Abstract
It is the main function of T cells to identify harmful antigens as quickly and precisely as possible. Super-resolution microscopy data has indicated that global clustering of the T cell receptor (TCR) occurs prior to T cell activation. Such pre-activation clustering has been interpreted as representing a potential regulatory mechanism that fine-tunes the T cell response. We found here that apparent TCR nanoclustering could be attributed to overcounting artifacts inherent to single-molecule-localization microscopy. Using complementary super-resolution approaches and statistical image analysis, we found no indication of global nanoclustering of the TCR on antigen-experienced CD4+ T cells under non-activating conditions. We also used extensive simulations of super-resolution images to provide quantitative limits for the degree of randomness of the TCR distribution. Together, our results suggest that the distribution of TCRs on the plasma membrane is optimized for fast recognition of antigen in the first phase of T cell activation.
Keywords: TCR nanoclusters, T cell activation, PALM, (d)STORM, STED
Adaptive immunity relies on the ability of T cells to rapidly and efficiently scan the surface of antigen-presenting cells (APCs). For specific antigen recognition, the T cell receptor (TCR) complex binds to cognate peptide-loaded major histocompatibility complex (pMHC) and translates binding events into a functional T cell response 1. In principle, rapid antigen scanning would be supported by randomly distributed TCRs, because such distribution would maximize the TCR-pMHC on-rate 2, 3. However, single-molecule localization microscopy (SMLM) indicated the nanoclustering of the TCR prior to activation in CD4+ and CD8+ primary T cells and in Jurkat T cells 4, 5, 6, 7. The TCR nanoclusters were reported to have an average size of 30-300 nm in diameter and harbor 7 – 30 TCR molecules. Various signaling proteins, such as Lck 8, LAT 4, 9, 10 and Slp-76 10 were described to associate with the TCR nanoclusters, in an activation-dependent manner. These observations prompted new models of T cell signaling by ascribing major regulatory functions to such clusters 2, 11. It is currently a common assumption that nanoclustering of the TCR is crucial to achieve the antigen sensitivity and specificity observed in T cells 11, 12.
Results from SMLM on the nanoscale distribution of the TCR 4, 5, 6, 7 were qualitatively consistent with older electron microscopy 13, 14 and biochemical data 15, with the advantage of being recorded under more natural conditions, partially even in living cells 4. SMLM is based on stochastically switching molecules between a fluorescent (“bright”) state and a non-fluorescent (“dark”) state. Experimental conditions are chosen to assure that most molecules are in the dark state, so that only few well-separated signals can be detected per image 16. The positions of these signals can be determined to an accuracy far below the diffraction limit, which is referred to as localization errors. Recording of thousands of images yields the positions of virtually all fluorophores. Ideally, such reconstructed images consist of the coordinates of all fluorescently labeled molecules and accurately reflect the subcellular distribution of the proteins of interest, below the diffraction limit. In reality, however, some molecules are counted multiple times due to reversible switching (a process known as “overcounting”), whereas other molecules are missed due to insufficient labeling or inactive fluorophores 17. Overcounting is bound to give rise to localization clusters, which emerge from the same single dye molecules counted multiple times, and severely complicates the interpretation of the localization maps observed. Analytical and experimental frameworks to address this problem have been suggested 17, 18, 19, 20.
Here, we applied label-density-variation SMLM to analyze the spatial distribution of the TCR on the plasma membrane of non-activated and activated primary CD4+ T cells. This method is based on deliberate variation of the label concentration and quantitative statistical image analysis of SMLM experiments. The results were verified by stimulated emission depletion (STED) microscopy 16, a complementary superresolution technique, which is not affected by overcounting artifacts. We used further image simulations and statistical image analysis to provide a quantitative estimate of the degree of randomness of the TCR distribution. Contrary to previous reports, our data did not support the existence of TCR nanoclusters in non-activated T cells. Our results were instead consistent with a random distribution of the TCR at the T cell plasma membrane, which appears best suited for rapid antigen scanning.
Results
SMLM of the TCR in CD4+ T effector cells
We used primary, antigen-experienced splenic CD4+ T effector (TEFF) cells from mice transgenic for the 5c.c7 TCR, which specifically recognizes a moth cytochrome c peptide bound to the MHC II molecule I-Ek (pMHC) 21. Cells were stimulated and cultivated in vitro for 7-9 days before being brought in contact with either non-activating or activating surfaces 22. For non-activating conditions, we used fluid supported lipid bilayers functionalized with the adhesion protein ICAM-1 (Supplementary Fig. 1a), a method used by previous studies reporting the nanoscale clustering of the TCR 4, 5. For antigen-specific T cell activation conditions, we used lipid bilayers functionalized with ICAM-1, and additionally with the co-stimulatory protein B7-1 and stimulatory pMHC loaded with moth cytochrome c peptide. Because the conditions used to maintain T cells in a resting state have generated controversy in the recent literature as to whether a true resting state can be observed when a T cell interacts with a flat surface 23, 24, we used live cell ratiometric calcium imaging via Fura-2 to check the activation state of T cells under identical conditions as for the imaging experiments (Supplementary Fig. 1b). We found that cells did not substantially activate on lipid bilayers bearing only ICAM-1. However, they did respond with rapid influx of Ca2+ when stimulated on lipid bilayers displaying ICAM-1, B-7 and pMHC. All other imaging experiments, unless otherwise indicated, were carried out after fixation of CD4+ TEFF cells to ensure the localization of fluorescent molecules with maximal positional accuracy, undisturbed by molecular diffusion.
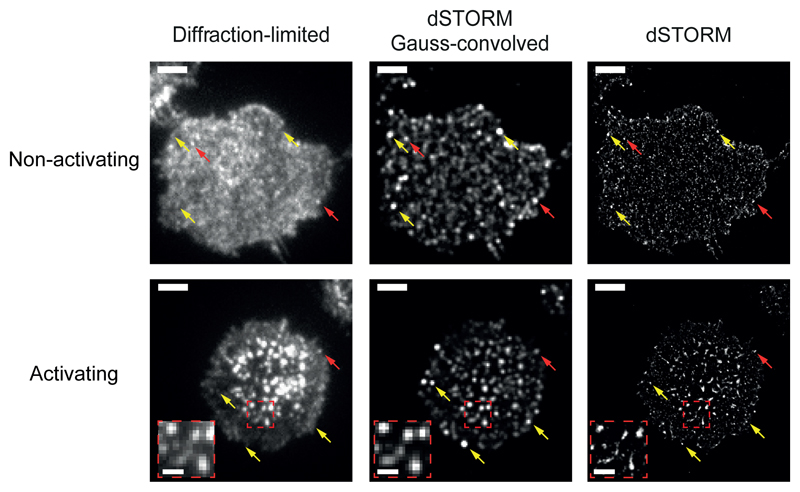
Random protein distributions appear clustered on SMLM imagesWe first performed dSTORM experiments on CD4+ TEFF cells plated on non-activating bilayers. To label the TCR we used a β-chain specific monoclonal antibody (clone H57) conjugated to AlexaFluor647 (AF647). Each experiment included the recording of a standard fluorescence microscopy image of a single T cell (referred to as “diffraction-limited” image), followed by dSTORM imaging and the reconstruction of localization maps. We could observe heterogeneities in the brightness of the diffraction-limited images (Fig. 1), which could be interpreted as an indication of a non-random protein distribution. However, these heterogeneities could also originate from the pixel-to-pixel fluctuations of the number of TCR complexes in combination with a stochastic labeling degree of the used antibody. Therefore, we compared the diffraction-limited images of T cells with images of localization maps convolved with the experimentally determined point-spread function (see Methods). If localization maps reflected the true spatial distribution of labeled proteins, the two images would be identical. However, there are bright spots in the reconstructed image which do not have a correspondence in the diffraction-limited image (Fig. 1), indicating the presence of overcounting artifacts across the image.
Figure 1. Blinking and multiple observations lead to over-representation of single molecules in SMLM images.
Diffraction-limited images (left), dSTORM localization maps (right), and back-calculated diffraction-limited images based on dSTORM localization maps (center) of fixed primary murine CD4+ TEFF cells labeled with 10μg/ml H57-AF647; images were recorded under non-activating (top) or activating (bottom) conditions. In the back-calculated image each xy-position of the dSTORM image was convolved with a Gaussian function characterized by its respective intensity and σ-width. Yellow arrows: features in the dSTORM and reconstructed images with no correspondence in the original diffraction-limited image. Red arrows: features that do have such a correspondence. Inserts (red dashed boxes) show zooms of regions in activated cells with pronounced microclustering, where high localization densities clearly correlated with high molecular densities. Scale bars: 3 μm in main images and 1 μm in enlarged regions; representative data (n=19 and n=16 biologically independent samples for activating and non-activating conditions, respectively).
Label-density-variation dSTORM reveals random TCR distribution
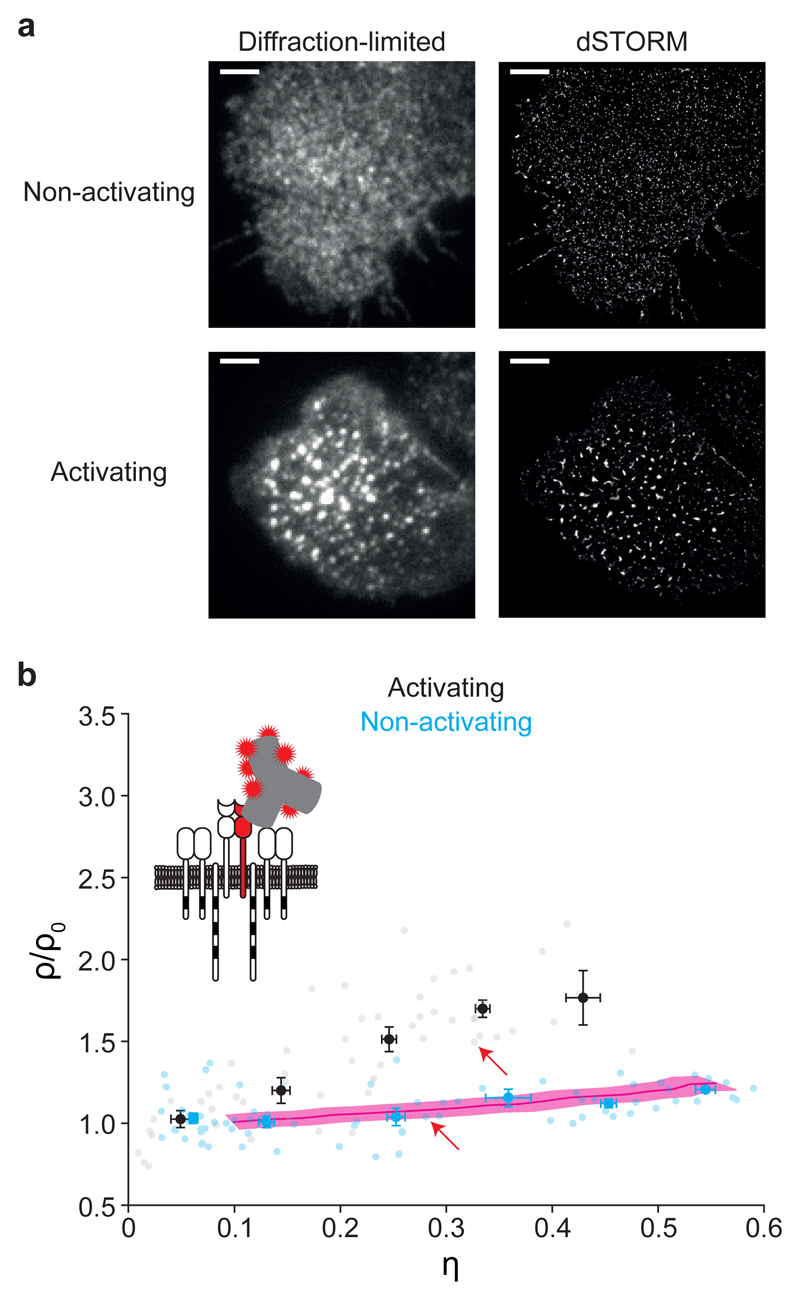
Label-density-variation SMLM was recently developed to discriminate true molecular clustering from overcounting artifacts 18. It exploits highly characteristic changes in the statistical properties of localization clusters when titrating the degree of labeling. In case of true nanoclustering, the number of localizations per detected localization cluster increases with increasing label concentrations. In case of a purely random protein distribution, the number of localizations per detected localization cluster only depends on the blinking properties of the probe, and hence remains unchanged with increasing label concentrations 18. In label-density-variation SMLM the relative area covered by clusters, η, and the normalized localization density within clusters, ρ/ρ0, are calculated to characterize localization clusters at different label concentrations. Here, we used label-density-variation SMLM to determine whether the deviations from a random localization distribution (Fig. 1) reflected nanoclustering of TCR, or whether it could be attributed to blinking of the dye molecules. For this, we labeled CD4+ TEFF cells with different concentrations of antibody specific for TCRβ, and analyzed the η and ρ/ρ0 of the detected localization clusters (Fig. 2 and Supplementary Fig. 1c). A random distribution is characterized by a rather flat curve indicated by the red line 18, which was calculated based on the experimentally determined blinking statistics of the used fluorescently labeled antibodies (Supplementary Fig. 2 and Methods). In contrast, clustering is indicated by an increase of the obtained ρ/ρ0 values versus η. For non-activated T cells, data did not show any deviations from the reference curve for a random distribution of molecules and were therefore fully consistent with a random TCR distribution (Fig. 2b). These results indicated that the localization clusters observed in SMLM (Fig 1 and Fig 2a) were due to overcounting of single H57-AF647-labeled TCR.
Figure 2. Label-density-variation dSTORM of TCRβ.
(a) Representative diffraction-limited microscopy images (left) and dSTORM localization maps (right) of fixed primary murine CD4+ TEFF cells labeled with H57-AF647 during interaction with non-activating (top) or activating (bottom) supported lipid bilayers (n=19 and n=16 biologically independent samples for activating and non-activating conditions, respectively); Scale bars: 3 μm. (b) Normalized ρ versus η plot derived from label-density-variation dSTORM of fixed primary murine CD4+ TEFF cells under non-activating (blue) and activating (black) conditions using H57-AF647 (0.05, 1, 5 and 10 μg/ml); data were binned based on η with a bin size of 0.1 and represented as means ± SEM; n=75 for non-activating and n=55 for activating conditions. Data for individual cells are shown in gray and light blue; red line with pink shaded region indicates reference line and its uncertainty, respectively, for a random distribution derived from simulations based on the experimentally determined blinking statistics of H57-AF647 (mean ± SEM; n=50 independent simulations); red arrows indicate data points corresponding to the cells shown in a; Cartoon to illustrate the labeling strategy.
To illustrate common pitfalls associated with overcounting blinking molecules, we simulated the localization maps for purely random distributions of proteins. For this, we assigned the experimentally-derived blinking statistics of the single dye-conjugated antibodies recorded in situ to randomly distributed molecule positions. Here we defined “random” as a uniform protein distribution following a Poisson process (Methods). We used the Ripley’s K function to illustrate the effect of fluorophore blinking in SMLM experiments. It quantifies the randomness of 2D-point-distributions by analyzing the number of points within a distance r of another point, and is commonly used to characterize clustering in SMLM 4, 8, 9. When plotted as a function of r, a constant value of L(r) - r would be indicative of a random point distribution, whereas clustered point distributions would yield a pronounced maximum approximately at the cluster radius 25. For control, we first performed simulations without fluorophore blinking. In this case, each molecule position resulted in a single localization. The localization maps showed scattered individual points, and L(r) - r yielded a constant value (Supplementary Fig. 3a). When the experimentally determined blinking statistics were included in the simulations, however, the images contained localization clusters. For each simulation, L(r) - r showed a pronounced maximum approximately at the size of the localization errors of the single-molecule signals (Supplementary Fig. 3b-j). These simulations illustrate that Ripley’s K function is sensitive to blinking-induced localization clusters that do not originate from molecular clustering. Hence, such localization clusters cannot be taken as indication of a non-random distribution of proteins.
Eventually, when analyzing the immunological synapse upon peptide-specific T cell activation, the TCR microclusters were readily detectable with diffraction-limited imaging, dSTORM imaging and the label-density variation approach (Fig. 1 and Fig. 2). As a control, labeling the CD3ε chain, which is non-covalently associated with the TCR, with an AF647-conjugated fluorescent antibody (clone KT3), yielded similar results, both under non-activating and activating conditions (Supplementary Fig. S4a, b and c). In conclusion, label-density-variation SMLM of the TCR in non-activated CD4+ TEFF cells indicates a random distribution of the TCR.
Label-density-variation PALM confirms random TCR distribution
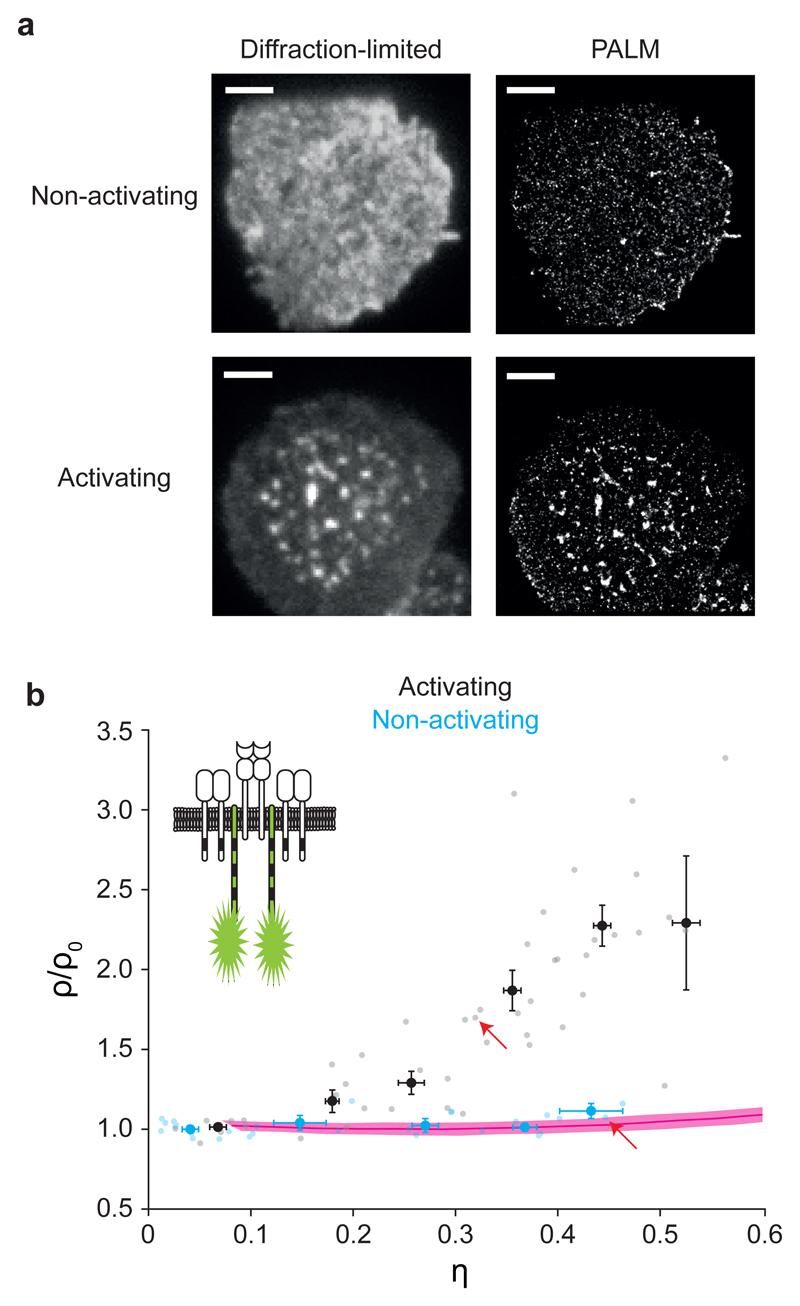
dSTORM is based on the repeated switching of dye molecules between a dark and a fluorescent state 26, 27, rendering blinking and associated overcounting an inherent complication. In contrast, PALM 28, 29 employs stochastic photoactivation, imaging and photobleaching of photo-switchable dyes, typically fluorescent proteins. PALM images yield one localization per protein molecule 4, but overcounting artifacts have been reported for photo-switchable proteins with PALM as well17, 18, 30. Photoswitchable cyan fluorescent protein 2 (PS-CFP2) is considered to be least prone to blinking and was hence used in pioneering studies on TCR organization 4. However, because a significant fraction (~65%) of molecules showed multiple detection events when observed at the single-molecule level (Supplementary Fig. 2) we tested whether overcounting using PS-CFP2 may have been misinterpreted as TCR nanoclusters. For the PALM experiments we ectopically expressed a PS-CFP2-CD3ζ fusion construct, which non-covalently associates with the TCR complex 4. We brought the cells in contact with non-activating or activating lipid bilayers and fixed them prior to imaging. Super-resolution images contained clusters of localizations (Fig. 3a), consistent with previous results 4, 5, 10. Taking advantage of the cell-to-cell variability in the PS-CFP2-CD3ζ expression levels, we carried out label-density variation PALM. The resulting ρ/ρ0 versus η curve was in agreement with a purely random TCR distribution indicated by the red reference curve, whereas label-density-variation PALM on activated CD4+ TEFF cells yielded ρ/ρ0 versus η curves that deviated from the reference curve (Fig. 3b).
Figure 3. Label-density-variation PALM of CD3ζ.
(a) Representative diffraction-limited microscopy images (left) and PALM localization maps (right) of fixed primary murine CD4+ TEFF cells expressing the fusion construct CD3ζ-PS-CFP2 and labeled with H57-scFv-AF647 (for diffraction-limited imaging) during interaction with non-activating (top) or activating (bottom) supported lipid bilayer (n=50 and n=30 for activating and non-activating conditions, respectively); Scale bars: 3 μm. (b) Normalized ρ versus η plot derived from label-density-variation PALM based on the intrinsic expression level variabilities of primary murine CD4+ TEFF cells expressing CD3ζ-PS-CFP2 under non-activating (blue) and activating (black) conditions; data were binned based on η with a bin size of 0.1 and represented as means ± SEM; n=30 for non-activating and n=50 for activating conditions. Data for individual cells are shown in gray and light blue; red line with pink shaded region indicates reference line and its uncertainty, respectively, for a random distribution derived from simulations based on the experimentally determined blinking statistics of PS-CFP2 (mean ± SEM; n=50 independent simulations); red arrows indicate data points corresponding to the cells shown in a; Cartoon to illustrate the labeling strategy.
Live cell PALM was suggested to be robust against overcounting artifacts, because the labeled molecules would move between the fluorescent blinks, and overcounts would not be detected on the same spot 4. We hence simulated a random distribution of PS-CFP2-labeled molecules diffusing by Brownian motion according to experimentally determined parameters. In addition, we used experimentally determined parameters for fluorophore blinking in the simulations. TCRβ in non-activated T cells has a mobility of D~0.047 μm2/s and a mobile fraction of 64% 31, 32. This rather low long-range TCR mobility results in PS-CFP2 molecules observable multiple times in close proximity to the initial observation. As a consequence, localization clusters were detected in the Ripley’s K analysis (Supplementary Fig. 3i). To experimentally validate the results of these simulations, we performed label-density-variation PALM on PS-CFP2-CD3ζ diffusing on the plasma membrane of non-activated live CD4+ TEFF cells. Data were analyzed, calculating ρ/ρ0 versus η, as for the analysis of fixed cell data. While localization clusters of PS-CFP2-CD3ζ were clearly visible in the images, the label-density variation analysis did not indicate any deviation from a random molecular distribution (Supplementary Fig. 4d and e). Taken together, the SMLM images did not provide any indication for the presence of substantial TCR nanoclusters at the plasma membrane of non-activated T cells.
Image simulations show quantitative limits of detection
Next, we assessed the sensitivity of label-density-variation SMLM for detecting nanoclusters. To this end, we performed comprehensive Monte Carlo simulations of SMLM images at different label densities, and calculated ρ/ρ0 versus η in the same way as for the experimental data. We simulated clusters at densities between 3 and 20 clusters per μm2 and with a radius between 20 nm and 150 nm, which covers values reported in the literature 4, 5. In addition, we included randomly distributed molecules outside the clusters, which account for a fraction of up to 20% non-clustered TCR 4, 5. To account for overcounting, we allocated multiple localizations to each simulated molecule, employing the empirically-determined blinking statistics of the probes used in our experiments. The blinking statistics were derived in situ, i.e. from label-density-variation SMLM samples at the lowest labeling concentration. Imaging conditions were kept identical to the ones we used for label-density-variation SMLM of the TCR. The surface density of the TCR was modeled according to typical values obtained in the experiments: we determined 59-81, 68-73 and 37-141 molecules/μm2 for H57-AF647, KT3-AF647 and CD3ζ-PS-CFP2, respectively.
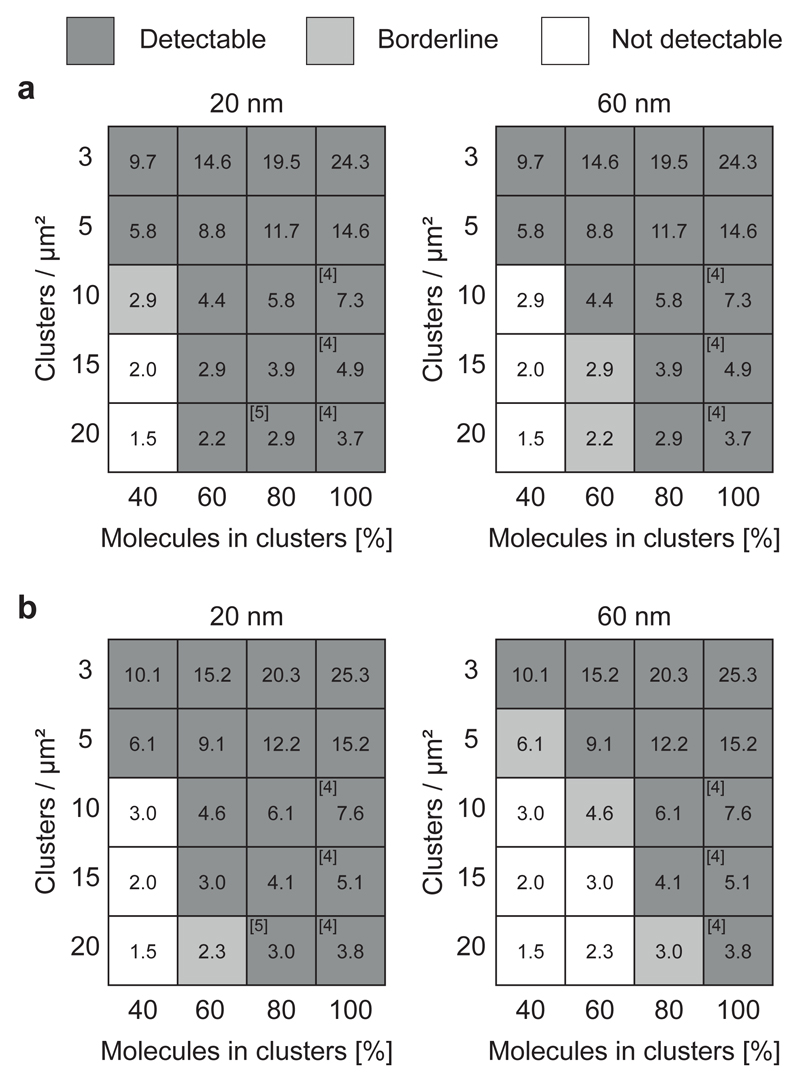
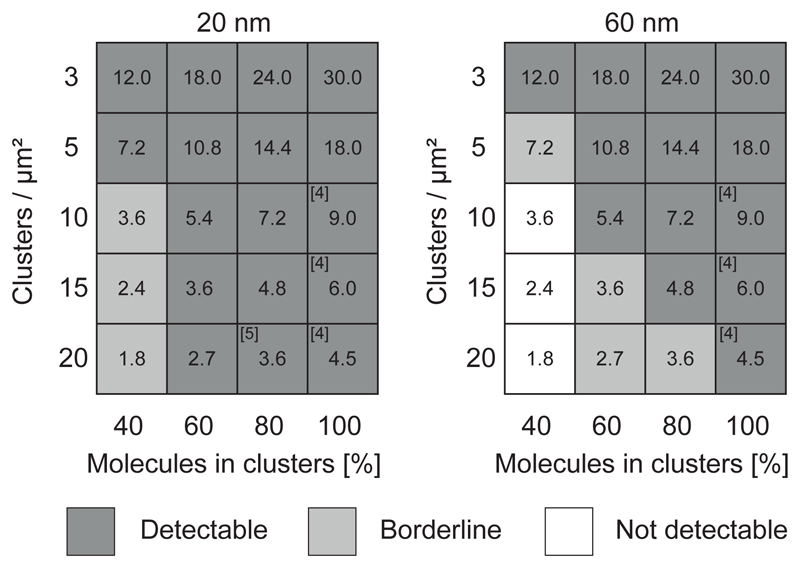
For each setting of cluster parameters, we classified the simulation results as clearly detectable, if the corresponding curves of ρ/ ρ0 versus η differed from the curves of a simulated random distribution of molecules. In a real experiment, non-random TCR distributions described by such cluster parameters would have been detectable. As we did not detect any difference from the random curve, these scenarios can be ruled out. ρ/ρ0 versus η curves of simulations, for which we found no difference between the clustered and random distribution, where classified as not detectable. In a real experiment, such TCR distributions would not result in differences from a random curve and would hence be missed by our approach. The classification scheme is described in Supplementary Fig. 5a. Together, label-density-variation SMLM experiments and sensitivity assessment based on simulations allow the conclusion that non-random TCR distributions down to 3 molecules per cluster at reported cluster sizes ≤ 80 nm can be ruled out (Fig. 4, Supplementary Fig. 5b and Supplementary Fig. 6).
Figure 4. Sensitivity of label-density-variation SMLM to detect nanoclustering.
Normalized ρ versus η plots were calculated for different simulated clustering scenarios and assessed for the difference from simulated random molecular distributions; detectable difference (dark gray), borderline (light gray) and not detectable difference (white) (see Supplementary Fig. 5a). Reference numbers indicate scenarios published in the literature; Simulations of nanoclusters with radii of 20 nm (left) or 60 nm (right) for 3, 5, 10, 15 and 20 clusters/μm2. The fraction of molecules inside clusters was varied between 40% and 100%. Average molecular densities were adjusted to (a) 73 molecules/μm2 based on H57-AF647 labeling of TCRβ (Fig. 2) and (b) 76 molecules/μm2 based on CD3ζ-PS-CFP2 PALM experiments (Fig. 3). Blinking statistics were based on experimental data of the respective fluorescent probes. Numbers in boxes indicate the average number of molecules per cluster.
STED microscopy confirms random distribution of TCR
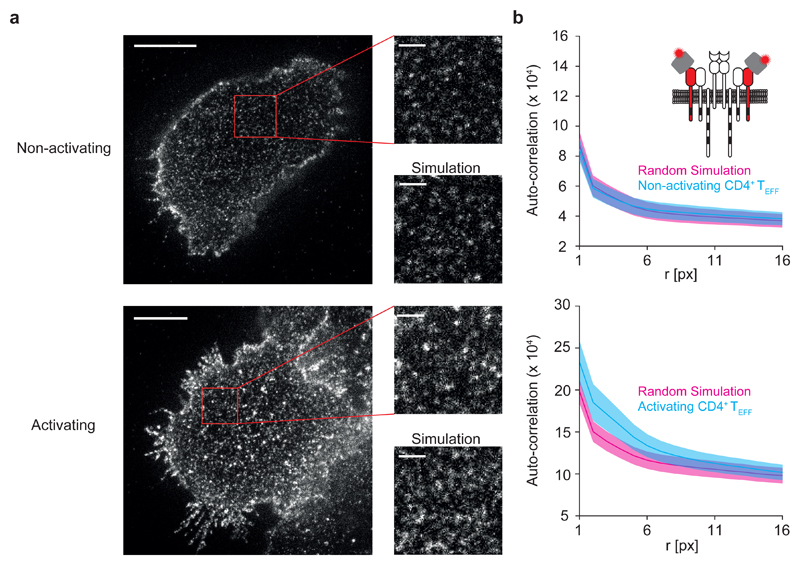
We then sought to validate our conclusion that the TCR was randomly distributed in non-activated CD4+ TEFF cells using a complimentary super-resolution imaging approach that is not susceptible to overcounting artifacts. In STED microscopy, resolution below the diffraction limit is achieved by reducing the width of the effective point-spread function to extremely small values using high-power stimulated emission depletion 16. Because the images are recorded in a single scan, cluster analysis from STED images is not affected by fluorophore blinking. We labeled CD3ε with an Abberior STAR 635P (AS635P)-conjugated single-chain variable fragment (scFv) derived from the antibody clone KT3 and analyzed fixed CD4+ TEFF cells under non-activating and activating conditions (Fig. 5a).
Figure 5. STED microscopy of CD3ε and image autocorrelation analysis.
(a) Representative STED microscopy images of fixed primary murine CD4+ TEFF cells labeled with KT3-scFv-AS635P during interaction with non-activating (top) or activating (bottom) supported lipid bilayers (n=2 biologically independent samples for both activating and non-activating conditions); Enlarged areas for each condition (right) are indicated by red boxes. Recorded and simulated images of randomly distributed labels are shown as indicated; for the simulations, the number of molecules per area and the point spread function were matched to the experimental data. Scale bars: 3 μm in full-size images and 500 nm in enlarged regions (b) Image autocorrelation analysis of STED microscopy data shown in (a); Autocorrelation curves of experimental data are shown in blue. At least five ROIs per cell were analyzed and averaged (means ± SEM; n= 5); Red line and pink shaded region indicates reference line and its uncertainty, respectively, for a random distribution derived from simulations based on the experimentally determined properties of the point spread function (mean ± SEM; n=50 independent simulations); Cartoon to illustrate the labeling strategy.
To objectively judge the randomness of the TCR distribution underlying the recorded STED images, we compared the STED images obtained from fixed CD4+ TEFF cells labeled with KT3-scFv-AS635P with simulated images obtained using the fitted single-molecule parameters of dye-conjugated scFvs recorded from sparsely labeled cells as input (Supplementary Fig. 7a and b). To this end, we employed image autocorrelation analysis, which allows the quantification of the spatial distribution of fluorophores in microscopy images 33. The autocorrelation function (ACF) quantifies the likelihood that two pixels separated by a distance r have similar brightness. ACF analysis yields a pronounced decrease of the curve at small r values as a consequence of the non-zero width of the point spread function. Clusters will appear as an additional, exponentially decaying term, with the length of the decay specifying the size of the clusters 34. The ACF amplitude scales with the inverse of the number of independent particles per pixel; a small surface density of molecules, but also the clustering of molecules, hence increases the ACF amplitude. ACF analysis of the STED images of fixed CD4+ TEFF cells labeled with KT3-scFv-AS635P yielded no difference in the curves compared to a simulated pure random distribution of TCR molecules (Fig. 5b and Supplementary Fig. 8a). Similar results were observed when we labeled the TCRβ chain with an AS635P-conjugated H57-scFv (Supplementary Fig. 8b).
To illustrate how true clustering would be visualized by STED microscopy, we simulated STED images of global TCR nanoclustering using previously reported parameters 4, 5 (i.e. densities between 3 and 20 clusters per μm2 with a radius between 20 nm and 150 nm and with fractions of up to 20% non-clustered TCR molecules). Compared to the experimentally measured data, such scenarios yielded substantially higher amplitudes and longer tails in the autocorrelation functions (Supplementary Fig. 7a-h). This led us to conclude that the presence of nanoclusters with densities between 3 and 20 clusters per μm2, radii between 20 nm and 150 nm and with fractions of up to 20% non-clustered TCR molecules could be ruled out.
In addition, we recorded STED images of activated CD4+ TEFF cells, and observed formation of microclusters that was not observable in the non-activated control cells. Concomitantly, ACF curves of the analyzed activated cells showed higher amplitudes and longer decays compared to the controls (Fig. 5b and Supplementary Fig. 8a). Of note, due to the rather small fraction of molecules present in microclusters (approximately 10%), the difference to a random distribution was not as pronounced as in the simulated cases of global nanoclustering (Supplementary Fig. 7g and h).
Finally, we determined the sensitivity of the autocorrelation method for the detection of nanoclustering in STED microscopy. We performed holistic simulations of STED images of random versus clustered scenarios for the same parameter settings as in Fig. 4, Supplementary Fig. 5 and Supplementary Fig. 6. For each setting of cluster parameters, we classified the simulation results as clearly detectable, if the corresponding ACF curve differed from the ACF curve of a simulated random distribution of molecules. In a real experiment, non-random TCR distributions described by such cluster parameters would have been detectable. As we did not detect any difference from the random curve, these scenarios can be ruled out. ACF curves of simulations, for which we found no difference between the clustered and random distribution, were classified as not detectable. In a real experiment, such TCR distributions would not result in differences from a random curve and would hence be missed by our approach. The classification scheme is described in Supplementary Fig. 9a. The simulations of STED microscopy data and their analysis with image autocorrelation showed similar sensitivity as label-density-variation SMLM (Fig. 6 and Supplementary Fig. 9b). Together these results suggest that STED imaging combined with autocorrelation analysis did not support the existence of substantial global TCR nanoclustering in non-activated CD4+ TEFF cells down to a sensitivity limit of 3 molecules per cluster at cluster sizes of ≤80 nm.
Figure 6. Sensitivity of STED image autocorrelation analysis to detect nanoclustering.
Image autocorrelation analysis was performed for different simulated clustering scenarios and assessed for the difference from simulated random molecular distributions; Detectable difference (dark gray), borderline (light gray) and not detectable difference (white) (Supplementary Fig. 9a). Reference numbers indicate scenarios published in the literature; Simulations of nanoclusters with radii of 20 nm (left) or 60 nm (right) for 3, 5, 10, 15 and 20 clusters/μm2. The fraction of molecules inside clusters was varied between 40% and 100%. Average molecular densities were adjusted to 75 molecules/μm2 based on KT3-scFv-AS635P labeling of CD3ε (Fig. 5). Parameters of the simulated point spread function were based on experimental data. Numbers in boxes indicate the average number of molecules per cluster.
Discussion
Here we found using label-density-variation SMLM that global nanoclustering of TCRs in non-activated T cells is not supported by commonly used superresolution microscopy data. We show that the TCR clusters reported previously originate from blinking artifacts inherent to SMLM. We corroborated our results using STED microscopy as a complementary super-resolution technique. It should be noted that occasional local deviations from a pure random distribution were detectable in our images, both in diffraction-limited and in superresolution microscopy. Such structures may include cell boundaries, filopodia or microvilli flattened out during the adhesion process 6, 35, or TCR located in endocytic vesicles close to the plasma membrane, which – in a 2D projection – would give rise to increased brightness and localization densities. In summary, our results indicate that state-of-the-art super-resolution microscopy techniques do not provide experimental evidence for the existence of global TCR nanoclustering. Together with recent data ruling out the existence of TCR oligomers down to the level of dimers 31, this data indicates that the TCR is distributed essentially at random across the T cell plasma membrane under non-activating conditions.
In the light of the current models of T cell activation, it appears that evolutionary processes have selected random TCR distributions over nanoclustering. This is in line with the central task of T cells to ensure rapid, sensitive and specific encounter of antigenic pMHC at the surface of APCs. Different concepts have been put forward to understand this process, such as TCR clustering 2, 11, pulling forces 21, 36, 37 or localized contacts between the T cell and the APC 35, covering different facets of the recognition process. However, from the perspective of the search for antigenic pMHC, TCR activation shows similarities to the situation of data-mining procedures for rare cases 38. During antigen recognition, T cells have to rapidly identify an extremely low number of relevant instances (here, antigenic pMHCs) over a vast excess of retrieved instances (here, non-activating pMHCs). Following common nomenclature in statistics, “precision” would define the percentage of times, in which antigenic pMHCs are correctly identified “sensitivity” defines the fraction of correctly detected antigenic pMHCs among all presented pMHCs For rare events, it’s difficult to optimize precision and sensitivity simultaneously. In such cases, a two-phase approach is expected to be optimal: in the first phase, called the “scanning phase”, low-precision rules are accepted, as long as the sensitivity remains high. In other words, erroneous signaling due to binding of non-activating pMHCs is accepted, as long as no antigenic pMHCs is missed. As such, at the scanning phase, the speed of pMHCs identification has to be optimized, even at the cost of limited precision. In the second phase, the “activation phase”, precision would be optimized, scrutinizing and, if necessary, revising antigen recognition. A classic example is the confrontation of T cells with very low numbers of antigenic pMHCs at the APC surface. Indeed, CD4+ TEFF cells can be stimulated with single pMHC molecules 39, potentially even at the level of single TCR molecules 40. Monomeric TCRs recruit ZAP70 to the initial contact sites during activation 31 and act as a catalytic unit for downstream signaling cascades 41, which lead to calcium signaling within seconds and the formation of TCR microclusters within minutes. In this context, calcium signaling could be interpreted as the readout of the scanning phase, while the TCR microcluster formation could be part of the recheck strategy during the activation phase. Other contributions for enhancing precision in the second phase may come from the abovementioned processes, such as force generation or dynamic T cell-APC contacts and may well involve other receptors as well. Notably, sustained TCR triggering over hours is required for maintenance of the immunological synapse and cytokine production 42, highlighting the relevance of recheck processes after the initial scanning phase.
Our data thus shed new light on how evolutionary pressure could have shaped the search strategy of T cells for cognate antigen. A random TCR distribution appears to be advantageous for the requirements of the scanning phase in resting T cells: maximizing the TCR-pMHC on-rates would optimize the likelihood of finding rare cognate antigens. Only during the activation phase, regulatory check and recheck mechanisms would come into play and modulate the activation process. Understanding the transition from purely random to clustered TCR distributions, as observed under activating conditions, will be a crucial aim for future studies.
Methods
Cell culture, DNA constructs, Antibodies and Reagents
All chemicals and cell culture supplies were from Sigma if not otherwise noted. Primary murine T cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1 kU/ml penicillin-streptomycin, 50 μM β-mercaptoethanol and 1 mM sodium pyruvate. The phoenix packaging cell line for retroviral infections was cultured in DMEM medium supplemented with 10 % FBS, 2 mM L-glutamine, 1 kU/ml penicillin-streptomycin. All cells were grown in a humidified atmosphere at 37 °C and 5% CO2.
For expression of the CD3ζ-PS-CFP2 fusion protein, we cloned the sequence of murine CD3ζ in frame with PS-CFP2 into the retroviral expression vector pIB2.
Alexa Fluor 647 (AF647)-conjugated TCRβ-specific antibody (clone H57-597); average degree of labeling of 7.4) was purchased from Biolegend (CatNo. 109218; LotNo. B206104). CD3ε-specific antibody (clone KT3) was from AbD Serotec/Bio-Rad Technologies (CatNo. MA1-80783; LotNo. 1603) and was conjugated to AF647 via NHS-ester chemistry following the supplier’s instructions. After removal of unreacted dye using Zeba desalting columns (Thermo Fisher Scientific) an average degree of labeling of 3.8 was determined.
T cell isolation and transduction
Primary T cells were isolated and treated as described elsewhere 42. Briefly, splenic T cells were isolated from 5c.c7 αβTCR transgenic mice and cultured in vitro in the presence of 1 μM moth cytochrome c (MCC) peptide (aa 88-103: ANERADLIAYLKQATK, T-cell epitope underlined, Elim Biopharmaceuticals) and IL-2 (added after 24 h) for 7-9 days. For retroviral transduction, we essentially followed protocols from the Nolan lab (Stanford University). Phoenix packaging cells were co-transfected with pIB2-CD3ζ-PS-CFP2 and pCL-eco using TurboFect (Invitrogen Life Technologies) on day 1 after T cell isolation, followed by two days of virus production. On day 3 after isolation, T cell blasts were infected by spin-infection in the presence of 10 μg/ml polybrene and 50 U/ml IL-2. Selection for positive cells was achieved by addition of 10 μg/ml blasticidin on day 4 after isolation. On day 6 after isolation dead cells were removed by a density-dependent centrifugation gradient using histopaque 1119. All experiments were conducted on days 7 – 9 after isolation.
Ethical compliance statement
All animal experimentation (related to breeding, sacrifice for T cell isolation) was evaluated by the ethics committee of the Medical University of Vienna and approved by the Federal Ministry of Science, Research and Economy, BMWFW (BMWFW-66.009/0378-WF/V/3b/2016). Animal husbandry and experimentation was performed under the national laws (Federal Ministry of Science, Research and Economy, Vienna, Austria) and ethics committee of the Medical University of Vienna and according to the guidelines of the Federation of Laboratory Animal 671 Science Associations (FELASA).
Protein expression and functionalization
IEk-MCC was prepared as described previously 21. For the generation of single-chain variable fragment (scFv) of TCRβ specific antibody (H57-scFv), mRNA was prepared from H57-597 or KT3 hybridoma (American Type Culture Collection) to serve as a template for 5′ rapid amplification of cDNA ends (RACE; Invitrogen). VH and VL antibody domains were fused as described in detail in Huppa et al 21 and mutagenized for site-specific modification using the Quikchange protocol (Stratagene). After refolding from inclusion bodies 43, scFv preparations were purified from aggregates on a S-200 size-exclusion column (GE Healthcare), site-specifically labeled for 2 h with AF647- or AS635P-maleimide in the presence of 50 μM tris(2-carboxyethyl) phosphine hydrochloride (TCEP; Pierce) and monomeric scFv–dye conjugates were again purified by size-exclusion chromatography (S-75; GE Healthcare). The label:protein stoichiometry was determined to be close to 1 in all cases.
Preparation of glass-supported lipid bilayers
All lipids were from Avanti Polar Lipids. Preparation of vesicles composed of 90% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 10% 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (18:1 DGS-NTA(Ni)) was done as described previously 21. Glass cover slides (#1.5, 24x60 mm, Menzel) were plasma cleaned for at least ten minutes and attached to 8-well LabTek chambers (Nunc), where the bottom had been removed. Glass slides were incubated for 10 min at room temperature with the vesicle suspension, followed by extensive rinsing with PBS. Supported lipid bilayers were functionalized with His10-ICAM-1 (Sinobiologicals) only or additionally with His12-pMHC and His10-B7-1 (Sinobiologicals) for 75 min, followed by extensive rinsing with PBS. Before addition of T cells, PBS was replaced with imaging buffer (HBSS + 2% FBS) by sequential dilution.
TCR labeling and sample preparation
All labeling steps were done on ice. Roughly 106 cells were washed in imaging buffer. For dSTORM experiments, we used full antibodies bearing multiple fluorophores. This is necessary, since due to high illumination powers used in dSTORM, not all fluorophores return to the active state within the imaging period. Multiple fluorophores per label increase the chance to detect most labels present in the sample. In a first step, unspecific binding sites were blocked with 5% BSA for 25 min, and then the cells were incubated with varying antibody concentrations (H57: 0.05, 1, 5 and 10 μg/ml; KT3: 0.02, 0.2, 2, 10 and 20 μg/ml) for 20 minutes. For experiments under non-activating conditions using KT3-AF647, blocking and antibody labeling was done after cell adhesion to the supported lipid bilayers and fixation, as labeling prior to cell seeding and adhesion induced Ca2+ influx. PALM and STED experiments were carried out with stoichiometrically labeled scFv, which replicates previously used labeling strategies. Labeling was done for 15 min. For ζ-PS-CFP2 PALM experiments, live T cells were labeled with 50 μg/ml H57-scFv-AF647 to follow microcluster formation before imaging PS-CFP2. For STED experiments we employed saturating concentrations of fluorescent scFv (5 μg/ml H57-scFv-AS635P or 50 μg/ml KT3-scFv-AS635P) or a mixture of fluorescent/non-fluorescent scFv to achieve labeling at single-molecule density (1:10 molar ratio).
After labeling, in all cases cells were washed twice with imaging buffer on ice before addition to the sample chambers. Cells were allowed to settle for 15 min under non-activating and 5 min under activating conditions. Cells were then fixed with 4% paraformaldehyde (PFA; Polysciences)/0.2% glutaraldehyde (GA) for 10 min at room temperature. For experiments under non-activating conditions using KT3-AF647, cells were extensively washed after labeling and again fixed for 10 min at room temperature to avoid detachment of the antibodies. Live cell PALM experiments were started 5 min after cell seeding without further preparations.
Single-molecule localization microscopy and tracking
A Zeiss Axiovert 200 microscope equipped with a 100x Plan-Apochromat (NA=1.46) objective (Zeiss) was used for imaging samples in objective-based total internal reflection (TIR) configuration. The setup was further equipped with a 640 nm diode laser (iBeam smart 640, Toptica), a 405 nm diode laser (iBeam smart 405, Toptica) and a 488 nm optically pumped semiconductor laser (Sapphire, Coherent). Acousto-optic modulators (AOM) were used to modulate intensity and timings using an in-house developed Labview software. For STORM experiments, we used a zt488/640rpc dichroic mirror (Chroma) and an FF01-538/685-25 emission filter (Semrock). For PALM experiments, instead of the emission filter we used a dual view system (Photometrix) with a 640dcxr dichroic mirror and emission filters FF01-525/45 and HQ 700/75m (Chroma). All data was recorded on a back-illuminated EM-CCD camera (Andor iXon DU897).
For SMLM images of fixed cells, 7,500 – 10,000 frames were acquired at 100 – 167 Hz, with illumination times of 2 - 3 ms. In live cell PALM experiments, 4,000 frames were recorded at 167 Hz (24 s total recording time). Imaging at 488 nm or 640 nm was done with 1.5 - 3 kW/cm2 and photoactivation was achieved with continuous 20 - 30 W/cm2 405 nm light. Importantly, the imaging parameters and sequence length were not changed within one set of experiments. dSTORM blinking buffer consisted of PBS (pH 7.4), 10% glucose, 500 μg/ml glucose oxidase, 40 μg/ml catalase and 50 mM cysteamine 27. PALM imaging was performed in imaging buffer.
Since the reliable detection of clusters requires immobilization of clusters during the acquisition time, we determined the residual mobility of antibody-labeled TCR upon fixation. We found only marginal fluctuations below the achieved localization errors.
We performed single-molecule tracking experiments on chemically fixed cells labeled with low concentrations of H57-scFv-AS635P. Image stacks were acquired at an illumination time of till = 10 ms and tdelay = 490 ms. Signal positions were determined as for SMLM. Tracking was then performed on the data using an in-house adaptation of the algorithm described in 44. Mean square displacement analysis was performed as described in 45 and fitted with the equation MSD = 4Dtlag + offset, where D specifies the lateral diffusion coefficient, tlag the analyzed time-lag, and offset the localization errors.
Quantitative analysis of single label blinking
To statistically quantify the blinking of single labels (H57-AF647, KT3-AF647 and CD3ζ-PS-CFP2), we analyzed cells recorded at low antibody concentrations or low expression levels of CD3ζ-PS-CFP2. All localizations appearing within a radius of 1 pixel were considered to be derived from one label molecule. We determined the first frame of appearance, the total number of detections per label (N), the time a label is detectable in consecutive frames (ton) and the time a label is not detectable (toff) (Supplementary Fig. 3a).
STED microscopy
STED measurements were done in ROXS buffer consisting of 2 mM Trolox, 1 mM methylviologen-dichloride hydrate, 50 μM glucose oxidase, 300 U/ml catalase and 5 %wt glucose 46. STED images were recorded on a custom-built microscope system, equipped with a 635 nm pulsed diode laser (LHD-D-C-635, PicoQuant) with < 100 ps pulse width. A Ti:Al2O3 laser (Mira900, Coherent) was tuned to 800 nm for stimulated emission depletion. The excitation and STED beam were fed into the objective (HC PL APO 100x/1.4 oil CS2, Leica) and emission was collected by the same objective. A dichroic mirror (zt 625-745 rpc, Chroma) was used to uncouple the emission light, which was then split into four identical channels by 50:50 beam splitters, filtered by band pass filters (685/70 ET, Chroma), coupled into multimode fibers and detected by avalanche photodiodes (SPCMAQR-13-FC, Perkin Elmer Optoelectronics). An additional short pass filter (ET750sp-sp, Chroma) was used to block the STED light. A 3-axis piezo stage (Tritor 102 Cap, Piezosystem Jena) was used to raster scan the T cell membrane. Signals were acquired with a time-correlated single-photon counting board in absolute timing mode (DPC-230, Becker & Hickl GmbH). Time gating was set to 0.8 to 7 ns in respect to the excitation pulse. Imaging for single-molecules as well as for fully labeled T cells was done with 50 fJ excitation and 1.6 nJ STED pulse energy at the back aperture of the objective and a pixel dwell time of 100 μs at a pixel size of 20 nm yielding a scanning time of 60-160 ms per line. To ensure that the chosen scanning speed was sufficiently fast to avoid diffusional spreading of the signals, we determined by single-molecule tracking the residual mobility of scFv-labeled TCR in fixed cells, yielding D = 1.6 x 10-5 ± 4 x 10-7 μm2/s. Together with the σ-width of about 40 nm for single-molecule signals (Supplementary Fig. 7), we estimate that the TCR diffuses 4-6 nm during the recording of a single-molecule signal, which is much smaller than the obtained resolution.
Calcium imaging and analysis
Roughly 106 cells were washed in imaging buffer and incubated with 5 μg/ml Fura-2-AM (Molecular Probes) for 20 min at room temperature. After washing, antibody or scFv labeling was done as for superresolution experiments. Fura-2-AM was excited using a monochromatic light source (Polychrome V, TILL Photonics), coupled to a Zeiss Axiovert 200M equipped with a 20x objective (Olympus) and an Andor iXon Ultra. Imaging was performed at 340 nm and 380 nm at illumination times of 50 and 10 ms, respectively. The total recording time was at least 10 minutes at 1 Hz.
ImageJ was used to generate the ratio images. Cells were segmented and tracked using a sum image of both channels using an in-house Matlab algorithm based on Gao Y. et al 44. Cellular positions and tracks were stored and used for intensity extraction based on the ratio image. Intensity traces were normalized to the starting value at time point zero; the data are displayed as medians ± standard error of the median.
Label-density-variation analysis
Single-molecule signals were fitted with a Gaussian intensity distribution by maximum likelihood estimation using the ImageJ plug-in ThunderSTORM 47 and filtered for intensity, σ and positional accuracy of the fit. dSTORM data were merged with a radius of 35 nm and a maximum off-time of 50 frames 18. PS-CFP2 data were merged with a maximum distance of 80 nm and a maximum off time of 1 frame. ρ/ρ0 versus η plots were obtained as described in detail in Baumgart F. et al 18. Briefly, we determined ρ0 by fitting the data with a polynomial of the form ρ=ρ0(1 + α × ηβ) with α=1.4 and β=4. In addition, to improve sensitivity random reference curves were generated for each probe corresponding to its specific blinking statistics: in each case, 50 titration curves were simulated with labeling efficiencies varied from 5% to 95% in 0.5% increments. For analysis of the simulations, the data points were pooled according to the simulated labeling efficiency into 20 equidistant bins (ranging from 0%-95%). From the mean values of these bins a reference line for randomly distributed data was generated. Confidence intervals (represented as SEM) of this simulated random curve were calculated reflecting the experimental situations shown in Fig. 2, Fig. 3, Supplementary Fig. 1, and Supplementary Fig. 4. To approximate the mean number of data points (n) typically found per bin in experimental data, we divided the total number of experimental data points by the number of bins (20). Calculated values for n were: n=3.7 for H57-AF647; n=2.9 for KT3-AF647 and n=1.45 for PS-CFP2.
Simulations for label-density-variation analysis
To test the sensitivity of the label-density-variation method, we simulated different clustering scenarios and compared them with simulated random distributions using in house-written Matlab code. Molecular densities were adjusted to values extracted from experimental data by dividing the number of localizations in fully labeled T cells by the mean number of localizations per label in sparsely labeled T cells, or, in case of PS-CFP2, T cells expressing low levels of CD3ζ-PS-CFP2. We determined 59-81, 68-73 and 37-141 molecules/μm2 for H57-AF647, KT3-AF647 and CD3ζ-PS-CFP2, respectively. Similar densities of H57-AF647 and KT3-AF647 labeling were a priori not expected, given the presence of two CD3ε subunits per TCRβ chain. We attribute this to incomplete labeling or steric hindrance of antibody binding in the case of KT3-AF647 (Brameshuber et al., Nature Immunology, accepted), and due to experimental differences in the staining and fixation procedures (see subsection “TCR labeling and sample preparation”).
Simulations were done in four steps. All parameters, if not otherwise stated, were randomized following a Poisson distribution with the indicated mean values.
First, we simulated the underlying protein distributions for regions of 10 x 10 μm, reflecting approximately the size of a typical cell. Clusters of proteins were placed randomly onto these regions, with adjustable number of clusters per μm2 (mean = 0, 3, 5, 10, 15 and 20) and σ-width (mean = 20, 40, 60, 80, 100 and 150 nm). We generated a probability mask for the whole region, where each cluster is represented by a Gaussian profile symmetrically truncated at 1 σ. Hence, the σ-width can be interpreted as the cluster radius. To avoid high protein densities in overlapping clusters, the probability map was thresholded at 0.9. Clusters were randomly filled with molecules (mean = 73, 70 and 76 molecules/μm2 for H57-AF647, KT3-AF647 and PS-CFP2, respectively), until the adjusted fraction of clustered molecules was reached (40, 60, 80, 100 %). The remaining molecules were distributed randomly in the areas outside of the clusters. A non-clustered scenario is naturally represented by the case of 0 clusters per μm2.
Second, labels were assigned randomly to the molecules; increasing the labeling probability from 5% to 95% allowed for the simulation of titration curves. For each titration step we simulated a new underlying spatial distribution of molecules.
Third, to simulate blinking, we assigned a number of detections to each label. This number was drawn from an empirical probability distribution recorded at low labeling concentrations in dSTORM experiments (H57-AF647 and KT3-AF647) or low expression levels of CD3ζ-PS-CFP2. Localization errors were simulated by spreading these detections using a Gaussian profile centered around the molecule position, with widths corresponding to the localization errors of the according experimental data calculated after single-molecule fitting 48, 49.
Fourth, to account for experimental errors, we included unspecifically bound labels at a mean density of 5 labels/μm2. We finally considered also false positive localizations by adding a random background of single localizations at a mean density of 10 localizations/μm2. Both values are realistic for the experimental settings.
10 or 50 titration curves were simulated for clustering random or random scenarios, respectively. Curves were analyzed as described in subsection “Label-density-variation analysis”.
For the analysis of KT3-AF647 data shown in Fig. 4b and Supplementary Fig. 6a we assumed a label stoichiometry of one antibody molecule per TCR complex. This assumption was corroborated by the experimental finding, that staining of CD3ε with full KT3 antibody after fixation yielded similar label densities as labeling of the monomeric β-chain with H57. Nevertheless, we also simulated scenarios assuming two KT3 antibody molecules per TCR complex, yielding essentially identical detection limits (data not shown). This was no surprise, as two antibodies per TCR complex would be mathematically equivalent to twice the amount of fluorophores per antibody; in our previous paper, we found no difference in the results, when performing the label-density-variation analysis of differently labeled antibodies 18.
Ripley’s K function analysis
Image analysis via Ripley’s K function was performed via in house-written Matlab code. As is common practice, we linearized the K-function and plotted 25.
STED autocorrelation analysis
To evaluate the degree of randomness in STED microscopy data, we calculated image autocorrelation functions for 5 different 2 x 2 μm regions of interest (ROIs) and averaged over all angles 20. ROIs from experimental samples were compared to simulated random distributions with experimentally determined parameters: STED images of T cells labeled at single-molecule density were used to characterize the point spread function (psf) of the imaging system by fitting single-molecule signals using the ImageJ plug-in ThunderSTORM 47. The background was determined from the mean fluorescence signal in non-cell regions next to the cells. TCR densities were estimated by dividing the background-corrected mean intensity in fully labeled cells by the average single-molecule intensity, yielding 40 - 150 molecules/μm2 in the case of H57-scFv-AS635P and 75 – 120 molecules/μm2 in the case of KT3-scFv-AS635P. Simulated images of random signal distributions were generated using the parameters derived from single-molecule signals, i.e. intensity (I), width (σ), background, and density. A log-normal distribution 50 was fitted to the intensity histogram (Supplementary Fig 7a). The dependence of σ on the intensity was fitted by the empirical function
| (Eq. 1) |
(Supplementary Fig. 7b). This dependence likely arose from bleaching of single molecules during the scanning process. To simulate single molecules, I was drawn from a log-normal distribution. For each molecule, σ(I) was calculated using Eq. 1, which was taken as the mean value of a normal distribution with constant standard deviation 6.24. A total of fluorescent counts (I) were then assigned for each molecule to 20 nm pixels using a Gaussian distribution with a radius σ. Finally, we included varying sources of noise in the simulations: (i) Line-scanning errors during the STED image acquisition were simulated by shifting each line horizontally for a randomized value of either -1, 0 or 1 pixels. These values were determined empirically from the STED images. (ii) A Poisson-distributed background was simulated with a mean intensity of 0.86, corresponding to the mean value determined in the experiments.
Simulations for STED autocorrelation analysis
To evaluate the sensitivity of the STED autocorrelation analysis, we compared simulated STED images of clustered and randomly distributed molecules using in house-written Matlab code. Molecular densities were matched to experimentally obtained values from T cells labeled with scFv-KT3-AS635P. We simulated underlying protein distributions in 2 x 2 μm ROIs as stated in the subsection “Simulations for label-density-variation analysis” and used these molecule positions to generate a STED image as described in “STED autocorrelation analysis”. For each scenario, we simulated five images and analyzed them as described in subsection “STED autocorrelation analysis”.
Supplementary Material
Acknowledgements
This work was supported by the Austrian Science Fund (FWF) projects P 26337-B21, P 25730-B21, P 30214-N36 (GJS), P27941-B28 (FB), as well as I953-B20 (MB), by the Vienna Science and Technology Fund (WWTF) LS13-030 (GJS, JBH). Funding was further provided by a Boehringer Ingelheim PhD Fellowship (RP) and a DOC Fellowship (24793) of the Austrian Academy of Sciences (AMA). We are grateful to Stefan Hell for providing infrastructure for the STED experiments.
Footnotes
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary.
Code Availability
All custom code used in the current study is available from the corresponding authors upon request.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding authors upon request.
Author Contributions
BR and FB performed and analyzed SMLM experiments. BR, MB and HT performed and analyzed STED experiments. MB contributed important ideas and performed initial PALM experiments. BR and AMA performed simulations and analysis algorithms. RP and FK produced key reagents. JBH provided key reagents, murine T cells and infrastructure as well as important ideas. BR, AMA, FB and GJS analyzed data. FB and GJS conceived the project. FB, BR and GJS wrote the manuscript.
Competing Interests
The authors declare no competing financial interests.
References
- 1.Huppa JB, Davis MM. T-cell-antigen recognition and the immunological synapse. Nat Rev Immunol. 2003;3:973–983. doi: 10.1038/nri1245. [DOI] [PubMed] [Google Scholar]
- 2.Chung W, Abel SM, Chakraborty AK. Protein Clusters on the T Cell Surface May Suppress Spurious Early Signaling Events. PLOS ONE. 2012;7:e44444. doi: 10.1371/journal.pone.0044444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein B, Wiegel FW. The effect of receptor clustering on diffusion-limited forward rate constants. Biophysical Journal. 1983;43:121–125. doi: 10.1016/S0006-3495(83)84330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lillemeier BF, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pageon SV, et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proceedings of the National Academy of Sciences. 2016;113:E5454–5463. doi: 10.1073/pnas.1607436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung Y, et al. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proceedings of the National Academy of Sciences. 2016;113:E5916–E5924. doi: 10.1073/pnas.1605399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu YS, Cang H, Lillemeier BF. Superresolution imaging reveals nanometer- and micrometer-scale spatial distributions of T-cell receptors in lymph nodes. Proceedings of the National Academy of Sciences. 2016 doi: 10.1073/pnas.1512331113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossy J, Owen DM, Williamson DJ, Yang Z, Gaus K. Conformational states of the kinase Lck regulate clustering in early T cell signaling. Nat Immunol. 2013;14:82–89. doi: 10.1038/ni.2488. [DOI] [PubMed] [Google Scholar]
- 9.Williamson DJ, et al. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat Immunol. 2011;12:655–662. doi: 10.1038/ni.2049. [DOI] [PubMed] [Google Scholar]
- 10.Sherman E, et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 2011;35:705–720. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schamel WW, Alarcon B. Organization of the resting TCR in nanoscale oligomers. Immunol Rev. 2013;251:13–20. doi: 10.1111/imr.12019. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Parajo MF, Cambi A, Torreno-Pina JA, Thompson N, Jacobson K. Nanoclustering as a dominant feature of plasma membrane organization. J Cell Sci. 2014;127:4995–5005. doi: 10.1242/jcs.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci U S A. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar R, et al. Increased Sensitivity of Antigen-Experienced T Cells through the Enrichment of Oligomeric T Cell Receptor Complexes. Immunity. 2011;35:375–387. doi: 10.1016/j.immuni.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Schamel WW, et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202:493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahl SJ, Hell SW, Jakobs S. Fluorescence nanoscopy in cell biology. Nature Reviews Molecular Cell Biology. 2017;18:685. doi: 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- 17.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Identification of clustering artifacts in photoactivated localization microscopy. Nat Methods. 2011;8:527–528. doi: 10.1038/nmeth.1627. [DOI] [PubMed] [Google Scholar]
- 18.Baumgart F, et al. Varying label density allows artifact-free analysis of membrane-protein nanoclusters. Nat Meth. 2016;13:661–664. doi: 10.1038/nmeth.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spahn C, Herrmannsdorfer F, Kuner T, Heilemann M. Temporal accumulation analysis provides simplified artifact-free analysis of membrane-protein nanoclusters. Nat Meth. 2016;13:963–964. doi: 10.1038/nmeth.4065. [DOI] [PubMed] [Google Scholar]
- 20.Sengupta P, Jovanovic-Talisman T, Lippincott-Schwartz J. Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat Protoc. 2013;8:345–354. doi: 10.1038/nprot.2013.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balagopalan L, Sherman E, Barr VA, Samelson LE. Imaging techniques for assaying lymphocyte activation in action. Nat Rev Immunol. 2011;11:21–33. doi: 10.1038/nri2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang VT, et al. Initiation of T cell signaling by CD45 segregation at 'close contacts'. Nat Immunol. 2016;17:574–582. doi: 10.1038/ni.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos AM, et al. Capturing resting T cells: the perils of PLL. Nature Immunology. 2018 doi: 10.1038/s41590-018-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiskowski MA, Hancock JF, Kenworthy AK. On the use of Ripley's K-function and its derivatives to analyze domain size. Biophys J. 2009;97:1095–1103. doi: 10.1016/j.bpj.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rust M, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 28.Betzig E, et al. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 29.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Annibale P, Scarselli M, Kodiyan A, Radenovic A. Photoactivatable Fluorescent Protein mEos2 Displays Repeated Photoactivation after a Long-Lived Dark State in the Red Photoconverted Form. The Journal of Physical Chemistry Letters. 2010;1:1506–1510. [Google Scholar]
- 31.Brameshuber M, et al. Monomeric TCRs drive T cell antigen recognition. Nature Immunology. 2018;19:487–496. doi: 10.1038/s41590-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dushek O, et al. Effects of Intracellular Calcium and Actin Cytoskeleton on TCR Mobility Measured by Fluorescence Recovery. PLOS ONE. 2008;3:e3913. doi: 10.1371/journal.pone.0003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson C, George SC. Theory and practical recommendations for autocorrelation-based image correlation spectroscopy. BIOMEDO. 2012;17:080801. doi: 10.1117/1.JBO.17.8.080801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengupta P, et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods. 2011;8:969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai E, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science. 2017;356 doi: 10.1126/science.aal3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klotzsch E, Schütz Gerhard J. Improved Ligand Discrimination by Force-Induced Unbinding of the T Cell Receptor from Peptide-MHC. Biophys J. 2013;104:1670–1675. doi: 10.1016/j.bpj.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu B, Chen W, Evavold Brian D, Zhu C. Accumulation of Dynamic Catch Bonds between TCR and Agonist Peptide-MHC Triggers T Cell Signaling. Cell. 2014;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maimon O, Rokach L. Data mining and knowledge discovery handbook. Springer; New York: 2005. [Google Scholar]
- 39.Huang J, et al. A Single Peptide-Major Histocompatibility Complex Ligand Triggers Digital Cytokine Secretion in CD4+ T Cells. Immunity. 2013;39:1–12. doi: 10.1016/j.immuni.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, Groves JT. Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. Elife. 2013;2:e00778. doi: 10.7554/eLife.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katz ZB, Novotna L, Blount A, Lillemeier BF. A cycle of Zap70 kinase activation and release from the TCR amplifies and disperses antigenic stimuli. Nat Immunol. 2017;18:86–95. doi: 10.1038/ni.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat Immunol. 2003;4:749–755. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- 43.Tsumoto K, et al. Highly efficient recovery of functional single-chain Fv fragments from inclusion bodies overexpressed in Escherichia coli by controlled introduction of oxidizing reagent—application to a human single-chain Fv fragment. Journal of Immunological Methods. 1998;219:119–129. doi: 10.1016/s0022-1759(98)00127-6. [DOI] [PubMed] [Google Scholar]
- 44.Gao Y, Kilfoil ML. Accurate detection and complete tracking of large populations of features in three dimensions. Opt Express. 2009;17:4685–4704. doi: 10.1364/oe.17.004685. [DOI] [PubMed] [Google Scholar]
- 45.Wieser S, Schütz GJ. Tracking single molecules in the live cell plasma membrane-Do's and Don't's. Methods. 2008;46:131–140. doi: 10.1016/j.ymeth.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Vogelsang J, et al. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew Chem Int Ed Engl. 2008;47:5465–5469. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- 47.Ovesný M, Křížek P, Borkovec J, Švindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plugin for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quan T, Zeng S, Huang Z. SPIE; 2010. Localization capability and limitation of electron-multiplying charge-coupled, scientific complementary metal-oxide semiconductor, and charge-coupled devices for superresolution imaging; p. 6. 2010. [DOI] [PubMed] [Google Scholar]
- 50.Mutch SA, et al. Deconvolving Single-Molecule Intensity Distributions for Quantitative Microscopy Measurements. Biophys J. 2007;92:2926–2943. doi: 10.1529/biophysj.106.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.