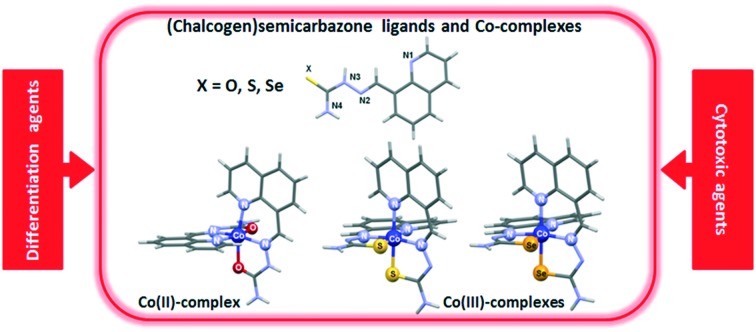
 A comparative study of (chalcogen)semicarbazones and their cobalt complexes was performed.
A comparative study of (chalcogen)semicarbazones and their cobalt complexes was performed.
Abstract
Cobalt complexes with semi- and thiosemicarbazones of 8-quinolinecarboxaldehyde have been synthesized and characterized by X-ray diffraction analysis. These novel complexes and a previously synthesized cobalt complex with a selenium-based selenosemicarbazone ligand showed myeloid differentiation activity on all trans retinoic acid resistant HL-60 acute myeloid leukaemia cells. They also showed varying levels of cytotoxicity on five human tumor cell lines: cervix carcinoma cells (HeLa), lung adenocarcinoma cells (A549), colorectal adenocarcinoma cells (LS-174), breast carcinoma cells (MDA-MB-361), and chronic myeloid leukaemia (K562) as well as one normal human cell line: fetal lung fibroblast cells (MRC-5). Leukaemia differentiation was most strongly induced by a metal-free oxygen ligand and the selenium ligand, whilst the latter and the cobalt(ii) complex with an oxygen ligand showed the strongest dose-dependent cytotoxic activity. In four out of five investigated tumor cell lines, it was of the same order of magnitude as cisplatin. These best compounds, however, had lower toxicity on non-transformed MRC-5 cells than cisplatin.
1. Introduction
Today cancer is a leading cause of death worldwide with 2030 projections estimated at 13.1 million deaths.1 The mainstay of treating widely dispersed (and therefore likely fatal) cancer is chemotherapy, which can be severely toxic, so the goal of much current research is to identify novel (safer and more effective) treatments for neoplastic conditions with the hope of decreasing morbidity and mortality. This has resulted in a large move towards targeted therapies for cancer, which aim to be less harmful to healthy proliferating cells in general. One such option is differentiation therapy, where malignant cells are stimulated to differentiate, thus maturing and entering natural apoptotic pathways.2,3 Leukaemia has been the main success story of differentiation therapy so far. This group of haematologic neoplasia is characterized by the unregulated proliferation of hematopoietic progenitor cells that fail to differentiate into mature cells leading to anaemia, immunocompromised state and bleeding disorders often times resulting in fatality. Acute myeloid leukaemia (AML) is the type with the worst prognosis, with 10 year survival rates of around 5% in older adults and around 50% in younger ones.4 Cytogenetic analysis is essential in prognosis and treatment of AML nowadays.5 Acute promyelocytic leukaemia (APL), one of the variants of AML, shows a translocation between the long arms of chromosomes, 15 and 17, leading to the production of an abnormal fusion protein (PML-RARα) made from the parts of 2 genes on the 2 separate chromosomes.6,7 This fusion protein interferes with DNA transcription, effectively blocking the differentiation of promyelocytes into myelocytes and causing the former's accumulation at the expense of functional fully differentiated white blood cells.8 This disease of young adults was almost invariably fatal, due to severe coagulopathy until the revolutionary introduction of ATRA (All Trans Retinoic Acid) as a treatment for APL.9 This drug, which is a ligand of the retinoic acid receptor α component of the fusion protein, when given in supra-physiological concentrations, is able to relieve the transcription block posed by the PML-RARα fusion protein and effectively induce differentiation.10 Together with a low dose of traditional chemotherapy, this has managed to achieve a cure rate of around 80%.11 The lack of toxic side-effects seen in standard chemotherapy is an added benefit.12 Unfortunately, ATRA works effectively only on the APL genetic subtype of acute myeloid leukaemia, although some efficacy has been seen on osteosarcoma and glioma cells or tumours.13,14 Due to this success story, extensive research is now being directed towards finding compounds which can cause differentiation in other types of immature leukemic cells.15 Initial testing of therapeutic candidates makes use of cell lines derived from leukemic patients which are easily available and can act as a shared standard across many research laboratories. Here we focus on the HL-60 cell line as our model.
(Chalcogen)semicarbazones, condensation products of (chalcogen)semicarbazides and carbonyl compounds, have been a subject of interest in coordination chemistry for many years. Versatile modes of coordination of this class of ligands have been reported.16–19 They can coordinate to metal ions bidentately via chalcogen donor and imine nitrogen atoms, but the coordination ability may be extended if a parent carbonyl compound possesses other suitable donor atoms. This general class of compounds has been shown to possess a wide range of bioactive properties, including antitumor activity.20–37 In fact (chalcogen)semicarbazones with activities comparable to standard anticancer drugs, like cis-diammindichloridoplatinum(ii) (cisplatin, CDDP), have been developed. A few comprehensive comparative studies of (chalcogen)semicarbazones and their complexes pointed out the importance of the chalcogen donor atom identity for biological activity.26,30,38 Among the three types of (chalcogen)semicarbazones, thiosemicarbazones have been studied to a greater extent than semicarbazones and selenosemicarbazones. Some results indicated that sulphur compounds are more active in comparison with oxygen analogues, which showed a rather limited spectrum of biological activities.24 These thiosemicarbazones are also more stable and safer to handle than their selenium analogues.37,39,40 Selenosemicarbazones and their metal complexes have been an area of our particular interest for years.31–47,41,42 Research suggests that the mechanisms of cytotoxicity of these compounds may include the production of reactive oxygen species and oxidative stress in the complexes with 2-quinolinecarboxaldehyde selenosemicarbazone.37
In the current study we used 8-quinolinecarboxaldehyde-based (chalcogen)semicarbazones as ligands (Scheme 1), providing detailed spectroscopic and structural characterization (X-ray diffraction, XRD) of the novel cobalt complexes with oxygen and sulphur-based ligands. Cobalt was chosen as the central metal ion since the coordination of (chalcogen)semicarbazones to cobalt often results in complexes with activities higher than the activity of standard anticancer drugs such as cisplatin.33,43–46 We analysed the effects of (chalcogen)semicarbazones and the corresponding cobalt complexes on the differentiation of high passage number (70+) HL-60 cells which are known to be poorly responsive to ATRA-induced differentiation.47 We further test the general cytotoxic effects of these compounds on a number of tumor cell lines and on normal cells and perform some analysis of the cell cycle perturbations induced.
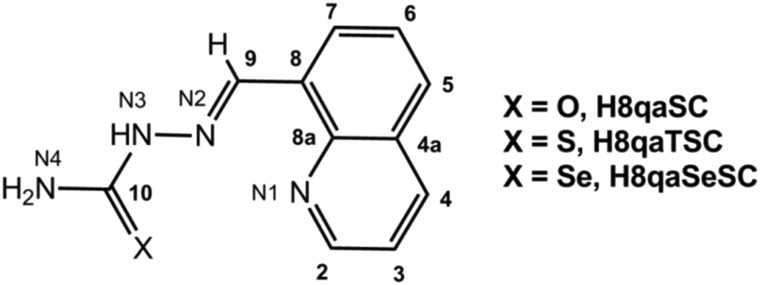
Scheme 1. Schematic structure and atomic numbering scheme of (chalcogen)semicarbazone ligands: H8qaSC, H8qaTSC and H8qaSeSC.
2. Results & discussion
2.1. General
(Chalcogen)semicarbazone cobalt complexes with H8qaSC and H8qaTSC ligands were synthesized by template reactions starting from metal salts, 8-quinolinecarboxaldehyde and corresponding (chalcogen)semicarbazide (mole ratio: 1 : 2 : 2, respectively). The same products were obtained also by direct reactions of metal salts and the ligands in a 1 : 2 molar ratio. The composition of the products was not affected by the change of the molar ratio of the reacting species. The complexes are soluble in MeOH, EtOH, MeCN, DMF and DMSO at room temperature. Molar conductivity measurements showed that the complex with the oxygen ligand H8qaSC is 1 : 2 electrolyte, while the complex with the sulfur ligand H8qaTSC is 1 : 1 electrolyte. Elemental analysis showed that the complex with the oxygen ligand contains two neutral H8qaSC ligand molecules, two chloride ions and two water molecules. This complex has a room-temperature magnetic moment of 1.80 μB, consistent with just one paramagnetic Co(ii) center, while the other complex is diamagnetic in nature and the central metal ion in the complex with the sulfur ligand is Co(iii). Based on these data, as well as spectroscopic data and X-ray structural analysis (vide infra) the general formula of the two novel complexes can be written as [Co(H8qaSC)2]Cl2·2H2O (1) and [Co(8qaTSC)2]ClO4·DMSO (2).
Synthesis of the cobalt(iii) complex with the oxygen ligand H8qaSC was unsuccessful even when a stream of air was passed through the reaction mixture for 3 h. Similarly, in the case of related 2-formyl, 2-acetyl, and 2-benzoylpyridine N(4)-cyclohexylsemicarbazones, cobalt(ii) complexes with neutral ligands were obtained, whereas with the analogous thiosemicarbazones, cobalt(iii) complexes with the anionic form of the ligands were obtained.48 It is anticipated that oxidation reaction of cobalt(ii) to cobalt(iii) species consists of two main steps: (1) the reversible formation of a dioxygen adduct (μ-peroxo-bridged cobalt compound) and (2) its irreversible decomposition to related cobalt(iii) complexes, where the redox rearrangement reactions can be classified as metal-centered oxidations and ligand-centered oxidative dehydrogenations.49,50 It is well known that the octahedral hexaaquacobalt(ii) ion is stable to aerial oxidation, but data on standard reduction potential for a variety of cobalt(iii) complexes showed the stabilization of the +3 oxidation state relative to the +2 as the ligands are changed from O- to N-donors.51 In the case of (chalcogen)semicarbazones derived from 8-quinolinecarboxaldehyde, it can be assumed that a combination of favorable thermodynamic and kinetic factors allows for facile synthesis of cobalt(iii) complexes with heavier chalcogens, as opposed to the cobalt(ii) complex with an O analogue.
2.2. Description of structures
Crystal data and structure determination results are summarized in Table 1. Single crystals of the ligand H8qaTSC were obtained by slow diffusion of ethanol into a DMSO solution of the ligand. The crystal structure of the ligand H8qaTSC has been previously determined,52 but it was not deposited in the Cambridge Crystallographic Data Centre (CCDC). The structure was re-investigated and it corresponds to that previously determined, except that we have obtained a better model with a slightly lower R factor value.
Table 1. Crystal data and structure refinement for the ligand H8qaTSC and complexes 1 and 2.
| H8qaTSC | 1 | 2 | |
| Crystal data | |||
| Empirical formula | C11H10N4S | C22H20CoN8O2·2Cl·2(H2O) | C22H18CoN8S2·ClO4·C2H6SO |
| Formula weight | 230.29 | 594.32 | 695.02 |
| Crystal system | Monoclinic | Monoclinic | Triclinic |
| Space group | P21/c | C2/c | P1[combining macron] |
| a, b, c (Å) | 8.9464 (9), 12.8728 (11), 9.7816 (7) | 21.752 (3), 10.0815 (9), 13.905 (2) | 10.3016 (6), 10.5421 (7), 14.5516 (10) |
| α, β, γ (°) | 90, 95.947 (8), 90 | 90, 124.39 (2), 90 | 80.662 (6), 89.829 (5), 65.733 (6) |
| V (Å3) | 1120.43 (17) | 2516.1 (8) | 1418.02 (17) |
| Z | 4 | 4 | 2 |
| μ (mm–1) | 0.27 | 0.94 | 0.97 |
| Crystal size (mm) | 0.20 × 0.18 × 0.12 | 0.15 × 0.12 × 0.09 | 0.09 × 0.08 × 0.08 |
| T min, Tmax | 0.697, 1.000 | 0.585, 1.000 | 0.955, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 16 892 | 18 343 | 20 909 |
| 2551 | 2881 | 6165 | |
| 2116 | 2365 | 4897 | |
| R int | 0.032 | 0.049 | 0.042 |
| (sin θ/λ)max (Å–1) | 0.650 | 0.650 | 0.639 |
| R[F2 > 2σ(F2)] | 0.039 | 0.042 | 0.046 |
| wR(F2) | 0.110 | 0.103 | 0.126 |
| S | 1.06 | 1.05 | 1.03 |
| No. of reflections | 2551 | 2881 | 6165 |
| No. of parameters | 157 | 188 | 389 |
| ΔΔ〉max, Δ, Δ〉min (e Å–3) | 0.21, –0.33 | 0.40, –0.26 | 0.53, –0.53 |
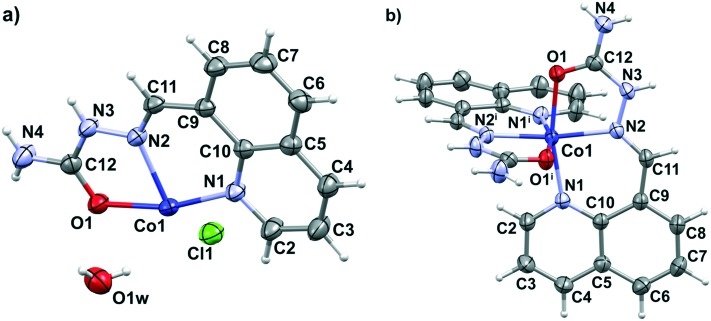
In the molecular structure of complex 1, a [Co(H8qaSC)2]2+ cation crystallizes with two chloride anions and two water molecules. The cobalt(ii) ion lies on a twofold rotation axis, hence the asymmetric unit of 1 consists of a half of the complex cation, one chloride ion and one water molecule (Fig. 1a). In the complex cation, the cobalt(ii) metal center is coordinated to two tridentate neutral H8qaSC ligands, giving rise to a chiral octahedral arrangement. Complex 1 is nonetheless a racemate, since it crystallizes in the centrosymmetric C2/c space group. Chelation occurs by means of quinoline (N1, N1i) and imine (N2, N2i) nitrogen atoms and by the oxygen donors (O1, O1i), where i = –x, y, 1.5 – z, in a mer geometry constrained by the ligand's planarity. Fig. 1b reports the molecular structure of [Co(H8qaSC)2]2+ in 1, while Table S1 (ESI‡) reports the coordination geometry. The coordinative bond lengths in 1 are similar to the respective bonds in analogous N-heteroaromatic semicarbazone cobalt(ii) complexes: bis{1-[(E)-2-pyridinylmethylidene]semicarbazide}cobalt(ii) diperchlorate monohydrate (CSD refcode ATUNEI)53 and bis[bis(2-pyridyl)ketone semicarbazonato-N,N′,N′′]cobalt(ii) dinitrate monohydrate (CSD refcode WEJNOO).54 The crystal packing (Fig. S1 and Table S2, ESI‡) is based on a 3D hydrogen bond network involving terminal NH2 groups, N–H groups, chloride ions and crystal water molecules.
Fig. 1. Asymmetric unit of complex 1 (a) and perspective view and labeling of the molecular structure of [Co(H8qaSC)2]2+ in 1 (b). Thermal ellipsoids are at the 50% probability level. Equivalent atoms are generated by the transformation i = –x, y, 1.5 – z.
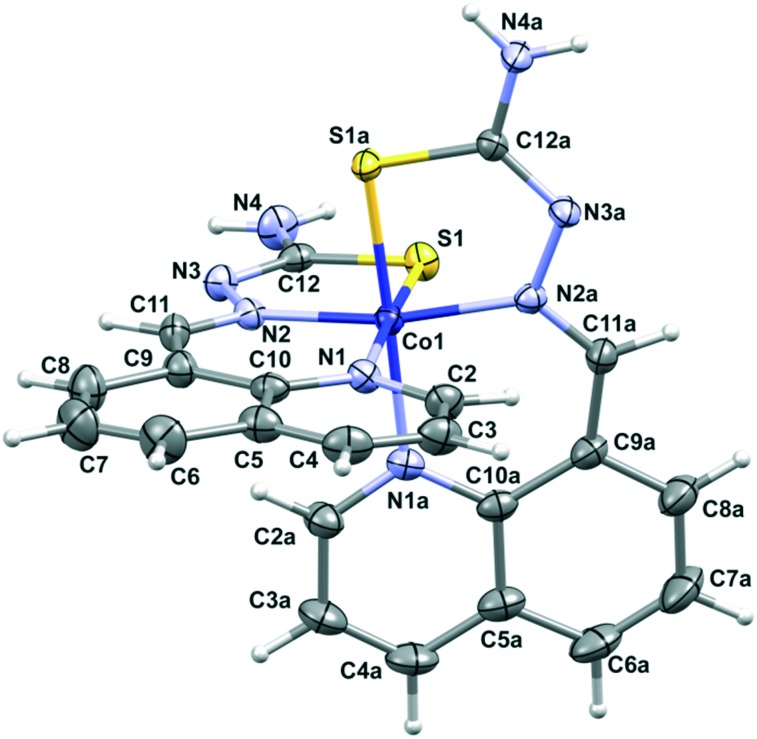
XRD shows that in the case of complex 2 two deprotonated H8qaTSC ligands are coordinated to the cobalt(iii) ion, while the outer sphere of the complex consists of a perchlorate ion and one DMSO solvent molecule. The octahedral bis-chelate cation [Co(8qaTSC)2]+ possesses a mer geometry. Since the complex crystallizes in the centrosymmetric P1[combining macron] space group, it is a racemic compound regardless of the chiral octahedral arrangement of cationic species in 2. Fig. 2 shows a perspective view of the complex cation in 2, while Table S1 (ESI‡) reports the most relevant bonding parameters. The monoanionic form of the H8qaTSC ligand coordinates the metal via the sulfur atom, the quinoline and the imine nitrogen atoms, with the formation of one six-membered and one five-membered chelate ring. All metal–donor atom bonds are close to the average corresponding bonds found in a search on quinoline thiosemicarbazone-cobalt systems performed through the Cambridge Structural Database (CCDC: 734053; 2016 release, v. 5.37 with updates: Nov15, Feb16).55 Besides electrostatic interactions between complex cations and counter ions, the crystal packing of complex 2 (Fig. S2 and Table S3, ESI‡) is based on hydrogen bonds and π–π stacking interactions of the quinoline rings. Complex 2 is isostructural with the cobalt(iii) complex with the selenium ligand [Co(8qaSeSC)2]ClO4·DMSO (3).56
Fig. 2. Perspective view and labeling of the molecular structure of [Co(8qaTSC)2]+ in 2. Thermal ellipsoids are at the 50% probability level.
2.3. Spectroscopic characterization
In the IR spectra of the ligands H8qaSC and H8qaTSC, there are sharp medium intensity bands in the region 3315–3155 cm–1 which are assigned to the ν(NH2) and ν(NH) stretching vibrations. The lack of a large systematic shift of these bands in the IR spectra of complexes 1 and 2 (Fig. S3 and S4, respectively, ESI‡) indicates no interaction between the terminal nitrogen atom and cobalt ion. The band at 1705 cm–1 in the spectrum of the ligand H8qaSC, which is ascribed to a ν(C O) vibration, is shifted to 1652 cm–1 in the spectrum of complex 1, suggesting the coordination of the oxygen atom. The coordination of azomethine nitrogen to cobalt in 1 is suggested by the shift of the ν(C N) toward lower frequency (1587 cm–1 in H8qaSC, 1550 cm–1 in 1). In the IR spectrum of complex 2 there is a systematic shift of the ν(C N) band to higher frequencies in comparison with that in the corresponding ligand (1603 cm–1 in H8qaTSC; 1637 cm–1 in 2). As it had been previously observed in other complexes with heavier (chalcogen)semicarbazones,19,31–33,41,42 upon coordination of the sulfur/selenium atom, the ν(C X) band (X = S, Se) was shifted to lower frequencies. Also, in the IR spectrum of complex 2, sharp and strong bands at 1015 cm–1 and ~ 1090 cm–1 can be observed, originating from the non-coordinated DMSO molecule and perchlorate ion, respectively. In complex 1, the broad absorption in the region above 3300 cm–1 was attributed to the lattice water.
1H and 13C NMR spectroscopy data (Fig. S5 and S6, respectively, ESI‡) confirm the tridentate coordination of the ligand H8qaTSC in its monoionic form in complex 2. The correlation of H–C2 with H–C9 and H–C7 in the 2D ROESY spectrum of complex 2 (Fig. S7, ESI‡), as previously noticed for the Co-selenosemicarbazone complex 3,56 is attributed to the octahedral geometry, which indicates that the geometry of complex 2 is preserved in the solution.
The electronic absorption spectra of the (chalcogen)semicarbazone ligands exhibited three bands in the region 350–220 nm, corresponding to the intra-ligand transitions associated with the azomethine, quinoline, and C X (X = O, S) portions of the ligands (Fig. S8, ESI‡). In the spectra of the complexes, the intense absorption bands attributed to the intra-ligand transitions within the coordinated ligand moiety and ligand-to-metal charge transfer transitions can be observed (Fig. S8, ESI‡). The aqueous solution behavior of complexes 1–3 with respect to hydrolysis was studied in DMSO/H2O 1 : 100 (v/v) solutions at ambient temperature over 24 h by UV-vis spectroscopy. Complexes 1–3 were stable, as can be seen from their electronic absorption spectra (Fig. S9, ESI‡).
2.4. Spectrophotometric assays of HL-60 differentiation induction and cytotoxic activity
As myeloid cells approach final maturation, they express and store oxidative burst enzymes capable of reducing soluble yellow nitroblue tetrazolium (NBT) into blue insoluble formazan crystals. The detection of this activity is therefore an indication of early myeloid maturation. Undifferentiated cells continue to proliferate, whilst those which entered differentiation will be reduced in number as more mature forms of myeloid cells stop proliferating as part of their maturation process. They also become capable of NBT reduction. The relative cell number can be assessed by (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (MTT) assay. Therefore, the NBT/MTT ratio is used as a screening method to gauge differentiation induction in this model cell line.57 Samples which showed good NBT/MTT ratios were then tested by morphologic assessment with Leishman's stain (vide infra).
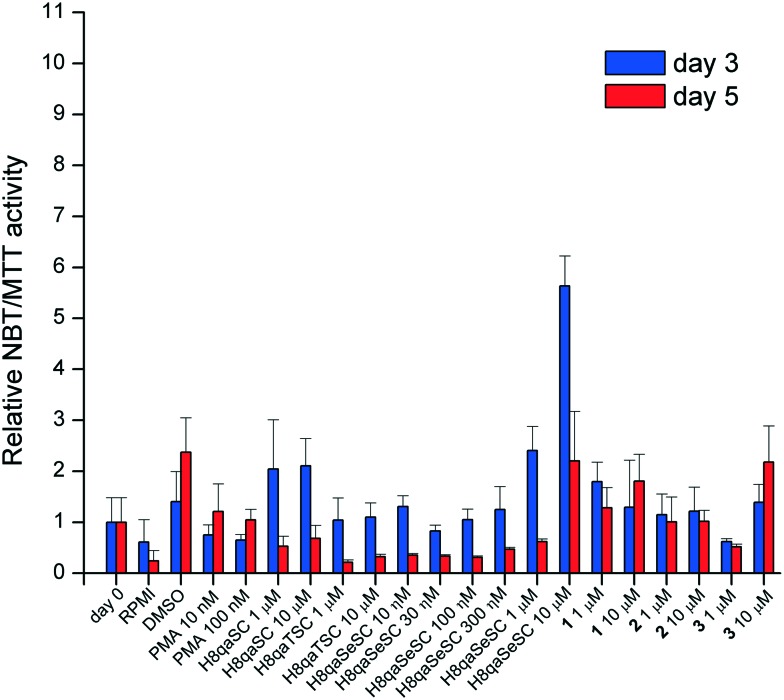
As can be seen in Fig. 3, the NBT/MTT ratio, a spectroscopic screen for the state of differentiation,57 shows that H8qaSC and its cobalt(ii) complex 1 and H8qaSeSC and its cobalt(iii) complex 3 appear to induce some differentiation (of greatly varying extent) whilst both the thiosemicarbazone ligand H8qaTSC and its cobalt(iii) complex 2 have relatively little effect. The selenium ligand H8qaSeSC, as well as the oxygen ligand H8qaSC, is effective at inducing differentiation at three days post-exposure even at 1 μM doses. After five days of exposure, H8qaSeSC and complex 1 and to a lesser extent 3 appear the better differentiating agents.
Fig. 3. Indicative differentiation (as expressed by the NBT/MTT ratio), relative to the same activity of undifferentiated cells (day 0) and without treatment after 3 or 5 days (indicated as RPMI – medium). Each value is a mean of three replicates and error bars indicate the range of values.
Data from the MTT assay acquired after 72 h incubation and used to calculate the NBT/MTT ratio were also employed to estimate the cytotoxic activity of the investigated compounds on HL60 cells. As it can be seen in Fig. S10A (ESI‡), ligands H8qaSC, H8qaSeSC and complex 3 were those whose activity reached IC50 concentrations. The IC50 value implies that the investigated treatment reduced the size of the treated population by 50% compared to non-treated control but does not obtain information on the particular mechanism responsible for this effect. In order to evaluate whether the cytotoxic activity or inhibition of proliferation was the underlying cause, cell cycle analysis on HL60 cells subjected to IC50 concentrations of H8qaSC, H8qaSeSC and 3 for 72 h was further performed to assess the percentages of cells accumulated at the sub-G1 subpopulation (Fig. S10B, ESI‡). Compound H8qaSeSC was the only one that notably, but still modestly, increased the percentage of dead cells, whereas H8qaSC and complex 3 barely raised the size of the sub-G1 fraction compared to control levels. These results clearly indicate that the investigated compounds did not exert cytotoxic activities on HL60 cells in a range of applied concentrations.
2.5. Differentiation results – morphology
The HL-60 cells exposed to the different chemicals being tested were also assessed for morphological evidence of differentiation after Leishman staining to visually confirm the effects of differentiation induction. Cells exposed to (chalcogen)semicarbazone ligands and their cobalt complexes showed varying pictures of differentiation, being clearly different from both positive controls DMSO and PMA, as well as from untreated HL-60 cells (Fig. 4). This may suggest an incomplete differentiation in certain cases, although some features of H8qaSeSC-induced differentiation, such as marked granule formation, are not even seen with the positive controls. It is interesting that they can be combined in future studies with DNA modifying agents to enhance their effect.58 Otherwise, as in the situation with APL, the combination with standard chemotherapy may result in the cells which are induced to differentiate being killed off resulting in a remission or even a cure.9
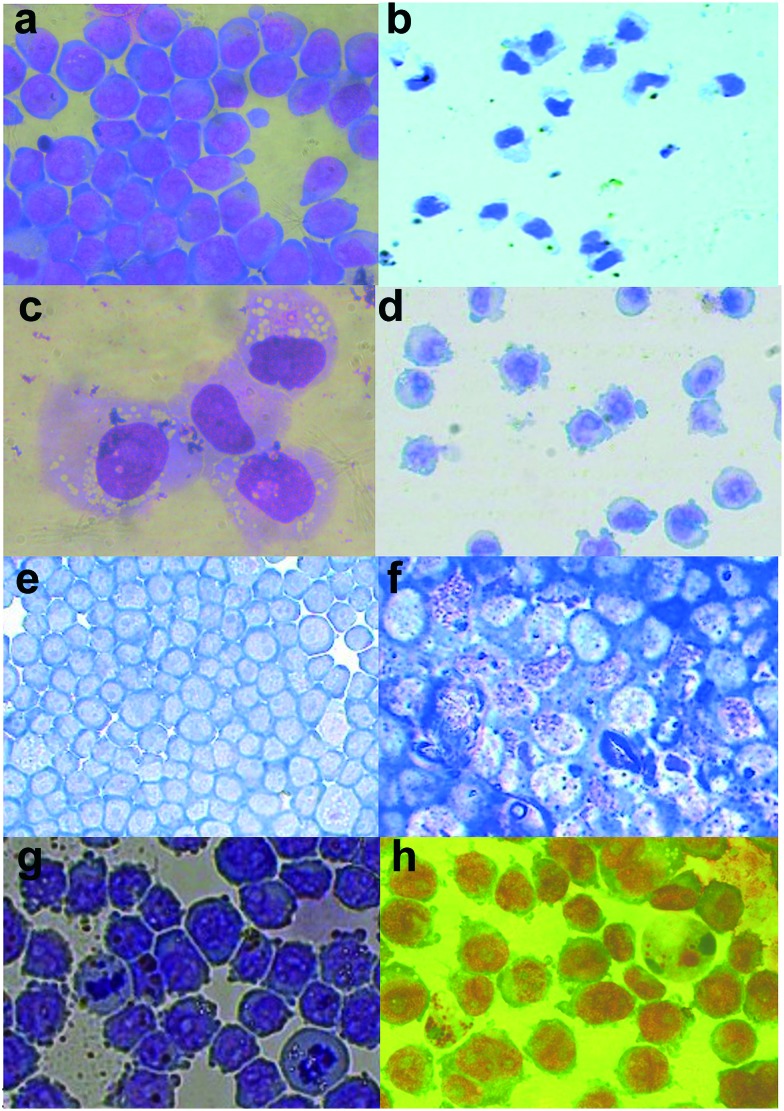
Fig. 4. Morphology of HL-60 cells upon exposure to (chalcogen)semicarbazones and corresponding complexes. Images are at 40× magnification. Plates a, b and c show the undifferentiated cells, and high power close ups of positive control differentiating agents, with DMSO day 5 differentiated (showing polymorphonuclear shaped nuclei) and PMA day 3 differentiated (showing more condensed nuclei and marked vacuolation), respectively. Plates d, f and h show H8qaSC (1 μM) at day 3, H8qaSeSC (10 μM) at day 3 and complex 3 (10 μM) at day 5, respectively, showing varying morphological evidence of differentiation. On the other hand, exposure to H8qaTSC (10 μM) at day 3 (plate e) and complex 1 (10 μM) at day 3 (plate g) show little change from the undifferentiated state.
The ligand H8qaSeSC which showed the strongest induction of NBT activity, through spectrophotometry, caused the development of numerous granules in the HL60 cells, indicating a strong differentiation towards granulocytes, together with the loss of nucleoli in the non-granular cells (Fig. 4f). The same compound has also been shown to induce some markers of differentiation in solid tumour cancer stem cells.56
Despite H8qaSC, H8qaTSC and H8qaSeSC having a rather similar structure with a sulphur or selenium atom substituting an oxygen atom in the parent compound H8qaSC, it appears that these small modifications greatly vary the differentiation-inducing activity of H8qaSC, with selenium enhancing it and sulphur removing it. Selenium is an important trace element in the body and the inclusion of this atom may mimic a selenium-containing natural factor. Zinc-finger transcription factor PLZF is known to abnormally repress gene activity in HL-60,59 and selenium is a known inhibitor of zinc-finger transcription factors.60,61 Thus inhibiting PLZF-dependent gene repression may result in differentiation.59,61 This may well be the reason for the stronger differentiation induced by H8qaSeSC as opposed to H8qaSC. The complexing of the selenium atoms with the cobalt in complex 3 may also then reduce this zinc-finger interaction and explain why the complex is less effective. The cobalt complex 3 resulted in some changes in nuclear chromatin condensation as well as of nuclear cytoplasmic ratios and development of numerous pseudopodia; however, the occasional granular cells also appeared. This, together with clearly lower NBT activity, suggests that the differentiation induced by the complex is less than that induced by the metal-free ligand H8qaSeSC, which may relate to the selenium-induced effects being suppressed by the very tight binding within the complex.
Exposure to the oxygen ligand H8qaSC shows smaller sized cells with reduced nuclear cytoplasmic ratios as well as evidence of nuclear chromatin condensation also indicating a partial differentiation. It is interesting to note that its complex 1 showed only minimal differentiation, unlike its strong cytotoxic activity to tumour cells (see below). From cell cycle analysis (vide infra), complex 1 appears to damage DNA resulting in sub G1 fragmentation of DNA, possibly due to the cobalt interfering with DNA repair proteins, including zinc-finger factors.62,63 However, one should point out that complex 1 toxicity occurs at doses above 40 μM, whilst effects on differentiation were tested at doses of 1 to 10 μM. Thus whilst the DNA damage induced at higher doses may be cytotoxic, the little amount induced at these low doses may be differentiating without being cytotoxic.64 As in the case of complex 3, in complex 1 too, the complexation appears to reduce the efficacy of the metal-free semicarbazone ligand as a differentiating agent, whilst in this case, markedly increasing its cytotoxicity.
On the other hand, the H8qaTSC and its cobalt(iii) complex 2 (latter not shown) both show little activity on the NBT screen and similarly show a morphology very similar to the undifferentiated cells, with large nuclear-cytoplasmic ratios, nucleoli and many mitoses. Again here, the stronger binding between the sulphur chalcogen donor atoms in the complex with cobalt(iii) may reduce its ability to interact with repair proteins causing DNA damage.
One should point out that since these are abnormal leukaemia cells (and not normal haematopoietic progenitors) being differentiated, the variability in the morphology creates difficulty in understanding the nominal stage of (normal haematopoietic) differentiation induced by the various agents. For this reason, three independent medical observers reviewed the slides to score the indicative features of the differentiated cells (Table S4, ESI‡).
2.6. Cytotoxic effects on tumour cell lines and non-transformed cells
The anti-proliferative activity of the (chalcogen)semicarbazone ligands, their cobalt complexes 1–3 and reference compound CDDP was determined by the MTT assay after 48 h treatment on five tumour cell lines: cervix carcinoma cells (HeLa), lung adenocarcinoma cells (A549), colorectal adenocarcinoma cells (LS-174), breast carcinoma cells (MDA-MB-361), and chronic myeloid leukaemia (K562), as well as one normal human cell line: fetal lung fibroblast cells (MRC-5). Growth inhibition effects of the investigated compounds, expressed in terms of IC50 values (Table 2), were determined from the cell survival diagrams (Fig. S11, ESI‡).
Table 2. In vitro cytotoxicity (IC50 in μM) a of the ligands and corresponding cobalt complexes 1–3 determined by the MTT assay after 48 h incubation.
| IC50 (μM) |
||||||
| HeLa | A549 | MDA-MB-361 | LS-174 | K562 | MRC-5 | |
| H8qaSC | 36.5 ± 3.9 | >100 | >100 | >100 | >100 | >100 |
| H8qaTSC | 75.6 ± 5.8 | >100 | >100 | >100 | >100 | >100 |
| H8qaSeSC | 6.6 ± 1.4 | 53.1 ± 2.8 | 9.2 ± 4.4 | 14.4 ± 2.3 | 4.0 ± 0.2 | 30.3 ± 3.6 |
| 1 | 17.2 ± 1.6 | 24.9 ± 3.2 | 45.5 ± 3.4 | 32.9 ± 4.5 | 37.6 ± 0.3 | 39.5 ± 1.0 |
| 2 | 46.2 ± 3.6 | >100 | >100 | >100 | >100 | >100 |
| 3 | 33.2 ± 2.2 | >100 | 36.3 ± 0.4 | >100 | 31.3 ± 5.9 | >100 |
| CDDP | 5.2 ± 0.3 | 26.2 ± 5.4 | 14.7 ± 1.2 | 22.4 ± 7.2 | 18.6 ± 3.3 | 12.1 ± 0.9 |
aValues represent the mean ± SD from three independent experiments.
In the investigated series of compounds, the selenosemicarbazone ligand H8qaSeSC had the highest cytotoxicity. It showed a strong cytotoxic effect on HeLa, K562, LS-174 and MDA-MB-361 cells, in the range of the activity of CDDP. In fact for a number of these cell lines it appeared considerably more toxic than this standard. It is worth mentioning that the ligand H8qaSeSC had a lower toxicity on normal cells (MRC-5), than on most of the investigated tumor cell lines. The toxicity of H8qaSeSC on these untransformed MRC-5 cells was also considerably less than that of CDDP. The two other ligands, H8qaSC and H8qaTSC, had a low cytotoxicity on all investigated cell lines, reaching the IC50 in the investigated range of concentrations only on HeLa cells. Among the complexes, complex 1 with the semicarbazone ligand H8qaSC showed the highest cytotoxicity, possibly due to the cobalt(ii) ion and its possible interaction with DNA repair proteins,65,66 which can also explain the DNA damage and sub-G1 cells seen in the cell cycle analysis. However, this toxicity was still considerably less than that of the metal-free selenium ligand on most cell lines. HeLa cells were the most sensitive to the action of complex 1, while the breast cancer cells (MDA-MB-361) were the most resistant. The cytotoxicity of this complex on the normal MRC-5 cells was two times lower than the activity on HeLa cells. The selenosemicarbazone complex 3 showed similar cytotoxicity on HeLa, MDA-MB-361 and K562 cells, but it was not cytotoxic to A549, LS-174 and normal cells (MRC-5) in the investigated concentration range. The thiosemicarbazone complex 2 had the lowest activity in the investigated series of complexes, reaching IC50 only on HeLa cells in the investigated concentration range.
The IC50 values (Table 2) indicate that the ligands generally show a cytotoxic activity in the following order: H8qaSeSC > H8qaSC > H8qaTSC, which is consistent with the literature data for related (chalcogen)semicarbazones.26,30 The order of activity for the complexes is: 1 > 3 > 2, where complexation increased the activity just in the case of complex 1. In this case cytotoxicity is most likely due to the metal, as the ligand H8qaSC is not active. Namely, it is known that cellular uptake of cobalt is genotoxic due to radical-mediated DNA damage and direct cobalt interference with DNA repair probably by substituting zinc ions from zinc-finger proteins.63,67 Also, cobalt(ii) ions themselves induce generation of reactive oxygen species in a Fenton-like reaction,68 and can replace magnesium(ii) ions in enzymatic physiological enzyme reactions, which strongly enhance DNA cleavage.69 It can be assumed that cobalt(ii) complex 1 is involved in oxidative damage of DNA, which is further supported indirectly by the results of cell cycle analysis (vide infra). Selectivity toward cancer cells as compared to the normal cell line was noticed for the selenosemicarbazone ligand H8qaSeSC, its cobalt(iii) complex 3, and cobalt(ii) complex 1 with the semicarbazone ligand.
2.7. Cell cycle analysis and mechanistic analysis of cell death
The effects of the ligands and complexes on cell cycle progression of the HeLa cells were examined by flow cytometry, after continual treatment for 24 and 48 h, using staining with propidium iodide (PI). Examination of the histograms of HeLa cells (Fig. S12, ESI‡) indicated that the ligands have different methods of action on dividing cells. H8qaSeSC as well as complexes 2 and 3 induce a reduction in G1 and an accumulation of cells in S and G2 phases of the cell cycle, indicating a G2/M arrest. This is a common feature of the toxic effects of a number of related chemical agents, including selenium containing compounds and cobalt organometallics.70–73 The toxicity of complex 3 may well have been due to a reduced effect of a cell cycle G2/M response as compared to the metal-free selenosemicarbazone ligand. On the other hand, complex 1 causes a clear increase in the sub-G1 fraction of cells, indicating a possible apoptosis or necrosis, probably secondary to DNA damage. As indicated above, this may be due to the function of the cobalt ion. The closer interaction of the sulphur and selenium atoms and the cobalt(iii) unlike the cobalt(ii) in complex 1 may reduce this cobalt toxicity in the other complexes. Some toxicity can occur with cobalt(iii) complexes,45 which may explain the cell cycle and microscopic changes seen in this case.
An initial assessment of mechanisms of cell death induced by these compounds was performed using Annexin-V and PI staining both with cytometry and microscopy. Results of the microscopy (Fig. S13, ESI‡) clearly confirm the patterns shown by the cell cycle analysis with H8qaSeSC and the complexes having the major effects and with complex 2 exhibiting effects more rapidly than the others. Further details of perturbations of the cell cycle and apoptosis induction (Fig. S14, ESI‡) are found in the ESI.‡
3. Conclusions
Two cobalt-(chalcogen)semicarbazone complexes have been synthesized and characterized by X-ray crystallography. All the complexes possess an octahedral geometry, with coordination of the ligands via quinoline and azomethine nitrogen atom and chalcogen donor atom. Preparation of all three cobalt-(chalcogen)semicarbazone complexes derived from 8-quinolinecaroxaldehyde made it possible to study the effect of metal ion complexation and chalcogen donor identity on the cytotoxic activity, but direct comparison can be made just for Co(iii) complexes 2 and 3. The strong differentiation-inducing ability of the semicarbazone and selenocarbazone ligands may provide future differentiation agents for use either alone or with standard chemotherapy or DNA modifying drugs. The plan is to proceed with testing these effects in the future on other leukaemia cell lines, using cytometry as an adjunct test, to see if the strong effects detected are replicated. Differentiation assessment will also be performed on certain solid tumour lines, particularly brain and bone tumour cells. The cobalt(ii) complex with the semicarbazone ligand shows strong anti-tumour cell activity and yet minimal toxicity on normal cells. Thus further investigation may yield novel chemotherapeutic agents for tumours with better safety profiles.
Supplementary Material
Acknowledgments
The authors acknowledge networking support by the COST Action CM1106 StemChem – “Chemical Approaches to Targeting Drug Resistance in Cancer Stem Cells”. The work was financially supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Grants OI 172055 and III 41026).
Footnotes
†The authors declare no competing interests.
‡Electronic supplementary information (ESI) available: Experimental section. Selected geometrical parameters (Table S1); hydrogen bond and π–π stacking interaction parameters (Tables S2 and S3); packing diagrams (Fig. S1 and S2); IR spectra of 1 and 2 (Fig. S3 and S4); NMR spectra of 2 (Fig. S5–S7) and UV-vis spectra (Fig. S8 and S9); feature scoring to indicate signs of differentiation (Table S4); concentration–response curves and cell cycle on HL60 cells (Fig. S10); cell survival diagrams (Fig. S11); cell cycle distribution (Fig. S12); fluorescence micrograph (Fig. S13); flow cytometry dot plot diagrams (Fig. S14). CCDC 1401684–1401686. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c6md00501b
References
- Brustugun O. T., Moller B., Helland A. Br. J. Cancer. 2014;11:1014–1020. doi: 10.1038/bjc.2014.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. J., Zhou G. B. IUBMB Life. 2012;64:671–675. doi: 10.1002/iub.1055. [DOI] [PubMed] [Google Scholar]
- Bozic I., Allen B., Nowak M. A. Trends Mol. Med. 2012;18:311–316. doi: 10.1016/j.molmed.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofran Y., Rowe J. M. Acta Haematol. 2014;132:292–297. doi: 10.1159/000360200. [DOI] [PubMed] [Google Scholar]
- Graubert T., Stone R. Semin. Hematol. 2014;51:322–329. doi: 10.1053/j.seminhematol.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Lavau C., Jansen J., Dejean A. Pathol. Biol. 1995;43:188–196. [PubMed] [Google Scholar]
- Randolph T. R. Clin. Lab. Sci. 2000;13:106–116. [PubMed] [Google Scholar]
- Grignani F., Ferrucci P. F., Testa U., Talamo G., Fagioli M., Alcalay M., Mencarelli A., Grignani F., Peschle C., Nicoletti I., Pelicci P. G. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- Fenaux P., Castaigne S., Dombret H., Archimbaud E., Duarte M., Morel P., Lamy T., Tilly H., Guerci A., Maloisel F. Blood. 1992;80:2176–2181. [PubMed] [Google Scholar]
- Chen S. J., Zhu Y. J., Tong J. H., Dong S., Huang W., Chen Y., Xiang W. M., Zhang L., Li X. S., Qian G. Q. Blood. 1991;78:2696–2701. [PubMed] [Google Scholar]
- Coombs C. C., Tavakkoli M., Tallman M. S. Blood Cancer J. 2015;5:e304. doi: 10.1038/bcj.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarezaie A., Khaledi F., Esfahani H. N., Ghaleghasemi T. M. J. Educ. Health Promot. 2014;3:64. doi: 10.4103/2277-9531.134778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco A., Carli M., Iacona I., Frascella E., Ninfo V., Rosolen A. Cancer. 2000;89:2661–2666. doi: 10.1002/1097-0142(20001215)89:12<2661::aid-cncr20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Liang C., Yang L., Guo S. Oncol. Lett. 2015;9:2833–2838. doi: 10.3892/ol.2015.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Schelker R., Klobuch S., Zaiss S., Troppmann M., Rehli M., Haferlach T., Herr W., Reichle A. Haematologica. 2015;100:e4–e6. doi: 10.3324/haematol.2014.115055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhye S. B., Kauffman B. G. Coord. Chem. Rev. 1985;63:127–160. [Google Scholar]
- Casas J. S., Garcia-Tasende M. S., Sordo J. Coord. Chem. Rev. 2000;209:197–261. [Google Scholar]
- Pelosi G. Open Crystallogr. J. 2010;3:16–28. [Google Scholar]
- Lobana T. S., Sharma R., Bawa G., Khanna S. Coord. Chem. Rev. 2009;253:977–1055. [Google Scholar]
- Moorthy N. S. H. N., Cerqueira N. M. F. S. A., Ramos M. J., Fernandes P. A. Mini-Rev. Med. Chem. 2013;13:1862–1872. doi: 10.2174/13895575113136660090. [DOI] [PubMed] [Google Scholar]
- Yu Y., Gutierrez E., Kovacevic Z., Saletta F., Obeidy P., Rahmanto Y. S., Richardson D. R. Curr. Med. Chem. 2012;19:2689–2702. doi: 10.2174/092986712800609706. [DOI] [PubMed] [Google Scholar]
- Kovacevic Z., Kalinowski D. S., Lovejoy D. B., Yu Y., Rahmanto S., Sharpe P. C., Bernhardt P. V., Richardson D. R. Curr. Top. Med. Chem. 2011;11:483–499. doi: 10.2174/156802611794785190. [DOI] [PubMed] [Google Scholar]
- Kalinowski D. S., Quach P., Richardson D. R. Future Med. Chem. 2009;1:1143–1151. doi: 10.4155/fmc.09.80. [DOI] [PubMed] [Google Scholar]
- Beraldo H., Gambino D. Mini-Rev. Med. Chem. 2004;4:31–39. doi: 10.2174/1389557043487484. [DOI] [PubMed] [Google Scholar]
- Yu Y., Kalinowski D. S., Kovacevic Z., Siafakas A. R., Jansson P. J., Stefani C., Lovejoy D. B., Sharpe P. C., Bernhardt P. V., Richardson D. R. J. Med. Chem. 2009;52:5271–5294. doi: 10.1021/jm900552r. [DOI] [PubMed] [Google Scholar]
- Kowol C. R., Eichinger R., Jakupec M. A., Galanski M., Arion V. B., Keppler B. K. J. Inorg. Biochem. 2007;101:1946–1957. doi: 10.1016/j.jinorgbio.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Serda M., Kalinowski D. S., Mrozek-Wilczkiewicz A., Musiol R., Szurko A., Ratuszna A., Pantarat N., Kovacevic Z., Merlot A. M., Richardson D. R., Polanski J. Bioorg. Med. Chem. Lett. 2012;22:5527–5531. doi: 10.1016/j.bmcl.2012.07.030. [DOI] [PubMed] [Google Scholar]
- Chaston T. B., Lovejoy D. B., Watts R. N., Richardson D. R. Clin. Cancer Res. 2003;9:402–414. [PubMed] [Google Scholar]
- Raja D. S., Bhuvanesh N. S. P., Natarajan K. Inorg. Chem. 2011;50:12852–12866. doi: 10.1021/ic2020308. [DOI] [PubMed] [Google Scholar]
- Agrawal K. C., Booth B. A., Michaud R. L., Moore E. C., Sartorelli A. C. Biochem. Pharmacol. 1974;23:2421–2429. doi: 10.1016/0006-2952(74)90233-0. [DOI] [PubMed] [Google Scholar]
- Gligorijević N., Todorović T., Radulović S., Sladić D., Filipović N., Gođevac D., Jeremić D., Anđelković K. Eur. J. Med. Chem. 2009;44:1623–1629. doi: 10.1016/j.ejmech.2008.07.033. [DOI] [PubMed] [Google Scholar]
- Todorović T. R., Bacchi A., Sladić D. M., Todorović N. M., BoŽić T. T., Radanović D. D., Filipović N. R., Pelizzi G., Anđelković K. K. Inorg. Chim. Acta. 2009;362:3813–3820. [Google Scholar]
- Bjelogrlić S., Todorović T., Bacchi A., Zec M., Sladić D., Srdić-Rajić T., Radanović D., Radulović S., Pelizzi G., Anđelković K. J. Inorg. Biochem. 2010;104:673–682. doi: 10.1016/j.jinorgbio.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Srdić-Rajić T., Zec M., Todorović T., Anđelković K., Radulović S. Eur. J. Med. Chem. 2011;46:3734–3747. doi: 10.1016/j.ejmech.2011.05.039. [DOI] [PubMed] [Google Scholar]
- Zec M. M., Srdić-Rajić T. V., Krivokuća A. M., Janković R. N., Todorović T. R., Anđelković K. K., Radulović S. S. Med. Chem. 2014;10:759–771. doi: 10.2174/1573406410666140327122009. [DOI] [PubMed] [Google Scholar]
- Zec M., Srdić-Rajić T., Konić-Ristić A., Todorović T., Andjelković K., Filipović-Ljeskovi I., Radulović S. Anti-Cancer Agents Med. Chem. 2012;10:1071–1080. doi: 10.2174/187152012803529682. [DOI] [PubMed] [Google Scholar]
- Filipović N., Polović N., Rašković B., Misirlić-Denčić S., Dulović M., Savić M., Nikšić M., Mitić D., Anđelković K., Todorović T. Monatsh. Chem. 2014;145:1089–1099. [Google Scholar]
- Revenko M. D., Prisacari V. I., Dizdari A. V., Stratulat E. F., Corja I. D., Proca L. M. Pharm. Chem. J. 2011;45:351–354. [Google Scholar]
- Castle T. C., Maurer R. I., Sowrey F. E., Went M. J., Reynolds C. A., McInnes E. J. L., Blower P. J. J. Am. Chem. Soc. 2003;125:10040–10049. doi: 10.1021/ja035737d. [DOI] [PubMed] [Google Scholar]
- Andaloussi M. B. D., Mohr F. J. J. Organomet. Chem. 2010;695:1276–1280. [Google Scholar]
- Todorović T. R., Bacchi A., Pelizzi G., Juranić N. O., Sladić D. M., Brčeski I. D., Anđelković K. K. Inorg. Chem. Commun. 2006;9:862–865. [Google Scholar]
- Todorović T. R., Bacchi A., Juranić N. O., Sladić D. M., Pelizzi G., BoŽić T. T., Filipović N. R., Anđelković K. K. Polyhedron. 2007;26:3428–3436. [Google Scholar]
- Manikandan R., Vijayan P., Anitha P., Prakash G., Viswanathamurthi P., Butcher R. J., Velmurugan K., Nandhakumar R. Inorg. Chim. Acta. 2014;421:80–90. [Google Scholar]
- Fan X., Dong J., Min R., Chen Y., Yi X., Zhou J., Zhang S. J. Coord. Chem. 2013;66:4268–4279. [Google Scholar]
- Manikandan R., Viswanathamurthi P., Velmurugan K., Nandhakumar R., Hashimoto T., Endo A. J. Photochem. Photobiol., B. 2014;130:205–216. doi: 10.1016/j.jphotobiol.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Ramachandran E., Thomas S. P., Poornima P., Kalaivani P., Prabhakaran R., Padma V. V., Natarajan K. Eur. J. Med. Chem. 2012;50:405–415. doi: 10.1016/j.ejmech.2012.02.026. [DOI] [PubMed] [Google Scholar]
- Fleck R. A., Athwal H., Bygraves J. A., Hockley D. J., Feavers I. M., Stacey G. N. In Vitro Cell. Dev. Biol.: Anim. 2003;39:235–242. doi: 10.1290/1543-706X(2003)039<0235:OONAHD>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- West D. X., Billeh I. S., Jasinski J. P., Jasinski J. M., Butcher R. J. Transition Met. Chem. 1998;23:209–314. [Google Scholar]
- Fallab S. and Mitchell P. R., Advances in Inorganic and Bioinorganic Mechanisms, vol. 3, Academic Press, London, 1984. [Google Scholar]
- Eaton D. R., O'Reilly A. Inorg. Chem. 1987;26:4185–4188. [Google Scholar]
- Blackman A. G., in Encyclopedia of Inorganic Chemistry, ed. R. Bruce King, Wiley, Chichester, Hoboken, New York, ch. Cobalt: Inorganic & coordination Chemistry, 2005, pp. 1–25. [Google Scholar]
- Chumakov Y. M., Biyushkin V. N., Bodyu V. G. J. Struct. Chem. 1985;26:929–934. [Google Scholar]
- Zhou J., Chen Z.-F., Tan Y.-S., Wang X.-W., Tan Y.-H., Liang H., Zhang Y. Acta Crystallogr., Sect. E: Struct. Rep. Online. 2004;60:m519–m521. [Google Scholar]
- Battaglia L. P., Berzolla P. G., Corradi A. B., Pelizzi C. J. Crystallogr. Spectrosc. Res. 1993;23:973–979. [Google Scholar]
- Allen F. H. Acta Crystallogr., Sect. B: Struct. Sci. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- Filipović N. R., Bjelogrlić S., Portalone G., Pelliccia S., Silvestri R., Klisurić O., Senćanski M., Stanković D., Todorović T. R., Muller C. D. MedChemComm. 2016;7:1604–1616. [Google Scholar]
- Stoica S., Magoulas G. E., Antoniou A. I., Suleiman S., Cassar A., Gatt L., Papaioannou D., Athanassopoulos C. M., Schembri-Wismayer P. Bioorg. Med. Chem. Lett. 2016;26:1145–1150. doi: 10.1016/j.bmcl.2016.01.048. [DOI] [PubMed] [Google Scholar]
- Valiuliene G., Stirblyte I., Cicenaite D., Kaupinis A., Valius M., Navakauskiene R. J. Cell. Mol. Med. 2015;19:1742–1755. doi: 10.1111/jcmm.12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig A., Blessing H., Schwerdtle T., Walter I. Toxicology. 2003;193:161–169. doi: 10.1016/j.tox.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Larabee J. L., Hocker J. R., Hanas J. S. J. Inorg. Biochem. 2009;103:419–426. doi: 10.1016/j.jinorgbio.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de The H. Nat. Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Beyersmann D., Hartwig A. Arch. Toxicol. 2008;82:493–512. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- Asmuss M., Mullenders L. H., Hartwig A. Toxicol. Lett. 2000;112–113:227–231. doi: 10.1016/s0378-4274(99)00273-8. [DOI] [PubMed] [Google Scholar]
- Wingert S., Thalheimer F. B., Haetscher N., Rehage M., Schroeder T., Rieger M. A. Stem Cells. 2016;34:699–710. doi: 10.1002/stem.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Dong J., Min R., Chen Y., Yi X., Zhou J., Zhang S. J. Coord. Chem. 2013;66:4268–4279. [Google Scholar]
- Baldwin E. L., Wilson Byl J. A., Osheroff N. Biochemistry. 2004;43:728–735. doi: 10.1021/bi035472f. [DOI] [PubMed] [Google Scholar]
- Chao H. and Hi L.-N., Metallotherapeutic Drugs and Metal-Based Diagnostic Agents: The Use of Metals in Medicine, ed. M. Gielen and E. R. T. Tiekink, John Wiley & Sons, England, 2005, ch. 11, pp. 201–218. [Google Scholar]
- Beyersmann D., Hartwig A. Arch. Toxicol. 2008;82:493–512. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- Baldwin E. L., Byl J. A., Osheroff N. Biochemistry. 2004;43:728–735. doi: 10.1021/bi035472f. [DOI] [PubMed] [Google Scholar]
- Zhao R., Xiang N., Domann F. E., Zhong W. Nutr. Cancer. 2009;61:397–407. doi: 10.1080/01635580802582751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N., Shi X., Zhao Y., Tian L., Wang D., Yang X. J. Agric. Food Chem. 2013;61:579–588. doi: 10.1021/jf3036929. [DOI] [PubMed] [Google Scholar]
- Choi A. R., Jo M. J., Jung M. J., Kim H. S., Yoon S. Eur. J. Pharmacol. 2015;764:63–69. doi: 10.1016/j.ejphar.2015.06.046. [DOI] [PubMed] [Google Scholar]
- Kowalski K., Hikisz P., Szczupak L., Therrien B., Koceva-Chyla A. Eur. J. Med. Chem. 2014;81:289–300. doi: 10.1016/j.ejmech.2014.05.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.