Abstract
Mutations in the E3 ubiquitin ligase Parkin (PARK2) and the protein kinase PINK1 (PARK6) are linked to autosomal-recessive juvenile Parkinsonism (AR-JP)1,2, and at the cellular level cause defects in mitophagy, the cellular process that organises destruction of damaged mitochondria3,4. Parkin is autoinhibited, and requires activation by PINK1, which phosphorylates Ser65 in ubiquitin and in the Parkin ubiquitin-like (Ubl) domain. Parkin binds phospho-ubiquitin, which enables efficient Parkin phosphorylation, however the enzyme remains autoinhibited with an inaccessible active site5,6. It is unclear how phosphorylation of Parkin activates the molecule.
We here follow the activation of full-length human Parkin by hydrogen deuterium exchange mass spectrometry, and reveal large scale domain rearrangement in the activation process, in which the phospho-Ubl rebinds to the Parkin core, and releases the catalytic RING2 domain. A 1.8 Å crystal structure of phosphorylated human Parkin reveals the binding site of the phosphorylated Ubl on the Unique Parkin Domain (UPD), involving a phosphate-binding pocket lined by AR-JP mutations. Strikingly, a conserved linker region between Ubl and UPD acts as an activating element (ACT) that contributes to RING2 release by mimicking RING2 interactions on the UPD, explaining further AR-JP mutations. Our data unveils how autoinhibition in Parkin is resolved, and suggests how Parkin ubiquitinates its substrates via an untethered RING2 domain. This opens exciting new avenues to design Parkin activators for clinical use.
Work in the last decade has shown how PINK1 and Parkin initiate mitophagy, and many steps in this process are mechanistically well understood3,4. It has further been suggested that targeted activation of either PINK1 or Parkin could increase mitochondrial turnover, and impede the progression of Parkinson’s disease. A detailed understanding of the underlying molecular mechanisms is hence essential.
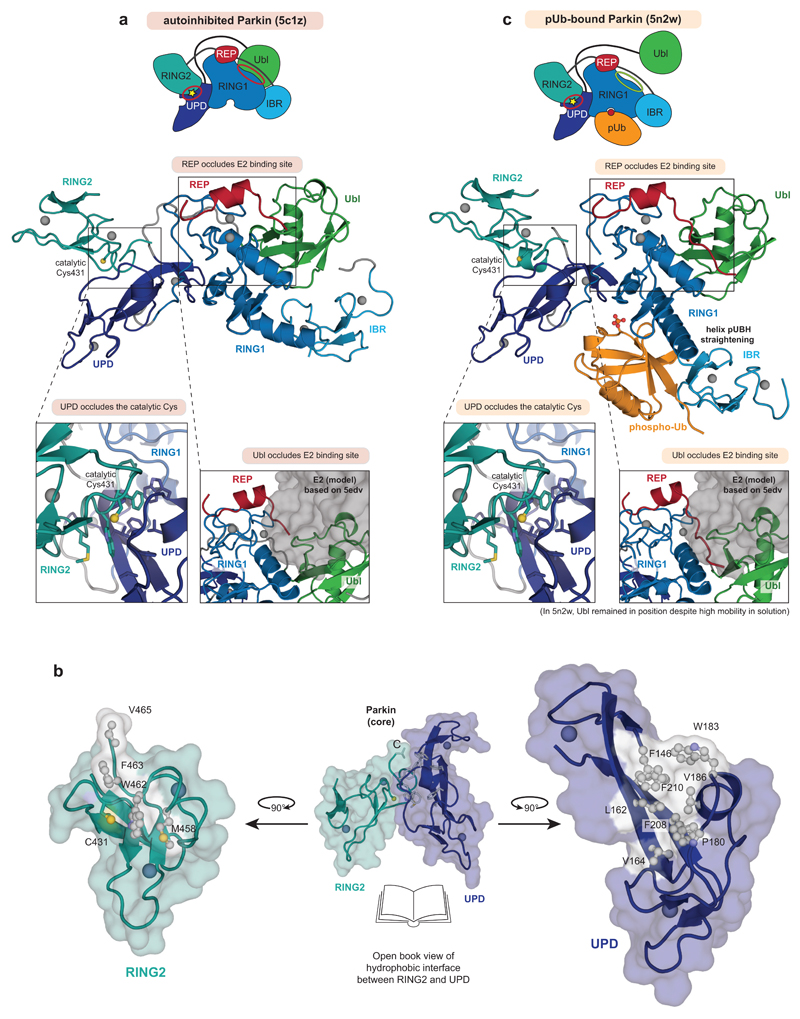
Parkin requires an elaborate activation mechanism. The first crystal structures of Parkin7–9 revealed several distinct mechanisms of autoinhibition (Fig. 1a, Extended Data Fig. 1a). Most strikingly, the active site Cys on the catalytic RING2 domain, which receives ubiquitin from the E2 enzyme, is obstructed by forming an interface with the UPD (also known as RING0)(Extended Data Fig. 1a). The RING2-UPD interface is highly hydrophobic7–9 (Extended Data Fig. 1b), and it is not clear how this intramolecular interaction can be opened.
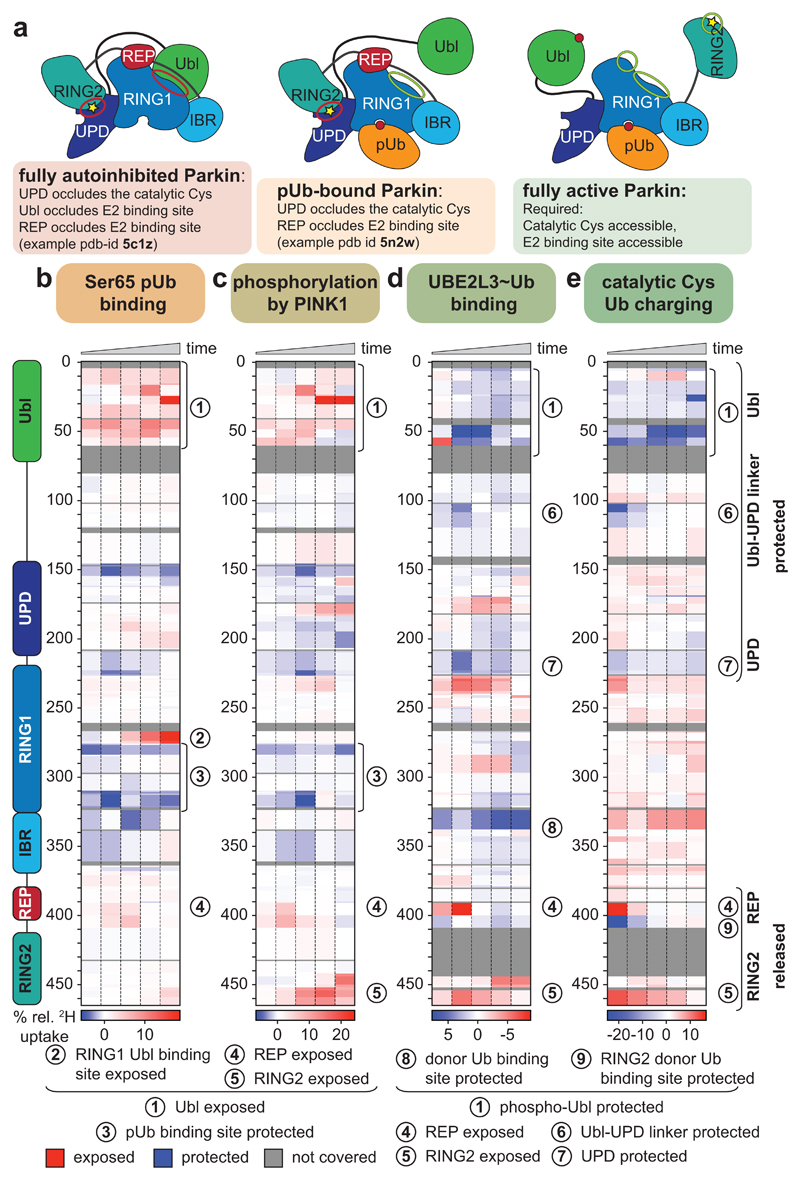
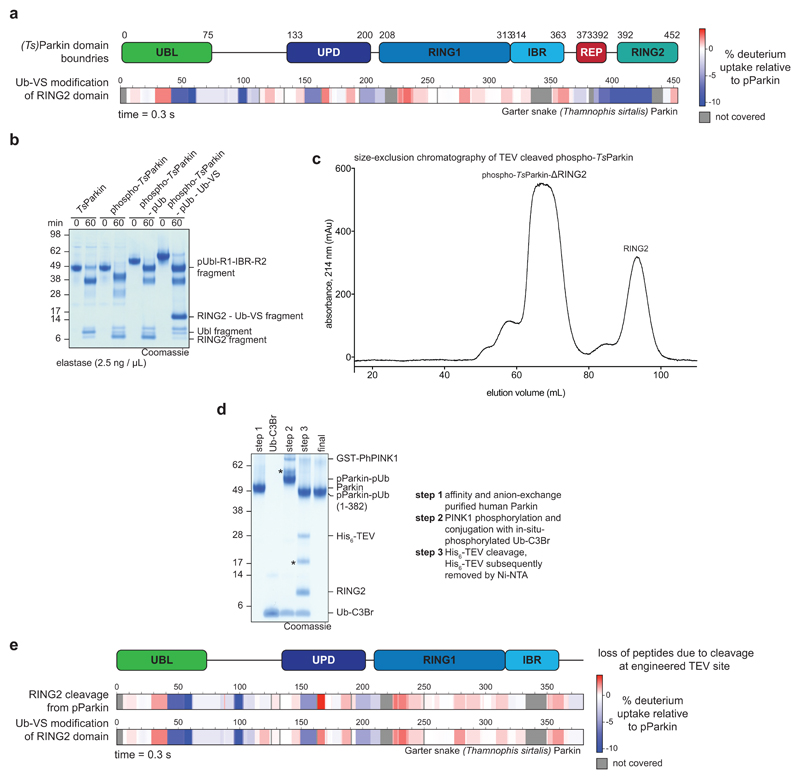
Figure 1. Domain rearrangements in Parkin, resolved by HDX-MS.
a, Cartoon of Parkin activation. Left, Parkin is autoinhibited by several mechanisms (red circles)7–9. Middle, phospho-ubiquitin binding to Parkin releases the Ubl domain, but most mechanisms of autoinhibition remain5,6. Right, after Ubl phosphorylation, Parkin is fully active (green circles), but a structure of active Parkin has not been reported. Also see Extended Data Fig. 1. b-e, HDX-MS difference map with the shortest peptides covering any given region, coloured from blue (more protected from exchange compared to previous state) to red (more accessible to solvent exchange), peptides for grey coloured regions could not be analysed (see Extended Data Fig. 3d). The five rows per sample indicate different time lengths for HD exchange (0.3 s, 3 s, 30 s, 300 s and 3000 s). All experiments were performed with human full-length Parkin, as technical triplicates. See Extended Data Fig. 2 and 3 for raw data and structural mapping, respectively. b, Difference between Parkin and Parkin bound to phospho-ubiquitin. c, Difference between Parkin:phospho-ubiquitin and phospho-Parkin:phospho-ubiquitin. d, Difference between phospho-parkin:phospho-ubiquitin, and phospho-Parkin:phospho-ubiquitin in complex with a non-dischargeable UBE2L3-Ub complex (see Online Methods). e, Difference between phospho-Parkin:phospho-ubiquitin and phospho-Parkin:phospho-ubiquitin charged with Ub-VS (see Online Methods).
Activation of Parkin is mediated by the mitochondrial outer membrane (MOM) Ser/Thr protein kinase PINK1, which phosphorylates Ser65 in ubiquitin (generating phospho-ubiquitin)10–14 and in the Parkin Ubl domain15–17. A current model for PINK1-mediated Parkin activation suggests that PINK1 phosphorylates ubiquitin attached to MOM proteins, and nanomolar affinity recruits autoinhibited, cytosolic Parkin to sites of PINK1 activity3,5,13,18–21. Phospho-ubiquitin binding induces conformational changes in Parkin that lead to the release of the Ubl domain from the Parkin core, and enable PINK1 to phosphorylate the Parkin Ubl domain3,5,7,13,18–23 (Fig. 1a, Extended Data Fig. 1c). Importantly, in structures of Parkin bound to phospho-ubiquitin5,22, Parkin is still autoinhibited: the E2 binding site remains blocked by the Repressor (REP) element, and RING2 and its catalytic Cys remain obstructed by the UPD (Fig. 1a, Extended Data Fig. 1c).
Indeed, Ubl phosphorylation is crucial to fully activate Parkin. A Parkin S65A mutant is not retained at mitochondria, is unable to trigger mitochondrial ubiquitination and mitophagy, and thus is physiologically inactive13,15,17,21,24. Biochemically, Parkin phosphorylation enhances activity more than phospho-ubiquitin binding13,21,25, and Parkin phosphorylation, but not phospho-ubiquitin binding, enables access of ubiquitin activity-based probes (Ub-ABPs) to the active site Cys5,21,26. How Ubl phosphorylation is able to activate Parkin, and in particular, how it is able to disrupt the RING2-UPD interface, has remained a mystery, and has led to various models of Parkin activation3,5,8,9,22,27.
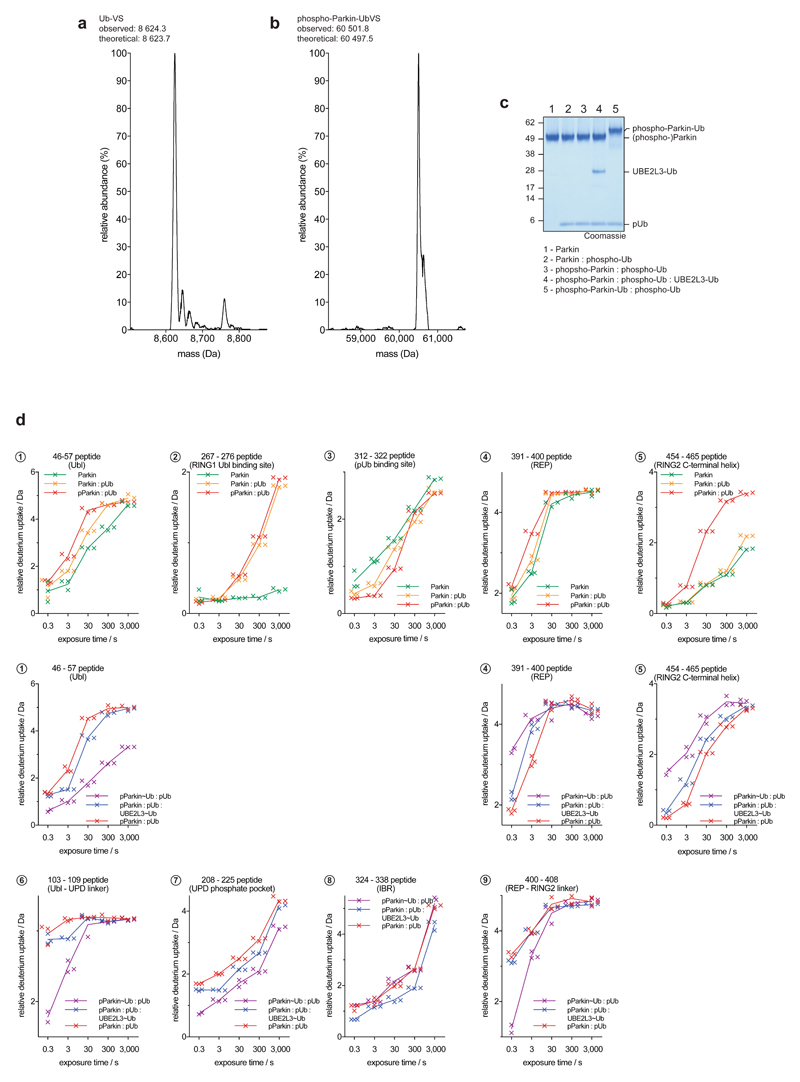
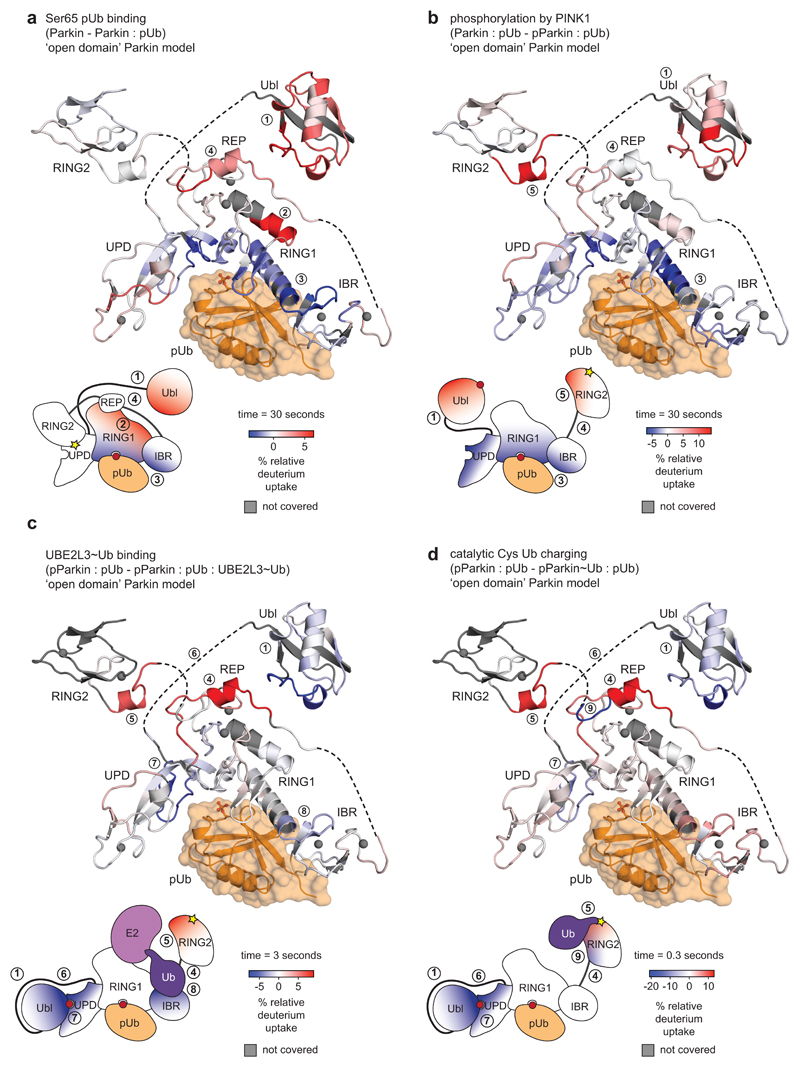
We reconstituted activation of full-length human Parkin by PINK1, and followed domain rearrangements by hydrogen deuterium exchange mass spectrometry (HDX-MS) 28(Fig. 1b-e, Extended Data Fig. 2, 3). HDX-MS reports on the relative rate of exchange of backbone amide hydrogens with deuterium based on the strength of hydrogen bonding and their solvent accessibility in the folded protein, and distinguishes peptides in a protein’s core (no/little exchange with solvent over time), with those at an exposed surface (high or increasing exchange with solvent over time). The power of the method lies in its ability to compare identical peptides between different states along an activation cascade, revealing peptides that become exposed and thus interfaces that are opened (red in Fig. 1b-e), and regions in the protein that become protected and form new interfaces (blue in Fig. 1b-e). For Parkin, this allowed us to confirm previously reported conformational changes upon phospho-ubiquitin binding5,22, whereby the Parkin Ubl is released and becomes exposed to solvent (1, 2 – numbers referring to Fig. 1b-e), the phospho-ubiquitin binding site becomes protected (3), while RING2, REP (4) and UPD are essentially unperturbed (Fig. 1b).
Phosphorylation of Parkin starts to show the first signs of REP and RING2 release (4, 5), especially at later time points, yet the phosphorylated Ubl also remains flexible and in exchange with solvent (1) (Fig. 1c, Extended Data Fig. 3b). The behaviour of the phospho-Ubl changes dramatically when a covalent, non-dischargeable E2~ubiquitin conjugate is added to the sample – now, the RING2 peptide at the UPD interface is solvent exposed (5), and the phosphorylated Ubl becomes protected (1), indicating formation of a new interface (Fig. 1d, Extended Data Fig. 2, 3c). Finally, charging of the RING2 catalytic Cys by ubiquitin was assessed using phosphorylated Parkin covalently modified with the Ub-ABP ubiquitin-vinylsulphone (Ub-VS)5,13(Online Methods, Extended Data Fig. 2a-c). ‘Charged’ phospho-Parkin reiterates the conformational changes observed in the phospho-Parkin E2~Ub bound sample (Fig. 1e), and revealed that the ubiquitin-modified RING2 had been fully released from the Parkin core (5). Overall, the HDX-MS experiments indicated considerable rearrangements of Ubl and RING2, with loss of old and formation of new intramolecular interfaces on the Parkin core (Fig. 1, Extended Data Fig. 3).
Surprisingly, a section of the linker between Ubl and UPD was protected together with phospho-Ubl re-binding (6) (Fig. 1d-e, Extended Data Fig. 2, 3). This region of Parkin, spanning amino acids (aa) 75-145, has remained unstudied as it is disordered in full-length Parkin7, and was removed in subsequent structures of human and rat Parkin18,19,22.
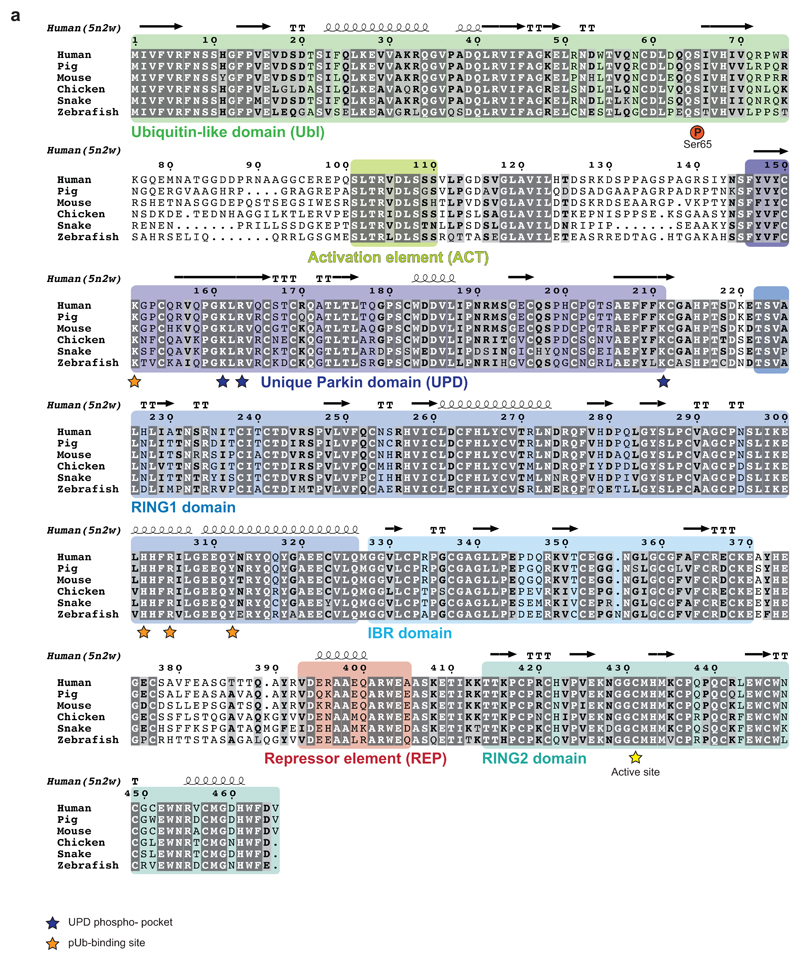
The Ubl-UPD linker contains two connected, short sections of highly conserved residues that are flanked by a variable number of unconserved residues (Extended Data Fig. 4). A minimal linker is present in Thamnophis sirtalis (Ts)Parkin (garter snake Parkin, sequence identity to human Parkin 73%, Extended Data Fig. 4), and TsParkin was used for comparative studies. HDX-MS revealed highly similar changes upon ubiquitin-charging in phosphorylated TsParkin when compared to human Parkin (Extended Data Fig. 5a, compare Fig. 1e). Moreover, limited proteolysis of full-length TsParkin revealed that autoinhibited, unphosphorylated TsParkin was cleaved first in the Ubl-UPD linker, whereas phosphorylated TsParkin was cleaved first in the IBR-RING2 linker, and no longer efficiently cleaved in the Ubl-UPD linker (Extended Data Fig. 5b). After cleavage of phospho-TsParkin, RING2 was no longer stably associated with the Parkin core (Extended Data Fig. 5c). Together these data again strongly suggested that the unstudied Ubl-UPD linker becomes ordered in activated Parkin, whereas REP and RING2 are dislodged, and RING2 becomes mobile.
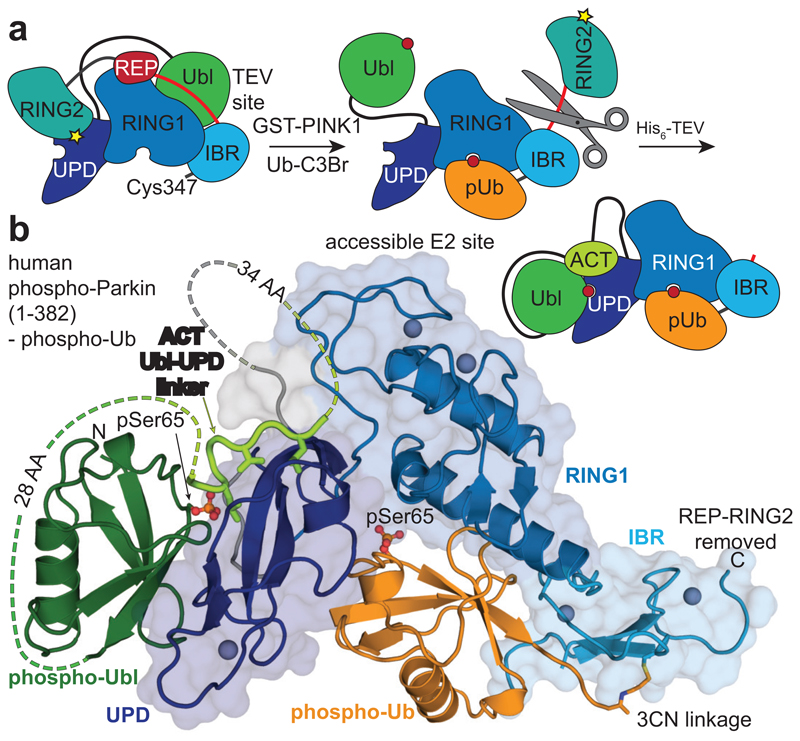
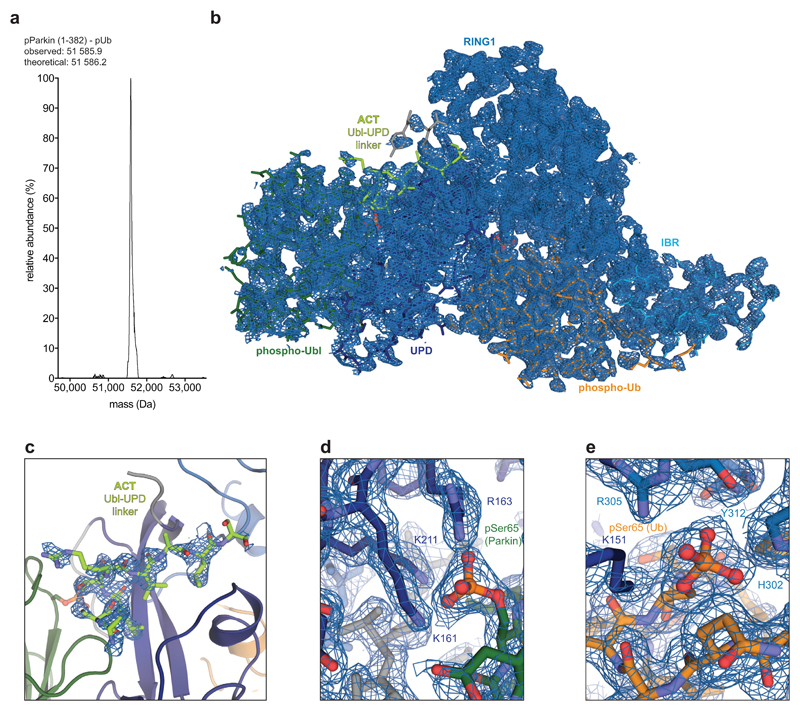
We realised that crystallographic analysis of active Parkin was likely impeded by a mobile RING2 domain, which inspired new construct design. Parkin is insoluble when expressed without the RING2 domain (data not shown), likely due to the exposed, hydrophobic UPD (Extended Data Fig. 1b). Hence, we engineered a TEV cleavage site into the IBR-RING2 linker (Fig. 2a, see Online Methods). This enabled removal of the RING2 domain upon phospho-ubiquitin binding and Ubl phosphorylation (Extended Data Fig. 5d). Importantly, UbVS-charged TsParkin and TsParkinΔRING2 displayed identical HDX-MS profiles, indicating that removal of the mobile RING2 has no effect on the remaining molecule (Extended Data Fig. 5e). For human Parkin, the resulting covalent phospho-ParkinΔRING2-phospho-ubiquitin (hereafter phospho-Parkin-pUb) complex was crystallised, and resulted in a 1.8 Å structure (Fig. 2b, Extended Data Table 1, Extended Data Fig. 6).
Figure 2. Structure of the phosphorylated Parkin core.
a, Schematic for obtaining a crystallisable phosphorylated Parkin core. Scissors indicate the introduction of a tobacco etch virus (TEV) protease cleavage site after the IBR domain (aa 382). b, Crystal structure at 1.80 Å of the human phosphorylated Parkin core lacking RING2, bound to phospho-ubiquitin. Phosphorylated residues are shown in ball-and-stick representation. A cartoon representation akin Fig. 2a is shown to the right. Also see Extended Data Fig. 6 and Extended Data Table 1.
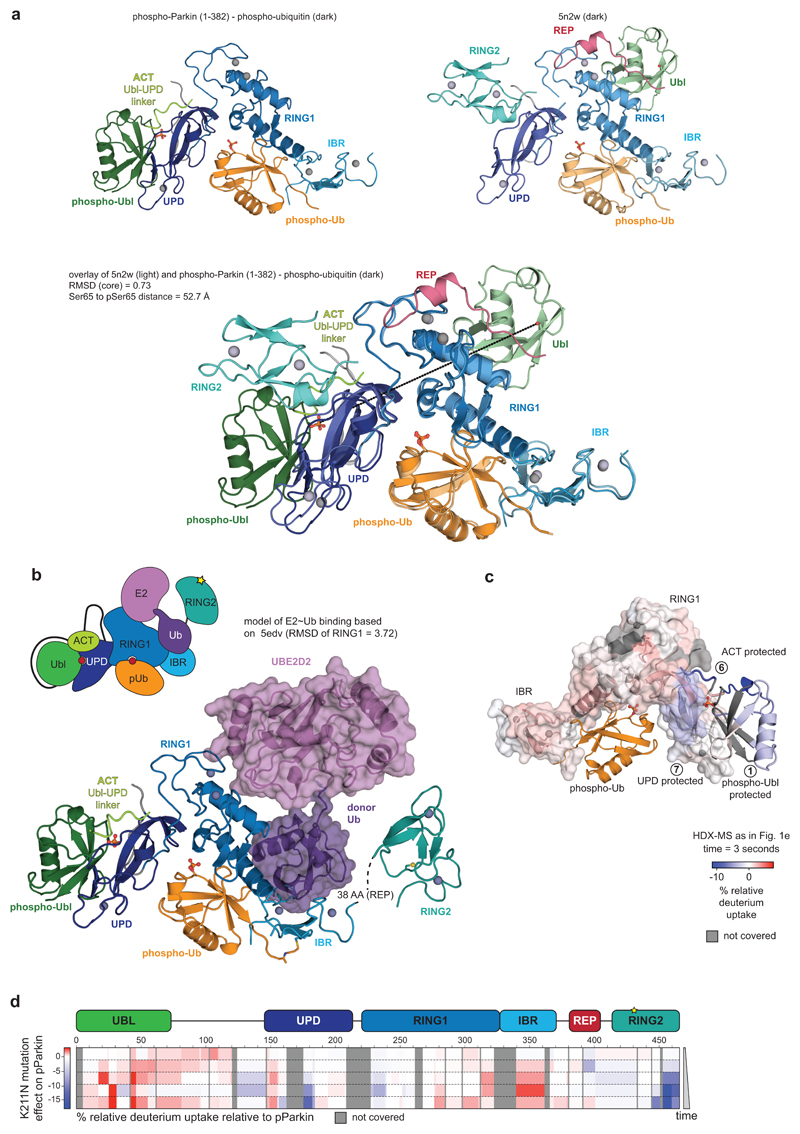
The structure of phospho-Parkin-pUb (Fig. 2a) revealed a near-identical organisation of the Parkin core (UPD-RING1-IBR) bound to phospho-ubiquitin, as compared to previous structures (RMSD 0.73 Å with human Parkin-pUb, pdb-id 5n2w, 22) (Extended Data Fig. 7a), and there no large conformational changes in individual domains. Modelling of an open E2~Ub conjugate structure (from 29) reveals sensible interfaces (Extended Data Fig. 7b) and corroborates the ubiquitin binding site observed in HDX-MS (Fig. 1d)22.
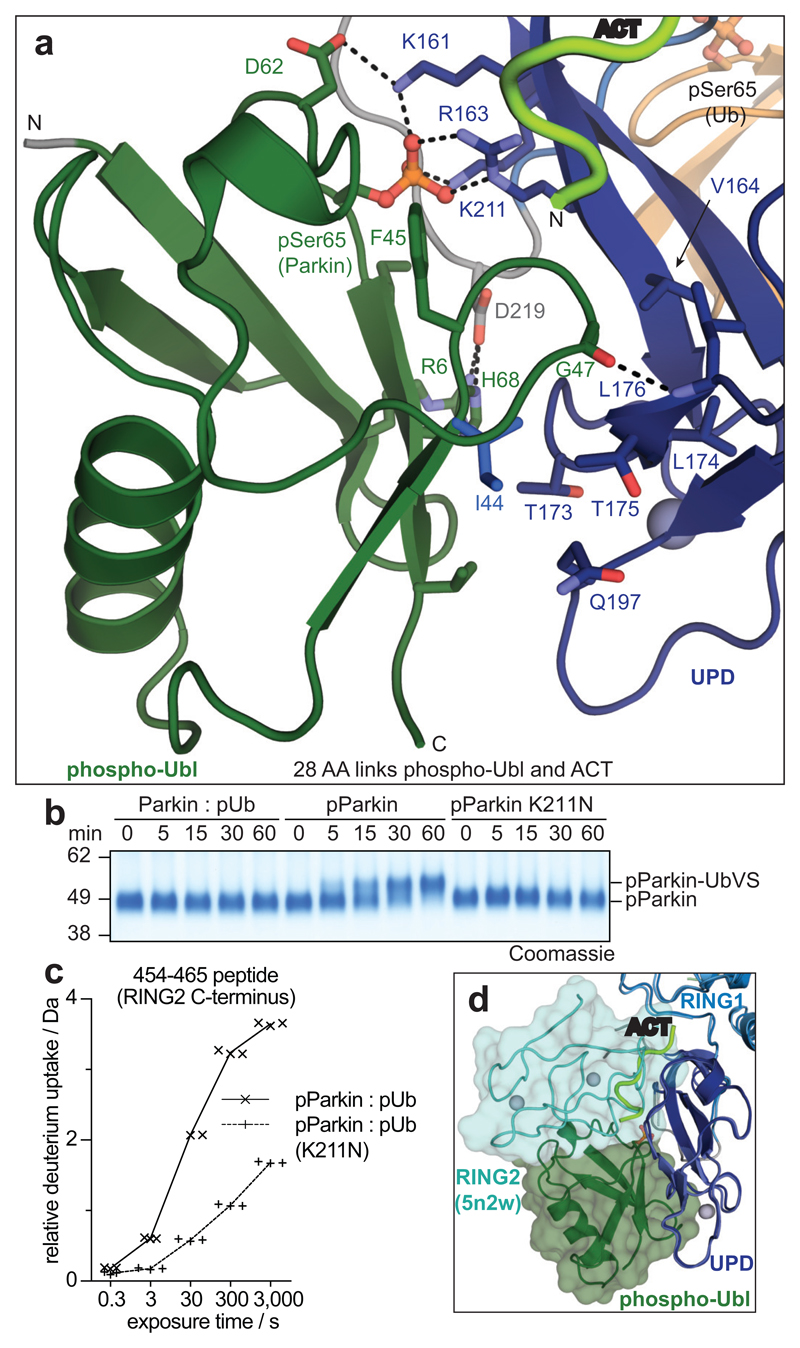
Importantly, the phosphorylated Ubl domain was bound to the UPD, and had moved by >50 Å compared to its position in autoinhibited Parkin (Fig. 2b, Fig. 3a, Extended Data Fig. 7a). The interface between phospho-Ubl and UPD is mediated via a common interaction site of ubiquitin-fold modifiers, the hydrophobic Ile44-patch of the Ubl, and engulfs the elongated UPD domain covering a surface of >800 Å2; this interface can be recapitulated in HDX-MS data (Fig. 1d, Extended Data Fig. 7c). Furthermore, the interaction places phosphorylated Ser65 into a positively charged pocket on the UPD (Fig. 3a). The phospho-acceptor pocket is lined by Lys161, Arg163 and Lys211, which contact the phosphate group forming four hydrogen bonds. We had previously noted this putative phosphate-acceptor binding site8, the importance of which is highlighted by two mutations found in AR-JP patients, K211N and K161N1,2, that also abrogate Parkin function in mitophagy13,30. Mechanistically, phosphorylated Parkin with a K211N mutation blocking the phospho-acceptor pocket, was no longer modified by Ub-VS (Fig. 3b)5. HDX-MS confirmed that phospho-Parkin-pUb K211N showed little signs of RING2 release, with strongest relative solvent protection in the C-terminus, where RING2 binds the UPD (Fig. 3c, Extended Data Fig. 7d). This indicated that the RING2 domain catalytic Cys remained inaccessible if the phospho-Ubl was unable to interact with its UPD binding site, and explained how AR-JP-causing K211N or K161N mutations produce Parkin variants unable to be activated by Ser65 phosphorylation.
Figure 3. The Parkin UPD - phospho-Ubl interaction.
a, Structural detail of the binding site between Parkin phospho-Ubl (green) and UPD (blue). Key residues are shown, and phospho-Ser65 is highlighted. Grey spheres indicate Zn atoms, and hydrogen bonds are shown as dotted lines. b, Ub-VS probe reactivity of the RING2 catalytic Cys residue with Parkin:phospho-ubiquitin, phospho-Parkin, or phospho-Parkin K211N. The experiment was done in duplicate with identical results, for gel source data, see Supplementary Fig. 1. c, HDX-MS analysis of phospho-Parkin:phospho-ubiquitin in comparison to phospho-Parkin K211N:phospho-ubiquitin. The C-terminal peptide profiles are compared, see Extended Data Fig. 7d for overall data. The RING2 C-terminus remains solvent protected in the phospho-Parkin K211N background. Technical triplicates are shown for all time points. d, Superposition of Parkin-pUb (5n2w, 6), and phospho-Parkin-pUb showing relative positions of the RING2 (cyan surface) and phospho-Ubl (green surface), respectively, on the UPD domain.
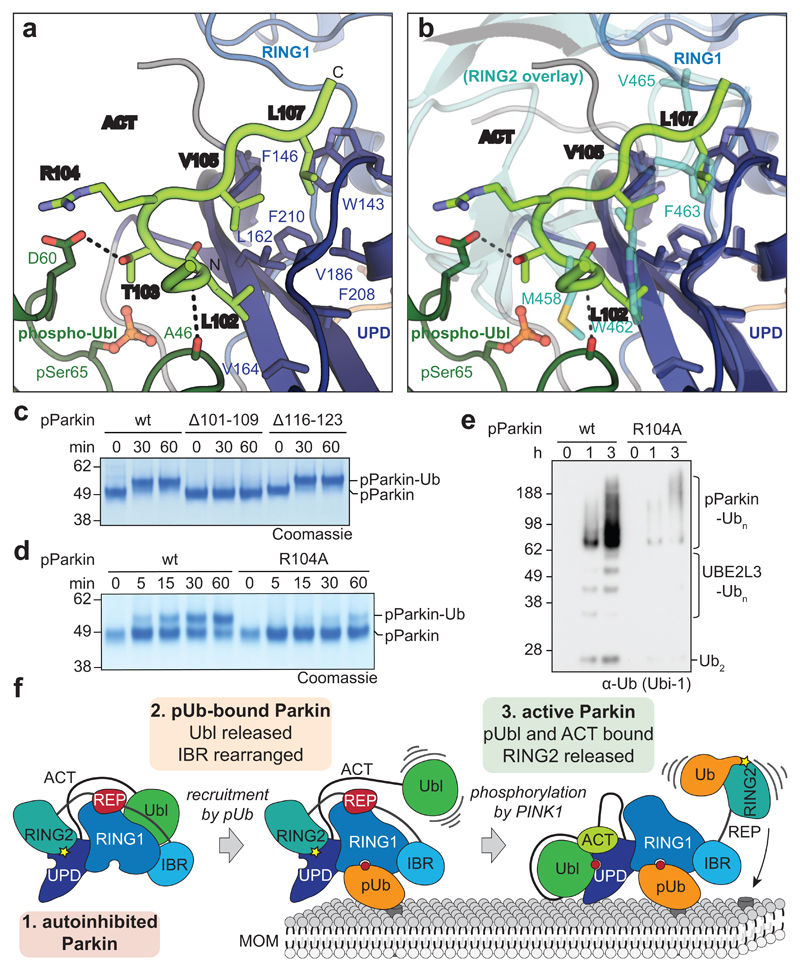
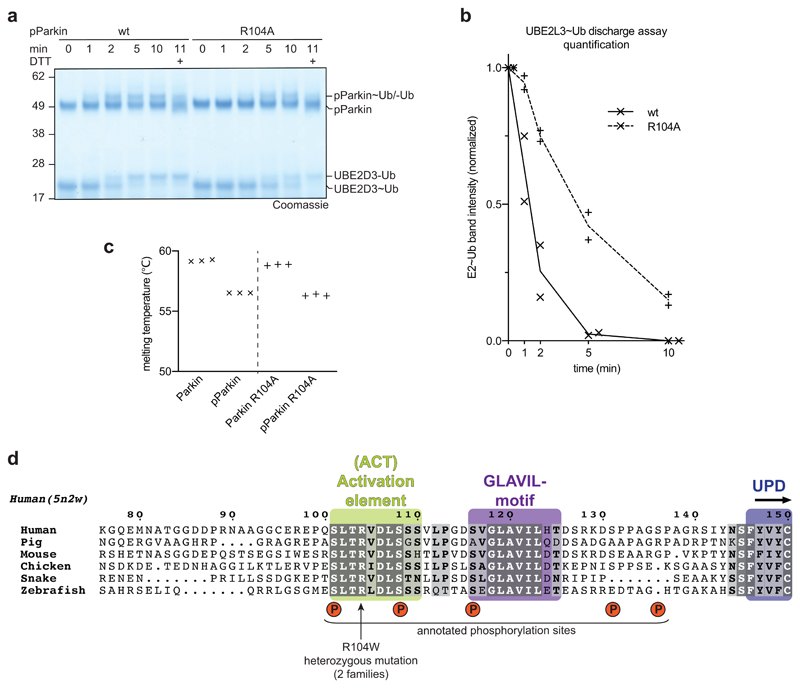
The position of the Ubl on the UPD overlaps only marginally with the position of RING2 in autoinhibited states of Parkin, and while binding of both would lead to steric clashes (Fig. 3d, Extended Data Fig. 7a), the hydrophobic RING2 binding site (Extended Data Fig. 1b) would remain unusually exposed upon opening of the RING2-UPD interface. In our structure, clear electron density for a stretch of residues was apparent at the RING2-binding site of the UPD (Extended Data Fig. 6c,d), which we could unambiguously assign to the sequence corresponding to the first conserved region of the Ubl-UPD linker (Fig. 4, Extended Data Fig. 4). In particular, residues Leu102, Val105 and Leu107 occupy pockets previously bound by RING2 residues Met458, Trp462 and Phe463 (Fig. 4a, b). This shields the hydrophobic patch on the UPD that opened by release of RING2. Indeed, similar to the K211N mutation, phospho-Parkin with deletion of the first set of conserved linker residues (Δ101-109), was unable to be charged by Ub-VS (Fig. 4c).
Figure 4. An Activating element (ACT) in Parkin.
a, Structural detail of the ordered Activating element (ACT) within the Parkin phospho-Ubl-UPD linker. Three hydrophobic ACT residues bind the hydrophobic UPD groove, and polar ACT residues contact the phospho-Ubl. b, Superposition of the ACT with RING2 (5n2w, 6, semi-transparent) in the same orientation as in a. Hydrophobic ACT residues mimic RING2 interactions. c, Ub-VS charging assay of phospho-Parkin, and phospho-Parkin variants lacking the ACT (∆101-109) or the second conserved hydrophobic linker sequence (∆116-123). Experiments were performed in duplicate with identical results, for gel source data, see Supplementary Fig. 1. d, Ub-VS charging assay as in c, for phospho-Parkin wild-type (wt) or R104A mutant. Patients with Parkin R104W suffer from AR-JP31. Experiments were performed in duplicate with identical results, for gel source data, see Supplementary Fig. 1. e, Activity of phospho-Parkin wild-type (wt) and R104A in vitro, with UBE2L3 as the E2 enzyme. The reaction was resolved by SDS-PAGE and Western blotted for ubiquitin. A representative gel of three independent experiments is shown. For source data, see Supplementary Fig. 1. f, Model of the sequential domain rearrangements required for full Parkin activation, extended from 3 (also see Fig. 1a). In autoinhibited Parkin, the Ubl, REP and RING2 assume inhibitory positions. Phospho-ubiquitin binding induces MOM localisation, repositioning of the IBR domain and release of the Ubl domain. Phosphorylation of Parkin allows the phospho-Ubl domain and ACT element to bind to the UPD, to replace and release the RING2 and REP, enabling MOM protein ubiquitination.
The linker provides additional contact points for the phospho-Ubl interface. Arg104 is located between two key hydrophobic residues, and contacts with its side chain the Ala46 loop in the phospho-Ubl. Importantly, Parkin R104W is an AR-JP patient mutation2,31, which we would predict to disrupt or misalign the observed hydrophobic interactions. Importantly, a phospho-Parkin R104A mutant was charged less efficiently by Ub-VS (Fig. 4d), showed slower E2~Ub discharge activity (Extended Data Fig. 8a, b), and reduced in vitro polyubiquitination activity (Fig. 4e), while its thermal stability remained unperturbed (Extended Data Fig. 8c).
Together, structural, biochemical and patient data confirmed the crucial importance of the first conserved stretch of the Ubl-UPD linker for Parkin activity, and defines a new activating element, which we term ACT, in this to-date understudied regulatory region of Parkin that also contains several phosphorylation sites (see further discussion in Extended Data Fig. 8d).
Our work resolves the activation mechanism of Parkin, finally visualising large-scale domain rearrangements. It is fascinating how the Parkin Ubl switches between an inhibitory position in the unphosphorylated molecule, to an activating position in phosphorylated Parkin. Our data is consistent with a model in which the phosphorylated Ubl and the here identified ACT element in the Ubl-UPD linker dislodge RING2 from its autoinhibited position, enabling it to be charged by E2~Ub, and ubiquitinate substrates in its vicinity, independently of the Parkin core (Fig. 4f). Notably, our model does not require Parkin dimerisation6,27.
Our structure of an activated Parkin core will inform drug discovery efforts that have set out to identify Parkin activators. With the realisation that the RING2-UPD interface opens and exposes a hydrophobic pocket, small molecules could be directed towards this interface. Such molecules may become particularly useful to restart mitophagy in AR-JP patients carrying Parkin variants that fail to be activated by PINK1 mediated Ubl phosphorylation.
Online Methods
Molecular Biology
cDNA of Thamnophis sirtalis (Ts) Parkin was obtained from GeneArt (Invitrogen) with codon-optimisation for bacterial expression and cloned into a pOPIN-K vector 32, using the In-Fusion HD Cloning Kit (Takara Clonetech). HsParkin and PhPINK1 constructs were also expressed from a pOPIN-K vector, while UBE2L3 was expressed from a pGEX6 vector. HsUBE1/PET21d was a gift from Cynthia Wolberger [Addgene plasmid # 34965, 33].
Site-directed mutagenesis was carried out using the QuikChange protocol with Phusion polymerase. A TEV cleavage site was introduced in the Parkin constructs using the NEB Q5 Site-Directed Mutagenesis Kit (NEB). In TsParkin residues 368 – 374 (KSPGATA) were replaced by the ENLYFQS TEV cleavage sequence, while in HsParkin residues 382 – 378 (EASGTTT) were replaced by the TEV cleavage sequence to yield cleavable constructs.
Protein purification
For Parkin expression, E. coli Rosetta2 pLacI cells (Novagen) were grown in 2xTY medium at 37 °C. At OD600 = 0.6 the temperature was reduced to 18 °C, expression induced at OD600 = 0.8 - 1.0 with 30 μM IPTG and the media supplemented with 200 μM ZnCl2, cells were harvested after overnight growth at 18°C and frozen at -20°C.
For Parkin purification, cells were resuspended in lysis buffer (300 mM NaCl, 10% (w/v) glycerol, 25 mM Tris (pH 8.5), 14.3 mM β-mercaptoethanol) supplemented with 2 mg/mL lysozyme, 0.2 mg/mL DNaseI and 80 μg/mL PMSF. The suspension was homogenised using an EmulsiFlex-C3 (Avestin) for two passes at ~15,000 psi and cleared by centrifugation at 46,000 g for 35 min at 4°C. The clarified lysate was applied to Amintra glutathione resin (Expedeon), resin washed with high salt buffer (25 mM Tris (pH 8.5), 500 mM NaCl, 10 mM DTT) and GST-fusion Parkin cleaved from the resin overnight at 4°C with GST-3C protease.
Samples were eluted and resin washed with no-salt buffer (25 mM Tris (pH 8.5), 10 mM DTT). All following purification steps were carried out on an Äkta Pure system (GE Healthcare). Pooled fractions were subjected to anion-exchange chromatography on a 6 mL Resource Q column (GE Healthcare) with a 0-25% linear gradient from buffer A (25 mM Tris (pH 8.5), 10 mM DTT, 50 mM NaCl) to buffer B (25 mM Tris (pH 8.5), 10 mM DTT, 1000 mM NaCl) over 15 column volumes. For phosphorylated Parkin, the resulting sample was phosphorylated using a 1:100 molar ratio of GST-PhPINK1 in phosphorylation buffer (10 mM ATP, 10 mM MgCl2, 200 mM NaCl, 50 mM Tris (pH 8.5), 10 mM DTT). PINK1 was subsequently removed by incubation with Amintra glutathione resin (Expedeon) and phosphorylated Parkin purified using anion exchange chromatography as above. Finally, samples were subjected to size-exclusion chromatography (HiLoad 16/600 Superdex 75 pg, GE Healthcare) into buffer C (25 mM Tris (pH 8.5), 10 mM DTT, 200 mM NaCl).
In short, HsUBE1 was purified as follows: An N-terminal GST-Ub fusion protein was expressed and lysed in β-mercaptoethanol-free lysis buffer and applied to Amintra glutathione resin (Expedeon). Upon washing, the resin was equilibrated with 50 mM Tris (pH 8.5) and 2 mM ATP. HsUBE1 β-mercaptoethanol-free clarified lysate was generated as above, supplemented with 10 mM ATP and 10 MgCl2 and incubated with the GST-Ub fusion-bound glutathione resin at room temperature for 30 min. The resin was then washed with DTT-free high salt buffer supplemented with 5 mM MgCl2. HsUBE1 was eluted in DTT-containing buffer and protein containing fractions were applied to anion-exchange and size-exclusion chromatography as above. UBE2L3, UBE2D3 and GST-PhPINK1 were purified as described previously34.
Generation of non-dischargeable E2-Ub complex
UBE2L3 (C86K) and ubiquitin were stored in charging buffer (25mM CAPSO (pH 9.5), 20 mM MgCl2, 150 mM NaCl). UBE2L3 (C86K) (450 μM) was incubated with Ub (900 μM) and HsUbe1 (2.5 μM) in charging buffer supplemented with 10 mM ATP at 37 °C overnight. The resulting mixture was applied to size-exclusion chromatography as above in buffer C. Fractions containing UBE2L3-Ub were pooled, concentrated and again applied to size-exclusion chromatography to remove free UBE2L3.
Ub-VS generation and Parkin coupling
Ub(1-75)-MesNa was prepared as described previously35. H-Gly-VS hydrochloride was a kind gift from Huib Ovaa and Bo Xin (Leiden University). Ub-MesNa, stored in buffer D at ~20 mg/mL was used to dissolve ~50 mg H-Gly-VS hydrochloride together with ~30 mg of N-hydroxysuccinamide (Fluka), acting as a catalyst. The pH was raised to 8.5 by addition of ~60 μL of 4 M NaOH and reaction incubated at 37°C for ~40 min. Reaction progress was monitored by LC-MS analysis. When the ratio of Ub(1-75)-VS to the hydrolysed Ub-MesNa product was ~1:1, with a minimum formation of the doubly coupled, Ub(1-75)-VS-VS species, the reaction was quenched by addition of 20 μL 12 M HCl (~30 min). The subsequent sample was diluted in 50 sodium acetate (pH 4.5) and applied to cation-exchange chromatography on a 1 mL MonoS column (GE Healthcare) with a 10-35% linear gradient between 50 mM sodium acetate (pH 4.5) containing 0 M and 1 M NaCl respectively. The resulting fractions were analysed by LS-MS and Ub(1-75)-VS containing fractions were pooled and applied to size-exclusion chromatography as above in buffer D.
For quantitative Parkin Ub-VS coupling, phospho-Parkin was purified as above where 10 mM DTT in buffer C was replaced with 5 mM TCEP. Phospho-Parkin and Ub-VS were mixed at a 1:3 molar ratio and incubated at room temperature. Reaction progress was monitored by LC-MS analysis and upon completion, the reaction was quenched by addition of DTT. The resulting sample was purified using size-exclusion chromatography (buffer C).
Mass-spectrometry analysis
LC-MS analysis was carried out on an Agilent 1200 Series chromatography system coupled to an Agilent 6130 Quadrupole mass spectrometer. Samples were eluted from a phenomenex Jupiter column (5 lm, 300 Å, C4 column, 150 × 2.0 mm) using an acetonitrile gradient + 0.2% (v/v) formic acid. Protein was ionised using an ESI source (3 kV ionisation voltage), and spectra were analysed in positive ion mode with a mass range between 400 and 2,000 m/z. Averaged spectra were deconvoluted using Promass (Novatia, LLC) and plotted using GraphPad Prism (version 7).
Limited proteolysis
TsParkin, phospho-TsParkin, phospho-TsParkin-pUb, and phospho-TsParkin-pUb charged with Ub-VS, were purified as described above. A 1 mg/mL protein solution was mixed with 5 μg/mL solution of elastase from the Proti-Ace Kit (Hampton research) and incubated for 1 h at room temperature. The reactions were quenched by addition of DTT- and iodoacetamide-containing LDS buffer and resolved on a 4-12% SDS NuPAGE gradient gels (Invitrogen) and stained with Instant Blue SafeStain (Expedeon).
Hydrogen-deuterium exchange mass spectrometry (HDX-MS)
Complexes were formed on ice and incubated for 30 mins giving a final Parkin concentration of 10 μM. Deuterium exchange reactions of Parkin and the different complexes were initiated by diluting the protein in D2O (99.8% (v/v) D2O ACROS, Sigma, UK) in 25 mM Tris (pH 8.5), 200 mM NaCl, 1 mM TCEP to give a final D2O percentage of ~95%. For all experiments, deuterium labelling was carried out at 23°C (unless otherwise stated) at five time points, 0.3 seconds (3 seconds on ice), 3 seconds, 30 seconds, 300 seconds and 3000 seconds, in technical triplicate. The labelling reaction was quenched by the addition of chilled 2.4% (v/v) formic acid in 2 M guanidinium hydrochloride and immediately frozen in liquid nitrogen. Samples were stored at -80°C prior to analysis.
The quenched protein samples were rapidly thawed and subjected to proteolytic cleavage with pepsin followed by reversed phase HPLC separation. Briefly, the protein was passed through an Enzymate BEH immobilised pepsin column, 2.1 x 30 mm, 5 μm (Waters, UK) at 200 μL/min for 2 min, the peptic peptides were trapped and desalted on a 2.1 x 5 mm C18 trap column (Acquity BEH C18 Van-guard pre-column, 1.7 μm, Waters, UK). Trapped peptides were subsequently eluted over 11 min using a 3-43% gradient of acetonitrile in 0.1% (v/v) formic acid at 40 μL/min. Peptides were separated on a reverse phase column (Acquity UPLC BEH C18 column 1.7 μm, 100 mm x 1 mm (Waters, UK) and detected on a SYNAPT G2-Si HDMS mass spectrometer (Waters, UK) over an m/z of 300 to 2000, with the standard electrospray ionisation (ESI) source with lock mass calibration using [Glu1]-fibrino peptide B (50 fmol/μL). The mass spectrometer was operated at a source temperature of 80°C and a spray voltage of 2.6 kV. Spectra were collected in positive ion mode.
Peptide identification was performed by MSe 36 using an identical gradient of increasing acetonitrile in 0.1% (v/v) formic acid over 11 min. The resulting MSe data were analysed using Protein Lynx Global Server software (Waters, UK) with an MS tolerance of 5 ppm.
Mass analysis of the peptide centroids was performed using DynamX software (Waters, UK). Only peptides with a score >6.4 were considered. The first round of analysis and identification was performed automatically by the DynamX software, however, all peptides (deuterated and non-deuterated) were manually verified at every time point for the correct charge state, presence of overlapping peptides, and correct retention time. Deuterium incorporation was not corrected for back-exchange and represents relative, rather than absolute changes in deuterium levels. Changes in H/D amide exchange in any peptide may be due to a single amide or a number of amides within that peptide.
Protein preparation for crystallisation
TEV-cleavable Parkin was purified as described above. An anion-exchange purified Parkin sample (step 1) was incubated with Ub-3CBr in a 1:4 molar ratio and GST-PhPINK1 in a 9:1 molar ratio in phosphorylation buffer (10 mM ATP, 10 mM MgCl2, 200 mM NaCl, 50 mM Tris (pH 8.5), 10 mM DTT) at a final Parkin concentration of ~70 μM, yielding a phosphorylated, phospho-ubiquitin conjugated, TEV-cleavable Parkin sample (step 2). GST-PhPINK1 was subsequently removed using Amintra glutathione resin (Expedeon). The sample was subjected to His6-TEV cleavage overnight at 4°C (step 3). His6-TEV was removed using Ni-NTA agarose (Qiagen), sample diluted in Buffer A and applied to anion-exchange and size-exclusion chromatography as described above.
Ub(1-75)-MesNa was prepared as described previously 35. Ub-MesNa, stored in buffer D (20 mM HEPES, 50 mM sodium acetate (pH 6.5), 75 mM NaCl) was incubated with 0.2 g/mL 3-bromopropylamine hydrobromide (Fluka) dissolved in PBS (pH 4.8) at 2:1 molar ratio with final Ub(1-75)-MesNa concentration of 445 μM. The coupling was carried out on ice for 30 min following addition of 50 μL 4 M NaOH to raise the pH to 10.5. The reaction was quenched by addition of 12 μL of 12 M HCl and sample buffer exchanged using a disposable PD-10 desalting column (GE Healthcare) into buffer C.
Crystallisation
Initial crystals were found from crystallisation experiments carried out at 18°C in a 96-well sitting drop vapour diffusion plates in the MRC format (Molecular Dimensions) by mixing 100 nL of 4 mg/mL protein solution with 100 nL reservoir solution. The crystallisation condition of 12.5% (w/v) PEG 1000, 12.5% (w/v) PEG 3350, 12.5% (v/v) MPD, 0.03 M of each sodium nitrate, disodium hydrogen phosphate, ammonium sulphate, 0.1 M MOPS/HEPES-Na (pH 7.5) was found from the MORPHEUS screen (Molecular Dimensions). Seeds were obtained from a fine screen and streak seeding was carried out in a hanging drop format from an 8 mg/mL protein solution. Larger crystals were obtained after 6 days in the original crystallisation condition. Crystals were soaked in mother-liquor supplemented with 10% (v/v) glycerol prior to vitrification in liquid nitrogen.
Data collection, phasing and refinement
Diffraction data was collected at the Diamond Light Source, beam line I-24 (0.9686 Å, 100 K), and processed using DIALS37. The crystal structure was determined by molecular replacement in Phaser 38using the structure of the human Parkin core (pdb-id 5n2w, 6) truncated after the IBR as well as a human Parkin Ubl structure (pdb-id 5c1z, 19). The structure was built at 1.80 Å, in multiple rounds of model building in Coot 39 and refinement in PHENIX40. Phenix ReadySet-derived geometry restraints for the 3CN warhead were used, with external restraints defining the linkage points. Final Ramachandran statistics: 98.9 % favoured, 1.1 % allowed, and 0 % outliers. Structural figures were generated using PyMol (www.pymol.org). Data collection and refinement statistics can be found in Extended Data Table 1.
Parkin activity assays
Ub-VS conjugation assays
Indicated Parkin variants stored in either DTT- or TCEP-containing buffer were incubated with Ub-VS that was prepared as described above. The reactions were quenched at indicated time points by addition of DTT- and iodoacetamide-containing LDS buffer and resolved on a 4-12% SDS NuPAGE gradient gels (Invitrogen) and stained with Instant Blue SafeStain (Expedeon)
Parkin assembly assays
phospho-Parkin wild-type or R104A (5 μM) buffer were incubated in ubiquitination buffer (30 mM HEPES (pH 7.5), 100 mM NaCl, 10 mM ATP, 10 mM MgCl2) with HsUBE1 (0.2 μM), UBE2L3 (2 μM) and Ub (20 μM). The reactions were quenched at indicated time points by addition of DTT- and iodoacetamide-containing LDS buffer and resolved on a 4-12% SDS NuPAGE gradient gels (Invitrogen) and transferred to a PVDF membrane (BioRad). Membranes were blocked in a 5% (w/v) milk solution in PBS-T (PBS + 0.1% (v/v) Tween-20) for 30 min and incubated overnight at 4 °C with a ubiquitin recognizing antibody (Ubi-1, NB300-130, Novus Biologicals) in 5% (w/v) BSA in PBS-T and 0.1% (w/v) sodium azide. The membrane was then washed with PBS-T, incubated for 1 h at room temperature with anti-mouse IgG-HRP (NXA931, GE Healthcare) in 5% (w/v) milk in PBS-T, washed in PBS-T and visualised using the Amersham Western Blotting Detection Reagent (GE Healthcare) and a ChemiDoc Touch Imaging System (BioRad).
E2~Ub discharge assays
The UBE2D3~Ub conjugate was generated by incubating UBE2D3 (20 μM) with HsUBE1 (20 nM) and Ub (80 μM) in ubiquitination buffer supplemented with 5 μM CaCl2 at 37°C for 10 min. To remove remaining ATP, 0.5 U of Apyrase (NEB) were added and the reaction incubated at 30°C for 30 min. The discharge reaction was studied by addition of 1 μM phospho-Parkin wild-type or R104A to a diluted charging reaction mixture (final UBE2D3 concentration was 9 μM). The reactions were quenched at indicated time points by addition of DTT-free LDS buffer, while a final sample was collected at 11 min in DTT-containing LDS buffer to assess the extent of isopeptide-linked UBE2D3-Ub species formation. Samples were resolved on a 4-12% SDS NuPAGE gradient gels (Invitrogen) and stained with Instant Blue SafeStain (Expedeon). The gel band intensity was quantified in ImageJ by isolating the specific intensity of the UBE2D3~Ub thioester band as indicated, subtracting the background of the final reduced sample and normalized within each reaction.
Thermal denaturation assays
Protein melting curves were recorded on a Corbett RG-6000 real time PCR cycler (30°C to 85°C with 7 s per 0.5°C). Samples contained 4 μM Parkin protein and 4x SYPRO Orange in ubiquitination buffer + 5 mM TCEP. Melting curves were obtained as the maxima of dF/dT versus T plots. All data were recorded in triplicate.
Extended Data
Extended Data Figure 1. Mechanisms of Parkin autoinhibition.
a, Structure of autoinhibited, full-length human Parkin (pdb-id 5c1z, 19) shown schematically (top, as in Fig. 1a) and in cartoon representation in the same colours. Two insets show the UPD-RING2 interface (with Cys431 shows in ball-and-stick representation), and the blocked E2 binding site (with the E2 position, modelled according to 5edv 29 shown as grey surface). Zn ions are shown as grey spheres. b, An ‘open-book’ view on the UPD-RING2 interface, with hydrophobic residues coloured white on each surface. c, Structure of phospho-ubiquitin bound to full-length Parkin (pdb-id 5n2w, 6) as in a. Phospho-ubiquitin binding leads to helix straightening, and IBR domain repositioning, which releases the Ubl domain for phosphorylation5,6. In the shown structures of unphosphorylated Parkin, the Ubl and REP (red) inhibit E2 binding, and the RING2-UPD interface is intact, with Cys431 being inaccessible. The Ubl-UPD linker was removed from both crystallised constructs6,22.
Extended Data Figure 2. Samples preparation for HDX-MS and selected raw data.
a, Representative LC-MS spectrum of the prepared Ub-VS probe (see Online Methods). Experiment has been performed in duplicate. b, Representative LC-MS spectrum of Ub-VS-reacted phospho-Parkin. Experiment has been performed in duplicate. c, Samples used in HDX-MS analysis. In HDX-MS, non-covalent complexes with phospho-ubiquitin were used. Covalent complexes are indicated with a dash (‘-‘), thioester-based covalent complexes by a tilde (‘~’), and non-covalent complexes by a colon (‘:’). This is representative of at least three independent experiments, for gel source data, see Supplementary Fig. 1. d, Relative deuterium uptake (in Da) is shown for exemplary selected peptides across the Parkin molecule, over the timecourse of the experiment. Each point for the technical replicate experiments is shown. Data points were taken at identical time points, but are offset on the x-axis for clarity.
Extended Data Figure 3. Graphical representation of HDX-MS data.
Data from HDX-MS experiments (Fig. 1b-e) was plotted onto a stylised ‘open domain’ model of Parkin, with identical colouring (blue, more protected from solvent exchange compared to previous state; red less protected from solvent exchange compared to previous state). Grey regions correspond to peptides that were not covered or could not be analysed due to modification.
Schematic domain representations indicate an average change of the corresponding interfaces across all time points. White regions indicate no change.
a, Parkin compared to Parkin:phospho-ubiquitin.
b, Parkin:phospho-ubiquitin compared to phospho-Parkin:phospho-ubiquitin
c, phospho-Parkin:phospho-ubiquitin compared to phospho-Parkin:phospho-ubiquitin in complex with UBE2L3-Ub isopeptide UBE2L3~Ub thioester mimetic (see Online Methods). This experiment confirmed a previously reported binding site for the E2-conjugated ubiquitin on the RBR6,29 (8).
d, phospho-Parkin:phospho-ubiquitin compared to Ub-VS-reacted phospho-Parkin:phospho-ubiquitin. Reaction with Ub-VS leads to modification of the catalytic Cys431 containing-peptide, generating non-identical peptides precluding comparison by HDX-MS. Low coverage of the RING2 domain can be explained by ubiquitin resistance to pepsin cleavage, leading to protection of the linked RING2 domain and subsequent peptide loss. To allow comparison, these peptides were also omitted form analysis of the UBE2L3 sample.
In c and d, the structure representation is deceiving since REP and RING2 are highly mobile and are no longer bound to the Parkin core. Indeed, the high HD exchange in the REP sequence in active Parkin (see Fig. 1d, e, peptide (4) in Extended Data Fig. 2d) indicates an additional loss of secondary structure in this helical element when REP and RING2 are released.
Extended Data Figure 4. A conserved linker between Ubl and UPD.
Sequence alignment of Parkin, with domains coloured corresponding to 5n2w 6 as in Extended Data Fig. 1. Phosphate binding pockets are labelled. The linker region between Ubl and UPD (aa 76-143) contains two strings of highly conserved residues. Residues upstream and downstream of the conserved region are unconserved both in sequence and linker length. Thamnophis sirtalis (Ts, garter snake) Parkin shows the smallest number of residues in the linker (upstream, 25 aa in human Parkin, 18 aa in TsParkin; downstream, 18 aa in human Parkin, 11 aa in TsParkin). See also Extended Data Fig. 8d.
Extended Data Figure 5.
a, HDX-MS experiment comparing phospho-TsParkin reacted with pUb-C3Br and phospho-TsParkin reacted with pUb-C3Br and Ub-VS with identical colouring (blue, more protected from solvent exchange; red less protected from solvent exchange; grey, not covered in all of the compared states, see Fig. 1). The experiment was performed in technical triplicate. The TsParkin profile is highly similar to the profile of human Parkin in an analogous state (Fig. 1e). Higher peptide resolution in this sample reveals protection of the RING2 interface by reacted Ub-VS, but importantly, the C-terminus of RING2 that binds to the UPD interface is surface exposed. Secondly, both phospho-Ubl and the Ubl-UPD linker are protected in activated Parkin. b, Limited proteolysis of TsParkin with Elastase, in different stages of activation. In unphosphorylated, autoinhibited TsParkin, the Ubl is cleaved off in the Ubl-UPD linker. In activated forms of TsParkin (phospho-TsParkin, phospho-TsParkin reacted with pUb-C3Br, phospho-TsParkin reacted with pUb-C3Br and Ub-VS), the RING2 is readily cleaved off, while the Ubl is not efficiently removed. this suggests that the Ubl-UPD linker is not accessible in activated forms of TsParkin. A representative gel from three independent experiments is shown. For gel source data, see Supplementary Fig. 1. c, A TEV cleavage site was introduced after the IBR domain, so that after activation by phospho-ubiquitin and Ubl-phosphorylation, the released RING2 domain can be removed. Once removed, RING2 is no longer stably associated with the remaining Parkin core. Shown is a gel filtration profile illustrating this point. A representative profile from three independent experiments is shown. d, SDS-PAGE analysis of sample preparation process (see Online methods). An asterisk (*) denotes ubiquitin probe (Ub-C3Br) reacted material that modifies the RING2 catalytic Cys, which explains the cleaved, probe-reacted RING2 band (* in step 3). A representative gel from three independent experiments is shown. For gel source data, see Supplementary Fig. 1. e, HDX-MS experiment on TsParkin, comparing phospho-TsParkin reacted with pUb-C3Br with either phospho-TsParkin reacted with pUb-C3Br and Ub-VS (bottom) or with RING2-TEV cleaved phospho-TsParkin reacted with pUb-C3Br (top), colouring as in a. Identical profiles were obtained, showing that RING2 removal has no effect on the activated core of Parkin. This further indicates that RING2 acts independently of the Parkin core upon full activation. Notably, in both comparisons, we observed concomitant protection of phospho-Ubl and the Ubl-UPD linker. The experiment was performed in technical triplicate.
Extended Data Figure 6.
a, LC-MS spectrum of crystallised human phospho-Parkin (aa 1-382) bound to phospho-ubiquitin. This is representative of two independent experiments. b, Composite omit map (generated with simulated annealing) shown for the single complex in the asymmetric unit. 2|Fo|-|Fc| electron density is shown at 1σ. c, Electron density as in b for the Ubl-UPD linker. d, Electron density as in b for the Ser65 phospho-Ubl binding site on the UPD linker. e, Electron density as in b for the Ser65 phospho-Ub binding site. Since we are missing electron density for disordered regions in Ubl-ACT and ACT-UPD linkers, there is a remote possibility that the phospho-Ubl may interact in trans with a neighbouring Parkin molecule, which we cannot exclude.
Extended Data Figure 7. The phospho-Ubl binding site on the UPD.
a, Side by side view of phospho-Parkin-pUb (left) and Parkin-pUb (pdb-id 5n2w, 6, right), and superposition of both (below). The green Ubl domain changes position by >50 Å. b, E2~Ub from the structure of HOIP RBR domain in complex with UBE2D2~Ub29, was modelled onto phospho-Parkin-pUb, by superposition of the RING1 domains of each complex. The E2-conjugated ubiquitin molecule in the ‘open’ conformation binds to the previously recognised cryptic ubiquitin binding interface on RING1/IBR 6. The contact points correlate with HDX-MS data (Fig. 1d, Extended Data Fig. 2, 3). c, HDX-MS data from Fig. 1e was plotted onto the phospho-Parkin-pUb structure with identical colouring (blue, more protected from solvent exchange; red less protected from solvent exchange; grey, not covered in all of the compared states, compare Fig. 1). Protected regions on UPD match the observed phospho-Ubl interface. d, HDX-MS experiments comparing a Parkin mutant with a mutation in the phospho-acceptor binding site on the UPD, (phospho-Parkin K211N:phospho-Ub) compared with phospho-Parkin:phospho-Ub, coloured as in c. The mutant is unable to protect the Ubl, and to release RING2 and REP. Experiments were done as technical triplicate.
Extended Data Figure 8. A regulatory role of the Parkin Ubl-UPD linker.
a,b, E2 discharge assay resolved on a Coomassie stained SDS-PAGE gel (a) and quantified from band intensities (b) for phospho-Parkin and phospho-Parkin R104A. This is representative of at least two independent experiments, for gel source data, see Supplementary Fig. 1. The mutation in the ACT element leads to lower discharge activity, suggesting that the residue is required to dislodge RING2 from the Parkin core. c, Parkin R104A is equally stable as compared to wild-type Parkin, in the unphosphorylated or phosphorylated form. Thermal denaturation experiments were performed as technical triplicate.
d, Sequence detail of the Ubl-UPD linker, which contains the here described ACT element. In the ACT element as bound to phospho-Parkin-pUb, the positions for two annotated (in PhosphoSitePlus) Parkin phosphorylation sites, Ser101 and Ser108, are resolved. Phosphorylation of Ser101 decreases Parkin activity41, which is likely explained by phosphorylation preventing phospho-Ubl and/or linker binding to the UPD. It is hence highly likely that phosphorylation of Parkin on these residues provides additional layers of Parkin regulation that remain to be uncovered in future work. As an example, Parkin phosphorylation by PKA was recently reported to be a mechanism of Parkin inhibition in beige-to-white adipocyte transition, although phosphorylation sites remained unclear 42.
Residues before the ACT element (aa 73-99), and after the ACT element, (aa 109-142) are disordered in our structure. The last ordered residue, Ser108, is tantalisingly close to the REP binding site as well as to the phospho-ubiquitin binding pocket, but disorder suggests that clear binding sites for other conserved linker residues, in particular for the Parkin GLAVIL motif, are not present. HDX-MS also does not reveal additional protection of the linker, even when the E2~Ub conjugate is bound, suggesting that the GLAVIL motif may not bind the E2 (Fig. 1, Extended Data Fig. 2, 3). On the other hand, there are at least three additional annotated phosphorylation sites, Ser116, Ser131 and Ser13615,41,43,44, suggesting that the second part of the linker may also be regulated. Phosphorylation on these residues could change its ability of the disordered parts of the linker to interact with Parkin in cis. For example, we would speculate that a phosphorylated Ser116 could e.g. reach the phosphate binding pocket occupied by phospho-Ser65 of ubiquitin. Alternatively, the remaining Ubl-UPD linker may be important for substrate recruitment, or involved in other, PINK1-independent mechanisms of Parkin activation.
Extended Data Table 1. Data collection and refinement statistics.
Values in parentheses are for highest-resolution shell.
| phospho-Parkin (1-382) - phospho-Ub | |
|---|---|
| Data collection | |
| Space group | P 32 2 1 |
| Cell dimensions | |
| a, b, c (Å) | 83.93, 83.93, 105.12 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 72.69 –1.80 (1.84 – 1.80) |
| Rmerge | 0.065 (0.773) |
| I / σI | 13.80 (2.40) |
| Completeness (%) | 100.00 (99.30) |
| Redundancy | 6.7 (6.7) |
| Refinement | |
| Resolution (Å) | 59.79 –1.80 |
| No. reflections / test set | 40229 / 2020 |
| Rwork / Rfree | 0.180 / 0.205 |
| No. atoms | |
| Protein | 3039 (398 aa) |
| Ligand/ion | 41 |
| Water | 165 |
| B-factors | |
| Protein | 45.05 |
| Ligand/ion | 66.19 |
| Water | 46.94 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.19 |
Values in parentheses are for highest-resolution shell.
Supplementary Material
Acknowledgments
We thank beamline scientists at Diamond Light Source (DLS) for support at beamlines I24, Huib Ovaa and Bo Xin (Leiden University) for H-Gly-VS hydrochloride, Jonathan Pruneda, Paul Elliott, and Martin Michel for help with crystallography and data collection, and members of the DK lab for reagents and discussions. Access to DLS was supported in part by the EU FP7 infrastructure grant BIOSTRUCT-X (contract no. 283570). The D.K. lab is supported by the Medical Research Council [U105192732], the European Research Council [724804], the Michael J. Fox Foundation and the Lister Institute for Preventive Medicine.
Footnotes
Data availability statement
Coordinates and structure factors have been deposited with the protein data bank under accession codes 6GLC. Uncropped versions of all gels are displayed in Supplementary Fig. 1. All reagents and data are available upon reasonable request from the corresponding author.
Author contribution
C.G. performed all experiments and analysed the data presented in this manuscript. S.M. and J.M.S. performed and analysed HDX-MS experiments. D.K. directed the project, analysed the data, acquired funding and wrote the manuscript with help from all authors.
Author information
The authors declare no competing interests.
References
- 1.Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- 2.Pickrell AM, Youle RJ. The Roles of PINK1, Parkin, and Mitochondrial Fidelity in Parkinson's Disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harper JW, Ordureau A, Heo J-M. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol. 2018;19:93–108. doi: 10.1038/nrm.2017.129. [DOI] [PubMed] [Google Scholar]
- 4.Pickles S, Vigié P, Youle RJ. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wauer T, Simicek M, Schubert AF, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–374. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A, et al. Parkin–phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity. Nat Struct Mol Biol. 2017;24:475–483. doi: 10.1038/nsmb.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trempe J-F, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 8.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riley BE, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nature Communications. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 11.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazlauskaite A, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ordureau A, et al. Quantitative Proteomics Reveal a Feedforward Mechanism for Mitochondrial PARKIN Translocation and Ubiquitin Chain Synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wauer T, et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34:307–325. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biology. 2012;2 doi: 10.1098/rsob.120080. 120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iguchi M, Kujuro Y, Okatsu K, Koyano F. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. Journal of Biological Chemistry. 2013;288:22019–22032. doi: 10.1074/jbc.M113.467530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiba-Fukushima K, et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauvé V, et al. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J. 2015;34:2492–2505. doi: 10.15252/embj.201592237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J. 2015;34:2506–2521. doi: 10.15252/embj.201592337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazlauskaite A, et al. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 2015;16:939–954. doi: 10.15252/embr.201540352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ordureau A, et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proceedings of the National Academy of Sciences. 2015;112:6637–6642. doi: 10.1073/pnas.1506593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A, Chaugule VK, Condos T. Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity. Nat Struct Mol Biol. 2017;24:475–483. doi: 10.1038/nsmb.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubert AF, et al. Structure of PINK1 in complex with its substrate ubiquitin. Nature. 2017;552:51–56. doi: 10.1038/nature24645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ordureau A, et al. Dynamics of PARKIN-Dependent Mitochondrial Ubiquitylation in Induced Neurons and Model Systems Revealed by Digital Snapshot Proteomics. Mol Cell. 2018;70:211–227.e8. doi: 10.1016/j.molcel.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S, Foote PK, Krist DT, Rice SE, Statsyuk AV. UbMES and UbFluor: Novel probes for ring-between-ring (RBR) E3 ubiquitin ligase PARKIN. Journal of Biological Chemistry. 2017;292:16539–16553. doi: 10.1074/jbc.M116.773200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pao K-C, et al. Probes of ubiquitin E3 ligases enable systematic dissection of parkin activation. Nat Chem Biol. 2016;12:324–331. doi: 10.1038/nchembio.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arkinson C, Walden H. Parkin function in Parkinson's disease. Science. 2018;360:267–268. doi: 10.1126/science.aar6606. [DOI] [PubMed] [Google Scholar]
- 28.Harrison RA, Engen JR. Conformational insight into multi-protein signaling assemblies by hydrogen–deuterium exchange mass spectrometry. Curr Opin Struct Biol. 2016;41:187–193. doi: 10.1016/j.sbi.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechtenberg BC, et al. Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature. 2016;529:546–550. doi: 10.1038/nature16511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biology. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 31.Chaudhary S, et al. Parkin mutations in familial and sporadic Parkinson's disease among Indians. Parkinsonism & Related Disorders. 2006;12:239–245. doi: 10.1016/j.parkreldis.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Berrow NS, et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berndsen CE, Wolberger C. A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal Biochem. 2011;418:102–110. doi: 10.1016/j.ab.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gladkova C, et al. An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1. EMBO J. 2017 doi: 10.15252/embj.201797876. e201797876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkinson KD, Gan-Erdene T, Kolli N. Derivitization of the C-terminus of ubiquitin and ubiquitin-like proteins using intein chemistry: methods and uses. Meth Enzymol. 2005;399:37–51. doi: 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
- 36.Silva JC, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 37.Waterman DG, et al. Diffraction-geometry refinement in the DIALS framework. Acta Crystallogr D Struct Biol. 2016;72:558–575. doi: 10.1107/S2059798316002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto A. Parkin Phosphorylation and Modulation of Its E3 Ubiquitin Ligase Activity. J Biol Chem. 2004;280:3390–3399. doi: 10.1074/jbc.M407724200. [DOI] [PubMed] [Google Scholar]
- 42.Lu X, et al. Mitophagy controls beige adipocyte maintenance through a Parkin-dependent and UCP1-independent mechanism. Science signaling. 2018;11 doi: 10.1126/scisignal.aap8526. eaap8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avraham E, Rott R, Liani E, Szargel R, Engelender S. Phosphorylation of Parkin by the Cyclin-dependent Kinase 5 at the Linker Region Modulates Its Ubiquitin-Ligase Activity and Aggregation. J Biol Chem. 2007;282:12842–12850. doi: 10.1074/jbc.M608243200. [DOI] [PubMed] [Google Scholar]
- 44.Durcan TM, et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 2014;33:2473–2491. doi: 10.15252/embj.201489729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.