Abstract
Brain tumours are among the most lethal and devastating cancers. Their study is limited by genetic heterogeneity and the incompleteness of available laboratory models. Three-dimensional organoid culture models offer innovative possibilities for modelling human disease. Here, we establish a 3D in vitro model, named neoplastic cerebral organoid (neoCOR), in which we recapitulate brain tumorigenesis by introducing oncogenic mutations in cerebral organoids via transposon- and CRISPR/Cas9-mediated mutagenesis. By screening clinically-relevant mutations identified in cancer genome projects, we define mutation combinations that result in glioblastoma-like and central nervous system primitive neuroectodermal tumour (CNS-PNET)-like neoplasms. We demonstrate that neoCORs are suitable to study aspects of tumour biology such as invasiveness, and to evaluate the effect of drugs in the context of specific DNA aberrations. neoCORs will provide a valuable complement to current basic and preclinical models for studying brain tumour biology.
Introduction
Malignant brain tumours are among the most devastating cancers with almost negligible survival rates1. Although these tumours have been studied in many experimental model systems, their survival rate has not improved in decades. There remains a need for the development of new experimental model systems to study human brain tumours.
Among currently available models, genetically engineered mouse models (GEMMs) are broadly used for both biological and preclinical investigations. GEMMs relatively accurately mimic the pathophysiological features of human brain tumours, but their application is limited by the genetic, morphological, and physiological differences between human and rodent brains2. Establishing GEMMs is also relatively expensive and time consuming, making them sub-optimal as a screening system for tumorigenic drivers from the numerous candidates identified by brain cancer sequencing projects3–5. Patient-derived xenografts (PDXs) represent, to a large extent, the heterogeneity of patient brain tumours, but are not suitable for studying tumour initiation. Furthermore, biopsy-derived PDXs take time to establish, and are financially infeasible for further drug testing6. Human brain cancer cell lines as well as cancer stem cells cultured in 2D have served as surrogate models for brain tumours, but do not recapitulate the 3D tumour environment7,8. Tumour sphere models generated from either tumour cell lines or cancer stem cells mimic a 3D structure, but lack the organ-like histology and the interaction between tumour and normal tissues9–11.
The recent development of in vitro organoid culture has opened new avenues for modelling diseases directly in human tissues. Recapitulating either organ regeneration from adult stem cells (ASCs)12 or organ development from pluripotent stem cells (PSCs)13, organoids can accurately resemble organ histology and physiology14,15. Organoids have been used to model various human diseases16, including cancer17. Human cerebral organoids recapitulate human brain development in vitro, and has been used to model human neurodevelopmental disorders18–21. Thus far, in vitro 3D organoid models have not yet been developed to study human brain tumour initiation, progression, and treatment.
Here, we report the development of 3D organoid models to study human brain tumour initiation, progression, and response to perturbation. We applied genome-editing techniques to introduce tumorigenic mutations into human cerebral organoids. These models allow us to test the tumorigenic capability of gain- and loss-of-function mutations, singly or in combination, in a systematic manner. We demonstrate that mutations found in cancer patients result, in our model system, in xeno-transplantable tumours that can be classified as central nervous system primitive neuroectodermal tumour (CNS-PNET) or glioblastoma (GBM). The neoplastic cerebral organoid (neoCORs) model provides a valuable tool to study fundamental brain tumour biology as well as to test potential drugs in a personalized setting.
Results
Clonal mutagenesis in cerebral organoids induces tumour overgrowth
A recent re-classification of brain cancer subtypes includes DNA aberrations as a defining feature22, highlighting the need for genetically defined human brain cancer models. Brain tumours are characterized by a variety of DNA aberrations that either cause oncogene overexpression and/or loss of tumour suppressor gene function3–5.
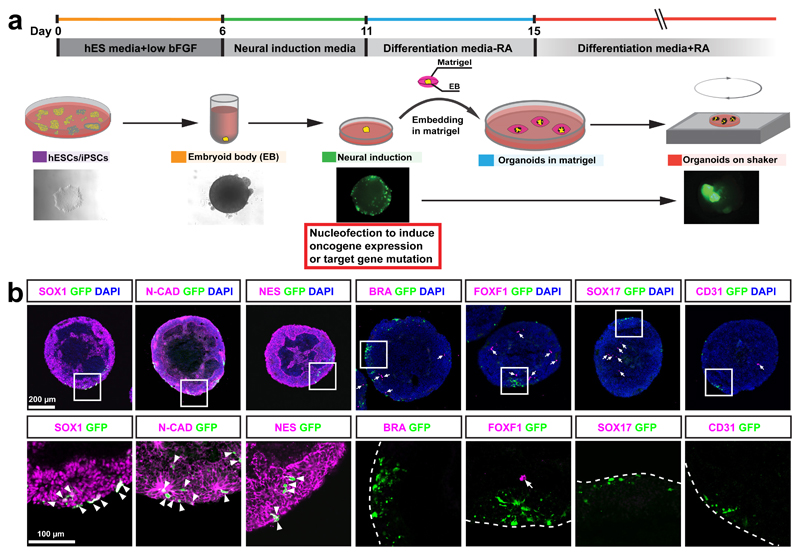
To recapitulate tumorigenic events in cerebral organoids, we combined Sleeping Beauty (SB) transposon-mediated gene insertion for oncogene-amplification with CRISPR/Cas9-based mutagenesis of tumour suppressor genes. We introduced combinations of plasmids encoding (1) the SB transposase for integration of IR-flanked expression elements into the genome, (2) GFP flanked by SB inverted repeats (IRs) for cell tracing, (3) any oncogene flanked by IRs for oncogene overexpression, and (4) plasmids expressing the Cas9 nuclease together with guide RNAs (gRNAs) for mutagenesis of tumour suppressor genes into cerebral organoids, by electroporation before matrigel embedding (Supplementary Fig. 1a). This strategy allows us the flexibility to introduce any combination of gain- and/or loss-of-function tumorigenic genes. At the end of neural induction stage of our cerebral organoid development protocol18 (Fig. 1a), neural stem and progenitor cells (NS/PCs), which are believed to be one of the cell of origins for many different subtypes of brain tumours23–30, are expanding on the surface of embryoid bodies (EBs). Immunostaining of both sectioned EBs and adherent culture of EBs 1 day after nucleofection of pCAG-GFP showed that 100% of GFP+ cells are SOX1+, CDH2+ (N-CADHERIN+), and NES+ NS/PCs (Fig. 1b). None of GFP+ cells are BRACHYURY+ (BRA+) or FOXF1+ mesodermal cells, or SOX17+ or PECAM1+ (CD31+) endodermal cells (Fig. 1b and Supplementary Fig. 1b-d). Thus, the electroporated plasmids are exclusively delivered into NS/PCs.
Figure 1. Introducing genome-editing constructs into neural stem/precursor cells (NS/PCs) of cerebral organoids.
(a) Schematic of cerebral organoid culture and nucleofection strategy. (b) The images show immunofluorescence staining for the indicated markers in EBs 1 day after nucleofection. Lower panel shows high-magnification images of nucleofected cells. Arrowheads point to nucleofected cells (GFP, green) that express NS/PC markers; arrows point to cells expressing mesodermal (BRA or FOXF1) or endodermal (SOX17 or CD31) markers. This experiment was performed twice independently with same results. EB, embryoid body; bFGF, basic fibroblast growth factor; hESCs, human embryonic stem cells; hiPSCs, human induced pluripotent stem cells; RA, retinoic acid; N-CAD: N-CADHERIN; NES: NESTIN; BRA: BRACHYURY. Scale bar: b, upper panel: 200 μm; lower panel: 100 μm.
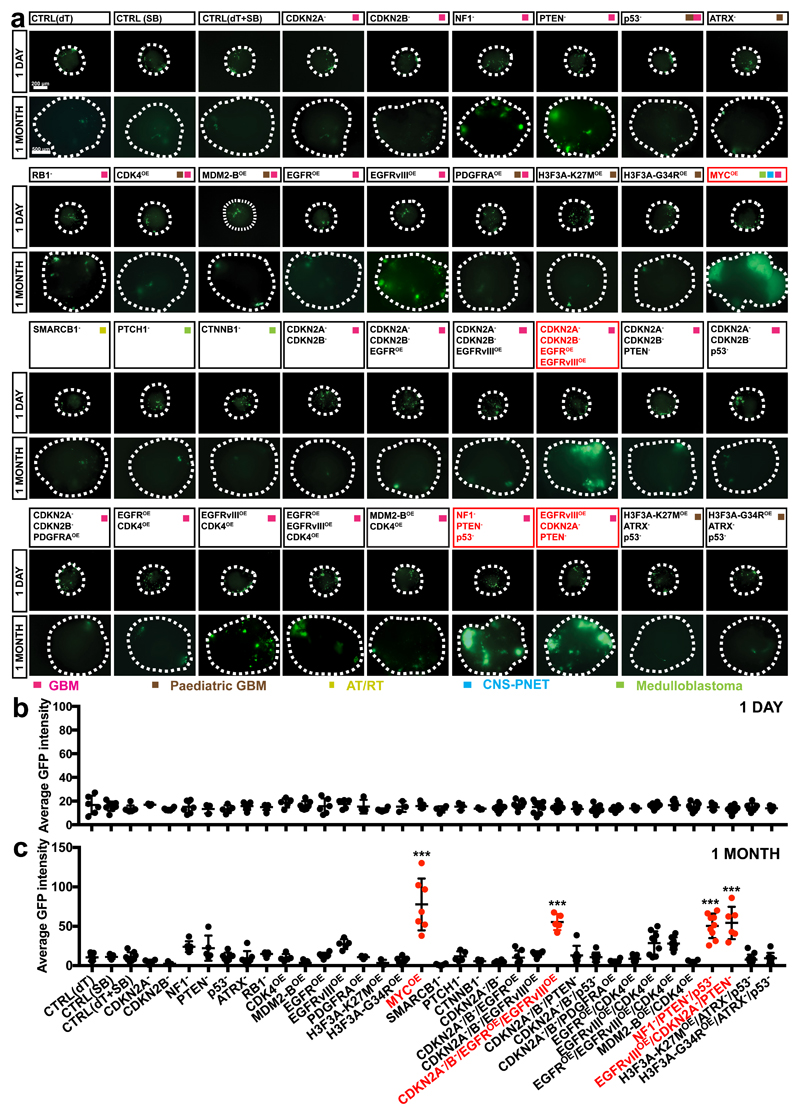
We tested whether tumorous overgrowth can be induced in cerebral organoids. We introduced into the organoids 18 single gene mutations or amplifications as well as 15 of the most common clinically-relevant combinations observed in brain tumours such as GBM3, paediatric CNS-PNET31, atypical teratoid/rhabdoid tumour (AT/RT)32, and Medulloblastoma5 (Supplementary Table 1). As most electroporated cells carry the CAG-GFP insertion, we used GFP intensity to quantify proliferation of cells carrying gene aberrations. One day after electroporation, EBs from all groups contained similar amounts of GFP+ cells (Fig. 2a,b). One month later, however, striking overgrowth of GFP+ cells was observed in organoids carrying the MYC-amplification (MYCOE), and in organoids carrying CDKN2A-/CDKN2B-/EGFROE/EGFRvIIIOE, NF1-/PTEN-/TP53- (p53-), and EGFRvIIIOE/CDKN2A-/PTEN- (Fig. 2a,c). As these combinations of gene aberrations are commonly found in GBM, we refer to them as GBM-1, GBM-2, and GBM-3, respectively.
Figure 2. Clonal mutagenesis in organoids induces tumor overgrowth.
(a-c) Immunofluorescence images (a) and quantification of the GFP fluorescence intensity (b,c) of organoids mutagenized with the indicated mutation combinations 1 day (b) and 1 month (c) after nucleofection. Organoids from four groups exhibited significant overgrowth at one month: MYCOE (n=7; adjusted p<0.0001 v.s. CTRL(SB)), CDKN2A-/CDKN2B-/EGFROE/EGFRvIIIOE (n=5; adjusted p<0.0001 v.s. CTRL(dT+SB)), NF1-/PTEN-/p53- (n=9; adjusted p<0.0001 v.s. CTRL(dT+SB)), EGFRvIII/PTEN-/CDK2A- (n=6; adjusted p<0.0001 v.s. CTRL(dT+SB)). This experiment was performed once. Statistical analysis was performed using one-way ANOVA with Tukey’s test. Data are presented as mean±SD; full details including all sample sizes are provided Source Data. ***, p<0.001. Scale bar: a, 1 day: 200 μm; 1 month: 500 μm.
We confirmed that the genome editing techniques actually altered the genome in tumour cells by analysing the expression of oncogenes and/or sequencing CRISPR-targeting regions. We observed that tumour cells carried the expected gene mutations/amplifications (Supplementary Fig. 2a-d). Thus, cerebral organoids can be used as a platform to test the tumorigenic capacity of different gene aberrations.
MYCOE and GBM-like neoCORs have distinct transcriptional profiles
To test whether brain tumour-like organoids resemble distinct brain tumour-subtypes, we performed transcriptome analysis on GFP+ cells isolated by FACS. Principal component analysis (PCA) of the top 500 variable genes between different groups identified three distinct clusters. Cluster 1 included all control (CTRL) organoids which harbour only CAG-GFP and a control gRNA targeting dTomato (Fig. 3a). Cluster 2 included the organoids carrying the MYCOE construct, while Cluster 3 contained the organoids carrying genetic aberrations found in GBM (GBM-1, GBM-2, GBM-3). Based on these clusters, we identified the differentially expressed genes (DESeq, adjusted p value <0.05) between Cluster 2 or Cluster 3 versus CTRL. As expected, the Venn diagram hypergeometric test showed that the majority of genes deregulated in the MYCOE group (Cluster 2) are distinct from those deregulated in the GBM-groups (Cluster 3) (Supplementary Fig. 3a). Further KEGG pathway analysis of differentially expressed genes (DESeq, adjusted p value <0.05) between Cluster 2 and Cluster 3 indicated upregulation of metabolic pathways and cell cycle genes in the tumour cells from Cluster 2 neoCORs, but also the Hippo, WNT, TGFβ, and TP53 signalling pathways that are known to be connected to MYC33–35 (Supplementary Fig. 3b). In addition, the MYCOE group showed upregulation of an epithelial differentiation signature, suggestive of a CNS-PNET-like neoplasm of neuroepithelial cellular origin. KEGG pathway analysis also confirmed a glioma signature in Cluster 3 neoCORs, and showed upregulation of the PI3K-AKT, RAP1, ERBB, HIF1A, NF-kappa B, and Estrogen signaling pathways, relevant for GBM36–40 (Supplementary Fig. 3b).
Figure 3. MYCOE and GBM-like neoCORs have distinct transcriptional profiles and cellular identities.
(a) Principle component analysis (PCA) of the top 500 variable genes between normal cells from CTRL organoids and tumour cells from different neoCOR groups. (b) The heatmap shows normalized expression levels for differentially expressed genes (adjusted absolute log2fc value >1 or <-1 and adjusted p value <0.05) between Cluster 2 and Cluster 3 (n=3 for Cluster 2 and n=7 for Cluster 3 from one experiment) selected from differentially expressed genes between human primary CNS-PNET and GBM tumours. The heatmap was created from log2(TPM) transformed data that was row (gene) normalized using the “Median Center Genes/Rows” and “Normalize Genes/Rows” functions to report data as relative expression between samples. (c) Representative immunofluorescence images of four-month-old organoids from CTRL, MYCOE, and GBM-1. The staining was performed from six independent experiments with the similar results. (d-i) Quantification of the indicated markers from panel c (magenta) and Supplemental Fig. 5 in CTRL and all neoCOR groups. Quantification was performed on organoids from three independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett’s test. Data are presented as mean±SD, with details of sample sizes and values, as well as adjusted p value in Source Data. *, p<0.05; **, p<0.01; ***, p<0.001. Scale bar: c: 100 μm.
To test the similarity of the tumour cells from neoCORs with primary tumours, we examined the transcriptome data from neoCORs for genes known to be differentially expressed between CNS-PNET and GBM patient tumours31. Hierarchical clustering revealed that neoCORs from the MYCOE group showed a strong CNS-PNET signature, while organoids from Cluster 3 exhibited upregulation of GBM genes (Fig. 3b). This data suggests that we have developed two distinct types of tumour overgrowth in human cerebral organoids, depending on the genetic aberrations induced: either a CNS-PNET-like or a GBM-like neoplastic growth.
MYCOE and GBM-like neoCORs have different cellular identities
In patients, CNS-PNETs are embryonic neuroepithelial neoplasms characterized by sheets of primitive neuroepithelial cells and frequent rosette formation41. These undifferentiated cells feature SOX2 positivity and high CD99 expression42. GBMs, on the other hand, are high grade astrocytic neoplasms featuring a more diverse morphology with glial cell predominance. The glial markers GFAP and S100β, as well as the proliferative marker Ki67 are diagnostic for GBM.
We analysed the expression of CNS-PNET and GBM markers in MYCOE and GBM neoCORs four months after nucleofection. In CTRL organoids, most GFP+ cells were HuC/D+ neurons (Fig. 3c,d and Supplementary Fig. 4a), while only a small portion of GFP+ cells were positive for SOX2 (Fig. 3c,e and Supplementary Fig. 4b) and Ki67 (Fig. 3c,f and Supplementary Fig. 4c), or the glial markers S100β (Fig. 3c,g and Supplementary Fig. 4d) and GFAP (Fig. 3c,h and Supplementary Fig. 4e). GFP+ cells located in the ventricular zone of cortical regions, expressed SOX2 and Ki67, while GFP+/HuC/D+ neurons were located in the basal cortical regions (Fig. 3c and Supplementary Fig. 4a-f).
In contrast, the MYCOE organoids contained few GFP+ cells that were HuC/D+ (Fig. 3c,d and Supplementary Fig. 4a), or expressed the glial markers S100β (Fig. 3c,g and Supplementary Fig. 4d) or GFAP (Fig. 3c,h and Supplementary Fig. 4e). Instead, most GFP+ cells were SOX2+ (Fig. 3c,e and Supplementary Fig. 4b), and nearly 50% expressed Ki67 (Fig. 3c,f and Supplementary Fig. 4c). In addition, most MYCOE/GFP+ cells expressed high levels of CD99 antigen (Fig. 3c,i and Supplementary Fig. 4f), further confirming their CNS-PNET-like cellular identities. GFP+ cells formed large sheets of cells and rosette structures (Fig. 3c and Supplementary Fig. 4a-f).
In the GBM-like groups, GFP+ regions were positive for S100β (Fig. 3c,g, and Supplementary Fig. 4d and 5) and GFAP (Fig. 3c,h, and Supplementary Fig. 4e and 5) indicating their glial identity while containing only a few HuC/D+ neurons (Fig. 3c,d, and Supplementary Fig. 4a and 5). Compared with CTRL organoids, we also detected more SOX2+ (Fig. 3c,e, and Supplementary Fig. 4b and 5) and Ki67+ (Fig. 3c,f, and Supplementary Fig. 4c and 5) cells, which are often observed in the central core of GBM tumours43. In addition, GFP+ regions in GBM-relevant groups showed elevated CD99 levels (Fig. 3c,i and Supplementary Fig. 4f and 5), a feature also reported for GBM tissues44. Tumour regions in the GBM-like organoids showed a disorganized architecture (Fig. 3c, and Supplementary Fig. 4a-f and 5).
We also examined one-month-old control organoids and neoCORs, which showed comparable cellular identities and histological features as four-month-old organoids (Supplementary Fig. 6a-e and 7a-e). Thus, neoCORs induced by distinct genetic aberrations recapitulate the cellular identities and partial histo-morphologic features CNS-PNET or GBM tumours.
NeoCORs retain viability and expand upon renal subscapular engrafting
The ability for self-renewal and immortality are two hallmarks of cancer cells. To examine if neoCORs exhibited these features in vivo, we implanted them into the renal subcapsular space of immuno-deficient mice (Supplementary Fig. 8a). In controls, 4 out of 5 organoids were resorbed within six weeks and the remaining organoid was reduced to only a tiny cluster of cells (Fig. 4a) that had lost cellularity and architectural detail (Fig. 4b). In contrast, 17 out of 20 neoCORs were retained and often expanded beyond the renal capsule (Fig. 4a and Supplementary Fig. 8b). Immunohistochemical analysis revealed many neuro-epithelial areas in organoids of the MYCOE group positive for the NS/PC marker SOX1 (Fig. 4c) and cell cycle marker Ki67 (Fig. 4d), but very few cells positive for the glial marker GFAP (Fig. 4e; arrow) or the neuronal marker MAP2 (Supplementary Fig. 8c; arrowhead), indicating their primitive, poorly differentiated state. Transplanted organoids from the MYCOE group proliferated massively (Fig. 4a and Supplementary Fig. 8b). They formed cell sheets and rosettes similar to CNS-PNET (Fig. 4b and Supplementary Fig. 8d-f). GBM groups instead displayed high expression of the glial marker GFAP, NS/PC marker SOX1, and Ki67 (Fig. 4c-e). GBM-1 and GBM-3 organoids displayed a glial neoplasm-like expansion (Fig. 4b), while GBM-2 showed glial neoplasm-like proliferation with additional cells of mature neuronal appearance reminiscent of glioneuronal tumours (Fig. 4b). Thus, neoCORs can engraft and expand in vivo and maintain their subtype identity upon renal transplantation into nude mice.
Figure 4. NeoCORs expand upon renal subcapsular xenografts.
(a) Brightfield and immunofluorescence images of the indicated renal subcapsular implants 1.5 months after implantation. (b) H&E staining of neoCORs under the renal capsule. Arrows indicate glial cells, arrowhead indicates a neuron. (c-e) Immunohistochemical staining of the indicated markers in implanted organoids. Scale bar: a, 500 mm; b-f, 200 μm and 50 μm (inset).
GBM-like neoCORs to study interaction between tumour and normal tissue
Compared to other in vitro brain tumour models, a distinct feature of the neoCOR model is that tumours are initiated by introducing genetic aberrations into a very small portion of cells in the cerebral organoid. This not only mimics human tumour initiation in vivo, but also results in a mixed structure containing both tumour and normal tissues. This allows us to use neoCORs to study important properties such as invasiveness.
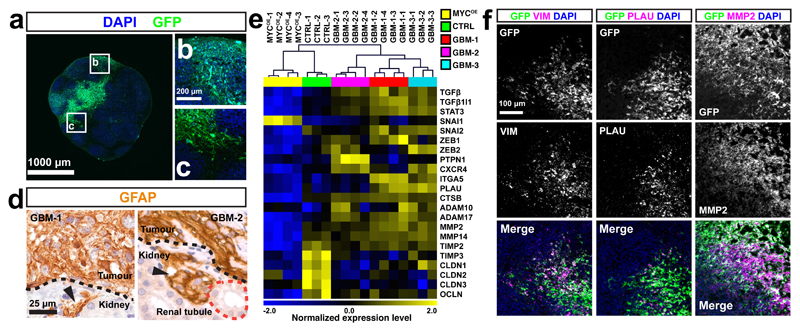
GBMs are known to extensively infiltrate into adjacent brain parenchyma, accompanied by anepithelial-mesenchymal transition (EMT) that confers invasive capabilities to tumour cells45,46. To assess whether neoCORs can be used to study this process, we evaluated the interface between tumor and normal cells in GBM-like neoCORs. We observed GFP+ tumour cells within normal regions (Fig. 5a-c). We also observed small foci of tumour cells that breached the renal capsule in the renal xenografts of GBM-group neoCORs (Fig. 5d).
Figure 5. GBM neoCORs exhibit features of GBM invasion.
(a-c) Representative images of the tumour-normal interface in GBM-1 neoCORs. Images are representative of at least three independent experiments. (d) Immunohistochemical staining of GFAP in GBM-like neoCORs. Images are representative of two independent renal implantations. Dotted black lines indicate the boundary between implanted neoCORs and murine kidney. Dotted red line indicates the renal tubule. Arrowheads indicate invaded tumour cells. (e) Hierarchical clustering analysis of GBM invasiveness-relevant genes from four-month-old organoids (n=3 for CTRL organoids; n=4 for MYCOE, n=4 for GBM-1, n=4 for GBM-2, and n=3 for GBM-3 neoCORs, from three independent cultures for each group). The heatmap was created from log2(TPM) transformed data that was row (gene) normalized using the “Median Center Genes/Rows” and “Normalize Genes/Rows” functions to report data as relative expression between samples. (f) Representative immunofluorescence staining of neoCORs from GBM-1 group for the indicated mesenchymal marker; GFP is also shown. Images are representative of two independent experiments. Scale bar: a, 1000 mm; b and c, 200 mm; d, 25 μm; f: 100 μm.
We compared the expression levels of invasion-related genes in tumour cells from four-month-old neoCORs of all three GBM groups and normal cells from the same age CTRL organoids, using RNA-seq analysis of FACS-sorted cells. Hierarchical clustering of samples based on reported GBM invasiveness-relevant genes47, including EMT-related transcriptional factors (TGFβ, TGFβ1I1, STAT3, SNAI2, ZEB1, ZEB2), migration-related receptor (CXCR4), extracellular matrix molecules (ITGA5), and proteases (PLAU, CTSB, ADAM10, ADAM17, MMP2, MMP14), showed that tumour cells from all GBM neoCORs clustered together and exhibited higher expression levels of invasiveness-relevant genes compared to normal cells from CTRL organoids and to tumour cells from the MYCOE group (Fig. 5e). In addition, tumour cells from all GBM groups exhibited downregulation of many genes inhibiting tumour invasion compared to normal cells in CTRL organoids, such as tissue inhibitors of matrix metalloproteinases (TIMP2, TIMP3), and tight junction components (CLDN1, CLDN2, CLDN3, OCLN)47 (Fig. 5e). Tumour cells from different GBM groups were further clustered into individual groups based on expression levels of various invasiveness-related genes. Most invasion-related genes were downregulated in MYCOE neoCORs compared to GBM groups (Fig. 5e), which correlates with the lower infiltration tendency of embryonic neoplasms compared to astrocytic neoplasms41.
Immunostaining of organoids for the mesenchymal marker vimentin (VIM), urokinase (PLAU), and matrix metalloproteinase 2 (MMP2), confirmed that tumour cells in GBM neoCORs expressed higher level of these invasiveness genes compared to the surrounding normal tissues (Fig. 5f).
NeoCORs are suitable for targeted drug testing
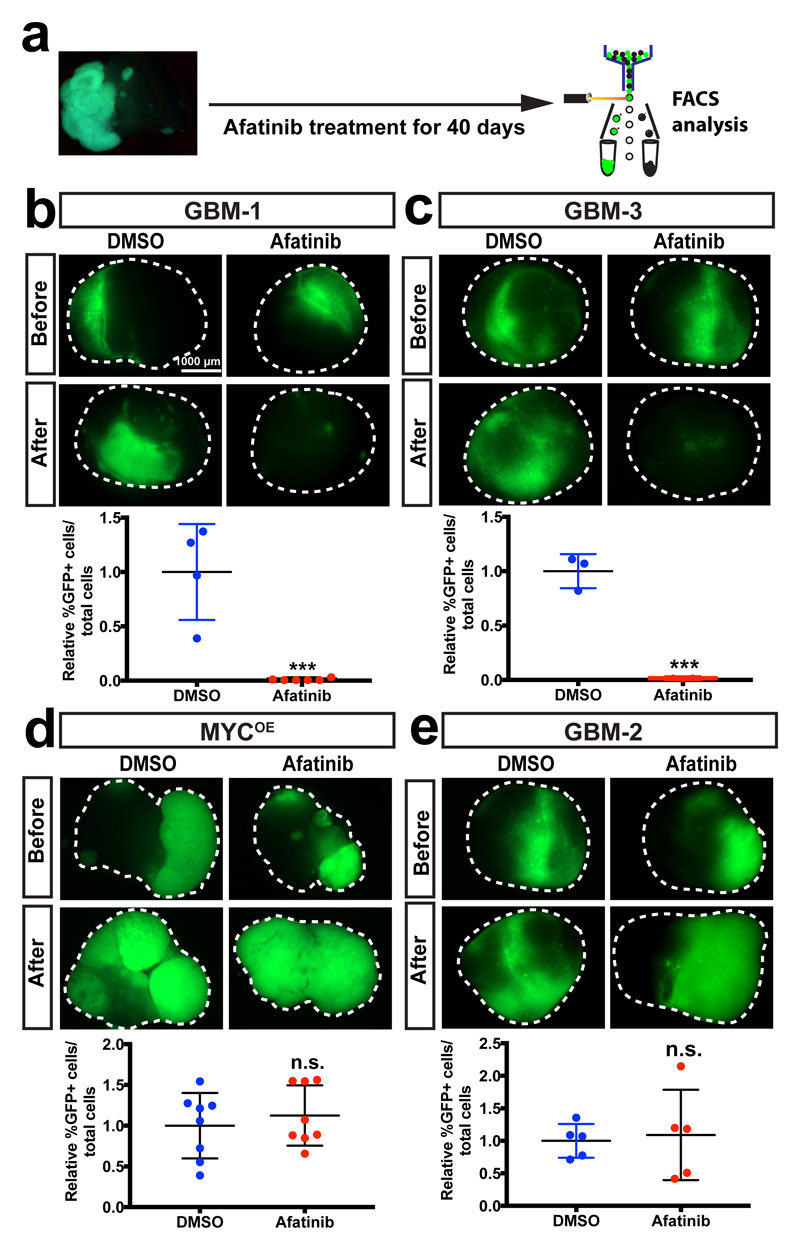
Since our approach initiates tumorigenesis by introducing defined gene aberrations, the neoCORs could be potentially used for targeted drug testing. To examine this, we assessed the effect of an EGFR inhibitor Afatinib, currently in a clinical trial for GBM (ClinicalTrials.gov NCT No.: NCT02423525), as a proof of principle (Fig. 6a). Forty days after treatment, Afatinib significantly reduced the number of tumour cells in GBM-1 and GBM-3 (Fig. 6b,c), but showed no effect on the MYCOE and GBM-2 groups (Fig. 6d,e), consistent with the fact that only GBM-1 and GBM-3 organoids show EGFR over-activation. Thus, neoCORs can be used to test the effect of chemical compounds on tumours originating from specific driver mutations.
Figure 6. NeoCORs are suitable for preclinical investigations.
(a) Schematic of the drug treatment and FACS analysis using neoCORs. (b-e) Images and FACS quantification of cells from neoCORs after the indicated treatment. The percentage of GFP+ cells from drug-treated groups was normalized to the percentage of GFP+ cells from DMSO-treated neoCORs. Afatinib diminished tumour cells in GBM-1 (b; n=6 from one experiment; p=0.0005) and GBM-3 (c; n=3 from one experiment; p=0.0004) neoCORs, but not in MYCOE (d; n=8 from one experiment; p=0.5261) and GBM-2 (e; n=5 from one experiment; p=0.7916) groups. The experiments were performed twice independently with similar results. Statistical analysis was performed using unpaired two-tailed Student’s t-test. Data were presented as mean±SD, details of sample size and values are provided in Source Data. ***, p<0.001. Scale bar: b-e, 1000 μm.
In an effort toward adapting this method for large scale screening, we modified the neoCOR system to include firefly luciferase for measurement of tumour size (Supplementary Fig. 9a). We applied five different EGFR inhibitors, including Afatinib, Erlotinib, and Gefitinib, which are approved for different types of cancers, and the experimental drugs Canertibib and Pelitinib, to GBM-1 organoids. Forty days after drug treatment, Afatinib and Erlotinib significantly reduced firefly luciferase activity (Supplementary Fig. 9b). Thus, these results suggested that our model could identify the efficacy of different compounds in the context of drug screening.
Discussion
By recapitulating genetic aberrations found in human brain cancer patients using genome-editing techniques in cerebral organoids, we have generated a new in vitro model system for human brain tumours, which we name neoCORs. These models exhibit many features of cancer, such as cellular identities, cancer pathway specific transcriptome profiles, and capability of in vivo expansion and invasion. We identified three mutant combinations that induce glial-orientated differentiation and abnormal overgrowth, indicating their glial neoplasm-like identities. By overexpressing oncogene MYC, we could generate neoCORs that showed histopathological features, cellular identities and transcriptome signatures very similar to those described for human CNS-PNET31,41, a tumour for which no successful animal or in vitro model exists48. It is interesting to note that amplification of MYC alone could initiate CNS-PNET-like neoplasm in cerebral organoids within a very short period, while normally it requires additional genetic events such as loss of p53 and much longer time in animal models, with low incidence28.
Unlike previous 3D culture models, such as brain tumour spheres9–11, neoCORs allow the functional analysis of genome aberrations within the same genetic background. By introducing genome aberrations in organoids started from patient iPSCs, neoCORs can be further used to test the susceptibility of individual patients to different combinations of driver mutations.
As opposed to brain tumour spheres as well as 2D glioblastoma cell cultures, neoCORs mimic in vivo structural organization, to a certain degree. They contain both tumour cells and normal cells within the same culture, so that interactions between transformed and non-transformed cells can be analysed. This feature of neoCORs not only makes them very useful to study essential tumour biology, but also valuable for preclinical investigation. For drug screening, this particular situation allows for an analysis of anti-tumour effects accompanied by a safety test in the same system. Like most organoid systems, neoCORs are limited by the lack of vasculature and therefore, certain features of GBM such as glomeruloid microvascular proliferations and perivascular palisading necrosis are not observable. Co-culture organoid systems like the ones generated for microglia49 and/or endothelial cells50 might help to overcome these limitations in the future.
Taken together, our results demonstrate the power of the neoCOR model system to further our knowledge human brain tumour biology by enabling screening of tumorigenic drivers. The system will complement other models, and clinical studies, in dissecting molecular mechanisms of tumour initiation, invasion and progression. It also opens the doors for validation of potential pharmacologic and biologic therapeutic approaches and exploratory drug discovery.
Online Methods
A step-by-step protocol is available as a Supplementary Protocol and has been submitted as an open resource to the Protocol Exchange51.
Plasmid constructs and materials
For overexpression constructs, based on the Sleeping Beauty Transposase System, the CMV promoter from pCMV(CAT)T7-SB100 (Addgene cat. No.: 34879)52 was replaced with CAG promotor from pCAGEN (Addgene cat. No.: 11160)53. IRDR-R and IRDR-L sequences from pT2/LTR7-GFP (Addgene cat. No.: 62541)54 were cloned into pCAGEN to produce pCAG-GS/IR. cDNAs used for overexpression were amplified from human cDNA and cloned into the MCS of pCAG-GS/IR. With the help of sleeping beauty transposase SB100X (pCAG-SB100X), CAG-GFP and CAG-oncogenes were integrated into the genome of cells in organoids. To introduce gene mutations, short guide RNAs of tumour suppressors were cloned into CRISPR/Cas9 vector pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene cat. No.: 42230)55. All cloning primers are listed in the Supplementary Table 2 and 3.
Mice
MF nu/nu mice were bred and maintained in the IMBA animal facility according to Austrian law. All animal experiments were performed under ethical animal license protocols from the Austrian Ministry of Science, Research, and Economics (BMWFW).
Human embryonic stem cell (hESC) culture
Feeder-free (FF) H9 hESCs were obtained from WiCell with verified normal karyotype and contamination free. FF H9 hESCs were cultured in a feeder-free manner on Matrigel (Corning, hESC-qualified Matrix)-coated plate with mTeSR medium (Stemcell Technologies). Feeder-dependent (FD) H9 hESCs were obtained from WiCell with verified contamination-free. FD H9 hESCs were cultured on CF-1-gamma-irradiated mouse embryonic stem cells (MEFs) (GSC-6001G, Global Stem) according to WiCell protocols. All cell lines were routinely checked for mycoplasma-negative. All stem cells were maintained in a 5% CO2 incubator at 37°C. Standard procedures were used for culturing and splitting hESCs as explained previously18. All hESCs were authenticated using Infinium PsychArray-24 Kit (Illumina).
Generation of cerebral organoids
Cerebral organoids were cultured as previous described18. Briefly, to make EBs, hESCs were trypsinized into single cells, and 9,000 cells were plated into each well of an ultra-low-binding 96-well plate (Corning) in human ES medium containing low concentration basic fibroblast growth factor (bFGF, 4 ng/ml) and 50 μM Rho-associated protein kinase (ROCK) inhibitor (Calbiochem). EBs were fed every three days for 6 days then transferred to neural induction media to form neuroepithelial tissues. After 5-7 days in neural induction media, EBs were embedded into droplets of matrigel (Corning) and cultured in differentiation medium without vitamin A (Diff-A). Finally, the EB droplets were transferred to 10 cm-dish containing differentiation medium with vitamin A (Diff+A) and cultured on an orbital shaker. Media were changed weekly.
Nucleofection of organoids to induce gene mutation/amplification
In order to initiate the brain tumours, we introduced the tumour suppressor mutations and/or oncogene amplifications on neuroepithelial cells at the end of neural induction culture, right before the Matrigel embedding. Briefly, 10-15 EBs were collected, resuspended in nucleofetion reagent (Nucleofector™ kits for human stem cells, Lonza) containing plasmids and transferred into nucleofection vials. Nucleofection was performed according to the manufacturer’s protocol. After electroporation, EBs were carefully transferred to 6 cm-dish containing neural induction medium, and cultured at 37°C incubator for four hours. Then nucleofected EBs were embedded into matrigel and cultured for organoids as described. The neoplastic cerebral organoids with significant overgrowth of GFP+ cells were selected for further investigations, in which the samples were randomly allocated.
Adherent cell culture of dissociated EBs
One day after nucleofection, the EBs were trypsinized at 37°C for 20 min to make single cell suspension. Then cells were plated on the poly-D-lysine- and laminin-coated coverglasses in neural induction medium with ROCK inhibitor, and cultured in a 5% CO2 incubator at 37°C. The further immunofluorescence staining and analysis were performed the day after.
RNA-sequencing and analysis
Organoids from control and neoplastic groups were collected 40 days and four months after nucleofection, and trypsinized with shaking at 37°C for half an hour. GFP+ cells were sorted according to the example gating strategy (Supplemental Fig. 10), and total RNA was isolated using RNeasy Micro kit (Qiagen) according to the manufacturer’s instruction. RNA concentration and quality were analysed using RNA 6000 Nano Chip (Agilent Technologies). Messenger RNA (mRNA) was enriched using SMART-Seq v4 Ultra Low Input RNA Kit (TaKaRa) according to manufacturer’s protocol. Libraries were prepared using NEB Next Ultra Directional RNA library Prep kit for Illumina (NEB). Barcoded samples were multiplexed and sequenced 50 bp SE on a HighSeq 2500 (Illumina). mRNA sample isolation, library preparation, and sequencing were performed at the VBCF NGS Unit (www.vbcf.ac.at).
The unstranded reads were screened for ribosomal RNA by aligning with BWA (v0.7.12) against known rRNA sequences (RefSeq). The rRNA subtracted reads were aligned with TopHat (v2.1.1) against the Homo sapiens genome (hg38). Microexon-search was enabled. Additionally, a gene model was provided as GTF (UCSC, 2015_01, hg38). rRNA loci are masked on the genome for downstream analysis. Aligned reads are subjected to TPM estimation with Kallisto (v0.43.0). Furthermore, the aligned reads were counted with HTSeq (v0.6.1; intersection-nonempty) and the genes were subjected to differential expression analysis with DESeq2 (v1.12.4).
Before the bioinformatics analysis, the expression of oncogenes according to the genome editing manipulation was checked, and one four-month-old sample from GBM-3 neoplastic cerebral organoid group was excluded for the further analysis because of the failure of introducing the overexpression of EGFRvIII.
PCA was performed using the top 500 variable genes between normal cells from CTRL organoids and tumour cells from different neoCOR groups. Venn diagram hypergeometric test was performed on differentially expressed genes between Cluster 2 or Cluster 3 versus CTRL, and KEGG pathway enrichment analysis were performed on differentially expressed genes between Cluster 2 and Cluster 3 with an adjusted absolute log2fc value >0.5 and adjusted p value <0.05. Venn diagram hypergeometric test was performed via R language. KEGG pathway enrichment was analysed using DAVID Bioinformatics (https://david.ncifcrf.gov)56. The heatmap of RNA-seq was generated using MeV57. For the heatmap of tumour-subtype gene profiling (Fig. 3c), the differentially expressed genes between Cluster 2 and Cluster 3 (adjusted absolute log2fc value >1 or <-1 and adjusted p value <0.05) were selected from the differentially expressed gene list (adjusted absolute log2fc value >1 or <-1 and adjusted p value <0.05) from human primary tumour transcriptome analysis31. For the heatmap of hierarchical clustering analysis of GBM invasiveness-relevant genes (Fig. 5e), differential expressed genes from any individual GBM groups versus CTRL organoids with an adjusted absolute log2fc value >0.5 and adjusted p value <0.05 were selected. The heatmap was created from log2(TPM) transformed data that was row (gene) normalized using the “Median Center Genes/Rows” and “Normalize Genes/Rows” functions to report data as relative expression between samples.
Verification of genome alteration introduced by SB and CRISPR/Cas9
To test whether the genome editing techniques actually altered the genome in tumour cells, GFP+ tumour cells were FACS sorted for genomic DNAs isolation for genotyping and for RNAs to verify the expression of oncogenes. RNAs were isolated using RNeasy Micro kit (Qiagen), and cDNA was synthesized according to previous description58. RT-PCRs for MYC, EGFR/EGFRvIII, and TBP were performed using the primers listed in Supplementary Table 4. Genomic DNAs were isolated using DNeasy Blood & Tissue Kits (Qiagen) according to the manufacturer’s instruction. The CRISPR/Cas9 targeted genome locus of tumour suppressor genes were amplified using primers listed in the Supplementary Table 5. The PCR products were inserted into T vector (Promega) according to the manufacturer’s instruction. Nighy-six colonies per gene were cultured for sequencing.
Renal subcapsular engrafting
All procedures were performed in accordance with institutional animal care guidelines and ethical license protocols. Briefly, adult MF1 nu/nu male mice (8 to 12 weeks) were anesthetized with ketamine solution. After disinfecting the surgical site with 70% alcohol, a 1.5-2 cm incision was made and the kidney was carefully exteriorized. The renal capsule was incised for 2-4 mm using a pipette tip, and a capsule pocket for the grafts was made using a blunted glass Pasteur pipette. Two-month-old organoids from each group were carefully implanted under the renal capsule, respectively. Then kidney was gently replaced back into the retroperitoneal cavity. During the exteriorization, the kidney was kept hydration by applying PBS with penicillin/streptomycin. The kidneys were collected one week and one and half months after xenograft for further analysis. The experiments were performed for three independently.
Immunofluorescence and immunohistochemistry
For immunofluorescence staining, tissues were fixed in 4% paraformaldehyde (PFA) at 4°C for overnight. The tissues were dehydrated in 30% sucrose overnight, embedded in Tissue-Tek (VWR), and then cryosectioned at 16 μm. Sections were blocked and permeabilized in 0.5% Triton X-100 and 4% normal donkey serum (NDS) in PBS at room temperature (RT). Sections were incubated at 4°C with primary antibody in 0.1% Triton-X-100 and 4% NDS in PBS. After washing three times for 10 min with PBS, sections were incubated with secondary antibodies in 0.1% Triton-X-100 and 4% NDS in PBS and DAPI consecutively for visualizing the immunostains. For immunofluorescence staining of adherent cell culture, cells on the coverglass were fixed in 4% PFA for 30 min, blocked and permeabilized in 0.5% Triton X-100 and 4% normal donkey serum (NDS) in PBS at room temperature (RT) for 30 min, incubated with primary and secondary antibodies for one hour at RT sequentially. Then the cells were incubated with DAPI for nuclei staining and mounted with DAKO Fluorescence Mounting Medium. The primary and secondary antibodies were used for immunofluorescence were listed in Supplementary Table 6 and 7. Images were captured using a confocal microscope (Zeiss LSM 780) and fluorescence microscope (Zeiss Axio Imager 2). Quantification of images from three independent preparations of neoplastic cerebral organoids was performed using Fiji. The experiments were performed using samples from three independent preparations.
For histologic and immunohistochemical staining, tissues were fixed in 4% paraformaldehyde overnight. Fixed tissues were rinsed in PBS, dehydrated by immersion in an ascending ethanol gradient (70%, 90%, and 100% ethanol), embedded in paraffin, and sectioned at a thickness of 2 to 5 μm. Sections were stained by a routine Hematoxylin and Eosin (H&E) protocol in a Microm HMS 740 automated stainer. Immunohistochemistry was performed using the Leica Bond III automated immunostainer. The primary and secondary antibodies used in this study were listed in Supplementart Table 6 and 7. Slides were reviewed with a Zeiss Axioskop 2 MOT microscope and images were acquired with a SPOT Insight digital camera. Slides were also scanned with a Pannoramic 250 Flash II Scanner (3D Histech). Digital slides were reviewed and images acquired with the Pannoramic Viewer software (3D Histech). Slides were reviewed by a board certified Veterinary Comparative Pathologist (A.K.).
For quantification of immunofluorescence staining, images from at least three organoids per group, which are collected from three independent experiments, were analysed. The detailed sample size was presented in the relevant figure legend and the associated Source Data.
Drug testing on neoplastic cerebral organoids
For drug testing, neoplastic cerebral organoids were first grown for two months, followed by drug treatment for 40 days. EGFR inhibitors Afatinib (http://www.selleckchem.com, cat. No.: S1011), Erlotinib (http://www.selleckchem.com, cat. No.: S7786), Gefitinib (http://www.selleckchem.com, cat. No.: S1025), Canertinib (http://www.selleckchem.com, cat. No.: S1019), and Pelitinib (Sigma-Aldrich, cat. #: 257933-82-7) (final concentration 1 μM) were applied, and DMSO was used as control. After drug treatment, neoplastic cerebral organoids were trypsinized for single cell preparation, followed by FACS analysis. Total cell numbers were counted to evaluate the cytotoxicity of the drugs. The drug testing experiment were performed twice independently.
Flow cytometry
Flow cytometry experiments were performed according to different experimental purposes. For RNA-sequencing analysis, organoids were trypsinized to make single cell suspension. GFP+ cells were collected using BD FACS AriaIII. Live cells were gated for sorting GFP+ cells for further RNA-seq analysis. For drug testing experiments, organoids were trypsinized to make single cell suspension. The proportion of GFP+ cells were analysed using BD LSR Fortessa2. Live cells were gated for analysis of GFP+ cell proportion. All cells were analysed for analysis of GFP+ cell proportion. Data were analysed using BD FACSDiva software. The example representing the gating strategy were presented in the Supplementary Fig. 10.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 7. Statistical analysis of quantification was performed using unpaired two-tailed Student’s t-test when comparing two groups; using one-way ANOVA with Tukey’s test or Dunnett’s multiple comparisons test when comparing multiple groups. Statistically significant threshold was accepted as p<0.05. No statistical method was used to predetermine the sample size. Sample sizes for experiments were estimated based on previous experience with a similar setup that showed significance. Experiments were not randomized, and were not blindly analysed. All the detailed sample size, statistical analysis, mean±SD, and adjusted p value for each experiment were provided in the relevant figure legends and in the associated Source Data.
Supplementary Material
Acknowledgements
We are grateful to all members of the Knoblich laboratory for discussions, and H. Gustafson and S. Wolfinger for technical support. We thank O. Wüseke and F. Bonnay for the comments on the manuscript. We thank T. Müller, P. Pasierbek, G. Petri, M. Weninger, G. Schmauss, and T. Lendl for FACS and help with imaging. We thank A. Piszczek, T. Engelmaier, J. Klughofer, and and M. Zeba for tissue processing and immunohistochemical staining. We thank the Next Generation Sequencing Facility for Next Generation sequencing. We thank Boehringer Ingelheim RCV GmbH & Co KG for providing EGFR inhibitors. We thank W. Hu for statistical advice. J.A. Bagley received funding from an EMBO postdoctoral fellowship, and funding from European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement (No.707109). Work in J.A. Knoblich laboratory is supported by the Austrian Academy of Sciences, the Austrian Science Fund (Z_153_B09), and an advanced grant from the European Research Council (ERC).
Footnotes
Life Sciences Reporting Summary
All the Life science study design, Statistical parameters, Software using in this study, Data availability, and specific materials were reported in the Life Sciences Reporting Summary.
Data Availability and Accession Code Available Statements
RNA-sequencing data are available at GEO with accession No.: GSE101577. The data that support the findings of this study are available from the corresponding author upon request.
Author Contributions
S.B. and J.A.K conceived the project and experimental design and wrote the manuscript. S.B. performed experiments and analyzed data. M.R, Z.G., and C.K. performed experiments. A.K contributed the histopathology data. T.R.B performed bioinformatics analysis of RNA-seq data. J.A.B contributed in RNA-seq analysis and quantification of immunostained tissues. J.A.K directed and supervised the project.
Competing Financial Interests
S.B. and J.A.K. have filed a patent application for using this method in future disease modelling and pre-clinical investigation.
References
- 1.Ostrom QT, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009-2013. Neuro-oncology. 2016;18:v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, et al. The Genetic Landscape of the Childhood Cancer Medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huszthy PC, et al. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro-oncology. 2012;14:979–993. doi: 10.1093/neuonc/nos135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu B, et al. Epigenetic Activation of WNT5A Drives Glioblastoma Stem Cell Differentiation and Invasive Growth. Cell. 2016;167:1281–1295.e18. doi: 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Z, et al. Zika virus has oncolytic activity against glioblastoma stem cells. J Exp Med. 2017;214:2843–2857. doi: 10.1084/jem.20171093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubert CG, et al. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Research. 2016 doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grotzer MA, Neve A, Baumgartner M. Dissecting brain tumor growth and metastasis in vitro and ex vivo. JCMT. 2016;2:149–14. [Google Scholar]
- 12.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 13.Kelava I, Lancaster MA. Stem Cell Models of Human Brain Development. Cell Stem Cell. 2016;18:736–748. doi: 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Clevers H, Loh KM, Nusse R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346 doi: 10.1126/science.1248012. 1248012–1248012. [DOI] [PubMed] [Google Scholar]
- 15.Lancaster MA, Knoblich JA. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science. 2014;345 doi: 10.1126/science.1247125. 1247125–1247125. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JZ, Hockemeyer D. Human stem cell-based disease modeling: prospects and challenges. Curr Opin Cell Biol. 2015;37:84–90. doi: 10.1016/j.ceb.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Neal JT, Kuo CJ. Organoids as Models for Neoplastic Transformation. Annu Rev Pathol Mech Dis. 2016;11:199–220. doi: 10.1146/annurev-pathol-012615-044249. [DOI] [PubMed] [Google Scholar]
- 18.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bershteyn M, et al. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell. 2017:1–29. doi: 10.1016/j.stem.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mariani J, et al. FOXG1-Dependent Dysregulation of GABA/ Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iefremova V, et al. An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome. Cell Rep. 2017;19:50–59. doi: 10.1016/j.celrep.2017.03.047. [DOI] [PubMed] [Google Scholar]
- 22.Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon C-H, et al. Pten Haploinsufficiency Accelerates Formation of High-Grade Astrocytomas. Cancer Research. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes & Development. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson P, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Momota H, Shih AH, Edgar MA, Holland EC. c-Myc and beta-catenin cooperate with loss of p53 to generate multiple members of the primitive neuroectodermal tumor family in mice. Oncogene. 2008;27:4392–4401. doi: 10.1038/onc.2008.81. [DOI] [PubMed] [Google Scholar]
- 29.Han Z-Y, et al. The occurrence of intracranial rhabdoid tumours in mice depends on temporal control of Smarcb1 inactivation. Nature Communications. 2016;7 doi: 10.1038/ncomms10421. 10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturm D, et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biegel JA, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Research. 1999;59:74–79. [PubMed] [Google Scholar]
- 33.Rogers HA, et al. WNT/β-catenin pathway activation in Myc immortalised cerebellar progenitor cells inhibits neuronal differentiation and generates tumours resembling medulloblastoma. British Journal of Cancer. 2012;107:1144–1152. doi: 10.1038/bjc.2012.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutter S, Bolin S, Weishaupt H, Swartling F. Modeling and Targeting MYC Genes in Childhood Brain Tumors. Genes. 2017;8:107–19. doi: 10.3390/genes8040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atkins M, et al. An Ectopic Network of Transcription Factors Regulated by Hippo Signaling Drives Growth and Invasion of a Malignant Tumor Model. Curr Biol. 2016;26:2101–2113. doi: 10.1016/j.cub.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 36.Gutmann DH, Saporito-Irwin S, DeClue JE, Wienecke R, Guha A. Alterations in the rap1 signaling pathway are common in human gliomas. Oncogene. 1997;15:1611–1616. doi: 10.1038/sj.onc.1201314. [DOI] [PubMed] [Google Scholar]
- 37.Clark PA, et al. Activation of Multiple ERBB Family Receptors Mediates Glioblastoma Cancer Stem-like Cell Resistance to EGFR-Targeted Inhibition. NEO. 2012;14:420–IN13. doi: 10.1596/neo.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer A, Schneider F, Vaupel P, Sommer C, Schmidberger H. Differential expression of HIF-1 in glioblastoma multiforme and anaplastic astrocytoma. Int J Oncol. 2012;41:1260–1270. doi: 10.3892/ijo.2012.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puliyappadamba VT, Hatanpaa KJ, Chakraborty S, Habib AA. The role of NF-κB in the pathogenesis of glioma. Mol Cell Oncol. 2014;1:e963478. doi: 10.4161/23723548.2014.963478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavares CB, et al. Expression of estrogen and progesterone receptors in astrocytomas: a literature review. Clinics (Sao Paulo) 2016;71:481–486. doi: 10.6061/clinics/2016(08)12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellison D, et al. Neuropathology. Elsevier Health Sciences; 2012. [Google Scholar]
- 42.Rocchi A, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J Clin Invest. 2010;120:668–680. doi: 10.1172/JCI36667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitz M, et al. Identification of SOX2 as a novel glioma-associated antigen and potential target for T cell-based immunotherapy. British Journal of Cancer. 2007;96:1293–1301. doi: 10.1038/sj.bjc.6603696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seol HJ, et al. Overexpression of CD99 Increases the Migration and Invasiveness of Human Malignant Glioma Cells. Genes & Cancer. 2012;3:535–549. doi: 10.1177/1947601912473603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwadate Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol Lett. 2016;11:1615–1620. doi: 10.3892/ol.2016.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Binder DK, Berger MS. Proteases and the biology of glioma invasion. J Neurooncol. 2002;56:149–158. doi: 10.1023/a:1014566604005. [DOI] [PubMed] [Google Scholar]
- 47.Paw I, Carpenter RC, Watabe K, Debinski W, Lo H-W. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trojanowski JQ, et al. In vivo and in vitro models of medulloblastomas and other primitive neuroectodermal brain tumors of childhood. Mol Chem Neuropathol. 1994;21:219–239. doi: 10.1007/BF02815352. [DOI] [PubMed] [Google Scholar]
- 49.Muffat J, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui X, et al. Hacking macrophage-associated immunosuppression for regulating glioblastoma angiogenesis. Biomaterials. 2018;161:164–178. doi: 10.1016/j.biomaterials.2018.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bian S, Knoblich JA. Genetic engineering to initiate tumorigenesis in cerebral organoids. Protocol Exchange. doi: 10.1038/protex.2018.071. [DOI] [Google Scholar]
- 52.Mátés L, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 53.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 55.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 57.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 58.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nature Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.