Abstract
A recent study identified a low frequency variant at CCND2 associated with lower risk of type 2 diabetes, enhanced insulin response to a glucose challenge, higher height and, paradoxically, higher BMI. We aimed to replicate the strength and effect size of these associations in independent samples and to assess the underlying mechanism. We genotyped the variant in 29,956 individuals and tested its association with type 2 diabetes and related traits. The low frequency allele was associated with a lower risk of type 2 diabetes (OR=0.53; p=2×10-13; 6,647 cases vs. 12,645 controls), higher disposition index (ß=0.07 log10; p=2×10-11; n=13,028) and higher Matsuda index of insulin sensitivity (ß=0.02 log10; p=5×10-3; n=13,118) but not fasting proinsulin (ß=0.01 log10; p=0.5; n=6,985). The low frequency allele was associated with higher adult height (ß=1.38 cm; p=6×10-9; n=13,927) but the association of the variant with BMI (ß=0.35 kg/m2; p=0.02; n=24,807), estimated in four population based samples, was less than in the original publication where the effect estimate was biased by analysing type 2 diabetes cases and non-diabetic controls separately. Our study establishes that a low frequency allele in CCND2 halves the risk of type 2 diabetes primarily through enhanced insulin secretion.
Introduction
A recent study used whole-genome sequencing and imputation techniques to identify one of the first robust associations between a low frequency variant (1.47% in Icelandic population) and type 2 diabetes(1). The effect of the G minor allele at rs76895963 was appreciably larger than that of known common variants (OR=0.53)(1). The G allele was associated with lower fasting glucose levels and higher insulinogenic index suggesting an effect on insulin secretion but paradoxically was associated with higher BMI (0.56 kg/m2)(1).
Genetic associations need testing in independent studies to ensure associations are not false positive results and to establish an effect size less biased by winner’s curse (regression to the mean). Once replicated, it is then important to test the underlying physiological mechanisms.
The apparently paradoxical association between the diabetes protective allele and higher BMI needs further explanation. Genetic associations that are paradoxical to epidemiological correlations have been described before and provide excellent targets for further investigation of biological mechanisms(2; 3). However, associations between known type 2 diabetes alleles and BMI can be biased by a form of “index event bias”, sometimes referred to as “truncation bias”, if datasets are restricted to cases or controls. This form of bias has likely led to associations between the risk allele at TCF7L2 and lower BMI in cases, because carriers of the risk allele do not need to be as overweight to develop diabetes(4).
We aimed to assess whether independent samples provide robust replication of the strength and effect sizes of the CCND2 associations and to investigate further the underlying mechanisms that result in a low frequency allele reducing the risk of type 2 diabetes but increasing height and BMI. We genotyped the CCND2 variant in 29,956 individuals and tested its association with risk of type 2 diabetes and with measures of insulin sensitivity and insulin secretion.
Research design and methods
We genotyped the low frequency CCND2 variant (rs76895963) in 23,359 individuals of European origin. Study characteristics and genotyping details are in Table 1. Call rates in all samples exceeded 95% with no evidence of departure from Hardy-Weinberg equilibrium (P > 0.05).
Table 1.
Summary details and relevant characteristics of the studies
| STUDY | N (N males / N females) | Age Mean (SD) years | Type 2 diabetes cases/controls | Fasting glucose Mean (SD) mmol/l | 2-hr OGTT Mean (SD) mmol/l | Matsuda ISI Mean (SD) mg/dl, mU/l | Insulinogenic index Mean (SD) pmol/mmol | Disposition index Mean (SD) | BMI Mean (SD) kg/m2 | Height Mean (SD) cm | Genotyping |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Avon Longitudinal Study of Parents and Children study (ALSPAC) data(6) | 6,597 (0 / 6,597) | 28.1 (4.8) | NA | NA | NA | NA | NA | NA | 22.9 (3.8) | NA | Fluorescence based competitive allele-specific assay (KASPar) at LGC Genomics (Hoddesdon, UK) |
| Genetics of Diabetes Audit and Research Tayside Study (GoDARTS)(12) | 13,512 (7,078 / 6,434) | 61.1 (10.6) | 6,145/5,045 | 4.9 (0.7) | NA | NA | NA | NA | 29.3 (5.8) | 167.9 (9.7) | Fluorescence based competitive allele-specific assay (KASPar) at LGC Genomics (Hoddesdon, UK) |
| Metabolic Syndrome in Men (METSIM)(5) | 8,102 (8,120/0) | 57.2 (7.1) | 1,602/6,500 | 5.7 (0.5) | 6.1 (1.7) | 6.9 (4.2) | 131.3 (220.3) | 739.2 (1566) | 26.8 (3.8) | 176.0 (6.4) | TaqMan Allelic Discrimination Assays (Applied Biosystems) |
| Relationship between Insulin Sensitivity and Cardiovascular disease (RISC)(13) | 1,285 (574 / 711) | 43.8 (8.3) | NA | 5.1 (0.6) | 5.7 (1.5) | 11.6 (6.1) | 96.9 (82.6) | 996.7 (990) | 25.5 (4.1) | 170.7 (9.4) | Fluorescence based competitive allele-specific assay (KASPar) at LGC Genomics (Hoddesdon, UK) |
We tested the association of the low frequency variant with risk of type 2 diabetes, diabetes-related intermediate traits (fasting glucose, 2-hour OGTT glucose, Matsuda index of insulin sensitivity, insulinogenic index, disposition index of beta cell function, proinsulin levels), BMI, fat percentage and height.
We used Matsuda index as a surrogate index of peripheral insulin sensitivity which is highly correlated (rho = 0.7) with the gold standard measure of insulin resistance (euglycemic-hyperinsulinemic clamp (M-value))(5).
We calculated Matsuda index of insulin sensitivity:
Insulinogenic index:
and insulin disposition index: Matsuda index of insulin sensitivity × Insulinogenic index.
To provide additional statistical power to estimate the effect of the variant on BMI, we genotyped the variant in 6,579 female participants with pre-pregnancy BMI data from Avon Longitudinal Study of Parents and Children study (ALSPAC)(6). Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. The ALSPAC study website contains details of all the data that is available through a fully searchable data dictionary(7).
To increase our statistical power to estimate the effect of the low frequency variant on Matsuda index, insulinogenic index and disposition index, we included 5,114 samples from the Inter99 study that was part of the original discovery(1; 8). The Danish study was approved by the Ethical Committee of the Capital Region of Denmark.
Diabetes-related intermediate traits were log10-transformed. We used age, sex, (and age2 for height and BMI) and, if applicable, measures required to correct for genetic background, as covariates. We assumed an additive genetic model.
Analyses of glycaemic traits, Matsuda index, insulinogenic index and disposition index were performed in non-diabetic individuals. For BMI, we limited analyses to studies most representative of the general population, with no or limited enrichment for or against type 2 diabetes. For the GoDARTs diabetes case control study we randomly selected a subset of cases to include with all the controls such that the “population” consisted of 5% type 2 diabetes and 95% controls. We also reanalyzed data from the population based studies from the original study but without separating diabetic from non-diabetic individuals and assessed the extent of enrichment for diabetes in the Decode population based study.
We performed fixed-effects inverse variance-weighted meta-analysis in R(9). Evidence of between-study heterogeneity was assessed using Cochran’s Q test and the I2 statistic(10).
Results
The CCND2 low frequency allele is associated with a lower risk of type 2 diabetes
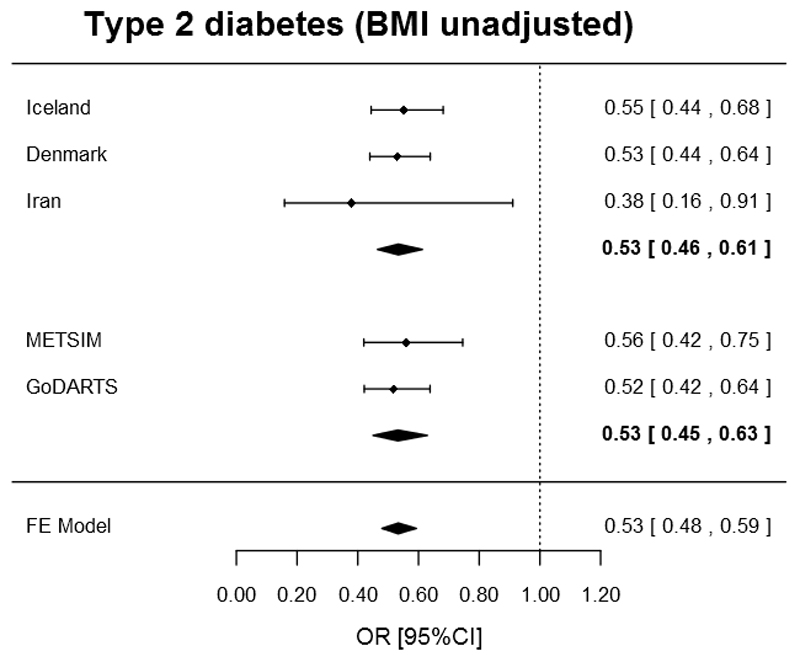
The frequency of rs76895963[G] was 1.97% in GoDARTS (Scottish), 2.15% in METSIM (Finnish), 1.51% in RISC (European-wide) and 2.04% in ALSPAC (South West UK). The rs76895963[G] allele was associated with a lower risk of type 2 diabetes with a very similar effect size to that described in the initial discovery study (OR unadjusted for BMI=0.53 [95% confidence interval: 0.45,0.63]; p=2 × 10-13, OR adjusted for BMI=0.49 [0.40,0.58]; p=2 × 10-14; 6,647 cases vs. 12,645 controls, Table 2 and Figure 1). Meta-analysis with 12,939 cases and 70,909 controls from the discovery studies revealed no evidence of heterogeneity of effect size across 5 studies (OR unadjusted for BMI=0.53 [0.48, 0.59]; p=1 × 10-30, OR adjusted for BMI=0.47 [0.42,0.53]; p=1 × 10-35; 19,586 cases vs. 83,554 controls; heterogeneity p=0.9 for both unadjusted and adjusted model, Table 2 and Figure 1)
Table 2.
Association of rs76895963 in CCND2 with type 2 diabetes, diabetes-related intermediate traits and anthropometric traits
| Trait/disease | Study | Effect size | 95% CI | P-value | N | I2 | Phet |
|---|---|---|---|---|---|---|---|
| Type 2 diabetes (BMI adjusted) (OR) |
Original study Current study Combined |
0.46 0.49 0.47 |
0.40, 0.54 0.40, 0.58 0.42, 0.53 |
6 × 10-23 2 × 10-14 1 × 10-35 |
12,939 vs. 70,909 6,647 vs. 12,645 19,586 vs. 83,554 |
0% 0% 0% |
0.7 0.6 0.6 |
| Type 2 diabetes (BMI unadjusted) (OR) |
Original study Current study Combined |
0.53 0.53 0.53 |
0.46, 0.61 0.45, 0.63 0.48, 0.59 |
8 × 10-19 2 × 10-13 1 × 10-30 |
12,939 vs. 70,909 6,647 vs. 12,645 19,586 vs. 83,554 |
0% 0% 0% |
0.7 0.7 0.9 |
| Fasting glucose (log) |
Original study Current study Combined |
-0.01 -0.02 -0.01 |
-0.02, -0.01 -0.02, -0.01 -0.02, -0.01 |
3 × 10-4 5 × 10-5 9 × 10-8 |
11,764 11,739 23,503 |
NA 0% 0% |
NA 0.9 0.8 |
| 2 hour OGTT (log) |
Original study Current study Combined |
-0.02 -0.05 -0.04 |
-0.04, 0.01 -0.08, -0.03 -0.05, -0.02 |
0.15 6 × 10-5 1 × 10-4 |
4,900 8,261 13,161 |
NA 0% 44% |
NA 0.6 0.2 |
| Matsuda index (log10)+ | Current study | 0.02 | 0.01, 0.03 | 5 × 10-3 | 13,118 | 0% | 0.8 |
| Disposition index (log10)+ | Current study | 0.07 | 0.05, 0.09 | 2 × 10-11 | 13,028 | 0% | 0.7 |
| Insulinogenic index (log10)+ | Current & Original study | 0.05 | 0.03, 0.07 | 8 × 10-6 | 13,181 | 0% | 0.5 |
| Fasting proinsulin (log10)* | Current study | 0.01 | -0.01, 0.02 | 0.5 | 6,985 | NA | NA |
| 30 min proinsulin (log10)* | Current study | -0.01 | -0.02, 0.01 | 0.3 | 6,947 | NA | NA |
| 120 min proinsulin (log10)* | Current study | -0.01 | -0.02, 0.00 | 0.1 | 6,978 | NA | NA |
| BMI (kg/m2) |
Population based studies** Current study *** Combined |
0.36 0.05 0.25 |
0.06, 0.65 -0.21, 0.30 0.08, 0.43 |
0.02 0.7 4 x 10-3 |
24,807 22,464 109,492 |
0% 0% 2% |
0.8 0.7 0.4 |
| Fat mass % (log10) | Current study | 0.00 | -0.01, 0.01 | 0.5 | 6,979 | NA | NA |
| Height (cm) |
Original study Current study Combined |
1.16 1.38 1.24 |
0.83, 1.50 0.92, 1.84 0.97, 1.51 |
6 × 10-12 6 × 10-9 2 × 10-19 |
78,236 13,927 92,163 |
0% 0% 0% |
0.7 0.6 0.8 |
Phet: Heterogeneity p value; NA: Not applicable because data from only one study was available.
Analysis of diabetes-related intermediate traits and height reported in the table were performed in non-diabetic individuals.
Values were adjusted for corresponding insulin measurements at same time-points during OGTT.
Results from population studies with no apparent enrichment for or against type 2 diabetes, including 3 studies from the original publication (Iranian TLGS study, Danish Inter99 study and Danish Health2006 study) and ALSPAC.
To avoid “index event bias” or “truncation bias” we used our population based studies (see method and discussion).
For Insulinogenic index we give the meta analysis results including data presented in the original paper from the Inter99 study. For Matsuda index and disposition index we give the meta analysis results including a new analysis of the Inter99 study, not previously presented.
Figure 1.
Forest plot of the association between the CCND2 rs76895963 low frequency allele and type 2 diabetes (unadjusted for BMI) in discovery and replication studies. The dashed line indicates null effect. The top, middle and bottom diamonds represent the effect size (center of diamond) and 95% confidence intervals (horizontal ends) from the discovery studies, replication studies and overall meta-analysis; respectively.
The CCND2 low frequency allele is associated with higher insulin secretion
The G minor allele was associated with lower fasting glucose (ß=-0.02 log [-0.02,-0.01]; p=5 × 10-5; n=11,739) and lower 2-hour OGTT glucose (ß=-0.05 log [-0.05,-0.02]; p=6 × 10-5; n=8,261). The combined meta-analysis estimated 0.01 log [-0.02,-0.01] lower fasting glucose levels (p=9 × 10-8; n=23,503) and 0.04 log [-0.05,-0.02] lower 2-hour OGTT glucose levels (p=1 × 10-4; n=13,161) per copy of type 2 diabetes protective allele (Table 2).
The type 2 diabetes protective allele was associated with improved ability to secrete insulin in response to a glucose challenge test: higher insulinogenic index (ß=0.06 log10 [0.03,0.09]; p=1 × 10-4; n=8,067; and ß=0.05 log10 [0.03,0.07]; p=8 × 10-6; n=13,181 including the original study, Table 2). The low frequency allele was associated with higher disposition index (ß=0.08 log10 [0.05,0.11]; p=1 × 10-7; n=8,050). Disposition index was not presented in the original study, but we analysed the Danish Inter99 study and meta-analysed with METSIM and RISC which provided an effect of 0.07 log10 [0.05,0.09] with higher disposition index (p=2 × 10-11, n=13,028; Table 2).
The G allele was not associated with any measures of proinsulin levels adjusted for corresponding insulin levels at same time-points during OGTT (Table 2).
The analysis of the CCND2 low frequency allele and the Matsuda index in METSIM and RISC produced a borderline result (ß=0.03 log10 [0,0.05]; p=0.05; n=8,134). A meta-analysis of all 13,118 non-diabetic individuals from METSIM, RISC and Danish Inter99 resulted in a small association with Matsuda index (ß=0.02 log10 [0.01,0.03]; p=5 × 10-3, Table 2).
The effect size of the CCND2 low frequency allele with height is consistent with the original study
The G minor allele was associated with higher adult height (ß=1.38 cm [0.92,1.84], p=6 × 10-9, n=13,927, Table 2). The combined meta-analysis including data from the original study estimated 1.24 cm [0.97,1.51] higher height per copy of the type 2 diabetes protective allele (p=2 × 10-19, n=92,163, Table 2).
The effect size of the CCND2 low frequency allele with BMI is lower than reported in the original study
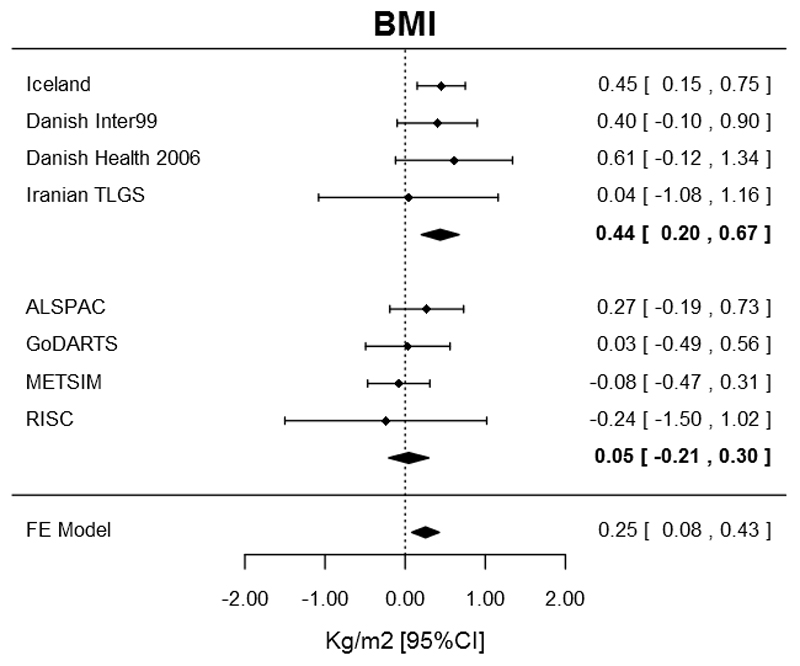
The original report found an association between the low frequency CCND2 allele and higher BMI (0.56 kg/m2) analysed separately in type 2 diabetic and non-diabetic individuals resulting in spurious associations due to index event biases. To further test the BMI association, we first showed that individuals from the Decode study with both CCND2 genotype and BMI available were slightly enriched for diabetic cases (Supplementary Figure 1). We showed that this type of enrichment in population studies results in a bias towards an association between the protective allele and lower BMI, because the diabetic cases tend to be heavier and carry less protective alleles than non-diabetic individuals (Supplementary Figure 2). We thus decided to focus our analysis of the BMI association to the four population studies with no apparent enrichment for or against type 2 diabetes, i.e. three studies from the original publication (the Iranian TLGS study (n=8,658), the Danish Inter99 study (n=6,228) and the Danish Health2006 study (n=3,324)) and ALSPAC (n=6,597). This resulted in a smaller effect of the variant on BMI than reported in the original publication (ß=0.36 kg/m2 [0.06,0.65]; p=0.02; n=24,807, Table 2). We observed no heterogeneity between the effect estimates between these four population based studies (heterogeneity p=0.8, Table 2). This analysis represented our least biased estimate of the effect size. When we combined results from the four studies included in the original publication (re-analysed including both type 2 diabetes cases and controls) with results from four studies added in this paper (with the GoDART study individuals sampled so as to include only 5% diabetic cases), the association with BMI was present but with lower effect size (ß=0.25 kg/m2 [0.08,0.43]; p=0.004; n=109,492; Table 2 and Figure 2) and with some evidence of heterogeneity (p=0.03).
Figure 2.
Forest plot of the association between the CCND2 rs76895963 low frequency allele and BMI including eight studies with no, or limited, ascertainment or enrichment for or against type 2 diabetes. These eight studies included four from the original paper, including Decode individuals and a sample of GoDARTs individuals made to consist of 5% diabetic cases. The dashed line indicates null effect. The top, middle and bottom diamonds represent the effect size (center of diamond) and 95% confidence intervals (horizontal ends) from the discovery studies, replication studies and overall meta-analysis; respectively.
We found no association with fat mass percentage (ß=0.00 [-0.01,0.01], p=0.5, n=6,979 non-diabetic individuals, Table 2).
Discussion
Our study provides robust replication of the relatively large protective effect of a low frequency variant at CCND2 against risk of type 2 diabetes, and its association with improved insulin secretion and higher height. The estimate of the effect size on risk of type 2 diabetes in our study was very close to that of the discovery studies and therefore confirms an unbiased estimate of the effect size – carriers of the low frequency allele are at approximately half the risk of type 2 diabetes compared to non-carriers. Our results, together with data from the original study, provide very strong evidence of the mechanism of diabetes protection. The associations with improved disposition index and insulinogenic index but smaller effects with the Matsuda index, in up to 13,181 individuals, show that the protective diabetes effect operates primarily through a mechanism of relatively favorable insulin secretory response to a glucose challenge and to lower blood sugar more effectively than non-carriers. The effect is unlikely to act through improved insulin processing, as we saw no association with proinsulin levels in the METSIM study, despite previous observations of associations between the TCF7L2 and other diabetes risk alleles in this study(2).
Our data suggest that the association between the CCND2 protective variant and higher BMI is lower than that previously reported. A re-analysis of previous data, together with new data, provided evidence of an association between the CCND2 protective allele and higher BMI but we observed a smaller effect and heterogeneity between studies. Determining the true biological effect of the variant on BMI was very difficult because of “index event” bias. The index “event” in this case was a classification of normoglycaemia, therefore people carrying a type 2 diabetes protective allele remain normoglycaemic at higher BMIs. Similar such likely biases have been observed between strong diabetes risk alleles and BMI where the risk allele at TCF7L2 was associated with lower BMI in type 2 diabetic individuals because individuals carrying a risk allele will develop diabetes at lower BMIs than non-carriers on average(4; 11). Index event bias means it is extremely difficult to determine whether or not diabetes risk alleles have biological effects on BMI.
In summary, we replicated the diabetes and height growth effects of the low frequency variant at CCND2 in 23,359 individuals. Our best estimate of the effect of the variant on BMI suggests that the effect is smaller than reported in the original publication due to index event bias. Further studies are needed to establish the size of the BMI association. Our data, together with the original finding, show a mechanism through improved insulin secretion which results in lower fasting glucose levels, lower 2-hour OGTT glucose levels and a lower risk of type 2 diabetes. Combining all data including 19,586 type 2 diabetes cases and 83,554 controls from the original study and our study provides evidence that carrying this variant reduces the risk of type 2 diabetes by approximately 50% relative to non-carriers.
Supplementary Material
Acknowledgements
We would like to thank the European Research Council (ERC: 323195, SZ-50371), Wellcome Trust (WT) and University of Exeter Medical School (UEMS). GoDARTS: We are grateful to all the participants in the GoDARTS study, the general practitioners, the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The study complies with the Declaration of Helsinki. We acknowledge the support of the Health Informatics Centre, University of Dundee for managing and supplying the anonymised data and NHS Tayside, the original data owner. The Wellcome Trust provides support for GoDARTS (Awards 072960/z/03/z, 099177/z12/z). MMcC is a Wellcome Trust Senior Investigator, Wellcome Trust: 090532, 098381, MRC: G0601261. METSIM was supported by grants from the Academy of Finland, University of Eastern Finland, and Sigrid Juselius Foundation.
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 092731) and the University of Bristol provide core support for ALSPAC. DAL is funded by SP/07 1008/24066. RMF is a Sir Henry Wellcome Postdoctoral Fellow (Wellcome Trust grant 085541/Z/08/Z). The Danish sub-study was supported by research grants from The Novo Nordisk Foundation Center for Basic Metabolic Research, an independent research center at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation (www.metabol.ku.dk), The Lundbeck Foundation (www.lucamp.org), The Danish Agency for Science, Technology and Innovation, the PhD School of Molecular Metabolism, University of Southern Denmark, and the Copenhagen Graduate School of Health and Medical Sciences. Funders had no influence on study design, data collection and analysis, decision to publish, or preparation of the manuscript. The Inter99 study was initiated by T. Jørgensen (principal investigator), K. Borch-Johnsen (co-principal investigator), H. Ibsen, and T.F. Thomsen. The steering committee comprises the former two and C. Pisinger. The Inter99 project was financially supported by research grants from the Danish Research Council, The Danish Centre for Health Technology Assessment, Novo Nordisk, Research Foundation of Copenhagen County, Ministry of Internal Affairs and Health, The Danish Heart Foundation, The Danish Pharmaceutical Association, The Augustinus Foundation, The Ib Henriksen Foundation, and the Becket Foundation. ATH is a Wellcome Trust Senior Investigator and a NHIR senior investigator.
Prof Timothy M Frayling is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions
Conceived and designed the study: HY TMF.
Analyzed the data: HY.
Genotyping/Phenotyping: RISC (EF AM MW) GoDARTS (CNAP ADM) METSIM (ML) ALSPAC (RMF SMR DAL GDS) Inter99 (TJ TH OP).
Statistical analysis: RISC (HY) GoDARTS (HY) METSIM (AS JV) ALSPAC (HY).
Commented on the manuscript: RMF CNAP GH DFG UP MIM ML.
Agreed with manuscript results and conclusions: AS RMF JV MNW WX ARW EF AM SMR DAL GDS TJ TH OP VS DFG GT UT KS ATH MW ADM MIM CNAP ML.
Competing Interests
No potential conflicts of interest relevant to this article were reported.
References
- 1.Steinthorsdottir V, Thorleifsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A, Helgadottir HT, Johannsdottir H, Magnusson OT, Gudjonsson SA, Justesen JM, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nature genetics. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 2.Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, Petrie JR, Travers ME, Bouatia-Naji N, Dimas AS, Nica A, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frayling TM, Hattersley AT. Physiology helps GWAS take a step closer to mechanism. Diabetes. 2014;63:1836–1837. doi: 10.2337/db14-0130. [DOI] [PubMed] [Google Scholar]
- 4.Stolerman ES, Manning AK, McAteer JB, Fox CS, Dupuis J, Meigs JB, Florez JC. TCF7L2 variants are associated with increased proinsulin/insulin ratios but not obesity traits in the Framingham Heart Study. Diabetologia. 2009;52:614–620. doi: 10.1007/s00125-009-1266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stancakova A, Javorsky M, Kuulasmaa T, Haffner SM, Kuusisto J, Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Davey Smith G. Cohort Profile: the 'children of the 90s'--the index offspring of the Avon Longitudinal Study of Parents and Children. International journal of epidemiology. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Available from http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/.
- 8.Jorgensen T, Borch-Johnsen K, Thomsen TF, Ibsen H, Glumer C, Pisinger C. A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99. European journal of cardiovascular prevention and rehabilitation : official journal of the European Society of Cardiology, Working Groups on Epidemiology & Prevention and Cardiac Rehabilitation and Exercise Physiology. 2003;10:377–386. doi: 10.1097/01.hjr.0000096541.30533.82. [DOI] [PubMed] [Google Scholar]
- 9.Viechtbauer W. Conducting meta-analyses in R with the metafor package. Journal of Statistical Software. 2010;36:1–48. [Google Scholar]
- 10.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 11.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature genetics. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 12.Doney AS, Fischer B, Leese G, Morris AD, Palmer CN. Cardiovascular risk in type 2 diabetes is associated with variation at the PPARG locus: a Go-DARTS study. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:2403–2407. doi: 10.1161/01.ATV.0000147897.57527.e4. [DOI] [PubMed] [Google Scholar]
- 13.Hills SA, Balkau B, Coppack SW, Dekker JM, Mari A, Natali A, Walker M, Ferrannini E. The EGIR-RISC STUDY (The European group for the study of insulin resistance: relationship between insulin sensitivity and cardiovascular disease risk): I. Methodology and objectives. Diabetologia. 2004;47:566–570. doi: 10.1007/s00125-004-1335-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.