 18α-GAMG exhibited strong anti-inflammatory activity through inhibiting the expression of iNOS, COX-2, and MAPKs, as well as activation of NF-κB.
18α-GAMG exhibited strong anti-inflammatory activity through inhibiting the expression of iNOS, COX-2, and MAPKs, as well as activation of NF-κB.
Abstract
Based on the SAR analysis of glycyrrhizin, 18α-glycyrrhetinic acid monoglucuronide (18α-GAMG) with strong inhibition against LPS-induced NO and IL-6 production in RAW264.7 cells was discovered. Western blotting and immunofluorescence results showed that 18α-GAMG reduced the expression of iNOS, COX-2, and MAPKs, as well as activation of NF-κB in the LPS-stimulated RAW264.7 cells. Further in vivo results showed that 18α-GAMG could significantly improve the pathological changes of CCl4-induced hepatic fibrosis.
Introduction
Licorice, the roots and rhizomes of the Glycyrrhiza species, is a natural sweetener and used as a traditional herbal medicine for the treatment of inflammation, gastric ulcers, liver disease, adrenal insufficiency, and tumours.1–3 Glycyrrhizin (glycyrrhizic acid, 18β-GA) is the major bioactive component in licorice with diverse pharmacological activities.4–7 Researches showed that 18β-GA can promote the maturation of murine dendritic cells (DCs) and can regulate interleukin (IL)-2, IL-10, IL-12, tumor necrosis factor-α (TNF-α), inducible nitric oxide synthase (iNOS), and cyclooxygenase (COX-2);8–11 additionally, it can down-regulate the production of IL-8 and eotaxin-1 in human lung fibroblast cells.12 Moreover, glycyrrhizin could prevent enteritis by reducing the nuclear factor-κB (NF-κB) p65 and p38 mitogen-activated protein kinase (p38MAPK) expression in rat, attenuated neuroinflammation and oxidative stress in the rotenone model of Parkinson's disease, and inflammatory response of isoflurane-induced cognitive deficits in neonatal rats.13–15 The anti-inflammatory effect of 18β-GA may be due to direct binding to high-mobility group box 1 protein, thus inhibiting its chemoattractant and mitogenic activities.16,17 These results indicated that 18β-GA could be used as immune modulators, which precisely regulate cellular immunity.
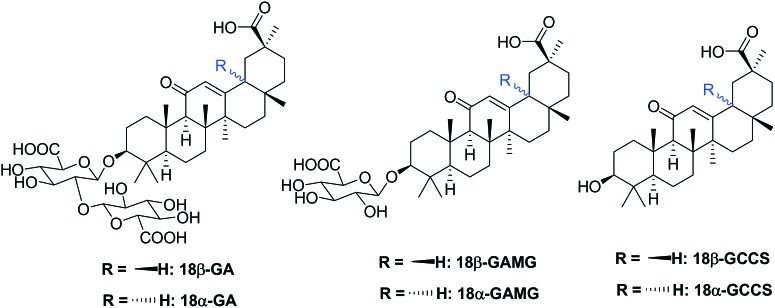
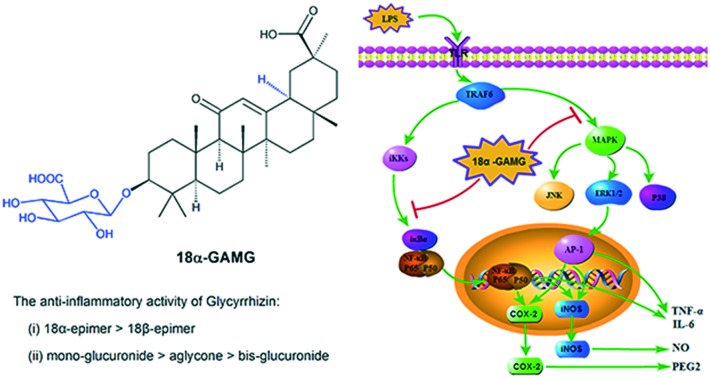
As a natural triterpene glycoside, 18β-GA contains one molecule of 18β-H-oleanane-type aglycone and two molecules of glucuronic acid (Fig. 1). 18β-Glycyrrhetinic acid (18β-GCCS), the aglycone of 18β-GA, is the active metabolite produced by the intestinal bacteria after the oral administration of 18β-GA.18 18β-GA and 18β-GCCS exhibited similar anti-inflammatory effects by inhibiting the production of LPS-induced nitric oxide (NO), prostaglandin E2 (PGE2), TNF-α, IL-6, IL-1β, and intracellular reactive oxygen species (ROS), reducing the expression of pro-inflammatory genes (iNOS and COX-2), and significantly blocking activation of transcription factors such as NF-κB and PI3K.19,20 18β-GA could be metabolized in the liver or be transformed via enzymolysis to 18β-glycyrrhetinic acid mono-glucuronide (18β-GAMG).21–23 18β-GAMG showed (or stronger) antitumor, antivirus, and anti-inflammatory activities similar to those of 18β-GA.24–27 18α-Glycyrrhizin (18α-GA), a D/E-trans-stereoisomer, exhibited anti-inflammatory and anticancer activities by inhibiting the activation of key arachidonic acid (AA) metabolism enzymes including phospholipase A2 (PLA2), cyclooxygenase (COXs), and 5-lipoxygenase (LOX) and release of AA pathway-generated inflammatory lipid mediators.28–30 Therefore, all glycyrrhizin analogues (Fig. 1, including metabolite and isomer) exhibited anti-inflammatory activities, which prompted us to analyze the structure–activity relationships (SAR) of 18β-GA.
Fig. 1. Chemical structures of the glycyrrhizin analogs.
In this study, we synthesized glycyrrhizin analogs with different glucuronide units and/or 18-α/β-stereoisomer to investigate the SAR and pharmacological mechanism of glycyrrhizin as an anti-inflammatory agent. Then, their anti-inflammatory activities in vitro and in vivo were further evaluated.
Results and discussion
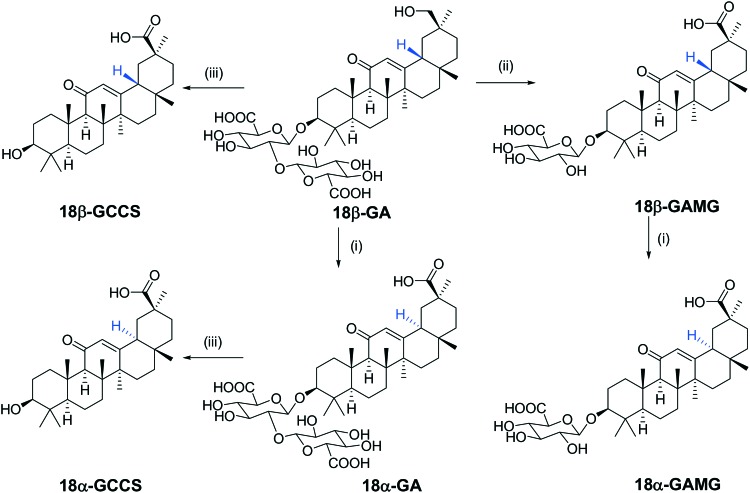
Glycyrrhizin analogs were synthesized and characterized via NMR and ESI-MS, and their detailed synthetic and structural information are provided in the ESI‡ (Scheme 1).
Scheme 1. Synthesis of the glycyrrhizin analogs. Reagents and conditions: (i) NaOH solution (5.0 M), 90 °C, and 12 h. (ii) β-Glucuronidase; and (iii) AcOH, 5 N HCl, and 100 °C.
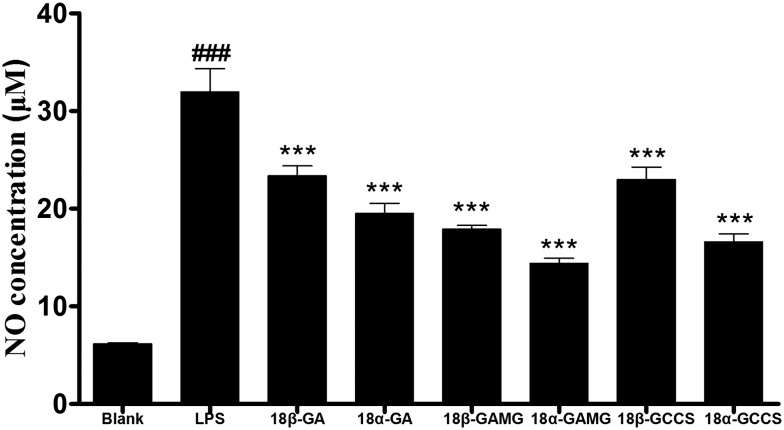
To evaluate the anti-inflammatory effect of the glycyrrhizin analogs, Griess reagent was used to detect the level of the LPS-induced NO release in the RAW264.7 cells. Excessive release of NO is regarded as an important factor in inflammatory responses.31 As shown in Fig. 2, after treatment with glycyrrhizin analogs, the increase in the LPS-induced NO release was significantly alleviated in the RAW264.7 cells. SAR analysis showed that (i) the anti-inflammatory activity of 18α-epimer of the oleanane-type aglycone was superior to that of 18β-epimer (18α-GA > 18β-GA, 18α-GAMG > 18β-GAMG, 18α-GCCS > 18β-GCCS) and (ii) the number of glucuronic acids at the C-3 position had an effect on the anti-inflammatory activity (mono-glucuronide > aglycone > bis-glucuronide, such as 18β-GAMG > 18β-GCCS > 18β-GA, 18α-GAMG > 18α-GCCS > 18α-GA). Glycyrrhizin analogs displayed preferable anti-inflammatory activity; among them, 18α-GAMG exhibited the strongest activity, and the NO inhibition rate exceeded 70% at a concentration of 40 μM.
Fig. 2. The inhibitory effects of the glycyrrhizin analogs on the NO production in the LPS-stimulated RAW264.7 cells. RAW264.7 cells were pretreated with glycyrrhizin analogs (40 μM) for 2 h and then in the presence or absence of LPS (1 μg mL–1) for 20 h. The results have been shown as means ± SD (n = 3) of at least three independent experiments. #p < 0.05, ##p < 0.01, ###p < 0.001 compared with the blank group; * p < 0.05, ** p < 0.01, *** p < 0.001 compared with LPS-stimulated group.
The cytotoxicity of the glycyrrhizin analogs was evaluated by the MTT assay in the RAW264.7 cells. As shown in Table S1,‡ glycyrrhizin analogs and LPS showed low toxicity, and the relative viabilities of the cells treated with them were more than 96%. These results indicated that glycyrrhizin analogs did not possess significant cytotoxic effects against the cells at the concentrations used herein.
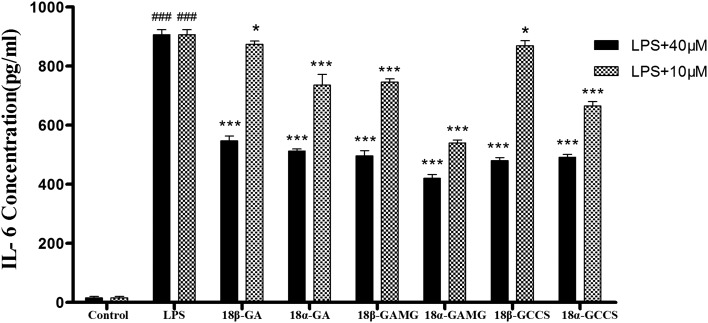
To further evaluate the effects of glycyrrhizin analogs on the LPS-induced IL-6 production, RAW264.7 cells were cultured with LPS (1 μg mL–1) in the presence of glycyrrhizin analogs for 20 h, and the levels of IL-6 in the supernatant were determined via ELISA. As shown in Fig. 3, the LPS-induced IL-6 production decreased after treatment with the glycyrrhizin analogs, and inhibitory effects were observed with the following conclusions: (i) for the inhibition of the IL-6 production, 18α-epimer was better than 18β-epimer (18α-GA > 18β-GA, 18α-GAMG > 18β-GAMG, 18α-GCCS ≈ 18β-GCCS); (ii) the number of glucuronic acids had an effect on the anti-inflammatory activity (mono-glucuronide > aglycone > bis-glucuronide, such as 18β-GAMG > 18β-GCCS > 18β-GA, 18α-GAMG > 18α-GCCS > 18α-GA). Thus, combining the anti-inflammatory activity and cytotoxicity of the glycyrrhizin analogs, 18α-GAMG was selected to further explore the mechanisms of the anti-inflammatory effect.
Fig. 3. The inhibitory effects of the glycyrrhizin analogs on the LPS-induced IL-6 production in the RAW264.7 cells. RAW264.7 cells were incubated with glycyrrhizin analogs (40 μM and 10 μM) for 2 h and then in the presence or absence of LPS (1 μg mL–1) for 20 h. The results have been shown as means ± SD (n = 3) of at least three independent experiments. #p < 0.05, ###p < 0.001 compared with the control group; * p < 0.05, *** p < 0.001 compared with LPS-stimulated group.
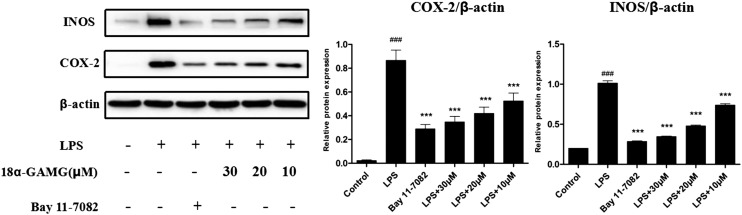
18α-GAMG was most effective in inhibiting the NO and IL-6 production. Thus, it was used to investigate the expression of the inflammation-related proteins.32,33 The expression of nitric oxide synthase (iNOS) and COX-2 was examined in the LPS-stimulated RAW264.7 cells. Western blotting results showed that 18α-GAMG strongly attenuated the expression of iNOS and COX-2 in the LPS-stimulated RAW264.7 cells in a dose-dependent manner (Fig. 4). These preliminary results demonstrated that 18α-GAMG may participate in signaling pathways activated by LPS in macrophages.
Fig. 4. Effects of 18α-GAMG on LPS-induced iNOS and COX-2 gene expression in the RAW264.7 cells. The cells were treated with different concentrations of 18α-GAMG and then in the presence or absence of LPS (1 μg mL–1) for 20 h. Bay 11-7082 is the NF-κB inhibitors (20 μM). The results have been shown as means ± SD (n = 3) of at least three independent experiments. #p < 0.05, ##p < 0.01, ###p < 0.001 compared with the control group; * p < 0.05, ** p < 0.01, *** p < 0.001 compared with LPS-stimulated group.
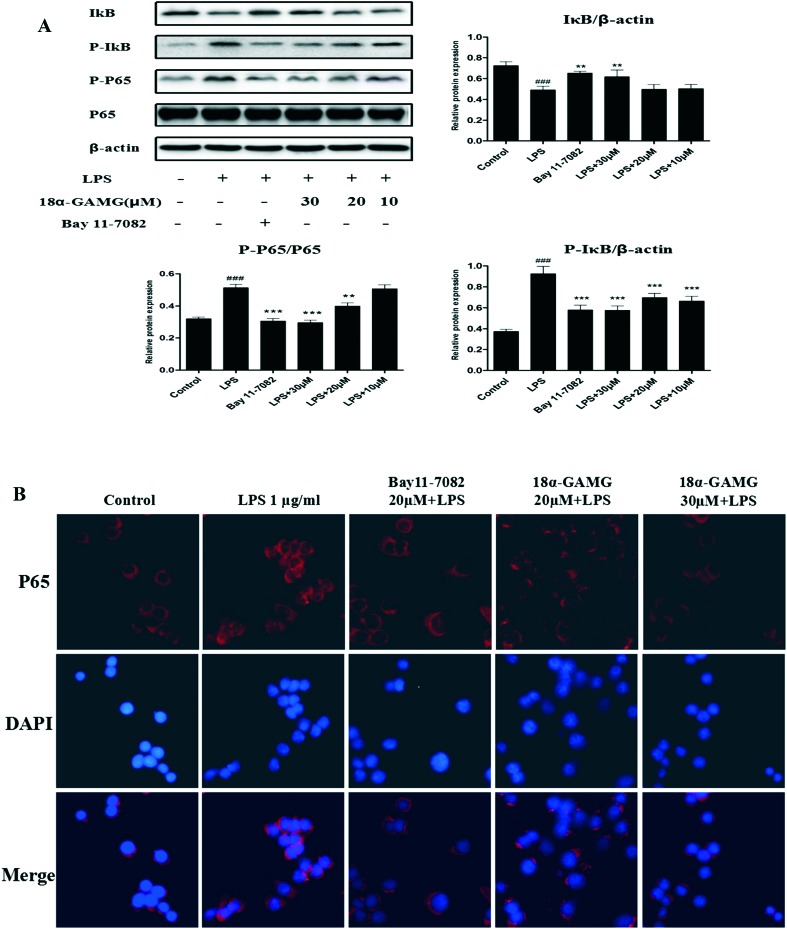
NF-κB is a well-known transcription factor that positively regulates inflammatory genes, such as iNOS, COX-2, and IL-6, in response to inflammatory stimuli.34 NF-κB activation is controlled by phosphorylation and degradation of IκB-α, a cognate regulatory subunit of NF-κB.35 Therefore, western blotting was used to examine the effects of 18α-GAMG on the NF-κB pathways in the LPS-stimulated RAW264.7 cells. As shown in Fig. 5A, LPS significantly increased the levels of phosphorylated NF-κB p65 and IκBα, and 18α-GAMG treatment could attenuate the activation of these proteins to varying degrees. Furthermore, the nuclear translocation of NF-κB was examined in the LPS-stimulated RAW264.7 cells. Immunofluorescence analysis showed that 18α-GAMG clearly inhibited NF-κB p65 nuclear translocation from the cytosol to the nucleus (Fig. 5B). These results further confirmed that 18α-GAMG might regulate the expression of pro-inflammatory proteins through the inhibition of the NF-κB signaling pathways.
Fig. 5. A: 18α-GAMG suppressed LPS-induced activation of the NF-κB signaling pathway in the RAW264.7 cells. RAW264.7 cells were treated with 18α-GAMG (10–30 μM) and LPS (1 μg mL–1) for 30 min. The levels of NF-κB p65 and IκB, and their phosphorylated forms were analyzed using western blotting. Bay 11-7082 is the NF-κB inhibitors (20 μM). The results have been shown as means ± SD (n = 3) of at least three independent experiments. #p < 0.05, ###p < 0.01, ###p < 0.001 compared with the control group; * p < 0.05, ** p < 0.01, *** p < 0.001 compared with LPS-stimulated group. B: 18α-GAMG clearly inhibited NF-κB p65 nuclear translocation. RAW264.7 were pretreated with 18α-GAMG for 1 h and then stimulated with LPS (1 μg mL–1) for 3 h.
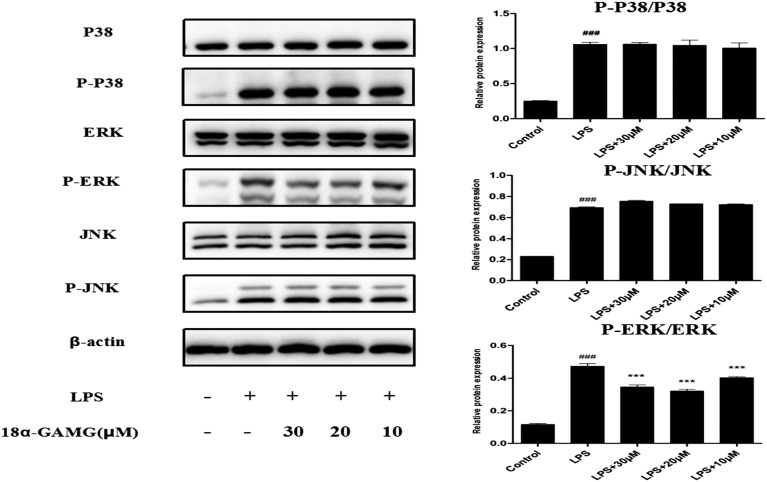
The mitogen-activated protein kinase (MAPK) transduction pathway is activated by NF-κB in mammalian cells.36,37 Inhibition of the activation of MAPK down-regulates the expression of the inflammatory mediators and thus improves the outcome of the experimental inflammatory diseases.38,39 To determine the role of 18α-GAMG in modulating MAPK activation in LPS-stimulated RAW264.7 cells, the expression of ERK, JNK, and p38 was examined. As expected (Fig. 6), the levels of phosphorylation of p38, JNK, and ERK increased after LPS-stimulated treatment for 30 min. 18α-GAMG dose-dependently (10, 20, and 30 μM) inhibited LPS-induced phosphorylation of ERK, but had little effect on the phosphorylation of JNK or p38 in RAW264.7 cells. These results suggested that the anti-inflammatory activity of 18α-GAMG might be associated with its negative effects on ERK activation.
Fig. 6. 18α-GAMG suppressed LPS-induced activation of the MAPK signaling pathway in the RAW264.7 cells. RAW264.7 cells were treated with 18α-GAMG (10–30 μM) and LPS (1 μg mL–1) for 30 min. The levels of JNK, ERK1/2, and p38 MAPK proteins, and their phosphorylated forms were analyzed using western blotting. The results have been shown as means ± SD (n = 3) of at least three independent experiments. #p < 0.05, ###p < 0.01, ###p < 0.001 compared with the control group; * p < 0.05, ** p < 0.01, *** p < 0.001 compared with LPS-stimulated group.
To further verify the anti-inflammatory activity of 18α-GAMG in vivo, a mice model of CCl4-induced hepatic fibrosis was established in this study to investigate its effect on hepatic fibrosis.40 Healthy C57BL6 mice (SPF, male, 20 ± 2 g) were purchased from the Experimental Animal Center of Anhui Medical University. Animals were housed in a temperature (22 ± 2 °C) and relatively humidity (50%)-controlled room under a 12 h light/dark cycle, given free access to food and water, and acclimatized for at least one week prior to use. All the animal experiments were performed in accordance with the Regulations of the Experimental Animal Administration issued by the State Committee of Science and Technology of China. Efforts were made to minimize the number of animals used and their suffering. Animals were maintained in accordance with the Guides of Center for Developmental Biology, Anhui Medical University for the Care and Use of Laboratory Animals, and in all the experiments, protocols approved by the institutions' subcommittees on animal care were used.
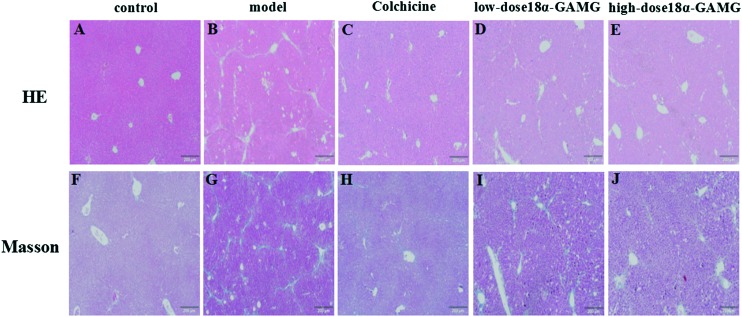
As shown in Fig. 7, in the control group, the structure of the liver was clear, and the size of hepatocytes was constant. The hepatic lobule was intact, without denaturation or necrosis (Fig. 7A and F). In the model group, the amount of blue collagen fibers was obviously increased. Fatty degeneration was apparent and ballooning degeneration of the hepatocyte can be seen in the model group (Fig. 7B and G). The extent of inflammatory cell infiltration, blue collagen fibers and fibrosis of liver in the colchicine group (0.1 mg kg–1), the high-dose 18α-GAMG group, and the low-dose 18α-GAMG were significant decreased, and the high-dose group was better than the low-dose group (Fig. 7C, D, E, H, I and J). These results showed that 18α-GAMG could significantly improve the pathological changes of the CCl4-induced hepatic fibrosis.
Fig. 7. Typical light photomicrographs of liver tissue showing the influence of 18α-GAMG on histological changes (H&E and Masson staining). (7A–E) represent H&E staining of the control group, model group, colchicine group (0.1 mg kg–1), low-dose 18α-GAMG group (100 mg kg–1), and high-dose 18α-GAMG group (200 mg kg–1), respectively. Fig. 7F–J represent the Masson staining of the control group, model group, colchicine group (0.1 mg kg–1), low-dose 18α-GAMG group (100 mg kg–1), and high-dose 18α-GAMG group (200 mg kg–1), respectively.
Conclusions
In conclusion, the clear structure–activity relationships of glycyrrhizin with anti-inflammatory activity was explained; among them, the glucuronide unit and the 18-α/β-stereoisomer were important factors. Among these compounds, 18α-glycyrrhetinic acid monoglucuronide (18α-GAMG) was found to exhibit strongest inhibition. Western blotting and immunofluorescence results showed that 18α-GAMG decreased the expression of iNOS, COX-2, and MAPKs, as well as the activation of NF-κB in the LPS-stimulated RAW264.7 cells. Overall, 18α-GAMG exerted its anti-inflammatory activity through the inhibition of NO generation as a result of inhibition of the NF-κB and MAPKs-related inflammatory signaling pathways. In addition, the in vivo results showed that 18α-GAMG could significantly improve the pathological changes of CCl4-induced hepatic fibrosis. Therefore, 18α-GAMG may be clinically useful for the reduction of inflammation in the future.
Conflict of interest
The authors declare no competing interests.
Supplementary Material
Acknowledgments
Financial support was provided by the National Natural Science Funding of China (21572003), Anhui University Natural Science Research Project (KJ2016A339), and University Project of Introduction and Cultivation of Leading Talents (gxfxZD2016044).
Footnotes
†Bo Li and Yongan Yang contributed equally to this work.
‡Electronic supplementary information (ESI) available: Experimental procedures for compound synthesis, characteristic data, 1H and 13C NMR spectra, HRMS assays for glycyrrhizin analogs. Details for the biological evaluation. Table of cytotoxicity analysis data. See DOI: 10.1039/c7md00210f
References
- Fiore C., Eisenhut M., Ragazzi E., Zanchin G., Armanini D. J. Ethnopharmacol. 2005;99:317–324. doi: 10.1016/j.jep.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monograph Altern. Med. Rev. 2005;10:230–237. [PubMed] [Google Scholar]
- Asl M. N., Hosseinzadeh H. Phytother. Res. 2008;22:709–724. doi: 10.1002/ptr.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K., Oh S. M., Kwon H. S., Oh Y. S., Lim S. S., Shin H. K. Biochem. Biophys. Res. Commun. 2006;345:1215–1223. doi: 10.1016/j.bbrc.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Cho H. J., Lim S. S., Lee Y. S., Kim J., Lee C. H., Kwon D. Y., Park J. H. Y. Food Chem. 2010;121:959–966. [Google Scholar]
- Yu J. Y., Ha J. Y., Kim K. M., Jung Y. S., Jung J. C., Oh S. Molecules. 2015;20:13041–13054. doi: 10.3390/molecules200713041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z. H., Li T., Tong Y. G., Chen X. J., Chen X. P., Wang Y. T., Lu J. J. Planta Med. 2015;81:1670–1687. doi: 10.1055/s-0035-1558227. [DOI] [PubMed] [Google Scholar]
- Hua H., Liang Z., Li W., Meng Y., Li X., Zhang Z., Lu C., Meng J., Shan F. Int. Immunopharmacol. 2012;12:518–525. doi: 10.1016/j.intimp.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Zhang Y. H., Isobe K., Nagase F., Lwin T., Kato M., Hamaguchi M., Yokochi T., Nakashima I. Immunology. 1993;79:528–534. [PMC free article] [PubMed] [Google Scholar]
- Wang H. L., Li Y. X., Niu Y. T., Zheng J., Wu J., Shi G. J., Ma L., Niu Y., Sun T., Yu J. Q. Inflammation. 2015;38:2269–2278. doi: 10.1007/s10753-015-0212-3. [DOI] [PubMed] [Google Scholar]
- Bordbar N., Karimi M. H., Amirghofran Z. Cell. Immunol. 2012;280:44–49. doi: 10.1016/j.cellimm.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Matsui S., Matsumoto H., Sonoda Y., Ando K., Aizu-Yokota E., Sato T., Kasahara T. Int. Immunopharmacol. 2004;4:1633–1644. doi: 10.1016/j.intimp.2004.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. M., Du G. Q. Mol. Med. Rep. 2016;13:3639–3646. doi: 10.3892/mmr.2016.4981. [DOI] [PubMed] [Google Scholar]
- Ojha S., Javed H., Azimullah S., Abul-Khair S. B., Haque M. E. Neurotoxic. Res. 2016;29:275–287. doi: 10.1007/s12640-015-9579-z. [DOI] [PubMed] [Google Scholar]
- Wang W., Chen X., Zhang J., Zhao Y., Li S., Tan L., Gao J., Fang X., Luo A. Neuroscience. 2016;316:328–336. doi: 10.1016/j.neuroscience.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Mollica L., Marchis F. D., Spitaleri A., Dallacosta C., Pennacchini D., Zamai M., Agresti A., Trisciuoglio L., Musco G., Bianchi M. E. Chem. Biol. 2007;14:431–441. doi: 10.1016/j.chembiol.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Du D., Yan J., Ren J., Lv H., Li Y., Xu S., Wang Y., Ma S., Qu J., Tang W., Hu Z., Yu S. J. Med. Chem. 2013;56:97–108. doi: 10.1021/jm301248y. [DOI] [PubMed] [Google Scholar]
- Takeda S., Ishthara K., Wakui Y., Amagaya S., Maruno M., Akao T., Kobashi K. J. Pharm. Pharmacol. 1996;48:902–905. doi: 10.1111/j.2042-7158.1996.tb05998.x. [DOI] [PubMed] [Google Scholar]
- Kao T. C., Shyu M. H., Yen G. C. J. Agric. Food Chem. 2010;58:8623–8629. doi: 10.1021/jf101841r. [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Kao T. C., Lo W. H., Yen G. C. J. Agric. Food Chem. 2011;59:7726–7733. doi: 10.1021/jf2013265. [DOI] [PubMed] [Google Scholar]
- Makino T., Okajima K., Uebayashi R., Ohtake N., Inoue K., Mizukami H. J. Pharmacol. Exp. Ther. 2012;342:297–304. doi: 10.1124/jpet.111.190009. [DOI] [PubMed] [Google Scholar]
- Makino T. Biol. Pharm. Bull. 2014;37:898–902. doi: 10.1248/bpb.b13-00997. [DOI] [PubMed] [Google Scholar]
- Feng S., Li C., Xu X., Wang X. J. Mol. Catal. B: Enzym. 2006;43:63–67. [Google Scholar]
- Kim D.-H., Hong S.-H., Kim B. T., Bae E.-A., Park H. Y., Han M. J. Arch. Pharmacal Res. 2000;23:172–177. doi: 10.1007/BF02975509. [DOI] [PubMed] [Google Scholar]
- Park H. Y., Park S. H., Yoon H. K., Han M. J., Kim D. H. Arch. Pharmacal Res. 2004;27:57–60. doi: 10.1007/BF02980047. [DOI] [PubMed] [Google Scholar]
- Yang Y. A., Tang W. J., Zhang X., Yuan J. W., Liu X. H., Zhu H. L. Molecules. 2014;19:6368–6381. doi: 10.3390/molecules19056368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. J., Yang Y. A., He X., Shi J. B., Liu X. H. Sci. Rep. 2014;4:7106. doi: 10.1038/srep07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltina L. A., Stolyarova O. V., Baltina L. A., Kondratenko R. M., Plyasunova O. A., Pokrovskii A. G. Pharm. Chem. J. 2010;44:299–302. doi: 10.1007/s11094-010-0454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A. V., Thirugnanam S., Dakshinamoorthy G., Samykutty A., Zheng G., Chen A., Bosland M. C., Kajdacsy-Balla A., Gnanasekar M. Int. J. Oncol. 2011;39:635–640. doi: 10.3892/ijo.2011.1061. [DOI] [PubMed] [Google Scholar]
- Xie C., Li X., Wu J., Liang Z., Deng F., Xie W., Zhu M., Zhu J., Zhu W., Geng S., Zhong C. Inflammation. 2015;38:1639–1648. doi: 10.1007/s10753-015-0140-2. [DOI] [PubMed] [Google Scholar]
- Aktan F. Life Sci. 2004;75:639–653. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- Tsatsanis C., Androulidaki A., Venihaki M., Margioris A. N. Int. J. Biochem. Cell Biol. 2006;38:1654–1661. doi: 10.1016/j.biocel.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Liew C. Y., Lam K. W., Kim M. K., Harith H. H., Tham C. L., Cheah Y. K., Sulaiman M. R., Lajis N. H., Israf D. A. Int. Immunopharmacol. 2011;11:85–95. doi: 10.1016/j.intimp.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Lawrence T. Cold Spring Harbor Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Ben-Neriah Y. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Hayden M. S. Nat. Rev. Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- Wang Z. S., Chen L. Z., Zhou H. P., Liu X. H., Chen F. H. Bioorg. Med. Chem. Lett. 2017;27:1803–1807. doi: 10.1016/j.bmcl.2017.02.056. [DOI] [PubMed] [Google Scholar]
- Vermeulen L., De Wilde G., Van Damme P., Vanden Berghe W., Haegeman G. EMBO J. 2003;22:1313–1324. doi: 10.1093/emboj/cdg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse K. H., Chow K. B., Leung W. K., Wong Y. H., Wise H. Neuroscience. 2014;267:241–251. doi: 10.1016/j.neuroscience.2014.02.041. [DOI] [PubMed] [Google Scholar]
- Czaja A. J. World J. Gastroenterol. 2014;20:2515–2532. doi: 10.3748/wjg.v20.i10.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.