Abstract
DNA in the eukaryotic nucleus is packaged in the form of nucleosomes, ~147 base pairs of DNA wrapped around a histone protein octamer. The position and histone composition of nucleosomes is governed by ATP dependent chromatin remodelers1–3 such as the 15 subunit INO80 complex4. INO80 regulates gene expression, DNA repair and replication by sliding nucleosomes, exchanging histone H2A.Z with H2A, and positioning +1 and -1 nucleosomes at promoter DNA5–8. A structure and mechanism for these remodeling reactions is lacking. Here we report the cryo-electron microscopy structure at 4.3Å resolution, with parts at 3.7Å, of an evolutionary conserved core INO80 complex from Chaetomium thermophilum bound to a nucleosome. INO80core cradles one entire gyre of the nucleosome through multivalent DNA and histone contacts. A Rvb1/2 AAA+ ATPase hetero-hexamer is an assembly scaffold for the complex and acts as stator for the motor and nucleosome gripping subunits. The Swi2/Snf2 ATPase motor binds to SHL-6, unwraps ~15 base pairs, disrupts the H2A:DNA contacts and is poised to pump entry DNA into the nucleosome. Arp5-Ies6 grip SHL-2/-3 acting as counter grip for the motor on the other side of the H2A/H2B dimer. The Arp5 insertion domain forms a grappler element that binds the nucleosome dyad, connects the Arp5 core and entry DNA over a distance of ~90Å and packs against histone H2A/H2B near the acidic patch. Our structure together with biochemical data8 suggest a unified mechanism for nucleosome sliding and histone editing by INO80. The motor pumps entry DNA across H2A/H2B against Arp5 and the grappler, sliding nucleosomes as a ratchet. Transient exposure of H2A/H2B by the motor and differential recognition of H2A.Z and H2A may regulate histone exchange during translocation.
Remodelers are grouped into INO80, SWI/SNF, CHD and ISWI families that collectively shape the nucleosome landscape on chromosomal DNA7,9. Although there might be fundamental differences in how remodelers slide, evict and edit nucleosomes1–3, it is suggested that a common ATP dependent DNA translocation of the motor domains underlies these distinct reactions3. Recent studies revealed how the SNF2 motor domain10 and Chd1 family proteins11,12 interact with the nucleosome, but there is very limited understanding on how stepwise DNA translocation results in its various large-scale reconfigurations. INO80 and the related SWR1 complex are megadalton large modular complexes13–15 that carry out intricate editing reactions. SWR1 incorporates H2A.Z16 while INO80 has been shown to exchange H2A.Z with H2A5,8. H2A.Z is a H2A variant found at promoter and enhancer elements and has important regulatory functions17. INO80 also slides nucleosomes and positions the -1 and +1 nucleosomes of genic arrays that flank nucleosome depleted promoter regions6–8. However, even nucleosome sliding requires extensive inter-subunit coordination18,19 and a clear mechanistic framework explaining these activities is not available. Intriguingly, biochemical evidence indicates that INO80 translocates and loops DNA at the H2A/H2B interface8, suggesting that sliding and editing are perhaps facets of a common, complex chemo-mechanical reaction.
To provide a structural mechanism for nucleosome recognition and remodeling by INO80 we performed cryo-EM analysis of an evolutionary conserved, recombinant 11 subunit INO80 from Chaetomium thermophilum (Ct) bound to a nucleosome (Fig. 1a-c). Our complex comprises the main ATPase Ino80, actin and actin related proteins Arp4,5,8, (INO80 subunits) Ies2,4,6, Taf14 and the AAA+ ATPases Rvb1 and Rvb2, but lacks the evolutionary less conserved subunits (in yeast INO80: Ies1, 3, 5, Nhp10) and the N-terminal part of Ino80 to which these subunits bind. Biochemical analysis shows a stoichiometric complex that stably binds and remodels nucleosomes (Ext. Data Fig. 1), consistent with activities of similar human14,18 and S. cerevisiae13 INO80 complexes. The nucleosome was assembled from human histones H2A, H2B, H3, H4 and a Widom 601 sequence with 50 bp (base pair) extranucleosomal DNA that matches the footprint identified for the entire S. cerevisiae (Sc) INO808.
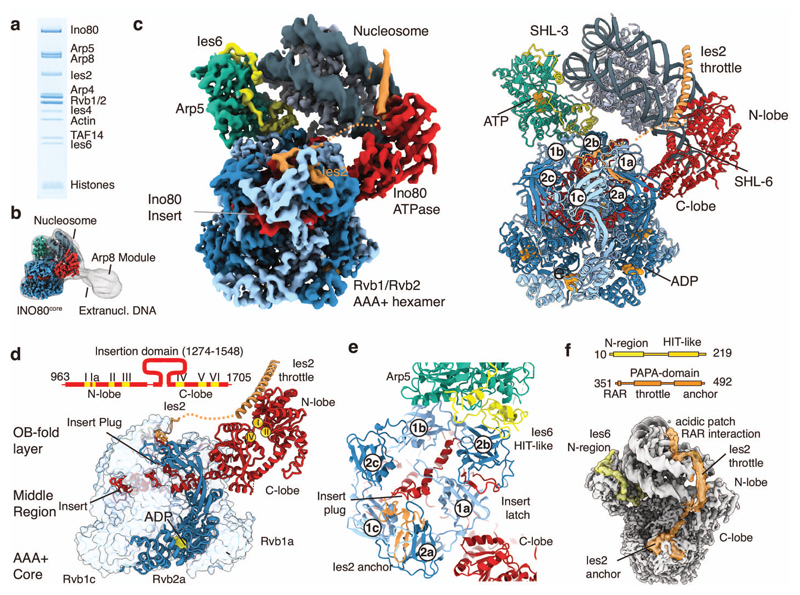
Figure 1. Structure of the INO80core nucleosome complex.
(a) Gel electrophoresis analysis of the purified, recombinant CtINO80 complex bound to nucleosome.
(b) Low resolution cryo-EM map showing extra density for the Arp8 module and extranucleosomal DNA. The high-resolution structure of INO80core is superimposed (see c-d).
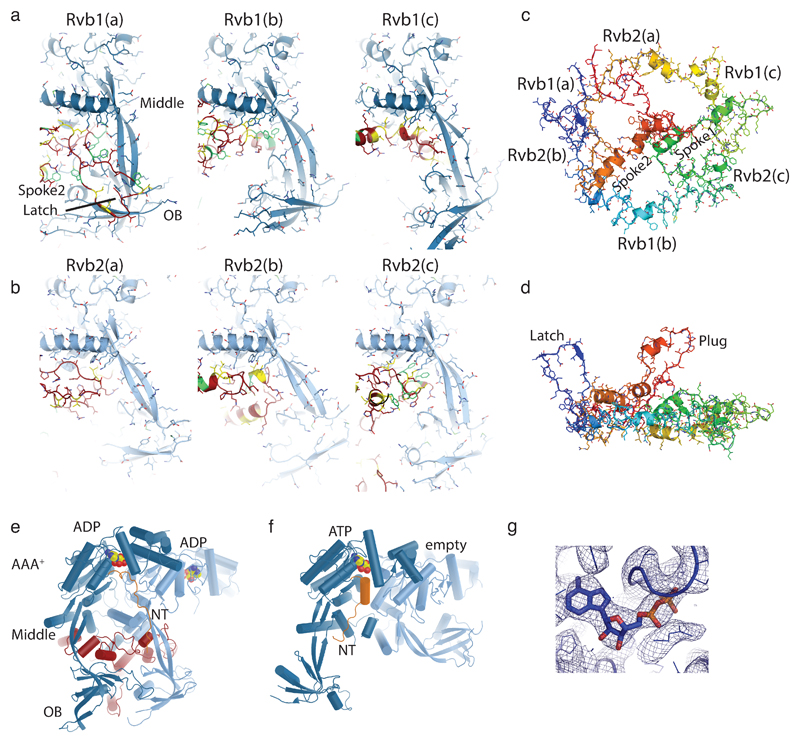
(c) (left) 4.3Å cryo-electron microscopy map reveals the architecture of INO80’s nucleosome remodeling core. Color code: nucleosome (steel), Ino80 (red), Ies2 (orange), Arp5 (green), Ies6 (yellow), three Rvb1 subunits (light blue), three Rvb2 subunits (dark blue). (right). Protein models obtained from interpretation of the cryo-EM map showing how INO80core binds the NCP. ADP and ATP molecules are indicated. The Rvb1/2 hexamer is assembled from three Rvb1/2 pairs, denoted “a”, “b” and “c” and organizes the nucleosome binding elements Arp5-Ies6, Ies2 and Ino80ATPase.
(d) Scheme of the Ino80ATPase showing the location of conserved helicase motifs (I-VI) and the INO80 family characteristic insert. The insert has a wheel-like structure that binds as client into the chamber of the three-layered Rvb1/2 hexamer. One Rvb1/2 subunit is shown as ribbon, the others as transparent surface.
(e) Details of the interactions of Arp5-Ies6, Ies2 and Ino80ATPase clients at the OB domain layer of Rvb1/2. Plug and latch of the Ino80insert recruit Ies2 and Arp5-Ies6 clients through direct interactions and/or orienting OB domains.
(f) Ies2 and Ies6 are extended proteins that functionally link Rvb1/2, nucleosome and motor or Arp5, respectively by multiple binding sites. Ies2 wraps around the nucleosome and binds the distal acidic patch. The domain architectures are shown above the map.
Cryo-electron microscopy and single particle reconstruction resulted in a map with a global resolution of 4.3Å and did not require crosslinking or the addition of nucleotides (Ext. Data Figs. 2,3, Table 1). The density reveals how a 590 kDa core module of INO80 (denoted INO80core, comprising Ino80, Arp5, Ies6, Ies2, Rvb1/2) recognizes and remodels the 200 kDa nucleosome core particle (NCP) (Fig. 1c). Focused refinement resulted in a 3.7Å map of the Rvb1/2-Arp5-Ies2-Ies6-Ino80 subcomplex (Ext. Data Figs. 2,3). We could build de novo atomic models for ATP-bound Arp5 actin-fold (Arp5core), Ies2, Ies6 and ADP-bound Rvb1/2 heterohexamer incorporating the complete Ino80ATPase insertion domain (Ino80insert). Pseudo-atomic models for the Ino80ATPase and the NCP were generated by flexible fitting of crystals structures and homology models (Fig. 1c). DNA visibly protrudes from the NCP, and a 20Å cryo-EM map, obtained from extensive 3D classification, indicates extranucleosomal binding of the 200 kDa Arp8 module (actin, Arp4, Arp8, Taf14, Ies4) (Fig. 1b), consistent with ChIP-Exo profiling that showed promoter DNA binding of Arp8 proximal to the +1 nucleosome in vivo20. However, the Arp8 module proved to be either unstable, or orientationally too heterogeneous to yield a high-resolution reconstruction at this stage.
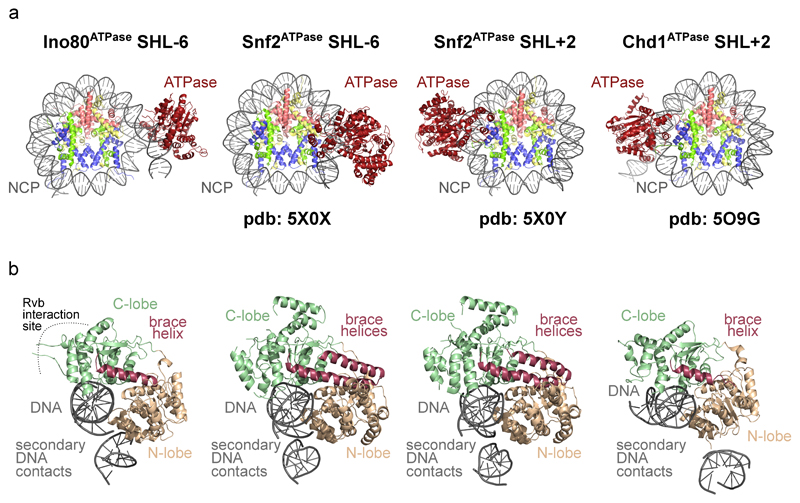
CtINO80core embraces one entire gyre of the nucleosome and binds in a multivalent fashion to nucleosomal DNA and histones (Fig. 1c). The overall mode of NCP recognition of INO80core matches closely the hydroxyl radical footprints of full ScINO808. The two main DNA contacts are to (superhelix location) SHL-6 by the Ino80ATPase motor8 and to SHL-2/-3 by Arp5-Ies6, latter explaining the second prominent NCP footprint of ScINO80. In addition, we observe contacts of Ino80ATPase and Ies2 to SHL2, of the ~325 amino acid large Arp5 insertion domain (denoted “grappler”) to the dyad, and of Grappler, Ies2 and Ies6 to the histone core (see below). Binding of SHL-6 by the Ino80ATPase motor differs from SHL-2 binding of Chd111,12 (Ext. Data Fig. 4) and ISWI remodeler21, indicating profoundly distinct remodeling mechanisms. The isolated Snf2 motor bound to SHL-2 but also to SHL-610. Therefore, clarification of mechanistic similarities and differences between INO80 and SWI/SNF require more complete structures of SWI/SNF remodelers.
The Rvb1/2 AAA+ ATPase is a prominent module of INO80 family remodelers and might act as an assembly chaperone22. We previously interpreted a low-resolution negative stain map as harboring the Rvb1/2 double-hexamer that forms in solution13, but our high-resolution structure now shows a single hexamer in nucleosome bound INO80, consistent with a recent structure of apo (human) hINO80core 23. However, by having the nucleosome bound state and the resolution to build atomic models for the clients, we can now reveal how Rvb1/2 specifically assembles INO80 and that it plays a key role in defining the functional arrangement of INO80 subunits for interaction with the NCP. The C-lobe of Ino80ATPase directly binds Rvb1/2 and contains a ~270 amino acid long insertion domain Ino80insert that adopts a wheel-like structure and sequentially binds to all six Rvb1/2 protomers in the central cavity (Ext. Data Fig. 5). Ino80 binding induces a marked asymmetry in the client binding OB domains inducing specific recruitment and positioning of motor, Ies2 and Arp5-Ies6 to grab the nucleosome from opposing sides (Fig. 1c).
Ino80insert does not bind to the individual Rvb1/2 units via a shared sequence or even a common structural fold, but the interactions are governed by different hydrophobic / aromatic elements, resembling models how bona fide chaperones may bind partially folded proteins24. Comparison with unliganded dodecameric Rvb1/225 reveals client induced conformational control (Ext. Data Fig. 5), consistent with a 16-fold stimulation of Rvb1/2’s ATP hydrolysis activity by Ino80 insertion peptides22. However, the observed post-hydrolysis ADP-state suggests that Rvb1/2 transforms into a more stable functional scaffold once the correct set of clients is assembled. A “latch” in Ino80insertion binds between OB folds 1a and 2b generating a distinct interaction site for Arp5-Ies6 at OBs 2a (Arp5) and 2b (Ies6). Notably, the C-terminal domain of Ies6 (Ies6CTD) resembles a degenerate Zn-HIT fold (that lost the zinc binding cluster), revealing how Zn-HIT domains can specifically bind Rvb1/2 in various complexes26. A “plug” closes the hole in the OB layer and directly binds Ies2, which wedges with a β-hairpin between OBs 2a and 1c (Fig. 1e). Intriguingly, Ies2 reaches all the way across from the Rvb1/2 OB layer via a flexible, but in length conserved linker, and pins the N-lobe to SHL2 (Figs. 1f,2a, Ext. Data Fig. 6 b,d). Ies2 further wraps around the nucleosome and binds the “acidic patch” at the INO80 distal side. Ies2 therefore links Ino80ATPase to Rvb1/2 and the nucleosome, showing why Ies2 acts a “throttle” for INO80’s remodeling activity18.
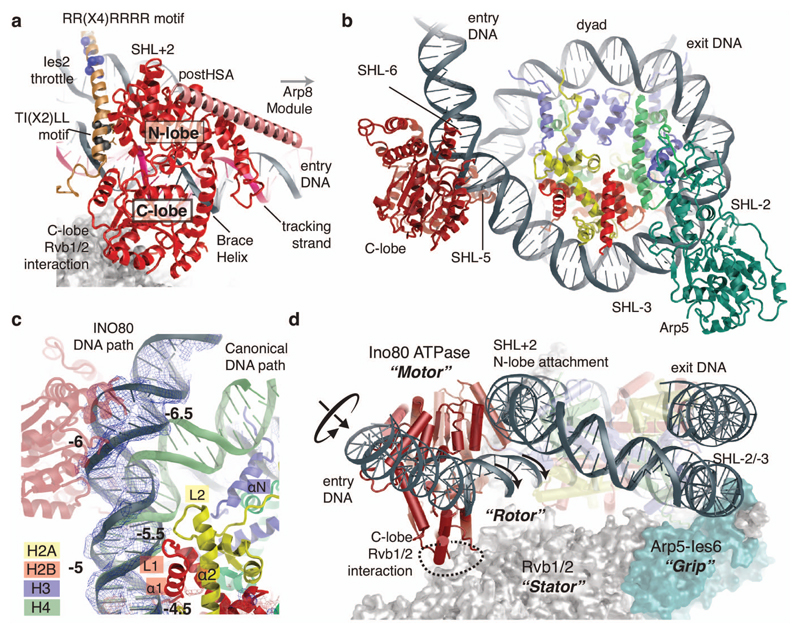
Figure 2. Ino80ATPase – Nucleosome Interaction.
(a) Details of the Ino80ATPase-Ies2 interaction with highlighted tracking-stand and notable conserved Ies2 sequence motifs. The postHSA (salmon) is provided as poly-alanine model (Ext. Data Fig. 6).
(b) The Ino80ATPase and Arp5 bind to opposing sides of the nucleosome, ~90Å apart (Rvb1/2 is not shown, for clarity).
(c) Binding of Ino80ATPase to exit DNA unwraps ~15bp from the nucleosome (blue with superimposed density), partially exposing H2A at SHL-5.5 and disrupting the H3 interaction at SHL-6.5. The canonical DNA path is shown in green for comparison. The unwrapped DNA is kinked through a widened minor groove at the interaction side with Ino80ATPase.
(d) Semi-schematic view showing how the Rvb1/2 hexamer positions the Ino80ATPase motor and Arp5 counter grip on opposing sides of the nucleosome. Rvb1/2 act as “stator” to prevent rotation of the “motor” with respect to the nucleosome, leading to rotation and translation of entry DNA instead.
Ino80ATPase is the motor of the remodeler. Conserved Swi2/Snf2 DNA binding motifs in both the N- and C-lobes engage with dsDNA, while the Swi2/Snf2 typical brace helix I reaches across both lobes, stabilizing their mutual orientation (Fig. 2a). The observed conformation suggests that the motor is poised to bind ATP and to translocate DNA by repetitive cycles of ATP binding and hydrolysis. Intriguingly, binding of Ino80ATPase at SHL-6 already unwraps ~15 bps of DNA from the entry site (Fig. 2b). Furthermore, DNA contacts to H2A L2 (loop 2) at SHL-5.5 and to H3 (helix αN) at SHL-6.5 are notably broken and the H2A/H2B dimer partially exposed. The full exposure of H2A also requires the disruption of H2A’s and H2B’s L1 and α1 DNA contacts, explaining why histone exchange additionally requires ATP driven DNA translocation8. Binding of SHL-6 to the Ino80ATPase goes along with a widening of the DNA minor groove, raising the possibility that INO80’s motor domain is influenced by DNA shape features (Fig. 2c). This could be interesting in determining nucleosome positioning at promoter regions7.
Swi2/Snf2 proteins translocate DNA by minor groove tracking27,28. The orientation of the Swi2/Snf2 motor at SHL-6 suggests that INO80ATPase pumps entry DNA into the nucleosome, consistent with INO80’s nucleosome centering activity (Fig. 2d, Ext. Data Fig. 1). An important, poorly understood feature of remodelers is how such stepwise translocation of the motor on DNA leads to large-scale reconfiguration of the nucleosome. Building up force on DNA in a processive manner through multiple consecutive steps requires arresting the motor with respect to the nucleosome. INO80’s motor is fixed by multiple interactions. Ies2 and a secondary DNA binding site pin the N-lobe to SHL2. Importantly, the C-lobe is held in place by Rvb1/2. Rvb1/2 therefore acts in conjunction with Arp5-Ies6 as a “stator”, enabling Ino80ATPase to apply force onto DNA (“rotor”) and to pump DNA into the nucleosome. This provides the means of conducting large-scale reconfigurations through multiple translocation steps.
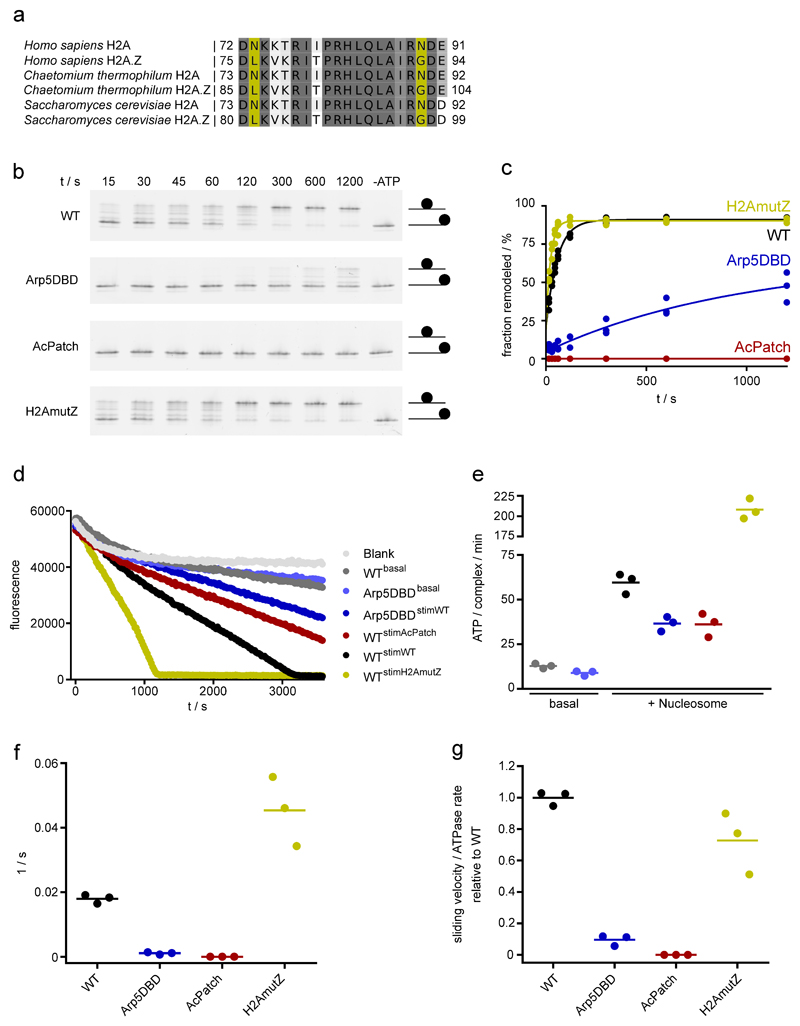
We identify here Arp5-Ies6 as a major nucleosome recognition module with multiple DNA and histone contacts of both Arp5core and Arp5’s 326 residue long insertion domain (Arp5insert) that forms a multi-armed “grappler” (Fig. 3a,b). The C-terminal Zn-HIT fold of Ies6 packs in between Rvb1/2 OB 2b and H2B αC (Fig. 3c,d), while the conserved N-region of Ies6 wraps around Arp5core at the nucleosome proximal DNA side and forms further contacts to Rvb1/2 OB 1b and H2B αC (Fig. 3c,d). Consequently, Arp5-Ies6 binds about 7-8 bases at SHL-2/-3, with both Ies6 and a DNA binding domain (DBD) of Arp5core (Fig. 3c). The DNA interaction explains hydroxyl radical footprints of full S. cerevisiae INO80 on nucleosomes that showed increased protection of SHL-2/-38. Of note, Arp5DBD is conserved from yeast to man and the structural equivalent of the “DNase I binding loop” of actin. Mutating conserved DNA binding arginines/lysines dramatically affected nucleosome sliding under conditions where INO80core still displayed robust ATPase activity (Fig. 3f; Ext. Data Fig. 7), comparable to Arp5 deletion in ScINO80 and hINO80core13,29. Taken together, we conclude that Arp5-Ies6 couples ATPase activity to nucleosome sliding, likely by gripping DNA and anchoring it to the histone octamer surface during ratchet translocation steps (see below).
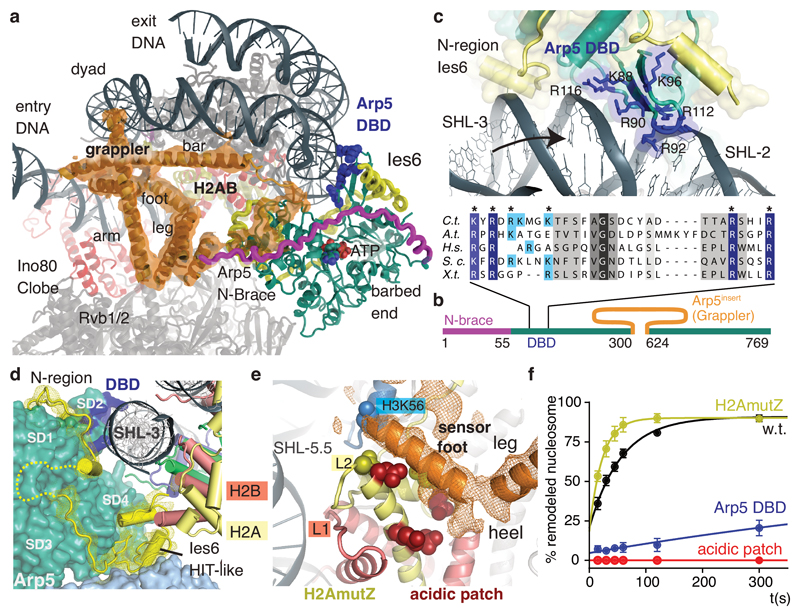
Figure 3. Multivalent nucleosome binding by Arp5.
(a) 4.7Å map showing the Arp5 insertion that forms a multi-armed grappler element (orange), along with the actin fold of Arp5 (green, with blue DNA binding domain (DBD)), the Arp5 N-terminal brace (magenta) and Ies6 (yellow). The grappler has multiple DNA and histone contacts and chemo-mechanically connects Arp5, dyad and H2A/H2B.
(b) Scheme of Arp5 domain structure with green actin-fold core domain and highlighted insertions.
(c) Detailed view of the DNA interactions by Arp5’s DBD and Ies6 along with a multiple sequence alignment showing conservation of DNA binding arginines/lysines in the DBD. Blue: residues mutated for functional analysis (panel e).
(d) The C-terminal HIT-like domain of Ies6 binds both H2A (yellow) and Rvb1 (light blue), while the N-terminal region wraps around Arp5core.
(e) Detailed view of the sensor foot and leg of the grappler (orange map and poly-alanine model). The sensor foot binds to the acidic patch of H2A/H2B and to H3 at K56, suggesting it is implicated in controlling histone variant exchange. Sites mutated (acidic patch: red; H2A.Z mimic: yellow) for the functional analysis in panel (e) are shown with side chains.
(f) Nucleosome sliding activities of INO80core and histone mutants. H2A.Z mimicking mutants (H2AmutZ) lead to increased sliding, while either mutating the H2A acidic patch or Arp5DBD abolishes or strongly reduces sliding under conditions where INO80 still displays robust ATPase. Shown are mean +/- standard deviation (n=3).
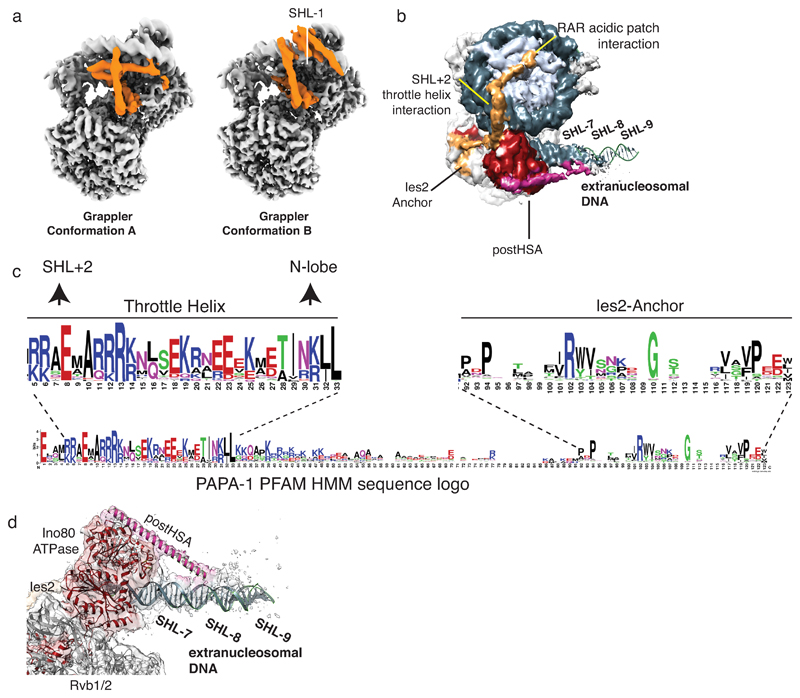
The grappler extends from subdomain 4 of Arp5’s actin fold and has a multi-armed structure with notable elements denoted arm, leg, foot, and bar. Masked 3D classification, leading to 4.7 Å (Fig. 3a) and 4.6Å (Ext. Data Fig. 6) maps, showed that the grappler adopts at least two conformations and allowed us to interpret the topology of its secondary structure with a poly-alanine model. The long N-terminal helix of the Arp5insert forms the “bar” that binds in a closed conformation of the grappler along the nucleosomal dyad and spans between Arp5core and entry DNA at SHL-7.5 over a distance of ~90Å. Importantly, the bar can adopt this binding mode as the entry DNA unwraps from the histone octamer due to binding of the Ino80 ATPase to SHL-6. The “arm” of the grappler stabilizes the bar at the dyad and connects it to the leg-foot element that packs against the H2A/H2B core at the acidic patch of the histone octamer (Fig. 3e). In an open conformation, the bar is released from the dyad, moves 45° to bind to SHL-1 and blocks the path of the exit DNA (Ext. Data Fig. 6). Therefore, we envision a switch-like behavior of Arp5 that is sensitive to the path of the entry and exit DNA.
Intriguingly, the foot backs H2A opposite of L2, as if to stabilize H2A to compensate for the broken DNA contacts due to unwrapping of entry DNA. Consequently, binding of the acidic patch on each side of the nucleosome plays an essential role for INO80: the grappler ensures the integrity of the histone octamer where the entry DNA unwraps, while Ies2 binds the acidic patch on the other side and acts as a throttle for Ino80 ATPase. In support, mutating the acidic patch targeting both interactions abrogates nucleosome sliding, even though it reduces ATPase rates only moderately (Fig. 3f). Of note, our structure predicts for a putative dimeric state of INO8029 competition of Ies2 and Arp5 grappler for the acidic patches on each side of the histone octamer, which might provide asymmetric control of the two Ino80 ATPases at SHL-6 and +6 to prevent simultaneous pumping of DNA in opposing directions.
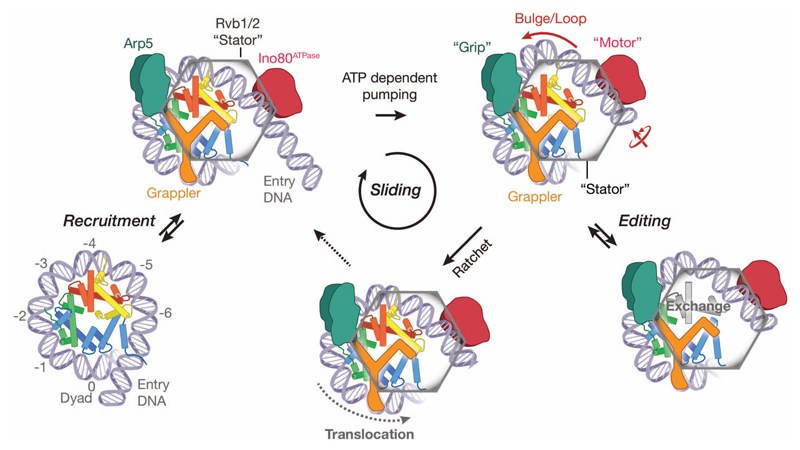
Together with biochemical studies8 our structure suggest a unified ratchet-like mechanism how INO80 slides and possibly edits nucleosomes (Fig. 4). We find that INO80core unwraps entry DNA and grips DNA and histones by multivalent interactions. The motor is positioned to pump DNA into the nucleosome against Arp5-Ies6, which could hold onto DNA until a sufficient force is generated by multiple small steps of the motor. Such groove tracking creates a DNA loop that is limited between the motor and the Arp5-Ies6 counter grip, persistently disrupting the H2A/H2B DNA interface8, and thus enables histone exchange until the pumped amount of DNA propagates across Arp5-Ies6 and grappler (i.e. the ratchet step). As a result, INO80 would move nucleosomes in larger steps (Fig. 4). 10-20bp step sizes are indeed observed8, but the precise stepping kinetics must await further analysis. During loop formation the grappler ensures structural integrity of the octamer by holding onto H2A/H2B, while our structure suggests in addition a function of its foot as a sensor during editing. The foot binds to H2A at a site where H2A differs in some amino acid residues from H2A.Z (Ext. Data Fig. 7). Introducing two H2A.Z mimicking mutations into H2A at this interface indeed increased sliding velocity (Fig. 3f), consistent with the observed faster sliding of H2A.Z nucleosomes by INO808,18. Intriguingly, we observe a direct contact of the “toe” of the sensor-foot with H3K56. Albeit controversial, acetylation of H3K56 has been shown to promote histone variant exchange by INO80 family remodelers30 and plays a pivotal role in DNA repair and replication as well as regulates gene expression homeostasis.
Figure 4. Model for INO80 nucleosome remodeling.
Unified model integrating structural (this work) and biochemical8 data. The functional architecture of INO80 with motor, grip and grappler suggests that processive nucleosome sliding proceeds via a ratchet mechanism. Transient generation of loops between the motor and the grip could expose H2A/H2B for editing. Direct binding of H2A/H2B by the grappler sensor-foot could regulate variant or modification specific editing (see text for details).
In summary, we provide first structural insights into the mechanism by which DNA translocation by a Swi2/Snf2 ATPase of a multisubunit remodeler governs large scale repositioning and editing reactions on nucleosomes. The “motor”, “stator” and multivalent “grip” of INO80 allows highly processive sliding without release and large-scale reconfigurations such as editing while keeping the remainder of the nucleosome intact. The proposed ratchet mechanism explains DNA loop formation resulting in large translocation steps, while providing also the means for ATP dependent H2A.Z -> H2A exchange8. Thus, our structure visualizes how nucleosome sliding and editing can be achieved by two facets of the same mechano-chemical cycle and how differential regulation might occur. Future studies need to address further how other modules that are not part of conserved INO80core function provide an additional layer of regulation e.g. in a promotor specific manner and will reveal how the principles discovered for INO80 apply to other remodeler families.
Methods
INO80 expression and purification
INO80 subunits were cloned and expressed using the MultiBac technology31. Genes coding for Ino80718-1848 - 2 x FLAG, Rvb1, Rvb2, Arp5, Ies2 and Ies6 were cloned in pACEBac1, pIDC and pIDK vectors, respectively and combined in a bacmid. Likewise genes coding for Ies4, Taf14, Arp8, actin and Arp4 were combined in a separate bacmid. Recombination steps were carried out in Escherichia coli XL1-Blue cells (Stratagene) or pirHC cells (Geneva Biotech) under addition of Cre recombinase (NEB). Baculoviruses were generated in Spodoptera frugiperda (SF21) insect cells (IPLB-Sf21AE). Trichoplusia ni High Five cells (Invitrogen) were co-infected with 1/100 of each baculovirus. Cells were cultured for 60 h at 27°C and harvested by centrifugation. For complex purification, cells were disrupted in lysis buffer (30 mM HEPES, pH 7.8, 300 mM NaCl, 10% glycerol, 20 µM ZnCl2, 0.25 mM DTT, 0.28 µg/mL Leupeptin, 1.37 µg/mL Pepstatin A, 0.17 mg/mL PMSF, 0.33 mg/mL Benzamidine) and gently sonified. Raw lysate was cleared by centrifugation at 30500 g and 4°C for 30 min. Supernatant was incubated with 4 mL Anti-FLAG M2 affinity gel (Sigma-Aldrich) for 1 h and washed with 75 mL lysis buffer and 50 mL wash buffer (30 mM HEPES, pH 7.8, 150 mM NaCl, 5% glycerol, 0.5 mM CaCl2, 20 µM ZnCl2, 0.25 mM DTT). The complex was eluted from the matrix by incubation with 8 mL elution buffer (wash buffer supplemented with 0.2 mg/mL FLAG peptide) for 20 min at 4°C. Next, the sample was loaded onto a Mono Q 5/50 GL column (GE Healthcare) and eluted by a gradient of increasing salt, resulting in a highly pure INO80 sample.
RALS measurement
Molecular weight of apo-INO80 was determined by right-angle light scattering (RALS). Size exclusion chromatography coupled light scattering was performed using an Äkta micro chromatography system equipped with a Superose 6 10/300 Increase column (GE Healthcare) and a right-angle laser light scattering device and refractive index detector (Malvern / Viscotek). BSA was used to calibrate the system. Evaluation was performed using the OmniSEC software (Malvern / Viscotek).
Purification of mononucleosomes
Canonical human histones and their mutants were purified by a combination of inclusion body purification and ion exchange chromatography as described previously32,33. In brief, histones were expressed in Escherichia coli BL21 (DE3) cells (Novagen) for 2 h after induction at 37°C. Cells were disrupted under non-denaturing conditions and inclusion bodies were washed with 1% Triton X-100. Inclusion bodies were resuspended in 7 M guanidinium chloride, dialyzed in 8 M urea and histones were purified by cation exchange chromatography. After refolding under low-salt conditions, anion exchange chromatography was performed as a final purification step. Histones were lyophilized for long-time storage. For octamer assembly, single histones were resuspended in 7 M guanidinium chloride, mixed at 1.2 fold excess of H2A and H2B and dialyzed against 2 M NaCl for 16 h. Histone octamers were purified by size exclusion chromatography using a Superdex 200 16/60 column (GE Healthcare) and were stored in 50% glycerol at -20°C. We used the Widom 601 DNA34 with 50 or 80 bp extranucleosomal DNA in the 0NX orientation35 for reconstituting mononucleosomes. DNA was amplified by PCR, purified using anion exchange chromatography and concentrated in vacuum. DNA and histone octamer were mixed at a 1.1 fold excess of DNA at 2 M NaCl and sodium chloride concentration was decreased to 50 mM over 17 h at 4°C. Finally, nucleosomes were purified by anion exchange chromatography, dialyzed to 50 mM NaCl, concentrated to 1 mg/mL and stored at 4°C.
Purification and vitrification of the INO80:0N50 complex
INO80 and 0N50 nucleosomes were mixed in a ratio of 2:1 and dialyzed to binding buffer (20 mM HEPES, pH 8, 60 mM KCl, 0.5% glycerol, 0.25 mM CaCl2, 20 µM ZnCl2, 0.25 mM DTT) for 1 h in slide-a-lyzer dialysis tubes (Thermo Fisher Scientific). The complex was purified by gel filtration using a Superose 6 3.2/300 column (GE Healthcare) and vitrified at a concentration of 1 mg/mL on Quantifoil R2/1 grids in the presence of 0.05% Octyl-β-glucoside using a Leica EM GP (Leica).
Electron microscopy and data collection
FEI Titan Krios transmission electron microscope was operated at 300 kV using a GIF quantum energy filter (slit width 20 eV) and a Gatan K2 summit direct electron detector. Two data sets of images (data set I and II) with a defocus ranging from 1.3 to 3.5 µM were collected at a calibrated pixel size of 1.34Å and 1.06Å and at a dose rate of 5.625 and 5.96 e-/Å2s, respectively. A total dose of 67.5 and 59.6 e-/Å2 was recorded during 12 and 10 seconds with a frame rate of 5 and 4 frames stored per second of dataset I and II, respectively. Data acquisition was carried out using SerialEM36 facilitated by a set of customized scripts allowing automated execution of low dose image acquisition including focus and drift determination as well as beam centering (Strauss et al., under development).
cryoEM data processing
Dose-fractionated image stacks were subjected to beam induced motion correction using MotionCor237. The first and the last frame were discarded and CTF parameters for each sum of remaining frames determined by CTFFIND438. Micrographs that exhibited too much drift, too much contamination or abnormal Fourier patterns were discarded. 1282 image stacks (dataset I at 1.34Å/pixel) and 3932 image stacks (dataset II at 1.06Å/pixel) were chosen for further processing, carried out by using MotionCor237 corrected sums that were filtered according to exposure dose. Particle selection, 2D classification, 3D classification as well as refinement was performed using RELION39 version 2.1.1b, unless stated otherwise. All resolutions reported below were determined by gold standard Fourier shell correlation 0.143 criterion. B-factors were automatically determined within RELION according to Reference40. Extended Data Figures 2a and 2b show an overview of the cryoEM processing scheme of dataset I and II. 2D class averages (Ext. Data Fig. 2d), used as 35Å low-pass filtered templates for the initial automated particle picking of dataset I, were calculated from 800 particles that were manually picked from a screening dataset acquired using a FEI Falcon II camera and a FEI Titan Halo transmission electron microscope at 300 kV. 6k semi-automatically picked particles from the same dataset were used to generate an 3D ab initio reconstruction in CryoSPARC41 (Ext. Data Fig. 2c), which served as a 40Å low-pass filtered reference for the first round of 3D classification in RELION. 2D and 3D classification (3DA1 and 3DA2, Ext. Data Fig. 2a) identified 18k particles corresponding to nucleosome bound INO80core complexes from 295k automatically picked particles. Since we refrained from crosslinking to stabilize complexes during sample and grid preparation, we observed at vitrified conditions a large number of disassembled complexes corresponding to free nucleosomes (class 1 of 3D-A2, Ext. Data Fig. 2a) or apo-INO80core complex (class 3 of 3D-A2, Ext. Data Fig. 2a). Severe orientational bias of particles in this dataset prevented meaningful refinement of the apo-INO80core complex beyond 8Å. In contrast, however, the identified set of 18k particles of nucleosome bound INO80core subjected to RELION refinement and subsequent solvent mask post-processing (B-factor of -100 for map sharpening) yielded a cryoEM map of the nucleosome complex at an overall resolution of 5.8Å. This map was used as a reference in order to determine a higher resolution structure using the larger dataset II recorded at higher magnification (1.06Å/pixel). In order to improve auto-picking of sparsely populated orientations of the complex we calculated 2D projections of the experimentally determined 5.8Å cryoEM map (Ext. Data Fig. 2e). To avoid false positives during particle picking we applied a 35Å low-pass filter to the projections before using them as templates and verified the quality of the automated picking procedure by visual inspection of the micrographs as well as by diagnostic 2D classifications in RELION. 252k particles derived from automated particle picking were subjected to successive rounds of 3D classification (3DB1, 3DB2 and 3DB3, Ext. Data Fig. 2b). Notably, an intermediate set of 144k particles yielded a cryoEM map of the INO80core complex at 3.9Å. Although inspection indicated still conformational or compositional heterogeneity within the region of the nucleosome, the particle density and signal to noise was sufficiently high to allow movie processing and particle polishing within Relion (using frames 1-30, running averages of 8 frames and a standard deviation of particles of 300Å). Subsequent refinement of the “polished” particles yielded a 3.7Å map that allowed de novo atomic model building and real space refinement of Ies2, Ies6, Arp5, Ino80insert and Arp5 (see below). 3D classification (3D-B2) yielded a class of 34k nucleosome bound particles. These particles were subjected to RELION refinement and solvent mask post-processing, yielding a cryoEM map of the complex at an overall resolution of 4.3Å (Ext. Data Fig. 2f). Finally, two classes showing different conformations of the “grappler” element were obtained by using a third 3D classification (3D-B3, Ext. Data Fig. 2b) in which the Euler angles derived from the previous refinement were kept fixed and a mask of the respective region of the complex was applied. Local resolution estimation and local resolution filtering was performed as implemented in RELION 2.1.1b.
Model building and refinement
In a first stage we performed rigid body docking in UCSF Chimera42 using available crystal structures of Xenopus laevis nucleosome with 601 Widom sequence (PDB 4R8P), crystal structures of Chaetomium thermophilum Rvb1 and Rvb2 (PDB 4WW4 and 4FM6) and homology models of Chaetomium thermophilum Arp5core residues 59-755 excluding insert residues 306-640 as well as Chaetomium thermophilum Ino80ATPase residues 964-1705 excluding insert residues 1274-1548. Homology models were build using SWISS-MODEL43. Available crystal structures of nucleosome-bound Snf2ATPase (pdb 5X0X) as well as a bound ATP-bound actin (PDB 1NWK) served as templates. Atomic model building of Ino80 insert (residues 1278-1544), Ies2 (residues 443-478), Ies6 (residues 10-52, 155-213) and Arp5 (15-107, 111-146, 153-300, 603-769) was performed using the 3.7Å map of INO80core and a combination of COOT44 and Moloc45. Model building and refinement was performed iteratively using restrained real space refinement in PHENIX 1.12. We used restraints for secondary structure, side chain rotamers, Ramachandran and Cβ restraints, while we restricted the resolution to 3.7 Å during refinement. At the final macrocycle grouped B-factor refinement for main chain and side chain was calculated. Statistics of the final refinement and the obtained structures are reported in Ext. Data Table 1. The obtained structures were subsequently used for interpretation of 4.3Å cryoEM map of the INO80core Nucleosome complex. To model regions with larger conformational deviations such as the nucleosomal DNA, the INO80 ATPase and regions at nucleosome interface of INO80core we employed a combination of flexible fitting and (re)building using a combination of Moloc45 and MDFF46. This procedure resulted in good fits of backbone atoms into the cryoEM map (Ext. Data Fig. 3c). The properties and limitations of the molecular models of the INO80core:NCP complex are briefly summarized in the following. Flexible fitting of nucleosomal DNA accounts for the large conformational change seen in the region between SHL-5.5 and SHL-7. Although additional unambiguous density corresponding to extranucleosomal DNA protrudes from the INO80core:NCP complex we did not attempt to build DNA beyond SHL-7 at this stage. The histone core required only minor adjustments. However, we do not observe density for H3 tail residues 37-44 at their canonical binding site above SHL 1. Interestingly, we observed instead unassigned density between the foot element of the grappler and the N-terminal H3 helix αN. Since this density can be also originate from grappler element of Arp5 (see below), we refrained from building the H3 tail at this stage. Ino80 residues 964-1274 and 1549-1705 were flexibly fitted into the density using MDFF and readily connected to the refined model of the insert region described above. The topology of the grappler element was unambiguously assigned to the Arp5 insert residues 306-640. However, model building was largely restricted to a poly-alanine model given the limited resolution of this element in the 4.6Å and 4.7Å subclasses (Ext. Data Fig. 3e). Similarly, we were able to build a poly-alanine model of Ino80 post HSA residues 820-855 and Ies2 residues 351-443 that includes the throttle helix bound to nucleosomal DNA (Ext. Data Fig. 6).
Electrophoretic-mobility shift assays
Electrophoretic mobility shift assays (EMSA) were used to monitor the interaction between INO80 and 0N50 mononucleosomes. Nucleosomes were labeled at the 5’-end of their extranucleosomal DNA with Fluorescein. 15 nM nucleosome was incubated with increasing concentrations of INO80 (0, 5, 10, 20, 40 nM) in EMSA buffer (25 mM HEPES, pH 8, 60 mM KCl, 7% glycerol, 0.25 mM DTT, 2 mM CaCl2) for 20 minutes on ice. Samples were analyzed at 4°C by native PAGE on a 3-12% acrylamide BIS-Tris gel (Invitrogen) and visualized using the Typhoon imaging system (GE healthcare).
Nucleosome sliding assays
0N80 mononucleosomes with 5'-Fluorescein-labeled extranucleosomal DNA were used for monitoring the sliding activity of INO80. 150 nM nucleosome was incubated with 50 nM INO80 in sliding buffer (25 mM HEPES, pH 8, 60 mM KCl, 7% glycerol, 0.10 mg/mL BSA, 0.25 mM DTT, 2 mM MgCl2) at 25°C. The reaction was started upon addition of 1 mM ATP and stopped at several time points (15, 30, 45, 60, 120, 300, 500, 1200 s) by addition of 0.2 mg/mL Lambda DNA (NEB). Nucleosome species were separated by native PAGE on a 3-12% acrylamide BIS-Tris gel (Invitrogen) and visualized using the Typhoon imaging system (GE healthcare). ImageJ was used to quantify gel bands and the fraction of remodeled band was plotted against the reaction time. Data describes a saturation curve and was fitted in Prism (GraphPad) using an exponential equation.
ATPase assays
A NADH-coupled based ATPase assay was used to determine the ATPase rate of INO80. 30 nM INO80 was incubated in assay buffer (25 mM HEPES, pH8, 50 mM KCl, 1 mM DTT, 2 mM MgCl2, 0.1 mg/mL BSA) with 0.5 mM phosphoenolpyruvate, 1 mM ATP, 0.1 mM NADH and 25 U/mL lactate dehydrogenase and pyruvate kinase (Sigma) at 25°C in a final volume of 50 µL. NADH concentration was monitored fluorescently over 1 h in non-binding, black, 384-well plates (Greiner Bio-One) using 340 nm for excitation and an emission of 460 nm with a Tecan Infinite M100 (Tecan). Where indicated, ATPase activity was determined in the presence of 150 nM nucleosome. ATP turnover was calculated using maximal initial linear rates, corrected for a buffer blank.
Figure Preparation
Figures were prepared with PyMol (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.), UCSF Chimera42 and UCSF ChimeraX47.
Extended Data
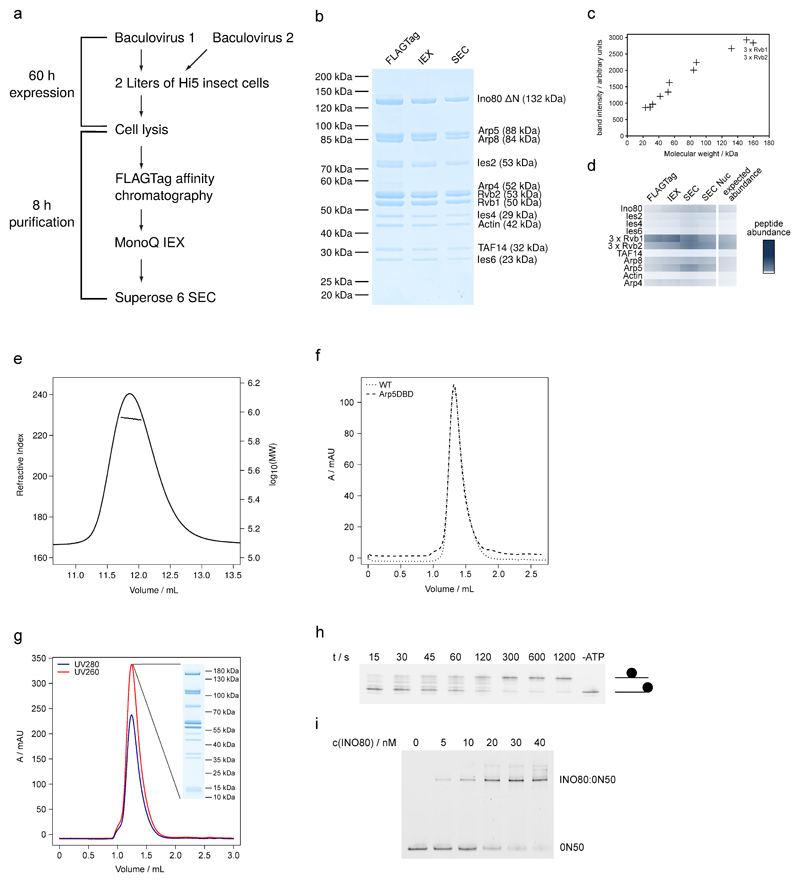
Extended Data Figure 1. Purification of apo-INO80, INO80:0N50 and sliding activity of INO80.
(a) Scheme of expression and purification of INO80.
(b) Documentation of INO80 purification by SDS PAGE (stained with simply blue). Protein identity was confirmed by mass spectrometry (data not shown).
(c) Quantification of band intensity from SDS PAGE (SEC sample) plotted against the molecular weight show stoichiometric presence of all subunits.
(d) Label free semi-quantitative mass spectrometry analysis of INO80core complexes after individual purification steps.
(e) RALS measurement of apo-INO80. Measured refractive index and calculated logarithmical molecular weight are plotted against the elution volume. The measurement yields a molecular weight of 880 kDa, confirming the integrity and correct stoichiometry of the purified complex.
(f) Comparison of the SEC elution profile of apo-INO80 and the Arp5DBD mutant on a Superose 6 3.2/300.
(g) Purification of the INO80:nucleosome complex. SEC elution profile of a Superose 6 3.2/300 is shown together with an analysis of the main peak fraction by SDS PAGE.
(h) Sliding of end-positioned 0N80 mononucleosomes by INO80. Native PAGE analysis of Fluorescein-labeled nucleosome is shown.
(i) Interaction of INO80 and mononucleosome monitored by electrophoretic-mobility shift assay.
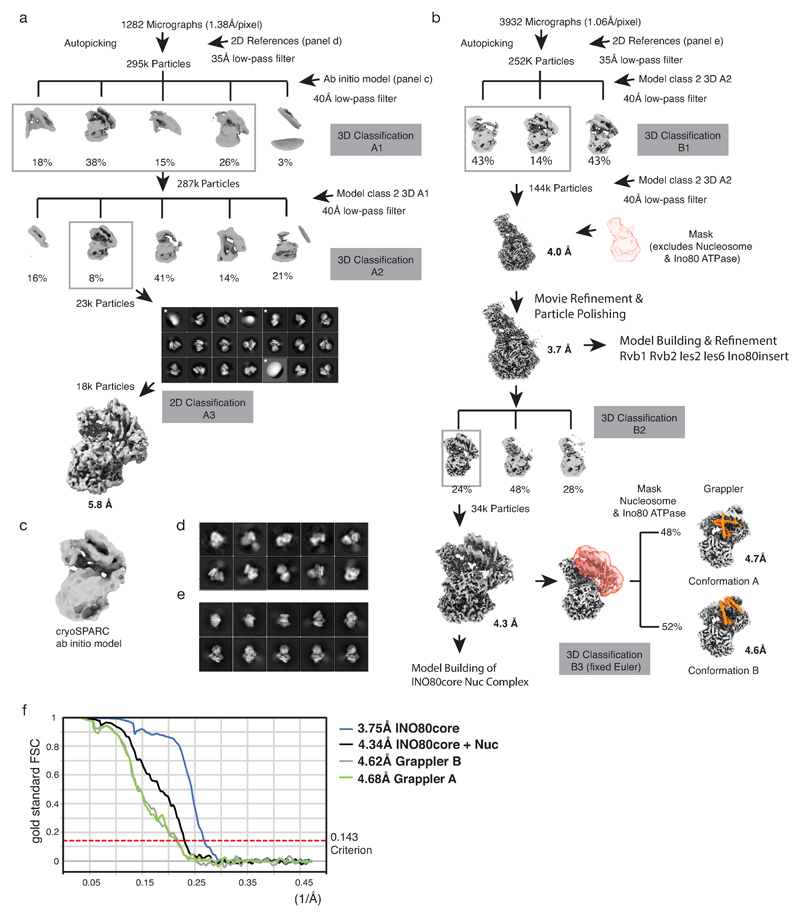
Extended Data Figure 2. Cryo EM data analysis.
(a) and (b). Panel (a) and (b) show the schemes of RELION39 classifications and refinements that were used to obtain cryoEM reconstructions of the INO80core:NCP complex. Panel (a) shows the outline of an initial classification scheme that used as a reference an cryoSPARC41 ab initio 3D reconstruction of the complex. Panel (b) describes the classification scheme that yielded the final cryoEM reconstructions. In both (a) and (b), boxed 3D classes were selected for further processing as indicated. 2D classes discarded for further processing are marked with an asterisk.
(c) ab initio 3D reconstruction by cryoSPARC
(d) Automated particle picking used as templates coarse 2D class averages from a small set of manually picked particles (see methods section). Shown are initial 2D class averages obtained from 800 manually picked particles from a screening dataset that were used to pick 6K particles semi-automatically for ab initio 3D reconstruction.
(e) projections of the experimentally determined 5.8Å cryoEM reconstructions obtain from the scheme in panel (a). These projections were low pass filtered to 35Å and used then as templates to improve automated picking of particles corresponding to sparsely populated orientations of the complex (see methods section). The quality of the automated particle picking was verified by visual inspection of micrographs as well as by diagnostic 2D classifications (not shown). Later 3D classifications in scheme (b) were facilitated by masks and fixed Euler angles from previous refinements as indicated (3D-B3).
(f) Gold standard Fourier shell correlation (FSC) curves of final maps (3.75, 4.34Å, 4.62Å and 4.68Å). The resolutions were determined using the 0.143 FSC criterion as indicated by the dotted line. Ext. Data Table 1 summarizes data collection and processing.
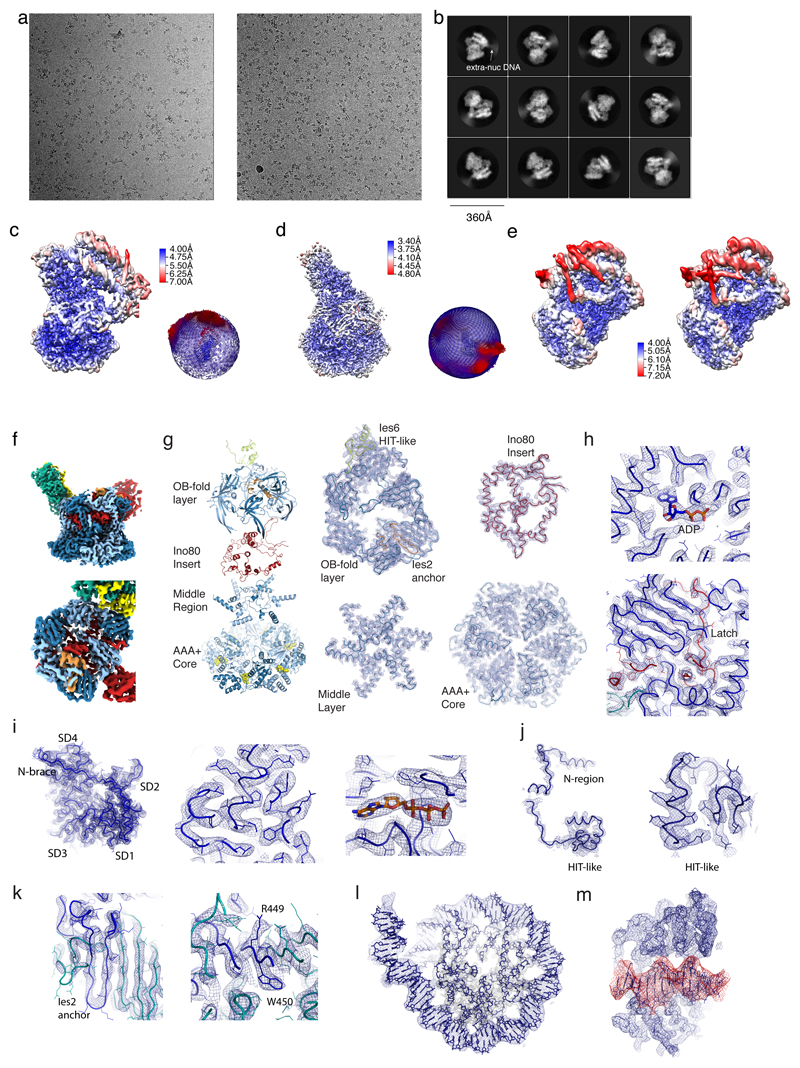
Extended Data Figure 3. Cryo EM data quality.
(a) Two representative micrographs of the set that was used to determine the structure of the INO80core:NCP complex.
(b) Typical 2D class averages of the INO80core:NCP complex. Note that dynamic extra-nucleosomal DNA (denoted as extra nuc DNA) visibly protrudes from the well-ordered core complex.
(c)-(e) The final 4.3Å (c, overall), 3.7Å (d, Rvb1/2-Arp5 mask) and 4.6Å and 4.7 Å (e, grappler conformations B and A) maps were analyzed by using ResMap48. Local resolution estimates are shown as a color-coded surface representation along with representations of angular distributions of particles contributing to the 4.3Å and 3.7Å maps.
(f)-(m) Representative examples of cryoEM map areas used for model building.
(f) The 3.7Å map using the color codes of Fig. 1c show the definition of Rvb1/2-client interactions.
(g) “Explosion” figure of the Rvb1/2 layers, along with corresponding regions of the 3.7Å map.
(h) Top: details showing a representative ATP/ADP binding site of Rvb1/2, with highlighted ADP, and showing the “latch” of the Ino80insert (red).
(i) Map at Arp5core, showing the N-terminal brace (left), with representative details of the actin core (middle) and the ATP binding site (right).
(j) Overview showing Ies6 (left) and details of its HIT-like domain (right).
(k) Map at the Ies2-Rvb1/2 interaction (left) with details showing an anchoring tryptophane.
(l) Map area at the NCP
(m) Map area at the Ino80 motor domain bound to SHL-6 (red).
Extended Data Figure 4. Comparison of nucleosome bound Swi2/Snf2-type ATPases.
(a) Interaction of Ino80ATPase (left, our study), Snf2ATPase bound to SHL6 (middle), Snf2ATPase bound to SHL2 and Chd1ATPase bound SHL2 (right) with nucleosome core particles (NCPs).
(b) Comparison of domain architectures reveals comparable arrangements of N- and C-lobes.
Extended Data Figure 5. Details of Rvb1/2-Ino80insert interactions.
(a) Close-up views of Rvb1 client cavities (blue), bound to the different interaction elements of Ino80insert (red, with yellow hydrophobic and green aromatic side chains).
(b) Same as (a), but depicting Rvb2 client cavities
(c) Ino80insert shown in rainbow coloring (red: N-terminus, blue C-terminus) to highlight the circular fold. Selected elements as well as the positions of the Rvb1/2 binding partners are annotated.
(d) Same as (c) but viewed from the side to highlight the protruding “plug” and “latch” elements
(e) Rvb1/2 pair (pair “c” from the hexamer in Fig. 1c) bound to Ino80insert in comparison with
(f) a Rvb1/2 pair from the unliganded dodecameric state (PDB: 4wvy). The comparison shows how client binding arranges the AAA+, OB and middle layers and displaces the N-terminal domain of Rvb1 from the client pocket, seen also for hINO80core 23. Both types of conformational changes impact on the ADP binding site (ADP, ATP: color coded spheres), suggesting how client interactions are allosterically coupled to ATPase activity of Rvb1/2.
(g) Exemplary view of the ADP coordination along with the superimposed map.
Extended Data Figure 6. Two conformations of the Grappler element and location of the post HSA domain.
Masked 3D classifications identified two conformations of the grappler element of INO80core and the postHSA domain of the INO80ATPase.
(a) Left: grappler conformation A as discussed in the main text. Right: “open” conformation B in which the bar interacts with SHL-1 of the nucleosome.
(b) Subclass showing the postHSA domain (magenta) at the INO80ATPase (red). postHSA protrudes towards extranucleosomal DNA. Ies2 is depicted in orange.
(c) Detailed view of the map around postHSA and extranucleosomal DNA with superimposed models.
(d) Hidden Markov Model (HMM) sequence logo of Ies2, showing high sequence conservation at key Ino80 and Rvb1/2 interaction sites.
Extended Data Figure 7. Analysis of the enzymatic activity of INO80.
(a): Sequence alignment of H2A and H2A.Z. Yellow: residues at the interface of H2A with the foot of the grappler differ in a species conserved fashion.
(b): Sliding of 0N80 mononucleosomes by INO80 analyzed by native PAGE. In the Arp5DBD mutant K88, R90, R92, K96, R112, R118 are mutated to alanines. AcPatch (E61A, E64A, D72A, D90A) and H2AmutZ (N73L, N89G) describe mutants of grappler-contacting residues of H2A (also see Fig. 3). Individual data points with exponential fit (n=3, technical replicates).
(c): Evaluation of INO80’s sliding activity. Band intensities of remodeled and unremodeled nucleosome species were quantified and the fraction of remodeled nucleosome plotted against time. Data points were fitted using an exponential equation.
(d): Raw data of ATPase assays (color code of panel c). Basal ATPase rates were determined for INO80 WT and the Arp5DBD mutant along with nucleosome-stimulated rates. Superscripted text indicates if and what type of nucleosome was used to stimulate ATPase activity.
(e): ATPase rates of INO80 with and without stimulation by nucleosomes. Rates were calculated from the linear area of the raw data and were corrected for a buffer blank (color code of panel c). Mean and individual data points (n=3, technical replicates).
(f): Initial sliding rates of INO80 and mutants (color code of panel c). Data were derived from exponential fits of individual sliding curves (panel c). Mean and individual data points (n=3, technical replicates).
(g): Quotient of the sliding rate (f) and ATPase rate (e) normalized to the WT. Mean and individual data points (n=3, technical replicates).
Extended Data Table 1. Cryo-EM data collection, refinement and validation statistics.
| #1 INO80core (EMD-4264) (PDB 6FHS) | #2 INO80core+nuc (EMD-4277) (PDB 6FML) | #3 INO80 Grappler A (EMD-4278) | #4 INO80 Grappler B (EMD-4280) | |
|---|---|---|---|---|
| Data collection and processing | ||||
| Camera | Gatan K2 | Gatan K2 | Gatan K2 | Gatan K2 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 59.6 | 59.6 | 59.6 | 59.6 |
| Defocus range (µm) | 1.3-3.5 | 1.3-3.5 | 1.3-3.5 | 1.3-3.5 |
| Pixel size (Å) | 1.06 | 1.06 | 1.06 | 1.06 |
| Symmetry imposed | Cl | Cl | Cl | Cl |
| Initial particle images (no.) | 251692 | 251692 | 251692 | 251692 |
| Final particle images (no.) | 144278 | 33937 | 16287 | 17263 |
| Map resolution (Å) | 3.75 | 4.34 | 4.68 | 4.62 |
| 0.143 FSC threshold | ||||
| Map sharpening B-factor (Å2) | -142 | -114 | -124 | -136 |
| Refinement | ||||
| Initial model used (PDB code) | models for refinement were build de novo or based on 4WW4 (Rvbl/2) and lNWK (actin) | models for refinement were build on 6FHS (INO80core) and 4r8p (nucleosome) | ||
| Model resolution (Å) | 3.82 | 4.47 | ||
| 0.5 FSC threshold | ||||
| Model resolution range (Å) | 360.4 - 3.7 | 360.4 – 4.3 | ||
| Map sharpening B factor (Å2) | -142 | -114 | ||
| Model composition | ||||
| Non-hydrogen atoms | 27184 | 43462 | ||
| Protein and DNA residues | 3481 | 5114 | ||
| Ligands | 7 | 7 | ||
| B factors (Å2) | ||||
| Protein and DNA | 59.6 | 112.9 | ||
| Ligand | 54.3 | 80.29 | ||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.008 | 0.010 | ||
| Bond angles (°) | 0.93 | 1.203 | ||
| Validation | ||||
| MolProbity score | 1.7 | 1.68 | ||
| Clashscore | 4.8 | 4.75 | ||
| Poor rotamers (%) | 0.1 | 0.13 | ||
| Ramachandran plot | ||||
| Favored (%) | 92.8 | 93.3 | ||
| Allowed (%) | 7.2 | 6.6 | ||
| Disallowed (%) | 0.0 | 0.1 | ||
Supplementary Material
Acknowledgements
We thank Philipp Korber and Elisa Oberbeckmann for help with biochemical analysis of INO80, Ed Hurt for help with Chaetomium thermophilum genome annotations, Otto Berninghausen for help with data collection, and Katja Lammens, Kilian Knoll, Rob Byrne, Gabriele Stöhr and Gregor Witte for discussions and technical help. We thank the LMU Protein Analysis Unit and the MPI of Biochemistry cryo-EM and biophysics core facilities for generous access and support, the SuperMUC for generous computing time. K.-P.H is supported by the Deutsche Forschungsgemeinschaft (CRC1064, RTG1721), the European Research Council (ERC Advanced Grant ATMMACHINE), the Gottfried-Wilhelm-Leibniz Prize and the Center for Integrated Protein Sciences Munich (CIPSM). S.E. acknowledges an EMBO long term fellowship, K.S. acknowledges funding by Quantitative Biosciences Munich (QBM).
Footnotes
Data availability
The electron density reconstruction and final model were deposited with the EM Data Base (accession codes EMD-4264, EMD-4277, EMD-4278, EMD-4280) and the PDB (accession codes 6HFS, 6FML). Uncropped images of the polyacrylamide gels are shown in the supplement.
Author Contributions
S.E. prepared cryoEM samples and performed structure determination. K.S. prepared nucleosomes, performed the biochemical analysis and participated in structure determination. S.E., D.K. and K.-P.H. build atomic models. K.L. characterized INO80 subcomplexes and participated in early stages of the EM analysis. M.S. operates the MPI Biochemistry cryo-EM facility, helped with EM data collection and provided general EM advice. M.M. helped with recombinant protein production. S.E. and K.-P.H. designed the overall study, analyzed the results and wrote the paper with contributions from all other authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 5.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udugama M, Sabri A, Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krietenstein N, et al. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell. 2016;167:709–721 e712. doi: 10.1016/j.cell.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahma S, et al. INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun. 2017;8:15616. doi: 10.1038/ncomms15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Li M, Xia X, Li X, Chen Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature. 2017;544:440–445. doi: 10.1038/nature22036. [DOI] [PubMed] [Google Scholar]
- 11.Sundaramoorthy R, et al. Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes. Elife. 2017;6 doi: 10.7554/eLife.22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnung L, Vos SM, Wigge C, Cramer P. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature. 2017;550:539–542. doi: 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tosi A, et al. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–1219. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, et al. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J Biol Chem. 2011;286:11283–11289. doi: 10.1074/jbc.M111.222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen VQ, et al. Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell. 2013;154:1220–1231. doi: 10.1016/j.cell.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 17.Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 18.Willhoft O, Bythell-Douglas R, McCormack EA, Wigley DB. Synergy and antagonism in regulation of recombinant human INO80 chromatin remodeling complex. Nucleic Acids Res. 2016;44:8179–8188. doi: 10.1093/nar/gkw509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Conaway RC, Conaway JW. Multiple modes of regulation of the human Ino80 SNF2 ATPase by subunits of the INO80 chromatin-remodeling complex. Proc Natl Acad Sci U S A. 2013;110:20497–20502. doi: 10.1073/pnas.1317092110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013;154:1246–1256. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangaraju VK, Bartholomew B. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol Cell Biol. 2007;27:3217–3225. doi: 10.1128/MCB.01731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou CY, et al. Regulation of Rvb1/Rvb2 by a Domain within the INO80 Chromatin Remodeling Complex Implicates the Yeast Rvbs as Protein Assembly Chaperones. Cell Rep. 2017;19:2033–2044. doi: 10.1016/j.celrep.2017.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aramayo RJ, et al. Cryo-EM structures of the human INO80 chromatin-remodeling complex. Nat Struct Mol Biol. 2017 doi: 10.1038/s41594-017-0003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koldewey P, Horowitz S, Bardwell JCA. Chaperone-client interactions: Non-specificity engenders multifunctionality. J Biol Chem. 2017;292:12010–12017. doi: 10.1074/jbc.R117.796862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakomek K, Stoehr G, Tosi A, Schmailzl M, Hopfner KP. Structural basis for dodecameric assembly states and conformational plasticity of the full-length AAA+ ATPases Rvb1 . Rvb2. Structure. 2015;23:483–495. doi: 10.1016/j.str.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Rivera-Calzada A, et al. The Structure of the R2TP Complex Defines a Platform for Recruiting Diverse Client Proteins to the HSP90 Molecular Chaperone System. Structure. 2017;25:1145–1152 e1144. doi: 10.1016/j.str.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 29.Willhoft O, et al. Crosstalk within a functional INO80 complex dimer regulates nucleosome sliding. Elife. 2017;6 doi: 10.7554/eLife.25782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013;340:195–199. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 32.Klinker H, Haas C, Harrer N, Becker PB, Mueller-Planitz F. Rapid purification of recombinant histones. PLoS One. 2014;9:e104029. doi: 10.1371/journal.pone.0104029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 34.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 35.Levendosky RF, Sabantsev A, Deindl S, Bowman GD. The Chd1 chromatin remodeler shifts hexasomes unidirectionally. Elife. 2016;5 doi: 10.7554/eLife.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Gerber PR, Muller K. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J Comput Aided Mol Des. 1995;9:251–268. doi: 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
- 46.Qi Y, et al. CHARMM-GUI MDFF/xMDFF Utilizer for Molecular Dynamics Flexible Fitting Simulations in Various Environments. J Phys Chem B. 2017;121:3718–3723. doi: 10.1021/acs.jpcb.6b10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2017 doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.