measurement methods cannot be equally weighted owing to variation in assay sensitivity, specificity, accuracy, and precision. Therefore, recent advances in epigenetic assays, mainly focusing on various scalable assays that not only target DNA methylation and histone modifications but also phenotype, are presented with the understanding that the relevance of each approach to specific science questions may depend upon study quality aspects and prospective design.
Molecular approaches.
DNA methylation.
DNA methylation, that is, 5-mC, can be measured by a variety of whole genome and site-specific approaches. Whereas traditional quantitative methods measure cytosine methylation using high-performance liquid chromatography (HPLC) or mass spectrometry, a more common practice uses a bisulfite conversion step before analysis by sequencing. During bisulfite conversion, DNA is treated with sodium bisulfite, which chemically converts unmethylated cytosines to uracil, a base read during DNA sequencing as thymine. By contrast, methylated cytosines are protected and will be read as cytosines. The bisulfite conversion and sequencing method is often considered the gold standard in validating methylation results from other assays such as MeDIP (see Table 4). The type of approach to be used depends on the cellular context and the purpose, that is, to examine global or site-specific methylation profiles. Of the DNA methylation approaches included, reduced representation bisulfite sequencing (RRBS) often offers a good compromise in terms of coverage and cost: It enriches for CpG islands and repetitive elements, but coverage is lower than in the other methods. Of note, RRBS and other methylation-detection methods have limitations in terms of coverage, sensitivity, cost, and sample size that might be solved by bisulfite sequencing combined with bead arrays (Bibikova et al. 2006), an adaptation of Illumina’s high-throughput GoldenGate® single nucleotide polymorphism (SNP) sequencing technology, which allows multiplexed detection of the methylation status of 1,536 CpG sites for 371 genes in each well of a 96-well plate using of genomic DNA. Finally, 5-hydroxymethyl cytosine (5-hmC), a 5-mC degradation intermediate that has some potential functions of its own (Hackett et al. 2013; Kafer et al. 2016), is an increasingly important object of study, and techniques to identify its presence by adapting bisulfite conversion methods have been developed (Field et al. 2015).
Table 4.
Epigenetic modification detection methods.
| Modification | Detection method | Description | Pros/cons |
|---|---|---|---|
| DNA methylation | Methylation array | Bisulfite-treated DNA is hybridized to a DNA microarray, and the ratio of methylated to unmethylated CpGs is assessed. Includes Illumina-based bead arrays. | Genome-scale, site-specific measurements. Low to medium throughput. Low to medium cost. |
| Methylated DNA immunoprecipitation (MeDIP) | DNA is fragmented, and methylated regions are immunoprecipitated with 5-mC capture beads. Captured DNA is eluted and sequenced. | Genome-scale, site-specific measurements biased towards hypermethylated regions. Low throughput. Medium to high cost. | |
| Whole genome bisulfite sequencing (WGBS) | Bisulfite-treated DNA is sequenced without any enrichment, thereby measuring the entire genome. | Genome-wide measurements of DNA methylation. Low throughput. High cost. | |
| Reduced representation bisulfite sequencing (RRBS) | Similar to WGBS, but DNA is restriction-enzyme digested to enrich for CpG-rich DNA. DNA is then bisulfite-treated and sequenced. | Genome-scale, biased for CpG-rich DNA. Low throughput. Medium cost. | |
| Nucleosome positioning and histone modifications | Chromatin immunoprecipitation with sequencing (ChIP-seq) | Antibodies specific to histone modifications (i.e., those associated with active or repressed chromatin) are used to immunoprecipitate crosslinked DNA, which is then subject to next-generation sequencing. | Genome-wide measurement of histone location and modification type. Low throughput. Medium cost. |
| Drop-ChIP | Micrococcal nuclease (MNase), detergents, DNA barcodes, and sample DNA are combined using a microfluidic method to form nucleosomal regions flanked by DNA barcodes. The chromatin is immunoprecipitated, amplified, and sequenced. | Genome-scale measurements with single-cell profiling of chromatin state. Low throughput. Medium to high cost. | |
| Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) | Hyperactive Tn5 transposase is integrated into accessible regions of the genome. Adaptors tag the locus and serve as templates for PCR amplification of sequences. | Genome-scale measurements with high reproducibility and resolution. Low throughput. Medium to high cost. | |
| Chromatin desilencing | CMV-driven GFP reporter | A plasmid encoding GFP is stably transfected and selected for absence of GFP expression. The CMV-driven promoter is highly active, and its silencing suggests integration into a repressive chromatin environment. Subsequent desilencing results in GFP expression. | Phenotypic measurement not specific for a genomic region. High-throughput assessment of chromatin desilencing not applicable to transgenerational effects. Low cost. |
| Transgenic let-858::GFP Caenorhabditis elegans | The transgene let-858::GFP encodes a nuclear-localized GFP fusion protein and is expressed in all somatic nuclei. Under normal conditions at baseline, GFP is epigenetically silenced. C. elegans DNA is unmethylated. GFP expression would suggest chromatin desilencing by histone modification independent of DNA methylation. | In vivo. Phenotypic measurement not specific for a genomic region. High-throughput assessment independent of DNA methylation. Low cost. | |
| Developmental and life-stage effects | Daphnia | Alternative invertebrate model that offers several phenotypic end points affected by epigenetic regulation. A useful tool for Tier 2 or follow-up screening evaluating the effects of dose and duration. | Invertebrate, developmental, and life-stage model with long culture time. Low to high throughput. Low cost. |
Note: CMV, cytomegalovirus; GFP, green fluorescent protein; PCR, polymerase chain reaction.
Chromatin accessibility and histone modifications.
Several different methods currently exist to assess other chromatin features in addition to DNA methylation. These methods can be divided into two broad categories: those that assess nucleosome positioning as a proxy for active/inactive chromatin and those that more specifically examine various types of histone modifications commonly associated with either transcriptional activity or repression (such as H3K4me3 or H3K9me3, respectively) (Bock and Lengauer 2008; Tsompana and Buck 2014) (Table 4). In most cases, the large amount of chromatin needed to accurately assess chromatin composition has precluded the development of high-throughput methods. However, the Drop-ChIP (Rotem et al. 2015) and Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-Seq) approaches may circumvent this problem by profiling at the single-cell or well level (Table 4) (Bao et al. 2015; Buenrostro et al. 2015). Drop-ChIP is an adaptation of the chromatin immunoprecipitation and sequencing (ChIP-seq) method and combines the use of microfluidic devices with nucleosome tagging and chromatin immunoprecipitation steps (Rotem et al. 2015). The tags allow single-cell profiling of their chromatin but also, via the aggregation of data from multiple cells of the same type, reconstruction of a high-resolution profile (Rotem et al. 2015). Drop-ChIP has not yet been adapted as a high-throughput platform, but it is tentatively possible to produce droplets of different cell types in a sequential manner that would be fused with different sets of bar codes, thereby creating a multiplexed assay. In contrast, ATAC-Seq shows a high degree of reproducibility and resolution for mapping accessible (i.e., open, chromatin-rich) regions, and requires little sample input. Thus, this method probes for genomic regions of lower nucleosome density and has been adapted for cells in culture in a 96-well format (Bao et al. 2015). Drop-ChIP and ATAC-Seq represent two practical and advantageous methods that require low material input and provide high-resolution chromatin profiling and mapping. These methods may be very useful for assessing epigenetic changes in response to multiple chemicals or in dose–response applications.
Phenotypic approaches.
In contrast to the direct-measure molecular approaches, phenotypic approaches have the advantage of directly screening chemicals for epigenetic effects. These methods could be used for chemical screening followed by measuring epigenetic marks through the assays described above. Several approaches are presented, in cell culture and in vivo, that may be leveraged for a chemical’s epigenetic impact.
Cell culture setting.
Martinez and colleagues describe the use of cytomegalovirus (CMV)-driven green fluorescent protein (GFP) reporter in mouse or human cell lines for chemical screening (Johnson et al. 2008; Martinez et al. 2006). In their assay scheme, a plasmid encoding GFP and the neo gene is transfected and is then selected for integration over several passages using G418 selection. Stable lines are then selected for both G18 resistance and absence of GFP expression, surmising that silencing of the highly transcriptionally active CMV promoter would represent an integration in a repressive chromatin environment. This approach was first validated using HDAC inhibitors and was then applied to small molecule screening (Johnson et al. 2008). By performing a screen against 69,137 small molecules, the authors were able to identify novel chemicals that acted as chromatin desilencers and to extract structure–activity relationships. This tantalizing approach is similar to the nematode assay described below and has the advantage of high scalability, but it does not allow the monitoring of epigenetic effects across generations.
Caenorhabditis elegans.
The nematode Caenorhabditis elegans is a free-living roundworm that has been extensively used as a model system for the study of a wide variety of cellular and developmental processes. Of particular relevance is that C. elegans lacks 5-mC, and it has been used for the identification of novel chromatin-regulating factors as well as for toxicity studies (Ferreira and Allard 2015; Lundby et al. 2016; Parodi et al. 2015). A transgenic let-858::GFP roundworm has recently been adapted in a fluorescence-based assay that evaluates chemical effects on removal of chromatin marks such as H3K4 methylation and the addition of repressive marks such as H3K9 methylation independent of DNA methylation in the germline (Table 4) (Kelly et al. 1997). The strain can therefore be used as a sensor to detect chemical exposures that lead to chromatin deregulation and desilencing. Worms are amenable to culture in 96-well plates and can be imaged in 96- or 384-well plates; therefore, the method is potentially scalable. Initial validation of the assay for chemical screening has been reported using known chemical inhibitors of chromatin-modifying enzymes such as valproic acid, a well-characterized histone deacetylase inhibitor (Lundby et al. 2016).
Daphnia.
The microcrustacean Daphnia collectively refers to a collection of species, most commonly D. magna and D. pulex, that have been used for ecological, environmental, and genetic studies (Harris et al. 2012). Daphnia offers several key advantages for epigenetic studies compared with other, perhaps more common, model organisms such as C. elegans and Drosophila melanogaster. First, Daphnia research over the last 150 y has generated a wealth of ecological data that encompass a deep understanding of its response to a variety of ecotoxicological conditions (Harris et al. 2012; Stollewerk 2010). Second, Daphnia shows a repertoire of epigenetic modifications that also include DNA methylation, a mark that has been shown to be responsive to environmental exposures (Robichaud et al. 2012; Vandegehuchte et al. 2009). Third, Daphnia species offer a wide variety of characteristics and phenotypes, such as egg type, growth, fertility and body morphs, that are highly dependent on environmental cues and on epigenetic marks. Variants in body morphs, such as Daphnia exhibiting a helmet or a neck tooth formation, are induced in response to the presence of predators, whereas variants in body length (e.g., growth) and reproduction (e.g., number of progeny) have been shown to be sensitive to chemical inhibitors of DNA methylation such as 5-azacytidine (5-aza) and the pesticide vinclozolin (Vandegehuchte et al. 2010). For example, 5-aza reduces both length and reproduction in the parental generation (F0) of D. magna as well as in their progeny (F1; Vandegehuchte et al. 2010). However, vinclozolin only affects growth in the F0 and does not affect reproduction (Harris et al. 2012; Vandegehuchte et al. 2010). With their small size, , Daphnia species are amenable to in vitro culture conditions and assays. However, the chemical characterization of their epigenetic regulation requires culture over several days (up to 7 d). Although slow, this alternative model is a valuable tool for the validation of epigenetic effects initially observed in other model systems.
The assays described above all have the potential to illuminate the study of environmental influences on the epigenome. The choice of strategy is highly dependent on the context of the study. For example, use of the C. elegans model may not offer the best context to study the epigenomic perturbation in a cell type that can be simply obtained from human or mouse. However, the high degree of conservation of epigenetic pathways and the ability to easily follow epigenetic modifications over several generations make this model highly suitable for transgenerational studies. The choice of platform is also dependent on the amount of material and the degree of data complexity required. For example, ATAC-seq requires little starting material and can provide a high-resolution map of genomic loci of higher chromatin accessibility. However, this method will not provide direct mechanistic insight into local epigenetic alterations such as changes in methylation or histone mark patterns. In those instances, RRBS or Drop-ChIP might be better options even if slightly more material is needed. Finally, cross-platform validation is likely to increase the reliability of the outcome. For example, one could start screening chemicals for a phenotypic readout, such as the use of the cell culture model described above, and follow the positive hits with ATAC-seq to assess chromatin relaxation across the genome.
Methodological Challenges.
To determine whether epigenetic changes associated with exposure might contribute to adverse health outcomes, a baseline epigenetic state is needed for comparison. Several large research consortia have focused on generating reference epigenomic maps from normal human cell and tissue types, including the National Institutes of Health (NIH) Roadmap Epigenomics Program (http://www.roadmapepigenomics.org/) (Roadmap Epigenomics Consortium et al. 2015) and the International Human Epigenome Consortium (http://ihec-epigenomes.org) (Stunnenberg et al. 2016). Reference maps such as these can be useful guides for interpreting exposure-associated epigenetic differences. These reference maps help to distinguish changes that may be directly linked to the exposure from changes that are more reflective of the relative ratios of specific cell types that make up a heterogeneous tissue sample. In essence, sample source and cell type heterogeneity (i.e., human genetic heterogeneity) are problematic in epigenomic studies because human samples often consist of blood (owing to ease of access) that is composed of mixed cell types. Each cell type has a unique epigenetic profile, and the relative proportion of each cell type in a sample may vary as a result of exposure (Stiegel et al. 2016). Reference resources and maps for a given cell type are typically collected from a limited number of people and provide no information about baseline epigenetic variation at a given locus. The implication is that high levels of underlying epigenetic variation could make it difficult to distinguish exposure-induced “signal” from background noise.
Discussion
Interpreting the Evidence
There is a clear challenge to linking epigenetic effects with outcomes in chemical risk assessment that supports the use of the AOP framework to organize epigenetic data into an inference-based biologically plausible framework. Integration of the evidence into conceptual frameworks, such as those depicted in Figure 1 and supported by Tables 2 and 3, improves interpretation of scientific information that, if not properly contextualized, is otherwise perceived as tenuous. For example, arsenic (and likely many other environmental toxicants) is not directly mutagenic, but it invokes epigenetic changes that may dysregulate the expression of tumor suppressors or oncogenes (Shinjo and Kondo 2015). Furthermore, epigenome reprogramming occurs during early embryonic and primordial germ cell development (Hackett and Surani 2013) and may restore epigenetic marks altered by prior exposure. However, exposures that occur during these developmentally sensitive windows may also lead to permanent reprogramming of the methylome, thus playing a role in the developmental origins of health and disease (Desai et al. 2013; Newbold et al. 2007), possibly including malignancy.
AOP evidence mapping exercises are broadly applicable to other environmental toxicants (e.g., metals, trichloroethylene and its metabolites, air pollution, benzene) that have been characterized as inducing epigenetic toxicity and have been reviewed (Bowers and McCullough 2017; Hodjat et al. 2017). The implication is that real-world exposures uniquely alter pharmacodynamics or molecular initiating effects that propagate across biology to achieve homeostasis, similar to a ripple in a pond, and converge at key events. These key events can be explicitly measured, used to infer outcome, and interpreted in the context of additional risk and modulating factors, such as genetic susceptibility, disease status, or nutrition. These concepts are critical for evaluating epigenetic modifications where the adversity depends on adaptive, adverse, or null effect types.
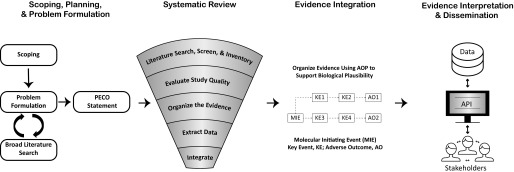
There is light at the end of the tunnel for epigenetic researchers hoping to intersect with the risk assessment community. Evidence-based mapping (ideally guided by a systematic literature search) can be used to help formulate the questions and to direct future research and resources (James et al. 2016) to areas where further scientific understanding of epigenetics is needed. Figure 2 outlines a general approach for evaluating an environmental protection goal where the state of the science is systematically analyzed using structured methods to identify existing studies that address the relevant questions (e.g., are epigenetic modifications adverse?). The relevant body of literature is systematically evaluated, considering the design, methods, conduct, and documentation irrespective of the study results. Assessing the study quality is an essential step to rule out bias before extracting results for interpretation (Ågerstrand et al. 2011; Beronius et al. 2014a, 2014b; Sterne et al. 2016). Even so, evaluating the significance of observational and experimental response data without proper biological context could result in meaningless and confusing interpretation. Organizing the evidence into a biologically based AOP framework can provide a roadmap from effect to outcome, help shape future research questions, and fill (or highlight) evidence gaps.
Figure 2.
Integration of the Adverse Outcome Pathway (AOP) framework into the risk-assessment process. The figure above depicts how the AOP framework could be used to organize study data supporting a scientific assessment. Although a systematic literature search was not performed in this example, a protocol including exposure and end point effects of interest (such as epigenetic modification) can be included in the scoping, planning, and problem formulation phase (column 1) and implemented during the systematic review (column 2). Relevant study evidence could then be assessed for quality and organized using an AOP framework (column 3) to improve interpretation. This workflow could build confidence in the assessment process by efficiently and transparently disseminating scientific information, perhaps through a public database accessible to stakeholders through an application programming interface.
Confidence in epigenetic data supported by an AOP framework may be enhanced by including weight of evidence and visuals. Weight of evidence guidance for the AOP framework has been previously described in the Organisation for Economic Co-operation and Development (OECD) (2016) Users’ Handbook supplement to the Guidance Document on the Reporting of Defined Approaches to be Used within Integrated Approaches to Testing and Assessment that identifies biological plausibility, empirical support, and uncertainties or inconsistencies that one key event will lead to another that can be cited from the literature. These evidence streams could be depicted by solid versus dashed arrows (Figure 1), the weight of an arrow or outline of a box, or some other type of quantitative visualization. A table with references, doses, study details, study types (epidemiologic, experimental, or controlled human exposure) could also complement the AOP. Ultimately, however, these qualitative AOP frameworks are more useful for hazard identification and would require translation into a framework that captures a quantitative understanding between exposure dose and effect. Nonetheless, organizing evidence using the AOP framework is an approach to clearly evaluating the biological evidence and should boost confidence during interpretation and information dissemination.
Conclusion
Toxicoepigenetics is a dynamic field that may have broad implications in human health and disease; however, there are challenges to be overcome before epigenetic data can be readily integrated into the chemical evaluation process. Organizing epigenetic information into AOP frameworks may help identify cause-and-effect relationships that help guide a science-based analysis of how a system will respond to epigenetic perturbations. The concept of harmonizing toxicological information using a broad toxicological framework to improve decision making is not new and was recently reviewed (Edwards et al. 2016). This approach could address the “basic problem in risk assessment … the incompleteness of data” (NRC 2009, p. 22). It is possible that epigenetic data can be layered with other diverse data streams and be used to inform events within the AOP framework so that there is sufficient evidence to conclude clear relationships between exposure, epigenetic modification, and adverse outcome. Yet, the success of the AOP framework and the pace at which epigenetic information is incorporated into the risk-assessment process depend on expert-driven AOP development and broad participation from the toxicoepigenetic community. The AOP paradigm may provide a platform for integrating toxicoepigenetic information with other biological pathways so that we may better understand if exposure-mediated epigenetic effects have an impact on adverse outcomes. This work breaks down barriers to incorporating epigenetic studies in science-based chemical risk assessment by describing a practical and integrative science-based approach.
Acknowledgment
The authors would like to thank D. Diaz Sanchez, J. Rogers, S. Dutton, L. D-Amico, and J. Vandenberg for their technical review and A. Jarabek for scientific advice.
Funding for this study came from the U.S. Environmental Protection Agency Office of Research and Development. The research described in this article has been reviewed by the National Center for Environmental Assessment of U.S. Environmental Protection Agency and approved for publication. The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
References
- Ågerstrand M, Küster A, Bachmann J, Breitholtz M, Ebert I, Rechenberg B, et al. 2011. Reporting and evaluation criteria as means towards a transparent use of ecotoxicity data for environmental risk assessment of pharmaceuticals. Environ Pollut 159(10):2487–2492, PMID: 21763042, 10.1016/j.envpol.2011.06.023. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, et al. 2010. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem 29(3):730–741, PMID: 20821501, 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 108(12):5003–5008, PMID: 21383194, 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ATSDR (Agency for Toxic Substances and Disease Registry). 2007. Toxicological Profile for Arsenic. Atlanta, GA:ATSDR; https://www.atsdr.cdc.gov/toxprofiles/tp2.pdf [accessed 16 November 2017]. [PubMed] [Google Scholar]
- Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, García-Vargas G, et al. 2013. Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. J Biochem Mol Toxicol 27(2):106–115, PMID: 23315758, 10.1002/jbt.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Rubin AJ, Qu K, Zhang J, Giresi PG, Chang HY, et al. 2015. A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol 16:284, PMID: 26683334, 10.1186/s13059-015-0840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beezhold K, Liu J, Kan H, Meighan T, Castranova V, Shi X, et al. 2011. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol Sci 123(2):411–420, PMID: 21750348, 10.1093/toxsci/kfr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. 2007. The complex language of chromatin regulation during transcription. Nature 447(7143):407–412, PMID: 17522673, 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125(2):315–326, PMID: 16630819, 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Beronius A, Hanberg A, Zilliacus J, Rudén C. 2014a. Bridging the gap between academic research and regulatory health risk assessment of endocrine disrupting chemicals. Curr Opin Pharmacol 19:99–104, PMID: 25238457, 10.1016/j.coph.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Beronius A, Molander L, Rudén C, Hanberg A. 2014b. Facilitating the use of non-standard in vivo studies in health risk assessment of chemicals: a proposal to improve evaluation criteria and reporting. J Appl Toxicol 34(6):607–617, PMID: 24481642, 10.1002/jat.2991. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. 2006. High-throughput DNA methylation profiling using universal bead arrays. Genome Res 16(3):383–393, PMID: 16449502, 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Kolano A, Xu GL. 2017. DNA demethylation pathways: additional players and regulators. Bioessays 39(1):1–13, PMID: 27859411, 10.1002/bies.201600178. [DOI] [PubMed] [Google Scholar]
- Bock C, Lengauer T. 2008. Computational epigenetics. Bioinformatics 24(1):1–10, PMID: 18024971, 10.1093/bioinformatics/btm546. [DOI] [PubMed] [Google Scholar]
- Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, et al. 2010. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect 118(6):763–768, PMID: 20061215, 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers EC, McCullough SD. 2017. Linking the epigenome with exposure effects and susceptibility: the epigenetic seed and soil model. Toxicol Sci 155(2):302–314, PMID: 28049737, 10.1093/toxsci/kfw215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkel J, Khan MH, Kraemer A. 2009. A systematic review of arsenic exposure and its social and mental health effects with special reference to Bangladesh. Int J Environ Res Public Health 6(5):1609–1619, PMID: 19543409, 10.3390/ijerph6051609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109(1): 21.29.1–21.29.9, PMID: 25559105, 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-García R, Sahu M, Abbey RJ, Laude J, Pham N, Blumberg B. 2013. Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ Health Perspect 121(3):359–366, PMID: 23322813, 10.1289/ehp.1205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S, Dasgupta UB, Guhamazumder D, Gupta M, Chaudhuri U, Lahiri S, et al. 2006. DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol Sci 89(2):431–437, PMID: 16251483, 10.1093/toxsci/kfj030. [DOI] [PubMed] [Google Scholar]
- Chervona Y, Hall MN, Arita A, Wu F, Sun H, Tseng HC, et al. 2012. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol Biomarkers Prev 21(12):2252–2260, PMID: 23064002, 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WA, Shao X, Morin A, Siroux V, Kwan T, Ge B, et al. 2017. Functional variation in allelic methylomes underscores a strong genetic contribution and reveals novel epigenetic alterations in the human epigenome. Genome Biol 18(1):50, PMID: 28283040, 10.1186/s13059-017-1173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304, PMID: 19355820, 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- de Conti A, Tryndyak V, Doerge DR, Beland FA, Pogribny IP. 2016. Irreversible down-regulation of miR-375 in the livers of Fischer 344 rats after chronic furan exposure. Food Chem Toxicol 98(Part A):2–10, PMID: 27371368, 10.1016/j.fct.2016.06.027. [DOI] [PubMed] [Google Scholar]
- Desai M, Beall M, Ross MG. 2013. Developmental origins of obesity: programmed adipogenesis. Curr Diab Rep 13(1):27–33, PMID: 23188593, 10.1007/s11892-012-0344-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Proudfoot NJ, Jopling CL. 2015. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol 22(4):319–327, PMID: 25730776, 10.1038/nsmb.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SW, Tan YM, Villeneuve DL, Meek ME, McQueen CA. 2016. Adverse outcome pathways-organizing toxicological information to improve decision making. J Pharmacol Exp Ther 356(1):170–181, PMID: 26537250, 10.1124/jpet.115.228239. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. 2007. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 104(40):15805–15810, PMID: 17890317, 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira DW, Allard P. 2015. Models of germ cell development and their application for toxicity studies. Environ Mol Mutagen 56(8):637–649, PMID: 25821157, 10.1002/em.21946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field SF, Beraldi D, Bachman M, Stewart SK, Beck S, Balasubramanian S. 2015. Accurate measurement of 5-methylcytosine and 5-hydroxymethylcytosine in human cerebellum DNA by oxidative bisulfite on an array (OxBS-array). PloS One 10(2):e0118202, PMID: 25706862, 10.1371/journal.pone.0118202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. 2012. The bromodomain interaction module. FEBS Lett 586(17):2692–2704, PMID: 22710155, 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11(13):1640–1650, PMID: 9224714, 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130(1):77–88, PMID: 17632057, 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, et al. 2013. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339(6118):448–452, PMID: 23223451, 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Surani MA. 2013. Beyond DNA: programming and inheritance of parental methylomes. Cell 153(4):737–739, PMID: 23663772, 10.1016/j.cell.2013.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43(8):768–775, PMID: 21706001, 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388, PMID: 23446346, 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Harrill AH, McCullough SD, Wood CE, Kahle JJ, Chorley BN. 2016. MicroRNA biomarkers of toxicity in biological matrices. Toxicol Sci 152(2):264–272, PMID: 27462126, 10.1093/toxsci/kfw090. [DOI] [PubMed] [Google Scholar]
- Harris KD, Bartlett NJ, Lloyd VK. 2012. Daphnia as an emerging epigenetic model organism. Genet Res Int 2012:147892, PMID: 22567376, 10.1155/2012/147892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39(3):311–318, PMID: 17277777, 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hodjat M, Rahmani S, Khan F, Niaz K, Navaei-Nigjeh M, Mohammadi Nejad S, et al. 2017. Environmental toxicants, incidence of degenerative diseases, and therapies from the epigenetic point of view. Arch Toxicol 91(7):2577–2597, PMID: 28516248, 10.1007/s00204-017-1979-9. [DOI] [PubMed] [Google Scholar]
- Hou LD, Zhang J. 2017. Circular RNAs: An emerging type of RNA in cancer. Int J Immunopathol Pharmacol 30(1):1–6, PMID: 28134598, 10.1177/0394632016686985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. 2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293(5531):834–838, PMID: 11452083, 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- James KL, Randall NP, Haddaway NR. 2016. A methodology for systematic mapping in environmental sciences. Environ Evid 5:7, 10.1186/s13750-016-0059-6. [DOI] [Google Scholar]
- Johnson RL, Huang W, Jadhav A, Austin CP, Inglese J, Martinez ED. 2008. A quantitative high-throughput screen identifies potential epigenetic modulators of gene expression. Anal Biochem 375(2):237–248, PMID: 18211814, 10.1016/j.ab.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer GR, Li X, Horii T, Suetake I, Tajima S, Hatada I, et al. 2016. 5-Hydroxymethylcytosine marks sites of DNA damage and promotes genome stability. Cell Rep 14(6):1283–1292, PMID: 26854228, 10.1016/j.celrep.2016.01.035. [DOI] [PubMed] [Google Scholar]
- Kan PY, Caterino TL, Hayes JJ. 2009. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol 29(2):538–546, PMID: 19001093, 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146(1):227–238, PMID: 9136012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapen D, Angrish M, Fortin M, Katsiadaki I, Lènoard M, Margiotta-Casaluci L, et al. 2018. Adverse outcome pathway networks I: development and applications. Environ Toxicol Chem, PMID: 29488651, 10.1002/etc.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong AP, Xiao K, Choi KC, Wang G, Chan MH, Ho CS, et al. 2012. Associations between microRNA (miR-21, 126, 155 and 221), albuminuria and heavy metals in Hong Kong Chinese adolescents. Clin Chim Acta 413(13–14):1053–1057, PMID: 22405870, 10.1016/j.cca.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128(4):693–705, PMID: 17320507, 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol 8(4):284–295, PMID: 17380162, 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Oren M. 2009. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 9(10):749–758, PMID: 19776744, 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Reinberg D. 2011. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 21(2):175–186, PMID: 21342762, 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17(4):207–223, PMID: 26948815, 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- Ludwig AK, Zhang P, Cardoso MC. 2016. Modifiers and readers of DNA modifications and their impact on genome structure, expression, and stability in disease. Front Genet 7:115, PMID: 27446199, 10.3389/fgene.2016.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby Z, Camacho J, Allard P. 2016. Fast functional germline and epigenetic assays in the nematode Caenorhabditis elegans. Methods Mol Biol 1473:99–107, PMID: 27518628, 10.1007/978-1-4939-6346-1_11. [DOI] [PubMed] [Google Scholar]
- Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK. 2012. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One 7(2):e31901, PMID: 22389676, 10.1371/journal.pone.0031901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. 2005. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6(11):838–849, PMID: 16261189, 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Martinez ED, Dull AB, Beutler JA, Hager GL. 2006. High-content fluorescence-based screening for epigenetic modulators. Meth Enzymol 414:21–36, PMID: 17110184, 10.1016/S0076-6879(06)14002-1. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338, PMID: 23446348, 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Newbold RR, Padilla-Banks E, Snyder RJ, Phillips TM, Jefferson WN. 2007. Developmental exposure to endocrine disruptors and the obesity epidemic. Reprod Toxicol 23(3):290–296, PMID: 17321108, 10.1016/j.reprotox.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC (National Research Council). 2009. “Science and Decisions: Advancing Risk Assessment.” Washington, DC: National Academies Press; https://www.nap.edu/catalog/12209/science-and-decisions-advancing-risk-assessment [accessed 9 June 2017]. [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co-operation and Development). 2013. “Guidance Document on Developing and Assessing Adverse Outcome Pathways. (Series on Testing and Assessment, No. 184).” Environment Directorate Paris, France:Organisation for Economic Cooperation and Development; http://search.oecd.org/officialdocuments/displaydocumentpdf/?cote=env/jm/mono(2013)6&doclanguage=en [accessed 9 June 2017]. [Google Scholar]
- OECD. 2016. “Guidance Document for the Use of Adverse Outcome Pathways in Developing Integrated Approaches to Testing and Assessment (IATA) Series on Testing & Assessment No. 260.” Paris, France:Environment Directorate OECD; https://one.oecd.org/document/ENV/JM/MONO(2016)67/en/pdf [accessed 9 June 2017]. [Google Scholar]
- Parodi DA, Damoiseaux R, Allard P. 2015. Comprehensive assessment of germline chemical toxicity using the nematode Caenorhabditis elegans. J Vis Exp (96):e52445, PMID: 25741987, 10.3791/52445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager JE, Moeller BC, Miller SK, Kracko D, Doyle-Eisele M, Swenberg JA, et al. 2014. Formaldehyde-associated changes in microRNAs: Tissue and temporal specificity in the rat nose, white blood cells, and bone marrow. Toxicol Sci 138(1):36–46, PMID: 24304932, 10.1093/toxsci/kft267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518(7539):317–330, PMID: 25693563, 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud NF, Sassine J, Beaton MJ, Lloyd VK. 2012. The epigenetic repertoire of Daphnia magna includes modified histones. Genet Res Int 2012:174860, PMID: 22567378, 10.1155/2012/174860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, et al. 2015. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 33(11):1165–1172, PMID: 26458175, 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. 2006. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9(6):435–443, PMID: 16766263, 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Shinjo K, Kondo Y. 2015. Targeting cancer epigenetics: linking basic biology to clinical medicine. Adv Drug Deliv Rev 95:56–64, PMID: 26494398, 10.1016/j.addr.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Tracey R, Guerrero-Bosagna C, Haque M, Nilsson EE. 2013. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med 11:228, PMID: 24228800, 10.1186/1741-7015-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. 2010. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA 16(6):1087–1095, PMID: 20423980, 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeester L, Rager JE, Bailey KA, Guan X, Smith N, García-Vargas G, et al. 2011. Epigenetic changes in individuals with arsenicosis. Chem Res Toxicol 24(2):165–167, PMID: 21291286, 10.1021/tx1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Step SE, Lim HW, Marinis JM, Prokesch A, Steger DJ, You SH, et al. 2014. Anti-diabetic rosiglitazone remodels the adipocyte transcriptome by redistributing transcription to PPARγ-driven enhancers. Genes Dev 28(9):1018–1028, PMID: 24788520, 10.1101/gad.237628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne JA, Hernán MA, Reeves BC, Savović J, Berkman ND, Viswanathan M, et al. 2016. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ 355:i4919, PMID: 27733354, 10.1136/bmj.i4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegel MA, Pleil JD, Sobus JR, Madden MC. 2016. Inflammatory cytokines and white blood cell counts response to environmental levels of diesel exhaust and ozone inhalation exposures. PLoS One 11(4):e0152458, PMID: 27058360, 10.1371/journal.pone.0152458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollewerk A. 2010. The water flea Daphnia–a 'new' model system for ecology and evolution? J Biol 9:21, PMID: 20478012, 10.1186/jbiol212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403(6765):41–45, PMID: 10638745, 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Stunnenberg HG, International Human Epigenome Consortium, Hirst M. 2016. The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell 167(5):1145–1149, PMID: 27863232, 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Tollefsen KE, Scholz S, Cronin MT, Edwards SW, de Knecht J, Crofton K, et al. 2014. Applying adverse outcome pathways (AOPs) to support integrated approaches to testing and assessment (IATA). Regul Toxicol Pharmacol 70(3):629–640, PMID: 25261300, 10.1016/j.yrtph.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Tracey R, Manikkam M, Guerrero-Bosagna C, Skinner MK. 2013. Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod Toxicol 36:104–116, PMID: 23453003, 10.1016/j.reprotox.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang V, Fry RC, Niculescu MD, Rager JE, Saunders J, Paul DS, et al. 2012. The epigenetic effects of a high prenatal folate intake in male mouse fetuses exposed in utero to arsenic. Toxicol Appl Pharmacol 264(3):439–450, PMID: 22959928, 10.1016/j.taap.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng WP, Chu HM, How SW, Fong JM, Lin CS, Yeh S. 1968. Prevalence of skin cancer in an endemic area of chronic arsenicism in Taiwan. J Natl Cancer Inst 40(3):453–463, PMID: 5644201. [PubMed] [Google Scholar]
- Tseng WP. 1977. Effects and dose–response relationships of skin cancer and blackfoot disease with arsenic. Environ Health Perspect 19:109–119, PMID: 908285, 10.1289/ehp.7719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompana M, Buck MJ. 2014. Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7(1):33, PMID: 25473421, 10.1186/1756-8935-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, et al. 2006. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 580(17):4214–4217, PMID: 16828749, 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- Tyler CR, Hafez AK, Solomon ER, Allan AM. 2015a. Developmental exposure to 50 parts-per-billion arsenic influences histone modifications and associated epigenetic machinery in a region- and sex-specific manner in the adult mouse brain. Toxicol Appl Pharmacol 288(1):40–51, PMID: 26193056, 10.1016/j.taap.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler CR, Weber JA, Labrecque M, Hessinger JM, Edwards JS, Allan AM. 2015b. ChIP-seq analysis of the adult male mouse brain after developmental exposure to arsenic. Data Brief 5:248–254, PMID: 26543888, 10.1016/j.dib.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahter M, Marafante E. 1988. In Vivo Methylation and Detoxification of Arsenic. Oxford, UK:Royal Society of Chemistry. [Google Scholar]
- Vandegehuchte MB, Kyndt T, Vanholme B, Haegeman A, Gheysen G, Janssen CR. 2009. Occurrence of DNA methylation in Daphnia magna and influence of multigeneration Cd exposure. Environ Int 35(4):700–706, PMID: 19249097, 10.1016/j.envint.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Vandegehuchte MB, Lemière F, Vanhaecke L, Vanden Berghe W, Janssen CR. 2010. Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp Biochem Physiol C Toxicol Pharmacol 151(3):278–285, PMID: 19961956, 10.1016/j.cbpc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL. 2015. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 16(3):178–189, PMID: 25650798, 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- Villeneuve D, Angrish M, Fortin M, Katsiadaki I, Lènoard M, Margiotta-Casaluci L, et al. 2018. Adverse outcome pathway networks II: network analytics. Environ Toxicol Chem, 10.1002/etc.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, et al. 2014a. Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol Sci 142(2):312–320, PMID: 25466378, 10.1093/toxsci/kfu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, et al. 2014b. Adverse outcome pathway development II: best practices. Toxicol Sci 142(2):321–330, PMID: 25466379, 10.1093/toxsci/kfu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken M, Knapen D, Vergauwen L, Hengstler JG, Angrish M, Whelan M. 2017. Adverse outcome pathways: A concise introduction for toxicologists. Arch Toxicol 91(11):3697–3707, PMID: 28660287, 10.1007/s00204-017-2020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, Galas DJ. 2010. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 38(20):7248–7259, PMID: 20615901, 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Humphries B, Xiao H, Jiang Y, Yang C. 2014. MicroRNA-200b suppresses arsenic-transformed cell migration by targeting protein kinase Cα and Wnt5b-protein kinase Cα positive feedback loop and inhibiting Rac1 activation. J Biol Chem 289(26):18373–18386, PMID: 24841200, 10.1074/jbc.M114.554246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhao Y, Smith E, Goodall GJ, Drew PA, Brabletz T, et al. 2011. Reversal and prevention of arsenic-induced human bronchial epithelial cell malignant transformation by microRNA-200b. Toxicol Sci 121(1):110–122, PMID: 21292642, 10.1093/toxsci/kfr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. 2012. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26(21):2392–2407, PMID: 23073843, 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. 2008. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31(4):449–461, PMID: 18722172, 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota J. 2000. Tumor progression and metastasis. Carcinogenesis 21(3):497–503, PMID: 10688870, 10.1093/carcin/21.3.497. [DOI] [PubMed] [Google Scholar]
- Zierold KM, Knobeloch L, Anderson H. 2004. Prevalence of chronic diseases in adults exposed to arsenic-contaminated drinking water. Am J Public Health 94(11):1936–1937, PMID: 15514231, 10.2105/AJPH.94.11.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]