 A series of novel β-carboline-based hydroxamate derivatives (8a–n) as HDAC inhibitors have been designed and synthesized.
A series of novel β-carboline-based hydroxamate derivatives (8a–n) as HDAC inhibitors have been designed and synthesized.
Abstract
A series of novel β-carboline-based hydroxamate derivatives (8a–n) as HDAC inhibitors have been designed and synthesized. Most of these compounds displayed potent histone deacetylase inhibitory effects and good antiproliferative activity with IC50s in the low micromolar range. One of the most potent compounds (8k) showed the strongest inhibition of the proliferation of human hepatocellular carcinoma (HCC) cells in vitro, with IC50 values lower than that of the currently approved HDAC inhibitor SAHA. Compound 8k also increased acetylation of histone H3 and α-tubulin, consistent with its potent HDAC inhibition. Importantly, 8k induced hypochromism by electrostatic interactions with CT-DNA, suggesting potential induction of DNA damage. Finally, 8k significantly induced HepG2 cell apoptosis by regulating apoptotic relative proteins expression. Together, our findings suggest that these novel β-carboline-based hydroxamate derivatives may provide a new framework for the discovery of novel antitumor agents for the intervention of human carcinoma cells.
Introduction
Many small molecules could interact with DNA through intercalation, surface binding, minor groove, or major groove binding.1 Studies on DNA interacting compounds in cancer chemotherapy have led to the development of many novel heterocyclic compounds as potential anticancer agents. In fact, anticancer drugs that target DNA are one of the most effective agents in clinical usage and have enhanced the survival rates of patients when used alone or in combination with other drugs.2 These agents exert their biological profile by non-covalent or covalent interactions in either the minor or major groove or between the base pairs (intercalation) of the double helix. As a result, a number of novel DNA interacting scaffolds such as anthracyclines, acridines, anthraquinones, distamycins, and β-carbolines have been developed in anti-cancer drug discovery efforts.3
β-Carbolines are alkaloids that contain a planar tricyclic pyrido-[3,4-b]indole ring structure. They are widely distributed in plants, marine creatures, and mammals, as well as human tissues and bodily fluids.4 Many naturally occurring β-carboline alkaloids and synthetic analogues containing the β-carboline subunit have been reported to inhibit cancer cell growth and induce apoptosis through multiple mechanisms such as DNA intercalation, inhibition of DNA topoisomerases I and II, CDK, PLK, and ABCG2.5–10 DNA intercalation is one of the important modes of action in clinical oncology and many drugs with this mechanism are currently used for the treatment of various cancers.11 Several β-carbolines including harmane, norharman, and β-carboline–benzimidazole conjugates have been reported to intercalate with DNA, alter DNA replication fidelity, and influence enzymatic activities in DNA repair processes due to the presence of a planar polycyclic aromatic pharmacophore, which is capable of stacking between DNA base pairs.5–7,12,13
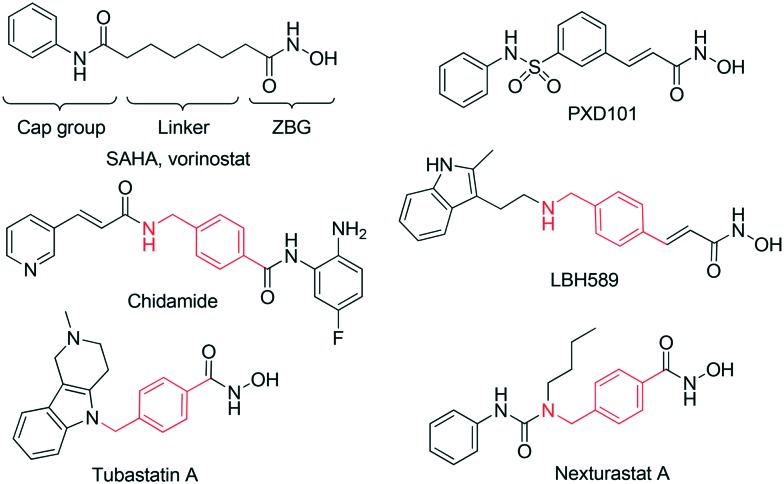
Inhibition of histone deacetylases (HDACs) has long been considered a promising therapeutic approach to the treatment of cancer. It has been shown that HDAC inhibitors (HDACi) significantly suppress cancer cell proliferation, angiogenesis, and metastasis, and induce apoptosis through multiple mechanisms, including changes in gene expression and alterations of both histone and non-histone proteins.14,15 Five HDACi have been approved for cancer treatment to date. Despite promising results in the treatment of cutaneous T-cell lymphoma, the two US FDA approved HDACi, Vorinostat (SAHA)16 and romidepsin,17 have not been proven effective in clinical trials involving solid tumors (Fig. 1).18,19 Other HDAC inhibitors that have recently been approved include belinostat (PXD101), panobinostat (LBH589), and chidamide (Epidaza), for the treatment of PTCL or multiple myeloma,20–22 although their effects on solid tumors have not been examined.
Fig. 1. Structures of clinically approved HDAC inhibitors in clinical trials, and some representative HDAC inhibitors with the benzylic linker.
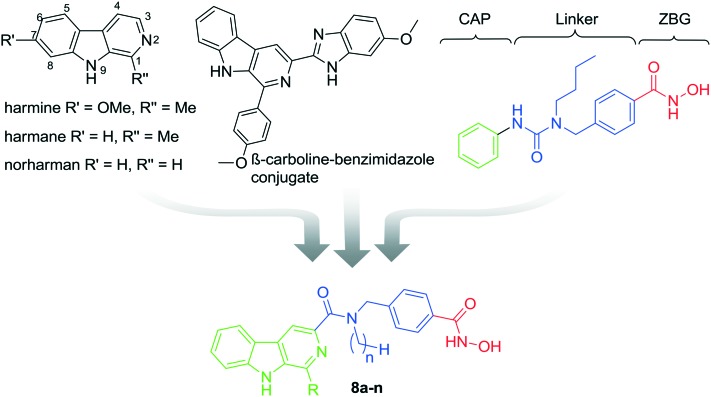
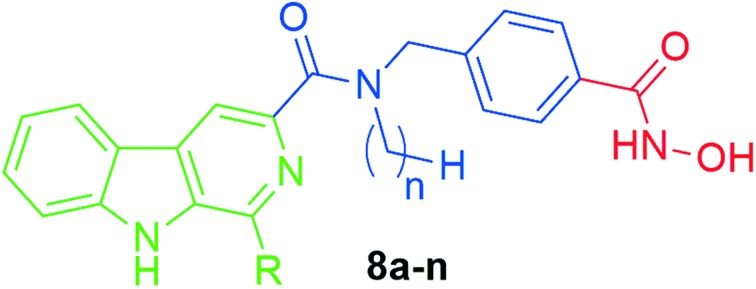
It has been widely reported that HDACi can synergistically enhance the inhibitory effect of other antitumor agents, which target tubulin, EGFR, DNA, or topoisomerase,23–30 in suppressing proliferation and inducing apoptosis in tumor cells. HDAC inhibitors have therefore been pursued in the design of multifunctional inhibitors to simultaneously interact with multiple targets with high potency and low toxicity.26–28 Given the DNA damage effects of β-carbolines, the combination of HDACi and β-carboline alkaloids may represent a promising strategy to achieve increased anticancer efficacy. A key feature of many potent and selective HDAC inhibitors is the presence of the benzylic linker built into the canonical inhibitor structure of the common CAP–linker–zinc binding group scaffold, as seen in nexturastat A, tubastatin A, and chidamide (Fig. 1). Thus, we aimed to integrate the key structural elements of β-carboline alkaloids and HDACi into one single structure to form novel β-carboline derivatives that may simultaneously inhibit DNA and HDAC and provide improved antitumor activity (Fig. 2). We herein report the synthesis and biological evaluation of a series of novel hydroxamic acid-based β-carboline derivatives (8a–n), as well as the investigation on their anti-tumor mechanisms in multiple cancer cell lines.
Fig. 2. The design of novel hydroxamate-based β-carboline derivatives 8a–n.
Results and discussion
Chemistry
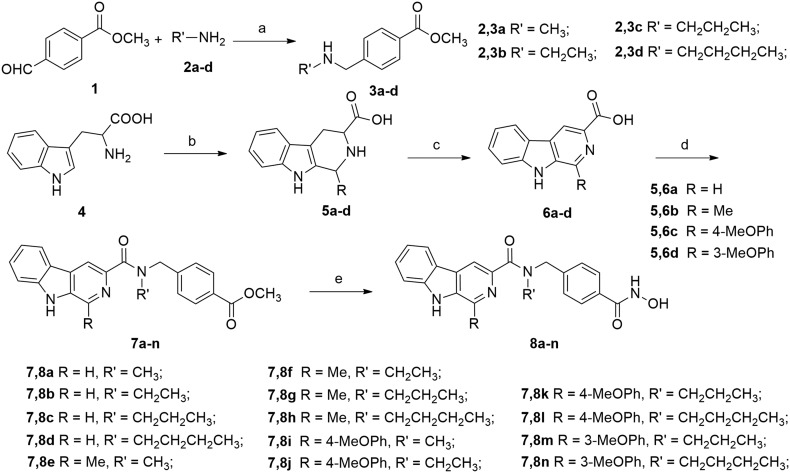
The synthetic route to compounds 8a–n is depicted in Scheme 1. The starting methyl 4-formylbenzoate 1 reacted with different alkyl amines in the presence of acetic acid to afford the imines, which were then reduced by NaBH4 to give the secondary amines 2a–d. Substituted β-carbolines 6a–d were prepared in a two-step sequence. First, commercially available l-tryptophan (4) was converted to 1-substituted-1,2,3,4-tetrahydro-β-carboline-3-carboxylic acid 5a–d in a Pictet–Spengler reaction with the treatment of differently substituted aldehydes (formaldehyde, acetaldehyde or 4-methoxyphenaldehyde). Intermediates 5a–d were then oxidized by KMnO4 in DMF to afford compounds 6a–d, which then reacted with secondary amines 3a–d in the presence of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDCI) and 4-dimethyl-amino-pyridine (DMAP) to produce amides 7a–n. Finally, intermediates 7a–n were treated with NH2OK to give the target compounds 8a–n. All target compounds were purified by column chromatography, and their structures were confirmed by 1H NMR, MS, and HRMS.
Scheme 1. Synthetic routes of target compounds 8a–n. Reagents and conditions: (a) (i) HOAc, THF, rt, 1–2 h, (ii) NaBH4, THF, rt, 2 h, 56–73%; (b) H+ or OH–, RCHO, reflux, 2–4 h, 81–86%; (c) KMnO4, DMF, rt, 6 h, 67–75%; (d) EDCI, DMAP, CH2Cl2, rt, 5–8 h; and (e) NH2OK, MeOH, rt, 10–15 h, 52–67%.
In vitro biological evaluations
All target compounds were first tested against a HeLa cell nuclear extract, a rich source of HDAC activity, for their HDAC inhibitory potencies,31 using SAHA, an FDA approved HDAC inhibitor, as the positive control. The IC50 values of compounds 8a–n against HDAC in the HeLa cell nuclear extract are listed in Table 2. Harmine showed an IC50 value greater than 10 μM. All target compounds showed HDAC inhibitory effects with IC50 values in the low micromolar range.
Table 2. IC50 values of compounds 5k–o and 11a–j against four CRC cell lines a .
| Compd. |
In vitro antiproliferative activity (IC50,
a
μM) |
|||
| HCT116 | HepG2 | SUMM-7721 | LOVO | |
| SAHA | 5.53 ± 0.68 | 6.26 ± 0.51 | 5.61 ± 0.57 | 6.75 ± 0.58 |
| Harmine | 46.7 ± 3.92 | 51.2 ± 4.76 | 55.3 ± 5.01 | 53.2 ± 4.35 |
| 8a | >12.5 | >12.5 | >12.5 | ND b |
| 8b | >12.5 | >12.5 | >12.5 | ND |
| 8c | >12.5 | >12.5 | >12.5 | ND |
| 8d | >12.5 | >12.5 | >12.5 | ND |
| 8e | >12.5 | >12.5 | >12.5 | >12.5 |
| 8f | >12.5 | >12.5 | >12.5 | >12.5 |
| 8g | 10.5 ± 0.93 | 9.83 ± 0.86 | 11.6 ± 1.28 | >12.5 |
| 8h | >12.5 | 11.7 ± 1.35 | >12.5 | >12.5 |
| 8i | 9.76 ± 1.09 | 10.5 ± 1.21 | >12.5 | >12.5 |
| 8j | 6.07 ± 0.58 | 8.34 ± 0.76 | 10.3 ± 0.87 | 9.24 ± 1.10 |
| 8k | 2.24 ± 0.21 | 2.15 ± 0.36 | 3.42 ± 0.31 | 5.07 ± 0.63 |
| 8l | 3.02 ± 0.26 | 3.37 ± 0.52 | 5.25 ± 0.60 | 4.76 ± 0.55 |
| 8m | 2.96 ± 0.30 | 3.64 ± 0.59 | 4.12 ± 0.36 | 5.31 ± 0.52 |
| 8n | 4.32 ± 0.41 | 5.16 ± 0.62 | 5.75 ± 0.58 | 7.63 ± 0.67 |
aThe inhibitory effects of individual compounds on the proliferation of cancer cell lines were determined by the MTT assay. The data are expressed as the mean ± SD of three independent experiments.
bND: not detected.
All the synthesized compounds 8a–n were then screened for their cancer cell growth inhibitory activity against human hepatocellular carcinoma cells (HepG2 and SMMC-7721) and human colon cancer cells (HCT116 and LOVO) using the MTT assay. β-Carboline analog harmine and SAHA were used as reference compounds. The IC50 values of 8a–n against the four human cancer cell lines are listed in Table 1. SAHA significantly inhibited the proliferation of all four types of cells, while harmine showed limited potency.
Table 1. Compounds 8a–n and their HDAC inhibition.
| Compd. | R | R′ | Nuclear extract (IC50, a μM) |
| SAHA | — | — | 0.48 ± 0.06 |
| Harmine | — | — | >10 |
| 8a | H | CH3 | 5.63 ± 0.65 |
| 8b | H | CH2CH3 | 4.78 ± 0.61 |
| 8c | H | CH2CH2CH3 | 2.82 ± 0.37 |
| 8d | H | CH2CH2CH2CH3 | 2.69 ± 0.30 |
| 8e | Me | CH3 | 3.06 ± 0.33 |
| 8f | Me | CH2CH3 | 1.75 ± 0.28 |
| 8g | Me | CH2CH2CH3 | 0.85 ± 0.31 |
| 8h | Me | CH2CH2CH2CH3 | 0.93 ± 0.22 |
| 8i | p-MeOPh | CH3 | 0.96 ± 0.13 |
| 8j | p-MeOPh | CH2CH3 | 0.83 ± 0.10 |
| 8k | p-MeOPh | CH2CH2CH3 | 0.26 ± 0.03 |
| 8l | p-MeOPh | CH2CH2CH2CH3 | 0.34 ± 0.05 |
| 8m | m-MeOPh | CH2CH2CH3 | 0.23 ± 0.04 |
| 8n | m-MeOPh | CH2CH2CH2CH3 | 0.37 ± 0.08 |
aThe data are expressed as the mean ± SD of three independent experiments.
These compounds were designed to evaluate the structure–activity relationships (SARs) at the C1 position of the β-carbolines, as well as the N-alkylation of the C3 amide. The results clearly demonstrate the impact of C1 substitution on both the HDAC inhibition and antiproliferation. For HDAC inhibition (Table 1), compounds 8a–d with R = H as in norhamane were less potent than 8e–h with R = Me as in harmane. The methoxyphenyl series (8i–n) were more potent than the previous two series. In cancer antiproliferative activity, a similar trend is present. Compounds 8a–d (R = H) and 8e–h (R = Me) all showed limited antiproliferative activities. Derivatives 8i–n with a methoxyphenyl group at the C1 position, as present in the previously reported β-carboline–benzimidazole conjugates,10 markedly inhibited human cancer cell growth, showing greater antiproliferative activity against the tested cells than harmine.
Within each subseries (R = H, Me, or methoxyphenyl), IC50 values in HDAC inhibition first increased then decreased with the elongation of the N-alkyl substitution, with the compounds bearing the propyl group being the most potent of each subseries. Compounds 8k and 8m were the most active analogues, with an IC50 value of 0.26 μM and 0.23 μM against the nuclear extract, respectively. In antiproliferation against the 4 cancer cells, the propyl analogs (8k and 8m) were also the most active compounds. Compound 8k exhibited anti-proliferative activities against HepG2 cells with an IC50 value of 2.15 μM, which is three-fold lower than SAHA (IC50 = 6.26 μM). Although 8k was slightly less potent than 8m in HDAC inhibition, it was more potent in anti-proliferation.
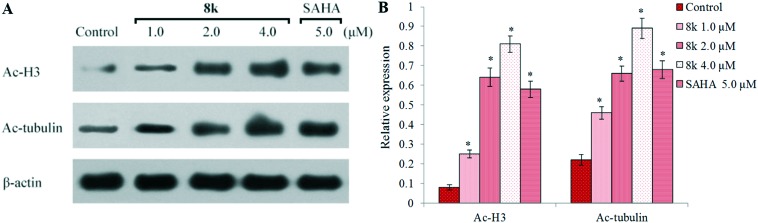
Acetylation of histone H3 and α-tubulin
Given that the inhibition of HDACs by β-carboline derivative 8k enhanced the tumor cell antiproliferative activity, we further investigated whether 8k induces acetylation of histones in HCC cells. The cells were treated with different concentrations of 8k and the levels of acetylation of histone H3 and α-tubulin were analyzed by western blotting assays using β-actin as the negative control (Fig. 3). HepG2 cells were incubated with the vehicle alone, SAHA (5.0 μM), or 8k (1.0, 2.0, and 4.0 μM) for 48 h. Compared to the control group, compound 8k demonstrated the ability to increase the expression of Ac-histone H3 and Ac-α-tubulin correlating with the 8k dose of treatment. The levels of Ac-histone H3 and Ac-α-tubulin in 8k treated groups (2.0 and 4.0 μM) were higher than or equivalent to the values from the 5.0 μM SAHA treated group, which was consistent with the results from the HDAC fluorimetric activity assay. The above results about Ac-histone H3 and Ac-α-tubulin clearly showed that 8k was not selective against HDAC1/2/3/6. Therefore, the selective profile of 8k against other representative HDAC isoforms, such as HDAC4 and HDAC8, was tested. However, 8k showed rather weak HDAC4 inhibition with IC50 values >10 μM and moderate HDAC8 inhibitory activity (IC50 = 0.76 μM), which were similar to SAHA.
Fig. 3. The expression of the acetylation for histone H3 and α-tubulin was examined by western blot analysis in vitro. (A) HepG2 cells were treated with 8k and SAHA for 48 h at the indicated concentrations, and the levels of protein expression were detected using anti-acetyl-histone H3, anti-acetyl-α-tubulin, and anti-β-actin antibodies, respectively. β-Actin was used as the loading control. Data shown are representative images of each protein for three separate experiments. (B) Quantitative analysis. The relative levels of Ac-H3 and Ac-α-tubulin used to control β-actin were determined by densimetric scanning. The data are expressed as means ± SD of three separate experiments. *P < 0.01 vs. control.
DNA binding studies by UV-visible spectroscopic analysis
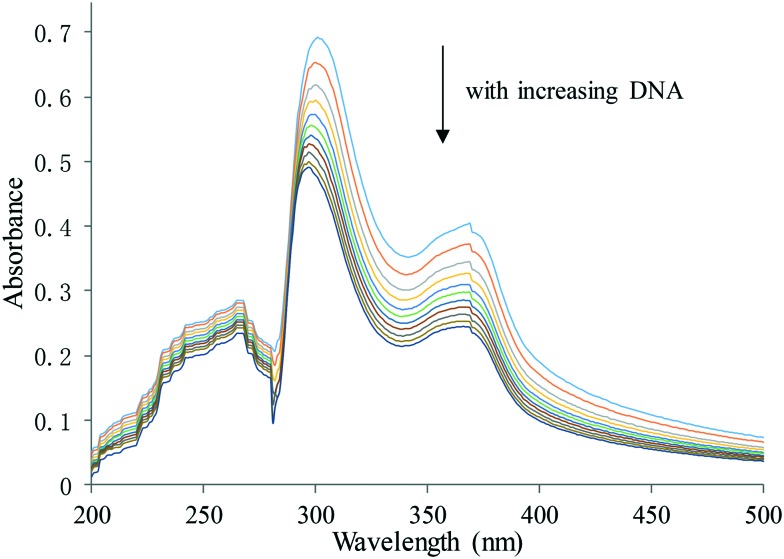
It is generally accepted that the planar structured β-carbolines can bind to DNA and induce DNA damage.11,12 The SAR results clearly showed the contribution of HDAC inhibition to the antitumor activities. To investigate whether the antitumor activities of these active β-carboline derivatives also arise partly from their interactions with DNA, UV-visible spectroscopic titration studies were performed. In general, hypochromism, or reduction in absorption, is observed when a small molecule binds to DNA through intercalation, resulting from a strong π–π stacking interaction between an aromatic chromophore from the molecule and the base pairs of DNA.24 In our study, the 8k–CT-DNA system showed absorption bands at 260 nm, 300 nm and 365 nm (Fig. 4). Upon addition of equal increments of CT-DNA to an 8k solution, the intensity of all 3 absorption bands gradually decreased. The maximum absorption was at 300 nm, and the strongest hypochromicity was also observed at this wavelength. A similar trend was observed in harmine, with the maximum absorption at 320 nm. However, the absorption slightly increased with SAHA upon addition of incremental CT-DNA (see the ESI‡). These results clearly indicate electrostatic binding of compound 8k to CT-DNA with intercalation.
Fig. 4. UV-visible absorption spectra of 8k in the presence of increasing amounts of CT-DNA. Arrows indicate the changes in absorbance with increasing concentration of DNA. [8k] = 20 μM.
Compound 8k induces apoptosis in HepG2 cells
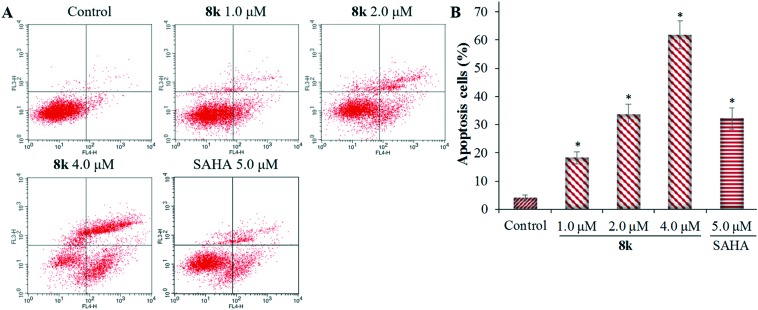
To determine whether the inhibitory effects of 8k on liver cancer cellular proliferation are accompanied by enhanced cancer cell apoptosis, FITC-annexin V/propidium iodide (PI) staining and flow cytometry assay were performed, and the percentages of apoptotic cells were determined. HepG2 cells were incubated with different concentrations of 8k or SAHA for 48 h. As shown in Fig. 5, 8k treated HepG2 cells exhibited a dose-dependent increase (P < 0.01) of apoptosis by 18.2%, 35.2%, and 61.8% at 1.0 μM, 2.0 μM, and 4.0 μM, respectively. The 61.8% induction of HepG2 cell apoptosis with 4.0 μM 8k was significantly higher than 5.0 μM SAHA (34.5% apoptotic cells).
Fig. 5. Compound 8k induces HepG2 cell apoptosis in vitro. HepG2 cells were incubated with the indicated concentrations of 8k or SAHA (5.0 μM) for 48 h, and the cells were stained with FITC-annexin V/PI, followed by flow cytometry analysis. (A) Flow cytometry analysis. (B) Quantitative analysis of apoptotic cells. Data are expressed as means ± SD of the percentages of apoptotic cells from three independent experiments. *P < 0.01 vs. control.
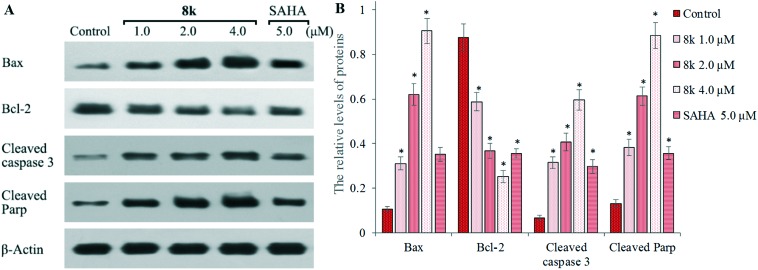
To further investigate the apoptosis induction of 8k, we examined the expression of apoptotic proteins Bax and Bcl-2, and the cleavage states of caspase-3 and PARP, in response to 8k treatment (Fig. 6). Sub-confluent HepG2 liver cancer cells were treated with or without 8k for 48 h, and then lysed and analyzed by western blot. β-Actin expression was used as an internal control. It was revealed that treatment with 8k dramatically increased the relative levels of pro-apoptotic Bax expression, but reduced the levels of anti-apoptotic Bcl-2 expression (Fig. 6A) in a dose-dependent manner. Furthermore, compound 8k resulted in more significant cleavage of both PARP and caspase-3 than the control group in Fig. 6B. Importantly, 8k treatment also induced more cleavage of PARP and caspase-3 than the SAHA treated group. Taken together, these results confirm that 8k treatment induced apoptosis associated with cleavage of caspase 3 and PARP in HepG2 cells.
Fig. 6. The expression of Bax, Bcl-2, cleaved caspase 3, PARP, and β-actin was examined by western blot analysis. (A) HepG2 cells were incubated with, or without, 8k and SAHA at the indicated concentrations for 48 h and the levels of protein expression were detected using specific antibodies. Data shown are representative images of each protein for three separate experiments. (B) Quantitative analysis of Bax, Bcl-2, cleaved caspase 3, and cleaved PARP. The relative levels of each protein compared to control β-actin were determined by densimetric scanning. Data are expressed as means ± SD from three separate experiments. *P < 0.01 vs. respective control.
Preliminary in vitro pharmacokinetic studies
In order to assess the drug-likeness of this class, we have conducted preliminary in vitro pharmacokinetic studies on compound 8k. Solubility is an important parameter to predict drug absorption and it is commonly accepted that insufficient drug solubility can lead to poor oral absorption. The most potent compound 8k of the series demonstrated moderate aqueous solubility (11.8 mg mL–1). We also evaluated the metabolic stability of 8k in incubation with rat liver microsomes (BD Gentest). The compound concentration was 1 μM and the final liver microsomal protein concentration was 0.5 mg mL–1. The percentage of remaining 8k was followed using LC-MS and the half-life (t1/2) was determined using the slope of the linear regression method. In this assay, 8k showed a t1/2 of 20.43 min, which is comparable to the reported t1/2 of SAHA (44 min) with a test concentration of 5 μM. These results are consistent with the fact that 8k and SAHA share similar sites of metabolism such as the hydroxamic acid moiety and the amide group.
Conclusion
In summary, we designed and synthesized a series of β-carboline-based hydroxamate derivatives (8a–n) and investigated their in vitro biological activities. Most of these compounds displayed significant histone deacetylase inhibitory effects and good antiproliferative activity with IC50 values in the low micromolar range. Their activities are clearly influenced by the substitution at the C1 position and the amide N-alkylation. Compound 8k with a C1-methoxyphenyl group and N-propyl was the most potent of the series and possessed greater antiproliferative activity against cancer cells than SAHA. In addition, compound 8k dose-dependently increased the acetylation of histone H3 and α-tubulin, as expected for HDAC inhibitors. The increased antiproliferative activity of 8k most likely resulted partly from induced DNA damage by the β-carboline structural moiety, as evidenced by the hypochromism observed in the DNA binding studies by UV-visible spectroscopic analysis. Further, 8k could significantly induce cell apoptosis by reducing the anti-apoptotic protein levels of Bcl-2 and up-regulating the pro-apoptotic proteins of Bax and the cleavage of caspase 3 and PARP. Finally, 8k showed reasonable solubility and metabolic stability. Our findings suggest that β-carboline-based hydroxamates such as 8k may hold promise as potential therapeutic agents for the intervention of human cancers.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the financial support by the Natural Science Foundation of China (Grant No. 81302628 and 81473089), the Project of “Jiangsu Six Peaks of Talent” (2014-SWYY-044 and SWYY-CXTD-008), China Pharmaceutical University for the Open Project Program of the State Key Laboratory of Natural Medicines (SKLNMKF201611), the China and Jiangsu Province Postdoctoral Science Foundation (2016 M590488 and 1601136B), the Jiangsu Government Scholarship for Overseas Studies (JS-2014-212), Jiangsu Key Training Programs of Innovation and Entrepreneurship for Undergraduates (201610304055Z), and the Applied Research Projects of Nantong City (MS12015060). The authors also thank the Priority Academic Programs Development (PAPD) of Jiangsu Higher Education Institutions for funding the project.
Footnotes
†The authors declare no competing interests.
‡Electronic supplementary information (ESI) available. See DOI: 10.1039/c6md00681g
References
- Hurley L. H. Nat. Rev. Cancer. 2002;2:188. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- Xu Y., Her C. Biomolecules. 2015;5:1652–1670. doi: 10.3390/biom5031652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momekov G., Bakalova A., Karavanova M. Curr. Med. Chem. 2005;12:2177. doi: 10.2174/0929867054864877. [DOI] [PubMed] [Google Scholar]
- Patel K., Gadewar M., Tripathi R., Tripathi S., Prasad K., Patel D. K. Asian Pac. J. Trop. Biomed. 2012;2:660–664. doi: 10.1016/S2221-1691(12)60116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaraiah N., Siraj K. P., Nekkanti S., Srinivasulu V., Sharma P., Senwar K. R., Sathish M., Vishnuvardhan M. V., Ramakrishna S., Jadala C., Nagesh N., Kamal A. Bioorg. Chem. 2015;59:130–139. doi: 10.1016/j.bioorg.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Wang K. B., Li D. H., Hu P., Wang W. J., Lin C., Wang J., Lin B., Bai J., Pei Y. H., Jing Y. K., Li Z. L., Yang D., Hua H. M. Org. Lett. 2016;18:3398. doi: 10.1021/acs.orglett.6b01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. H., Zhao M., Qian K. D., Lee K. H., Morris-Natschke S., Peng S. Q. Eur. J. Med. Chem. 2009;44:4153–4161. doi: 10.1016/j.ejmech.2009.05.006. [DOI] [PubMed] [Google Scholar]
- He L., Liao S. Y., Tan C. P., Ye R. R., Xu Y. W., Zhao M., Ji L. N., Mao Z. W. Chemistry. 2013;19:12152–12160. doi: 10.1002/chem.201301389. [DOI] [PubMed] [Google Scholar]
- Han X., Zhang J., Guo L., Cao R., Li Y., Li N., Ma Q., Wu J., Wang Y., Si S. PLoS One. 2012;7:e46546. doi: 10.1371/journal.pone.0046546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler A., Stefan K., Wiese M. J. Med. Chem. 2016;59:6121. doi: 10.1021/acs.jmedchem.6b00035. [DOI] [PubMed] [Google Scholar]
- Martinez R., Chacon-Garcia L. Curr. Med. Chem. 2005;12:127. doi: 10.2174/0929867053363414. [DOI] [PubMed] [Google Scholar]
- Kamal A., Rao M. P. N., Swapna P., Srinivasulu V., Bagul C., Shaik A. B., Mullagiri K., Kovvuri J., Reddy V. S., Vidyasagar K., Nagesh N. Org. Biomol. Chem. 2014;12:2370–2387. doi: 10.1039/c3ob42236d. [DOI] [PubMed] [Google Scholar]
- Lin Y. C., Chen Y. F., Tseng L. S., Lee Y. H., Morris-Natschke S. L., Kuo S. C., Yang N. S., Lee K. H., Huang L. J. Eur. J. Med. Chem. 2016;110:98. doi: 10.1016/j.ejmech.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. J., Seto E. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- Gryder B. E., Sodji Q. H., Oyelere A. K. Future Med. Chem. 2012;4:505–524. doi: 10.4155/fmc.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S., Easley C., Kirkpatrick P. Nat. Rev. Drug Discovery. 2007;6:21–22. doi: 10.1038/nrd2227. [DOI] [PubMed] [Google Scholar]
- Grant C., Rahman F., Piekarz R., Peer C., Frye R., Robey R. W., Gardner E. R., Figg W. D., Bates S. E. Expert Rev. Anticancer Ther. 2010;10:997–1008. doi: 10.1586/era.10.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico M., Bagella L. J. Biomed. Biotechnol. 2011;2011:475641. doi: 10.1155/2011/475641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Cang S., Ma Y., Petrillo R. L., Liu D. J. Hematol. Oncol. 2010;3:5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. A. Am. J. Health-Syst. Pharm. 2014;71:1328. doi: 10.2146/news140056. [DOI] [PubMed] [Google Scholar]
- Garnock-Jones K. P. Drugs. 2015;75:695–704. doi: 10.1007/s40265-015-0388-8. [DOI] [PubMed] [Google Scholar]
- Gu R., Liu T., Zhu X., Gan H., Wu Z., Li J., Zheng Y., Dou G., Meng Z. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2015;1000:181–186. doi: 10.1016/j.jchromb.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang J., Tong L., Luo Y., Su M., Zang Y., Li J., Lu W., Chen Y. Bioorg. Med. Chem. 2013;21:3240–3244. doi: 10.1016/j.bmc.2013.03.049. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Bao R., Tao X., Wang J., Atoyan R., Qu H., Wang D. G., Yin L., Samson M., Forrester J., Zifcak B., Xu G. X., DellaRocca S., Zhai H. X., Cai X., Munger W. E., Keegan M., Pepicelli C. V., Qian C. Cancer Res. 2010;70:3647–3656. doi: 10.1158/0008-5472.CAN-09-3360. [DOI] [PubMed] [Google Scholar]
- Sato T., Suzuki M., Sato Y., Echigo S., Rikiishi H. Int. J. Oncol. 2006;28:1233–1241. [PubMed] [Google Scholar]
- Sarcar B., Kahali S., Chinnaiyan P. J. Neurooncol. 2010;99:201–207. doi: 10.1007/s11060-010-0127-7. [DOI] [PubMed] [Google Scholar]
- Budman D. R., Tai J., Calabro A., John V. Invest. New Drugs. 2011;29:1224–1229. doi: 10.1007/s10637-010-9467-6. [DOI] [PubMed] [Google Scholar]
- Ling Y., Xu C. J., Luo L., Cao J., Feng J., Xue Y., Zhu Q., Ju C., Li F., Zhang Y. A., Ling X. J. Med. Chem. 2015;58:9214. doi: 10.1021/acs.jmedchem.5b01052. [DOI] [PubMed] [Google Scholar]
- Zhao X., Tan Q., Zhang Z., Zhao Y. Med. Chem. Res. 2014;23:5188. [Google Scholar]
- Zhao Y., Hui J., Zhu L. Med. Chem. Res. 2013;22:2505. [Google Scholar]
- Zhang Y., Feng J., Jia Y., Wang X., Zhang L., Liu C., Fang H., Xu W. J. Med. Chem. 2011;54:2823. doi: 10.1021/jm101605z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.