 The reduction–rebridging strategy is a powerful method for the preparation of stable and homogeneous antibody–drug conjugates (ADCs).
The reduction–rebridging strategy is a powerful method for the preparation of stable and homogeneous antibody–drug conjugates (ADCs).
Abstract
The reduction–rebridging strategy is a powerful method for the preparation of stable and homogeneous antibody–drug conjugates (ADCs). In this communication, we describe the development of the arylene-dipropiolonitrile (ADPN) functional group for the rebridging of reduced disulphide bonds and its application in the preparation of potent and selective ADCs.
Antibody–drug conjugates (ADCs) are efficient therapies that combine the targeting specificity of monoclonal antibodies with the potency of small molecular drugs. Four ADCs have been approved by the FDA and more than 65 are currently in clinical trials, illustrating the potential of these powerful targeted therapies.1 Recent research has shown that a number of parameters, such as the drug-to-antibody ratio (DAR),2 site of conjugation3 and linker chemistry,4,5 can influence the efficacy of ADCs. Currently marketed ADCs are heterogeneous mixtures of species with different DARs and conjugation sites. The heterogeneity complicates the development of ADCs, since species with different DARs and conjugation sites have different pharmacokinetic and toxicity profiles.6 Therefore, reducing the heterogeneity of ADCs can yield more efficient and predictable therapies.7 One of the possible ways to reduce the ADC heterogeneity is the site-specific introduction of amino acids or peptide sequences that can be further modified using enzymatic or chemical conjugation approaches.8 However, this approach requires the use of costly and laborious techniques for protein engineering in order to produce the modified antibodies.
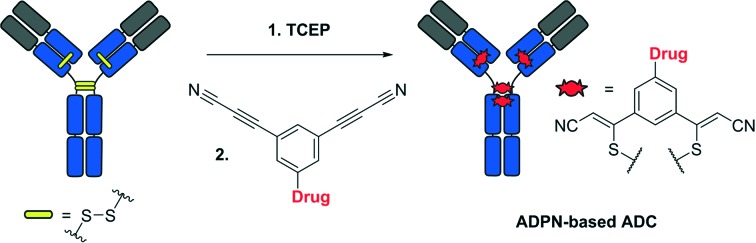
An elegant alternative strategy is reduction–rebridging, which exploits the higher accessibility of interchain disulphide bonds located in the hinge region of monoclonal antibodies. These disulphide bonds can be easily reduced under mild conditions without disrupting the intrachain disulphide bonds or the secondary structure of the antibody. Further rebridging can be achieved through a reaction with 3,4-disubstituted maleimides,9–11 bissulfones,12 dibromopyridazinediones13–16 or alkynes under UV irradiation.17 In addition to offering a higher homogeneity, ADCs prepared using the reduction–rebridging strategy retain the covalent links between the chains of the antibody, which improves the stability of their secondary structures.
Recently, we have described the use of the 3-arylpropiolonitrile (APN) moiety for the preparation of ADCs with improved plasma stability.18–20 The S–Csp2 bond formed upon conjugation of the APN group with cysteine residues has shown excellent stability in the presence of other amino acid residues, such as lysine. In this paper, we discuss the development and application of the 3,3′-arylene-dipropiolonitrile (ADPN) functional group for the preparation of ADCs using the reduction–rebridging approach (Fig. 1).
Fig. 1. Reduction–alkylation strategy (left) and rebridging strategy (right) for the preparation of ADCs.
We hypothesized that the introduction of a second propiolonitrile functional group into the aromatic ring would provide bifunctional reagents suitable for rebridging reduced disulphide bonds. In this pilot study, the potential of ADPN as a novel bioconjugation functional group was explored with ADCs consisting of trastuzumab, monomethyl auristatin E (MMAE) and our β-galactosidase-responsive self-immolative linker (Fig. 1).17–19 Following selective internalization inside HER2-positive cancer cells, these ADCs are designed to be activated by lysosomal β-galactosidase leading to the release of MMAE, thereby restoring its cytotoxic activity in a stringently controlled fashion.
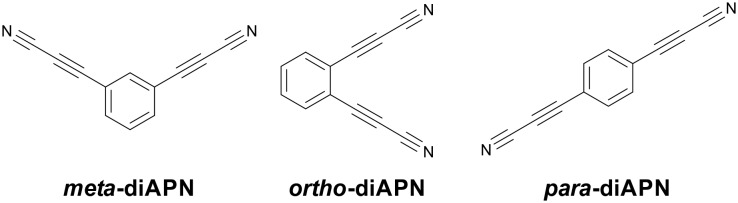
For our initial study, we have prepared meta, ortho and para isomers of ADPN (Fig. 2). The products were synthesized following a previously described procedure.20 Briefly, corresponding phenyl diiodides underwent Sonogashira coupling with propargyl alcohol to provide intermediates that were oxidized in the presence of ammonia to yield the isomers of ADPN.
Fig. 2. Structures of the model ADPN isomers used in the initial studies.
In order to choose the most suitable isomer for disulphide rebridging, we carried out a comparative study using trastuzumab (obtained from a specialty pharmacy and purified by ultrafiltration) as a model antibody. This monoclonal IgG1 antibody consists of two heavy chains (H) and two light chains (L), linked together by four interchain disulphide bridges. Rebridging of the reduced antibody can yield the total antibody (HHLL) as well as HHL, HL and HH fragments.
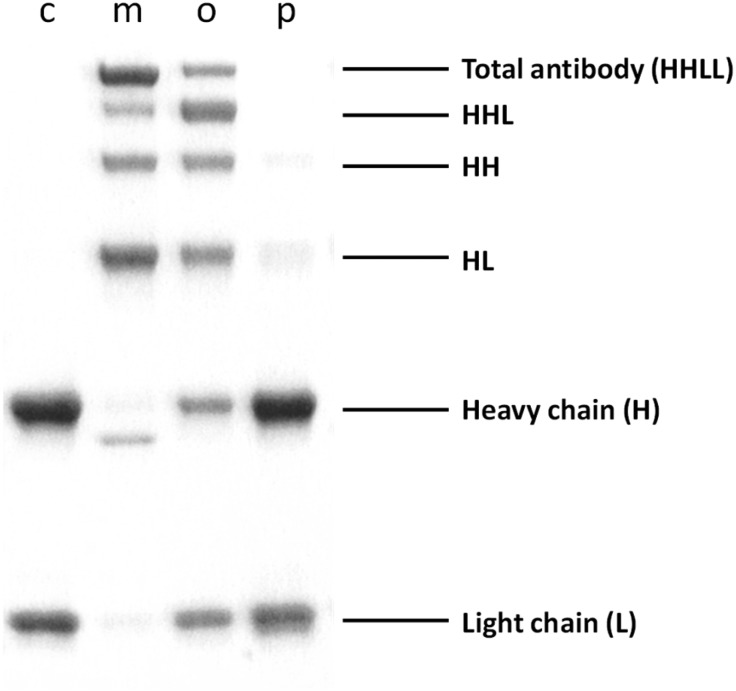
We incubated the reduced trastuzumab with each ADPN isomer for 12 h. The conjugates were purified by size-exclusion chromatography and analysed using SDS-PAGE under reducing conditions (Fig. 3). This allowed the identification of antibody fragments that were covalently bound by ADPN isomers. Surprisingly, para-ADPN yielded only trace amounts of the rebridged antibody fragments. Ortho-ADPN demonstrated higher efficiency in the rebridging, but substantial amounts of free heavy and light chains were still present. This lack of efficiency of para- and ortho-ADPN may be explained by the fact that the electron-withdrawing substituents in these positions reduce the hydrolytic stability of the propiolonitrile functional group, making it less compatible with the aqueous buffers used for conjugation. Meta-ADPN appeared to be the most efficient rebridging reagent, since SDS-PAGE revealed no free heavy or light chain fragments.
Fig. 3. Reducing SDS-PAGE analysis of the rebridged trastuzumab. Lane c – reduced trastuzumab. Lanes m, o and p – reduced trastuzumab rebridged with meta-, ortho- and para-ADPN, respectively.
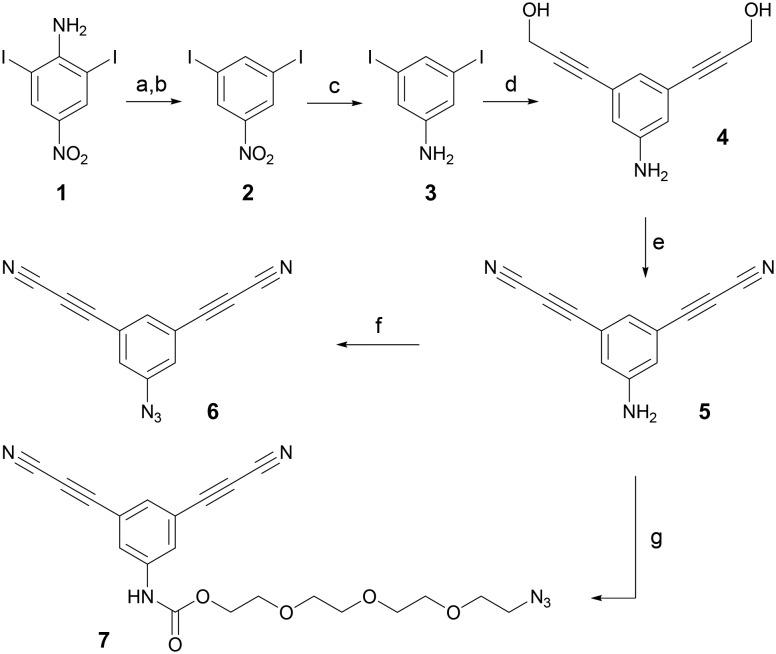
Encouraged by these results, we proceeded to the synthesis of cytotoxic payloads containing meta-ADPN for antibody rebridging. First, we prepared the trifunctional precursor containing meta-ADPN and an azide group, suitable for coupling various drugs (Scheme 1).
Scheme 1. Synthesis of the trifunctional compound 6. a) NaNO2, H2SO4, 0 °C, 2 h; b) CuSO4·5H2O, EtOH, reflux, 2 h, 36%; c) SnCl2·2H2O, NaBH4, EtOH, reflux, 0.5 h, 93%; d) propargyl alcohol (3 eq.), NEt3 (4 eq.) Pd(PPh3)2Cl2 (2 mol%), CuI (4 mol%), DMF, r.t., 14 h, 96%; e) MnO2 (30 eq.), MgSO4 (30 eq.), NH3 (8 eq., 2 M isopropanol solution), THF, r.t., 2 h, 11%; f) NaNO2 (1.1 eq.), NaN3 (2 eq.), HCl (15%), H2O, 0 °C, 5 min, 96%.
Thus, commercially available 2,6-diiodo-4-nitroaniline (1) was diazotized by NaNO2 in H2SO4 and reduced by heating in EtOH in the presence of CuSO4 to give 2. The nitro group in 2 was reduced by SnCl2 and NaBH4 in EtOH to provide product 3. Double Sonogashira coupling of 3 with propargyl alcohol yielded intermediate 4, which was oxidized by MnO2 in the presence of NH3 and MgSO4 to give product 5. Finally, an azido group was introduced by reaction of 5 with NaNO2/HCl followed by NaN3, giving the trifunctional precursor 6. Another trifunctional compound, 7, containing a short PEG linker, was prepared from 5 by reaction with triphosgene followed by N3-PEG-OH.
In order to prepare payloads for antibody rebridging, we used the galactoside (8) that was previously described by our group (Scheme 2).19,20 This has already been employed with success for the preparation of β-galactosidase-responsive ADCs that showed significant anticancer activity in vivo. Thus, the cytotoxic payload 9 was synthesized by coupling compounds 6 and 8 using the CuAAC reaction.
Scheme 2. Synthesis of cytotoxic payloads for antibody rebridging. a) Cu(MeCN)4PF6 (1.5 eq.), CH2Cl2/MeOH (1 : 0.2), r.t., 18 h, 75%; b) Cu(MeCN)4PF6 (1.5 eq.), CH2Cl2/MeOH (1 : 0.2), r.t., 6 h, 17% after purification by preparative reverse-phase HPLC.
Since the galactoside trigger must be readily accessible to lysosomal β-galactosidase for inducing efficient drug release, we also investigated the preparation of payload 10. Indeed, in payload 9, the short distance between the bulky antibody and the enzyme substrate could hamper the recognition of the carbohydrate moiety by β-galactosidase. Therefore, to prevent this possible issue, we synthesized payload 10, which contains a short PEG linker, by coupling compounds 7 and 8 under the same conditions as those used for the preparation of 9.
Trastuzumab was used as a model antibody for the preparation of new ADCs. The ADCs were prepared following the optimized reduction–rebridging protocol (ESI‡). Briefly, the interchain disulphide bonds of the antibody were reduced by incubation with 5 eq. of TCEP in PBS at 37 °C. The reduced antibody was subsequently incubated with 9 or 10 for 12 h at 25 °C. The resulting rebridged ADCs (T-ADPN-Gal-MMAE prepared from 9 and T-ADPN-PEG4-Gal-MMAE prepared from 10) were purified by size-exclusion chromatography. According to native mass spectrometry analysis, the average DAR values were 4.5 for T-ADPN-Gal-MMAE and 4.0 for T-ADPN-PEG4-Gal-MMAE. Although the major species in the DAR distribution of each ADC corresponded to DAR 4, small amounts of DAR 5 species were also observed (Fig. S1‡). The formation of DAR 5 species may occur if the reduced disulphide reacts with two different molecules of ADPN-based payload. Further optimization of the DAR distribution may be achieved through fine-tuning of the antibody and payload concentrations.
The in vitro cytotoxicity of the new ADCs was evaluated on SK-BR-3 HER2-positive and MDA-MB-231 HER2-negative cancer cell lines, obtained from the American Tissue Culture Collection (ATCC, Manassas, VA, USA). The FDA-approved trastuzumab emtansine (T-DM1, obtained from a specialty pharmacy and purified by ultrafiltration) was used as the benchmark in our studies. Both rebridged ADCs showed strong activity on HER2-positive cancer cells (Fig. S1‡), with IC50 values comparable with those of T-DM1 (Table 1). Furthermore, they were approximately 200 times less toxic for MDA-MB-231 cells, demonstrating their selectivity for killing HER2-positive cancer cells. Interestingly, there was no significant difference between conjugates with and without the PEG linker, suggesting that the hinge region of the antibody is sufficiently accessible for β-galactosidase to perform efficient drug release.
Table 1. IC50 values of the ADPN-based ADCs compared to those of T-DM1.
| ADC | IC50 (nM)SK-BR-3 (HER2+) cell line | IC50 (nM)MDA-MB-231 (HER2–) cell line |
| T-ADPN-Gal-MMAE | 0.022 | 4.39 |
| T-ADPN-PEG4-Gal-MMAE | 0.019 | 3.43 |
| T-DM1 | 0.032 | 19.03 |
Conclusions
In conclusion, we have developed a new functional group for the rebridging of reduced disulphide bonds. The ADPN functional group was successfully combined with a β-galactosidase-cleavable linker and MMAE to yield a series of novel ADC payloads. The resulting payloads were used for the preparation of potent and selective ADCs via the rebridging of reduced trastuzumab.
Conflicts of interest
The authors declare no competing interest.
Supplementary Material
Acknowledgments
This work was supported by CNRS, University of Strasbourg, Région Alsace, La Ligue contre le Cancer (comités Deux-Sèvres et Vienne), “Agence Nationale de la Recherche” (ANR) and the French Proteomic Infrastructure (ProFI; ANR-10-INBS-08-03). The International Center for Frontier Research in Chemistry (icFRC) is also acknowledged for financial support. A. E. acknowledges the «Association Nationale de la Recherche et de la Technologie» (ANRT) and Syndivia for funding his PhD fellowship. We thank Dr. Wojciech Krezel for the assistance with the in vitro experiments.
Footnotes
†The authors contributed equally to this work.
‡Electronic supplementary information (ESI) available. See DOI: 10.1039/c8md00141c
References
- Beck A., Goetsch L., Dumontet C., Corvaïa N. Nat. Rev. Drug Discovery. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- Sun X., Ponte J. F., Yoder N. C., Laleau R., Coccia J., Lanieri L., Qiu Q., Wu R., Hong E., Bogalhas M., Wang L., Dong L., Setiady Y., Maloney E. K., Ab O., Zhang X., Pinkas J., Keating T. A., Chari R., Erickson H. K., Lambert J. M. Bioconjugate Chem. 2017;28:1371–1381. doi: 10.1021/acs.bioconjchem.7b00062. [DOI] [PubMed] [Google Scholar]
- Strop P., Liu S.-H., Dorywalska M., Delaria K., Dushin R. G., Tran T.-T., Ho W.-H., Farias S., Casas M. G., Abdiche Y., Zhou D., Chandrasekaran R., Samain C., Loo C., Rossi A., Rickert M., Krimm S., Wong T., Chin S. M., Yu J., Dilley J., Chaparro-Riggers J., Filzen G. F., O'Donnell C. J., Wang F., Myers J. S., Pons J., Shelton D. L., Rajpal A. Chem. Biol. 2013;20:161–167. doi: 10.1016/j.chembiol.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Lyon R. P., Setter J. R., Bovee T. D., Doronina S. O., Hunter J. H., Anderson M. E., Balasubramanian C. L., Duniho S. M., Leiske C. I., Li F., Senter P. D. Nat. Biotechnol. 2014;32:1059–1062. doi: 10.1038/nbt.2968. [DOI] [PubMed] [Google Scholar]
- Burke P. J., Hamilton J. Z., Pires T. A., Setter J. R., Hunter J. H., Cochran J. H., Waight A. B., Gordon K. A., Toki B. E., Emmerton K. K., Zeng W., Stone I. J., Senter P. D., Lyon R. P., Jeffrey S. C. Mol. Cancer Ther. 2016;15:938–945. doi: 10.1158/1535-7163.MCT-16-0038. [DOI] [PubMed] [Google Scholar]
- Hamblett K. J., Senter P. D., Chace D. F., Sun M. M. C., Lenox J., Cerveny C. G., Kissler K. M., Bernhardt S. X., Kopcha A. K., Zabinski R. F., Meyer D. L., Francisco J. A. Clin. Cancer Res. 2004;10:7063–7070. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- Pillow T. H., Tien J., Parsons-Reponte K. L., Bhakta S., Li H., Staben L. R., Li G., Chuh J., Fourie-O'Donohue A., Darwish M., Yip V., Liu L., Leipold D. D., Su D., Wu E., Spencer S. D., Shen B.-Q., Xu K., Kozak K. R., Raab H., Vandlen R., Lewis Phillips G. D., Scheller R. H., Polakis P., Sliwkowski M. X., Flygare J. A., Junutula J. R. J. Med. Chem. 2014;57:7890–7899. doi: 10.1021/jm500552c. [DOI] [PubMed] [Google Scholar]
- Agarwal P., Bertozzi C. R. Bioconjugate Chem. 2015;26:176–192. doi: 10.1021/bc5004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais M., Nunes J. P. M., Karu K., Forte N., Benni I., Smith M. E. B., Caddick S., Chudasama V., Baker J. R. Org. Biomol. Chem. 2017;15:2947–2952. doi: 10.1039/c7ob00220c. [DOI] [PubMed] [Google Scholar]
- Behrens C. R., Ha E. H., Chinn L. L., Bowers S., Probst G., Fitch-Bruhns M., Monteon J., Valdiosera A., Bermudez A., Liao-Chan S., Wong T., Melnick J., Theunissen J., Flory M. R., Houser D., Venstrom K., Levashova Z., Sauer P., Migone T., van der Horst E. H., Halcomb R. L., Jackson D. Y. Mol. Pharmaceutics. 2015;12:3986–3998. doi: 10.1021/acs.molpharmaceut.5b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes J. P. M., Morais M., Vassileva V., Robinson E., Rajkumar V. S., Smith M. E. B., Pedley R. B., Caddick S., Baker J. R., Chudasama V. Chem. Commun. 2015;51:10624–10627. doi: 10.1039/c5cc03557k. [DOI] [PubMed] [Google Scholar]
- Badescu G., Bryant P., Bird M., Henseleit K., Swierkosz J., Parekh V., Tommasi R., Pawlisz E., Jurlewicz K., Farys M., Camper N., Sheng X., Fisher M., Grygorash R., Kyle A., Abhilash A., Frigerio M., Edwards J., Godwin A. Bioconjugate Chem. 2014;25:1124–1136. doi: 10.1021/bc500148x. [DOI] [PubMed] [Google Scholar]
- Robinson E., Nunes J. P. M., Vassileva V., Maruani A., Nogueira J. C. F., Smith M. E. B., Pedley R. B., Caddick S., Baker J. R., Chudasama V. RSC Adv. 2017;7:9073–9077. [Google Scholar]
- Maruani A., Smith M. E. B., Miranda E., Chester K. A., Chudasama V., Caddick S. Nat. Commun. 2015;6:6645. doi: 10.1038/ncomms7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. T. W., Maruani A., Richards D. A., Baker J. R., Caddick S., Chudasama V. Chem. Sci. 2017;8:2056–2060. doi: 10.1039/c6sc03655d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. T. W., Maruani A., Baker J. R., Caddick S., Chudasama V. Chem. Sci. 2016;7:799–802. doi: 10.1039/c5sc02666k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebenow N., Dilmaç A. M., Greven S., Bräse S. Bioconjugate Chem. 2016;27:911–917. doi: 10.1021/acs.bioconjchem.5b00682. [DOI] [PubMed] [Google Scholar]
- Kolodych S., Koniev O., Baatarkhuu Z., Bonnefoy J.-Y., Debaene F., Cianférani S., Van Dorsselaer A., Wagner A. Bioconjugate Chem. 2015;26:197–200. doi: 10.1021/bc500610g. [DOI] [PubMed] [Google Scholar]
- Koniev O., Kolodych S., Baatarkhuu Z., Stojko J., Eberova J., Bonnefoy J.-Y., Cianférani S., Van Dorsselaer A., Wagner A. Bioconjugate Chem. 2015;26:1863–1867. doi: 10.1021/acs.bioconjchem.5b00440. [DOI] [PubMed] [Google Scholar]
- Kolodych S., Michel C., Delacroix S., Koniev O., Ehkirch A., Eberova J., Cianférani S., Renoux B., Krezel W., Poinot P., Muller C. D., Papot S., Wagner A. Eur. J. Med. Chem. 2017;142:376–382. doi: 10.1016/j.ejmech.2017.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.