Abstract
Background:
Human health assessments synthesize human, animal, and mechanistic data to produce toxicity values that are key inputs to risk-based decision making. Traditional assessments are data-, time-, and resource-intensive, and they cannot be developed for most environmental chemicals owing to a lack of appropriate data.
Objectives:
As recommended by the National Research Council, we propose a solution for predicting toxicity values for data-poor chemicals through development of quantitative structure–activity relationship (QSAR) models.
Methods:
We used a comprehensive database of chemicals with existing regulatory toxicity values from U.S. federal and state agencies to develop quantitative QSAR models. We compared QSAR-based model predictions to those based on high-throughput screening (HTS) assays.
Results:
QSAR models for noncancer threshold-based values and cancer slope factors had cross-validation-based of 0.25–0.45, mean model errors of 0.70–1.11 units, and applicability domains covering of environmental chemicals. Toxicity values predicted from QSAR models developed in this study were more accurate and precise than those based on HTS assays or mean-based predictions. A publicly accessible web interface to make predictions for any chemical of interest is available at http://toxvalue.org.
Conclusions:
An in silico tool that can predict toxicity values with an uncertainty of an order of magnitude or less can be used to quickly and quantitatively assess risks of environmental chemicals when traditional toxicity data or human health assessments are unavailable. This tool can fill a critical gap in the risk assessment and management of data-poor chemicals. https://doi.org/10.1289/EHP2998
Introduction
Of the tens of thousands of chemicals in commerce or on various manufacturing inventories, only hundreds have comprehensive toxicological data or have undergone some form of human health risk assessment (Guyton et al. 2009; Judson et al. 2009). Most assessments that support risk-based environmental health decision making rely on epidemiologic data, experimental animal data, or both, to quantitatively estimate risk and develop toxicity values or exposure thresholds. Such assessments, including the U.S. Environmental Protection Agency’s (EPA’s) Integrated Science Assessments and Integrated Risk Information System (IRIS) toxicological reviews, are highly data-, time-, and resource-intensive. However, a need exists for reliable approaches to systematically estimate human health risks for all, rather than for just a few, chemicals. Accordingly, the National Academies recommended the “development of default approaches to support risk estimation for the large number of chemicals lacking chemical-specific information” (NRC 2009).
There are a number of examples of how experimental data from short-term studies in animals or nonmammalian systems, such as the lethal dose 50% () or mutagenicity determinations, can be used to develop chronic toxicity estimates. For example, Zeise et al. (1984) developed an approach to estimate the cancer potency of a chemical when only the of that chemical is known. That study showed that a quantitative estimate of chronic cancer hazard from acute lethality information can be used to provide a conservative estimate of a “worst-case scenario” to guide decision making based on the assumption that the initial stages of chemical absorption and distribution would be comparable for both lethality and carcinogenicity (Zeise et al. 1984). Follow-up work extended this approach to estimate cancer risk based on data from short-term mutation and toxicity tests (Travis et al. 1990). Such approaches are sensible because information is available for thousands of chemicals (https://chem.nlm.nih.gov/chemidplus/), but only a few hundred of these have been tested in a 90-d toxicity study or in a 2-y cancer bioassay.
Complementary methodologies with great utility for supporting a variety of risk-based decision contexts include high-throughput assessments that use in vitro data, systems biology approaches, and pharmacokinetic modeling (Judson et al. 2011; Krewski et al. 2014). Several large-scale screening programs now generate toxicity hazard data for thousands of chemicals in hundreds of assays (Kavlock et al. 2012; Tice et al. 2013). These data are now used for various decision contexts from replacement of animal tests (Judson et al. 2015) to estimating potential human exposure levels (Wambaugh et al. 2013, 2015) to predictions of acute toxicity (Zhu et al. 2009) or cancer hazard class (Sedykh et al. 2011). Similarly, approaches to how in vitro (Wetmore 2015) or short-term in vivo (Thomas et al. 2013) toxicity studies can be used for risk-based evaluations have been recently proposed, and their applications have been illustrated in a National Academies report (National Academies of Science, Engineering, and Medicine 2017).
In parallel to the efforts in high-throughput toxicity testing, quantitative structure–activity relationship (QSAR) tools for predicting chemical metabolism (Maltarollo et al. 2015; Pinto et al. 2016) or for generating categorical predictions, such as classifying cancer and noncancer hazards (Jolly et al. 2015; Low et al. 2014; Mansouri et al. 2016; Rusyn et al. 2012), are already widely used in various decision contexts. However, there are few computational tools to generate quantitative (e.g., dose–response) estimates, information that is highly relevant for decision making and risk management. Specifically, there is a need for predictions of the continuous values used in decision making above and beyond the prediction of a chemical as a “hazard” versus a “nonhazard.” Therefore, this study aimed to address this significant gap through the development of the Conditional Toxicity Value (CTV) Predictor. The CTV Predictor is a compendium of QSAR models and a web portal that predicts, based on the chemical’s structure, an array of toxicity values that are often used in risk management decisions. The term “conditional” is used to distinguish the QSAR-based prediction of a toxicity value from toxicity values derived using human, animal, and other traditional data streams. The CTV-predicted values include the reference dose (RfD) and concentration (RfC), the oral slope factor (OSF), the inhalation unit risk (IUR), the cancer potency value [CPV; a California EPA (CalEPA)-specific OSF], and various estimates of the point-of-departure (POD). CTV predictions rely on a comprehensive database of existing toxicity values and experimental data and incorporate Organisation for Economic Co-operation and Development (OECD) principles for model building and external cross-validation. All of the data, models, and results are made publicly available for use by interested stakeholders.
Methods
Compounds and Toxicity Values
As part of previously published work (Wignall et al. 2014) and further described in Table 1 and Table S1, we compiled a database of publicly available, peer-reviewed human health toxicity values as of June 2011 as found in specific U.S. EPA sources [IRIS, Office of Pesticide Programs (OPP), Superfund Regional Screening Level Tables (RSLs)], or from the California EPA (Office of Environmental Health Hazard Assessment). Superfund RSL Tables reference toxicity values from the abovementioned sources as well as from Centers for Disease Control and Prevention (CDC)/Agency for Toxic Substances and Disease Registry (ATSDR) toxicological profiles, U.S. EPA Provisional Peer Reviewed Toxicity Values (PPRTV), U.S EPA Health Effects Assessment Summary Tables (HEAST), and other U.S. EPA offices (Wignall et al., 2014). The toxicity values extracted for this effort included chronic values only, specifically RfDs [including chronic oral minimal risk levels (MRLs)], RfCs (including chronic inhalation MRLs and reference exposure levels), OSFs, IURs, and CPVs (CalEPA values only, for the purpose of exploring a non-Federal data set derived independently from other processes). We also collected no observed adverse effect levels (NOAELs) associated with the RfDs for POD modeling. It is possible that a source referenced above contains additional values (e.g., acute MRLs), but they were excluded from our efforts (e.g., insufficient number of values for model development, not chronic values).
Table 1.
List of the established toxicity values and chemical descriptors that were used for modeling in this study.
| Toxicity value name (abbreviation) [units] | Mean (90% CI) [min–max] | Number of compoundsa | Global domain of applicability coverageb |
|---|---|---|---|
| Oral exposure noncancer | |||
| Reference dose (RfD) [] | 7.3 (5.4, 9.6) [3.6–14.5] | 671 | 86.1% |
| RfD No observed adverse effect level (RfD NOAEL) [] | 4.8 (2.8, 6.9) [1.8–8.4] | 487 | 90.4% |
| RfD Benchmark dose (RfD BMD) [] | 4.0 (1.8, 6.1) [1.1–6.8] | 137 | 81.8% |
| RfD Benchmark dose lower limit (RfD BMDL) [] | 4.4 (2.1, 6.5) [1.3–7.9] | 137 | 81.8% |
| Oral exposure cancer | |||
| Oral slope factor (OSF) [] | 5.0 (2.8, 7.5) [1.5–10.7] | 302 | 86.7% |
| Cancer potency value (CPV) [] | 5.2 (3.0, 7.6) [2.5–10.7] | 225 | 81.6% |
| Inhalation exposure (noncancer and cancer) | |||
| Reference concentration (RfC) [] | 6.8 (3.9, 9.4) [3.1–12.9] | 152 | 61.7% |
| Inhalation unit risk (IUR) [] | 4.5 (2.2, 6.9) [] | 150 | 71.8% |
Note: CI, confidence interval.
Depending on the toxicity value, the number of compounds with an established number varied and was determined as described in “Methods.” See Table S1 for the chemical names, toxicity values, and their source (i.e., the federal or state agency that derived each value).
Percent of compounds from the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) chemical database of 32,464 compounds (Mansouri et al. 2016) within the global domain of applicability for the compounds with the corresponding toxicity values, based on Chemistry Development Kit (CDK) molecular descriptors and a Z-score cutoff . For a more restrictive Z-score cutoff , the percentages are 70%, 78%, 65%, 65%, 69%, 67%, 43%, and 54%.
When available, we also gathered the experimental dose–response data that were used as the basis for the PODs, the toxicity values, or both. Using U.S. EPA guidance for dose–response modeling, as implemented by Wignall et al. (2014), we calculated benchmark doses (BMDs) and their lower confidence limits (BMDLs) from the dose–response data supporting the RfDs; we also included these PODs for QSAR modeling.
For a given chemical, if a specific toxicity value was available from more than one source (e.g., RfD from both U.S. EPA IRIS and OPP), then the toxicity value used for QSAR modeling was selected according to the following hierarchies (first choice to last choice), modeled after the practice of the U.S. EPA’s Superfund program:
-
•
RfD and NOAEL: IRIS OPP PPRTV ATSDR CalEPA HEAST
-
•
RfC, OSF, and IUR: IRIS PPRTV ATSDR CalEPA HEAST
CPVs were collected from CalEPA only, and BMDs and BMDLs used for modeling were derived as described by Wignall et al. (2014). A separate model was also developed for CalEPA CPVs because some organizations prefer those models on account of their applying more broadly the potential for increased cancer susceptibility for exposures at early life stages. Toxicity value units were converted from milligrams to moles and then transformed. To maintain a consistent “directionality” in which higher numbers are more potent, -transformed RfDs, RfCs, and PODs were then multiplied by after transformation. These values were used as the input for QSAR modeling.
Chemical structures of the compounds of interest (those with toxicity values described above) were curated using a protocol described by Fourches et al. (2010) to ensure standardization, removal of redundant entries, and filtering out of compounds inappropriate for modeling (i.e., using the approaches described below, descriptors cannot be calculated for organometallics, inorganics, or mixtures).
Molecular Descriptors and Chemical Space
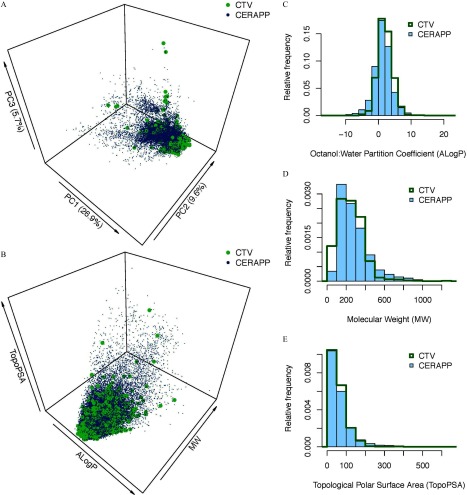
As described by Tropsha (2010), a chemical structure is mathematically represented using “descriptors” for the purposes of modeling. The open-source Chemistry Development Kit (CDK), as implemented in the R (version 3.3.2; R Core Team) package “rcdk” (Guha 2007), is one specific descriptor set that we used to calculate molecular descriptors for each compound with a toxicity value. These descriptors were generated based on compound structure as represented by simplified molecular-input line-entry system (SMILES) notation (Anderson et al. 1987) and represent features calculated from the chemical structure, such as octanol:water partition coefficient (logP), molecular weight (MW), and topological polar surface area (TPSA). If a descriptor could not be calculated for a chemical, it was imputed using the median value of that descriptor from other chemicals. Descriptors that could not be calculated for any chemical were dropped; each retained descriptor was available for at least 30% of the compounds. For comparison, CDK descriptors were also generated for the 32,464 structures in the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) “prediction” data set (Mansouri et al. 2016). This set of chemicals was designed to represent the compendium of chemicals to which humans may be exposed. To provide an overview of the diversity of the structures and the overall chemical space, the chemical structures in each list were analyzed using principal component analysis, as well as using the easily interpretable descriptors logP, MW, and TPSA (Figure 1). The global domain of applicability (or the chemical diversity represented by the model) was assessed by comparison with the CERAPP data set based on the distribution of pairwise Euclidean distances (descriptors were first standardized so that the range in the data set was 0–1), with the following Z-score cutoffs: (less restrictive) or (more restrictive).
Figure 1.
Chemical space coverage of 886 Conditional toxicity value (CTV) chemicals compared to 32,464 structures in the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) “prediction” data set, based on chemical descriptors generated from the Chemistry Development Kit (CDK). (A) Three-dimensional (3-D) plot of coverage comparison based on first three principal components (PC); (B–E) Coverage comparisons based on easily interpretable descriptors octanol:water partition coefficient (ALogP), molecular weight (MW), and topological polar surface area (TopoPSA). Panel (B) is a 3-D plot, and panels (C–E) are histograms comparing the distribution of compounds for each descriptor. The 3-D plots were created in R (version 3.3.2; R Core Team).
QSAR Modeling Approach
After the steps above were performed, each CTV compound of interest had a set of molecular descriptors describing the chemical structure and was associated with one or more quantitative “activity” values (the toxicity values described above). Using this data set, we explored an array of machine learning algorithms for generating statistical models to predict toxicity values (Tropsha 2010). The algorithms were trained and optimized based on our collected data set. In the testing phase, we evaluated the prediction accuracy of a wide array of chemical descriptors [i.e., CDK, ISIDA fragments (Ruggiu et al. 2010), Dragon (Mauri 2006)] and modeling approaches (i.e., random forest, support vector machines, k-nearest neighbors). Different modeling approaches performed similarly (data not shown); notably, no significant advantages in prediction accuracy were afforded by chemical structure descriptors that use proprietary software and thus would not be completely transparent. We therefore used publicly available molecular descriptors and software, specifically, the CDK molecular descriptors along with the machine-learning method of random forest (using the R package “randomForest” version 4.6-14, Liaw and Wiener, 2002), to construct QSAR models for predicting each toxicity value. Random forest is a method based on generating an “ensemble” of decision trees. Each “tree” is a predictive model that uses the molecular descriptors as predictors of the toxicity value, using a random subset of the “training” data (i.e., the toxicity values), where the default size of the random sample is a fraction (0.632) of the data set. Each overall model consists of the average prediction across a large number (in our case, 1,000) of individual “trees.” The random forest algorithm has an additional advantage in that it limits the number of descriptors used in each model (Fernández-Delgado et al. 2014) to balance against the number of compounds in the training set. Models developed in this project were validated using OECD principles (OECD 2007), which recommend techniques such as external cross-validation (i.e., the ability of the model to accurately predict the value of interest is judged on its ability to predict values for a subset of chemicals that were not used in model building). Specifically, measures of goodness-of-fit, robustness, and predictivity included a 5-fold external cross-validation–derived value, an indicator of the fraction of variance explained by the model (Golbraikh and Tropsha 2002), y-randomization (building models with the toxicity values randomly reordered), and the prediction error (i.e., distribution of residuals using cross-validation predictions). To facilitate a mechanistic interpretation, the frequency of use of each descriptor across each of the 1,000 individual “trees” was used as a surrogate for the importance of that descriptor in building the overall model, and the ability of the top two descriptors to discriminate between “high” and “low” potency compounds was examined visually. For comparison, alternative models were developed in which every chemical was assigned the mean value of the overall distribution of toxicity values, and prediction error for this approach was assessed.
CTV Predictor Web Portal
We developed the CTV predictor web portal (http://toxvalue.org), which allows users to obtain toxicity values for chemicals of interest. Specifically, a user can either draw a chemical structure, which will then generate SMILES using the JSME Molecular Editor (version 2013-10-13), or enter search terms. In either case, the portal sends a search query to ChemSpider (http://www.chemspider.com/) and retrieves the top-ranked result. After verifying the identity of the chemical, the user can select which toxicity values to predict. For each toxicity value, the web portal first searches the database to see if a regulatory value is available from one of the sources discussed above, or if a QSAR prediction is needed. The portal gives a warning if the chemical selected is inappropriate for modeling (i.e., inorganic, contains metals or metalloids, or of descriptors imputed). The web portal then returns a table containing the retrieved or predicted toxicity values. Predicted toxicity values include the (one-tail) lower and upper 95% confidence bounds based on the cross-validation residuals.
Comparison with HTS-Based Toxicity Values
To compare the CTV results against other methods, we evaluated CTV predictions along with HTS assay–based oral equivalent dose (OED) estimates from Wetmore (2015) in relation to “gold standard” regulatory NOAEL, BMDL, or RfD values. Note that the BMDL values included those recalculated by Wignall et al. (2014). Specifically, for each type of toxicity value, we compared predictions for the subset of compounds for which HTS-based values were available, evaluating the degree to which CTV- and HTS-based toxicity values correlated with the gold standard regulatory toxicity values. For HTS-based toxicity values, we used the fifth percentile OED () based on HTS assay results and in vitro–to–in vivo extrapolation (IVIVE) reported by Wetmore et al. (2015). We evaluated predictive power using a linear model on the values (slope, p-value, and adjusted ) as well as with respect to the median absolute deviation (also on values). Among the 163 compounds from Wetmore (2015), 36 had regulatory NOAELs, 14 had regulatory BMDLs, and 51 had regulatory RfDs from our database.
Results
Compounds and Toxicity Values
Our search and curation yielded a comprehensive data matrix containing regulatory agency–derived toxicity values and corresponding points of departure on 886 unique environmental chemicals (termed CTV compounds throughout). The chemical list, Chemical Abstracts Service (CAS) numbers, SMILES codes, and toxicity values are provided in Table S1. The largest data set is for oral exposure noncancer toxicity value RfD and its corresponding points of departure (NOAEL and BMD/L); inhalation exposure cancer and noncancer values were among the smaller data sets (Table 1). Only 40 chemicals have toxicity values for both oral and inhalation exposures and both cancer and noncancer hazards. For all types of toxicity values, the range of potency (in molar units) of the chemicals included in each data set spanned many orders of magnitude (Table 1), indicating that the database is balanced in terms of the range of toxicity values included, or “activity,” for deriving quantitative predictions.
Molecular Descriptors and Chemical Space
It is evident from comparing CDK molecular descriptor space to CERAPP compounds that the CTV compounds are chemically diverse. From the results of principal component analysis, there appears to be great overlap between CTV and CERAPP compounds (Figure 1A; see also Figure S1). A similar overlap is evident when major physicochemical properties frequently used in the pharmaceutical industry to triage compounds (OECD 2007) were considered together (Figure 1B) or independently (Figures 1C–E). The domain of applicability for most toxicity values was as high as 90%; only data sets for inhalation exposure–derived toxicity values had lower () coverage (Table 2). These results suggest that the chemicals with existing toxicity values are representative of the larger “universe” of the chemicals in the environment and on various chemical inventories.
Table 2.
Summary of the modeling outcomes.
| Toxicity value | Consensus model a | Model prediction absolute errorb | Mean value prediction absolute errorc | p-valued |
|---|---|---|---|---|
| RfD | 0.41 | 0.77 (0.06, 1.80) | 0.96 (0.08, 2.53) | |
| RfD NOAEL | 0.45 | 0.70 (0.06, 1.82) | 0.93 (0.05, 2.37) | |
| RfD BMD | 0.31 | 0.88 (0.13, 2.08) | 1.08 (0.09, 2.34) | 0.0098 |
| RfD BMDL | 0.28 | 0.93 (0.07, 2.19) | 1.13 (0.13, 2.44) | 0.0098 |
| OSF | 0.33 | 0.92 (0.07, 2.45) | 1.19 (0.12, 2.60) | |
| CPV | 0.25 | 0.97 (0.07, 2.53) | 1.19 (0.15, 2.65) | 0.0008 |
| RfC | 0.42 | 1.11 (0.12, 2.71) | 1.49 (0.20, 3.54) | 0.0015 |
| IUR | 0.42 | 0.93 (0.07, 2.69) | 1.29 (0.06, 2.85) |
Note: BMD, benchmark dose; BMDL, benchmark dose lower confidence limit; CPV, cancer potency value; IUR, inhalation unit risk; NOAEL, no observed adverse effect level; OSF, oral slope factor; QSAR, quantitative structure activity relationship; RfC = reference concentration; RfD = reference dose.
is the fraction of the variance explained by each model, estimated by 5-fold cross-validation. In all cases, the accuracy of the model built with original data was significantly higher than that of models built using y-randomized data sets ().
The mean and confidence interval (90%) of absolute error for the external prediction of each compound’s toxicity value against the QSAR model prediction under 5-fold cross-validation.
The mean and confidence interval (90%) of absolute error for the external prediction of each compound’s toxicity value against the mean value of the compounds with that particular toxicity value.
Kolmogorov-Smirnov statistics for the difference between model and “mean value” prediction absolute errors.
QSAR Modeling
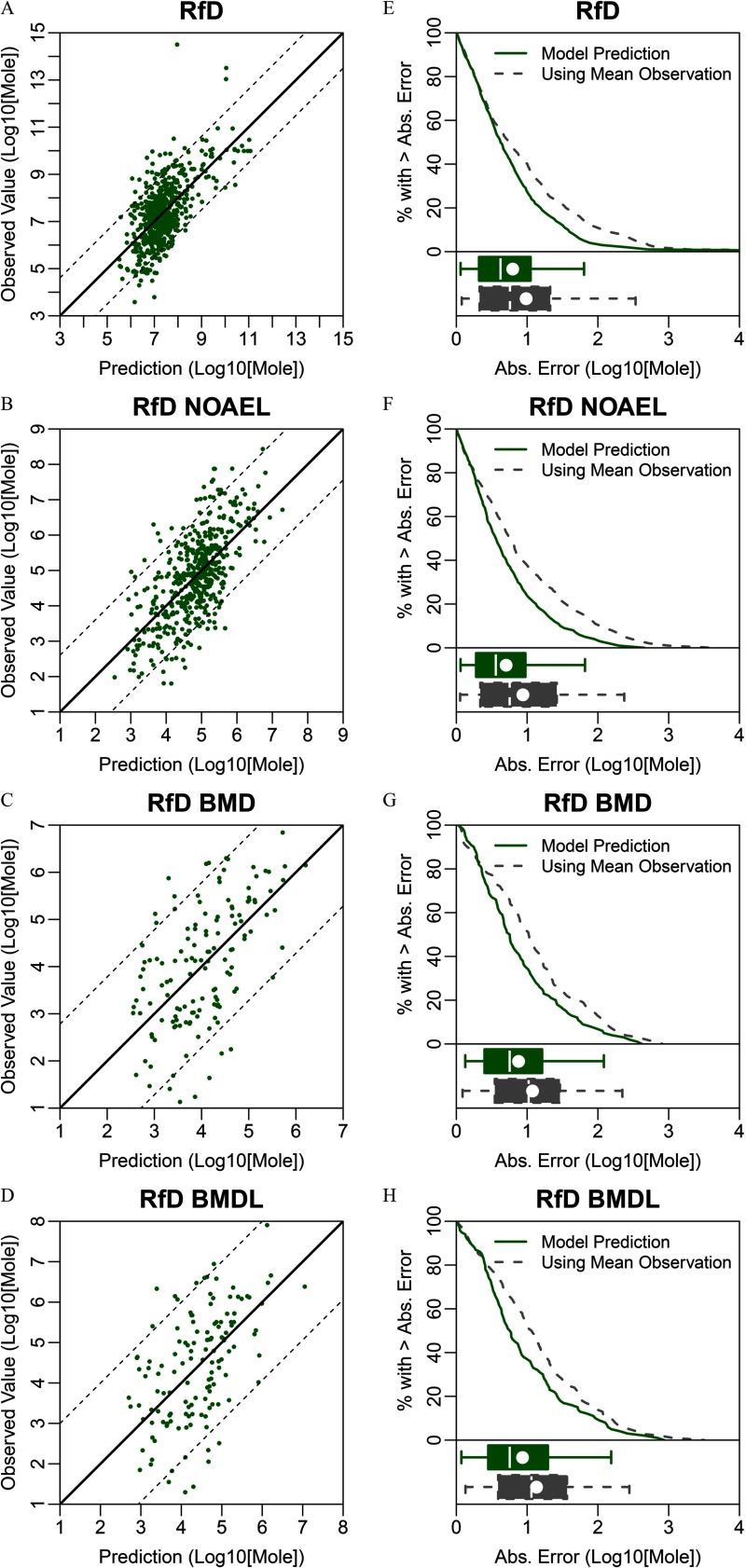
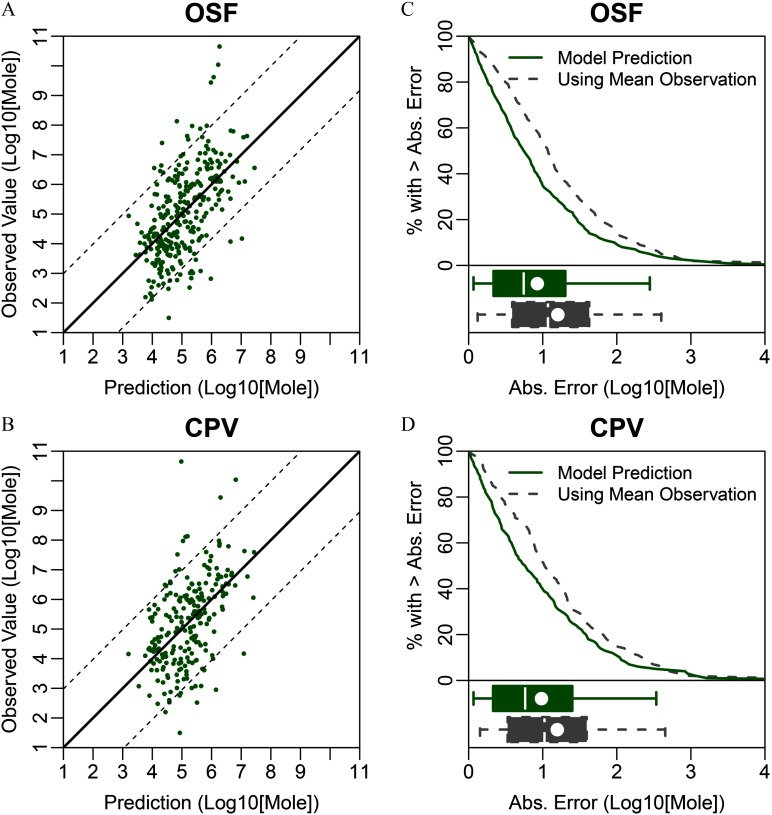
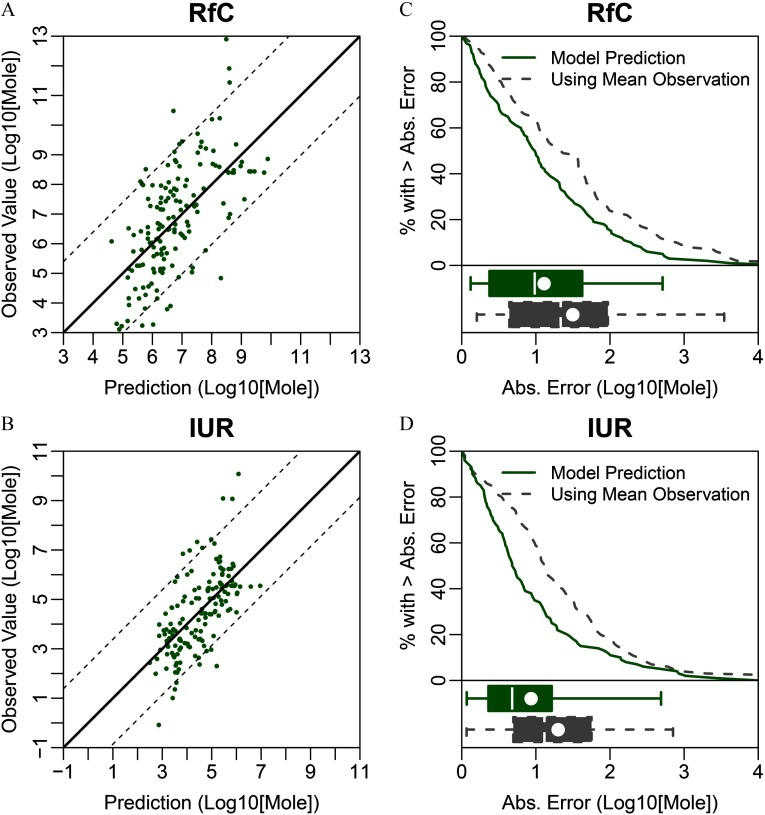
Predictive QSAR models were developed for eight oral and inhalation cancer and noncancer toxicity values. The range of values varied from 0.25 to 0.45 (Table 2). The accuracy of prediction for all models developed in this study was significantly () higher than that of models developed for each data set using y-randomized data. We observed that the range of predicted values was compressed on the margins in all data sets, indicating the challenges of predicting compounds with the lowest and highest toxicity values [Figures 2–4 (left panels)]. Absolute prediction errors for each model had means ranging from 0.70 to 1.11 (Table 2). We compared these errors to the alternative option for predicting an unknown toxicity value by using the mean in each data set as a prediction for each toxicity value [Figures 2–4 (right panels)]. We found that for all models, the QSAR model predictions had smaller mean absolute error and were highly significantly () more accurate than this alternative (Table 2). Additionally, it should be noted that regulatory toxicity values themselves may differ across federal and state agencies, which place limits as to how precise any predictions might be. For example, of the chemicals with RfDs used in modeling had at least one alternative RfD in our database developed by another federal or state entity (e.g., owing to different PODs and/or uncertainty factors). The median absolute deviation of the alternative RfDs from those used in modeling was 0.59, which is only slightly smaller than the median absolute error of 0.77 for CTV’s RfD predictions.
Figure 2.
Conditional toxicity value (CTV) prediction errors based on 5-fold cross-validation for the RfD and associated NOAEL, BMD, or BMDL. (A–D) Scatter plots of QSAR-predicted versus “observed” regulatory toxicity values, with the dotted lines showing the range encompassing 90% of the predictions. (E–H) Comparison of the distributions of absolute prediction errors for CTV (solid line) and an alternative model using the observed mean value for every prediction (dashed line). Box plots show the corresponding interquartile region (box), median (line in box), mean (dot in box), and 95% confidence interval (whiskers). Note: Abs., absolute; BMD, benchmark dose; BMDL, benchmark dose lower confidence limit; NOAEL, no observed adverse effect level; QSAR, quantitative structure–activity relationship; RfD, reference dose.
Figure 3.
Conditional toxicity value (CTV) prediction errors based on 5-fold cross-validation for the oral slope factor (OSF) and cancer potency value (CPV). (A–B) Scatter plots of QSAR-predicted versus “observed” regulatory toxicity values, with the dotted lines showing the range encompassing 90% of the predictions. (C–D) Comparison of the distributions of absolute prediction errors for CTV (solid line) and an alternative model using the observed mean value for every prediction (dashed line). Box plots show the corresponding interquartile region (box), median (line in box), mean (dot in box), and 95% confidence interval (whiskers). Note: Abs., absolute; QSAR, quantitative structure–activity relationship.
Figure 4.
Conditional toxicity value (CTV) prediction errors based on 5-fold cross-validation for the reference concentration (RfD) and inhalation unit risk (IUR). (A, B) Scatter plots of QSAR-predicted versus “observed” regulatory toxicity values, with the dotted lines showing the range encompassing 90% of the predictions. (C, D) Comparison of the distributions of absolute prediction errors for CTV (solid line) and an alternative model using the observed mean value for every prediction (dashed line). Box plots show the corresponding interquartile region (box), median (line in box), mean (dot in box), and 95% confidence interval (whiskers). Note: Abs., absolute; QSAR, quantitative structure–activity relationship.
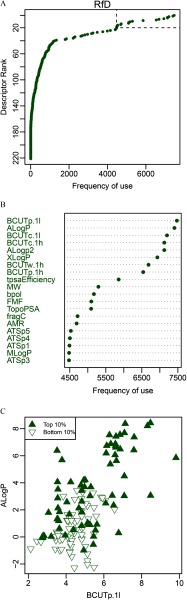
A mechanistic interpretation of a QSAR model, if possible, is encouraged for regulatory purposes (CDC 2014). Therefore, we examined the relative contribution of the individual descriptors, judged from the frequency of the descriptor’s use in the model, to the accuracy of the model predictions. Figure 5 (top panel) shows an example of this analysis for the RfD toxicity value (see the Supplemental Material, Figures S2–S4 for analyses for all other toxicity values). Top-20 descriptors are exemplified in Figure 5 (middle panel). Note that descriptors may be used more than once in each tree (e.g., partitioning the data at different cut points and/or in different branches of the tree). Even though the modeling relied on the multivariate data matrix, there is some clustering of higher- and lower-potency chemicals based on pairwise comparisons of descriptors (Figure 5, lower panel). For example, in the case of the RfD, compounds with lower values (i.e., less toxic, owing to transformations described in “Methods”) tend to have lower ALogP and BCUT.p-1l values. The separation is, of course, imperfect, because a two-dimensional “slice” cannot be expected to recapitulate the full multivariate model.
Figure 5.
Example of mechanistic interpretation of QSAR model for the RfD. (A) Ranks of the molecular descriptors by their frequency of use in the model. The top twenty are denoted by dashed lines. (B) The top twenty descriptors shown separately, with the descriptor names. (C) comparison of the descriptor values for the top two descriptors between the highest and lowest potency RfDs. Note: QSAR, quantitative structure activity relationship; RfD, reference dose. Definitions of each molecular descriptor can be found online: https://cdk.github.io/cdk/1.5/docs/api/org/openscience/cdk/qsar/descriptors/molecular/package-summary.html.
CTV Predictor Web Portal
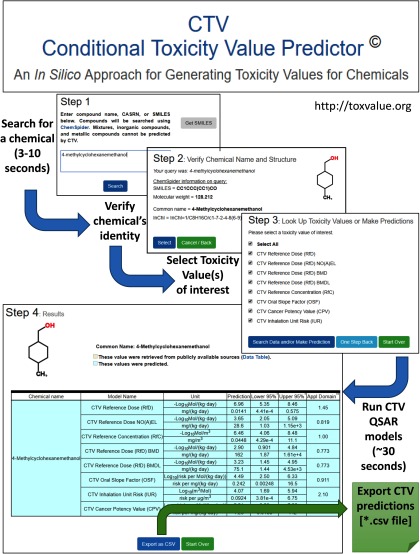
To enable access to the QSAR models developed in this study, we developed a public web portal (http://toxvalue.org) for both retrieving existing toxicity values (collected as described in “Methods”) and predicting values for data-poor chemicals. Screen shots of each step are shown in Figure 6. All steps, from entering a search term to obtaining a CSV file of the results, can be completed in . In the example shown in Figure 6, the chemical 4-methylcyclohexanemethanol, which was accidentally spilled into the Elk River in West Virginia on 9 January 2014, is used. This chemical lacked any regulatory toxicity values, hampering the response and the communication of potential risks. Thus, in the days after the spill, the CDC established a screening level concentration for less-than-lifetime exposure of , which corresponds to a daily dose of approximately (TERA 2014). Approximately five months after the spill, a short-term RfD of was derived by a group of consultants commissioned by the state of West Virginia (Bhhatarai et al. 2016; Dimitrov et al. 2016; Patlewicz et al. 2016). Note that neither of these values, however, underwent formal external peer review or regulatory endorsement; therefore, they were not included in our database. However, CTV is able to make a prediction about the RfD, based on the QSAR models we have developed, with the result being a predicted RfD [90% confidence interval (CI)] for this compound of 0.014 (, 0.58) . Although this value is for chronic exposures, it is within a factor of 7 of the value derived by the CDC and within a factor of 5 of the value derived by the state consultants. This example illustrates how CTV could be used to derive a screening level when few or no toxicological data are available and be applied in an emergency situation.
Figure 6.
Screen shots illustrating use of the Conditional Toxicity Value (CTV) Predictor web portal (http://toxvalue.org). Steps include searching for a chemical, verifying its identity, selecting toxicity values of interest, running quantitative structure–activity relationship (QSAR) models, and exporting predictions to a comma-separated-values file. The entire process can be completed in approximately 60 s. CTV is a public web portal maintained by two of the authors (IR and WAC).
Comparison with HTS-Based Toxicity Values
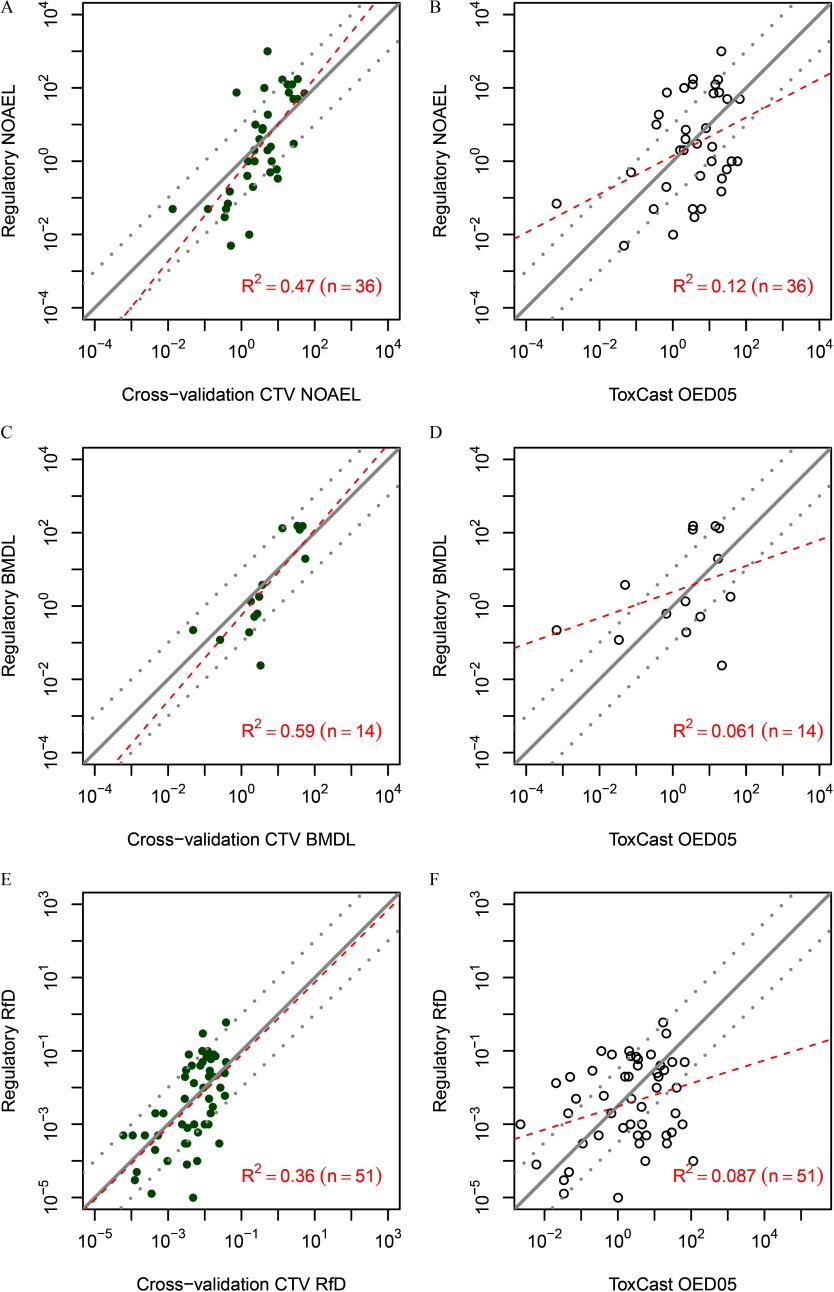
Comparisons between CTV predictions and HTS-based toxicity values are shown in Table 3 and Figure 7. The CTV predictions had smaller absolute deviations from the regulatory values than predictions based on HTS assays and IVIVE (Table 3). A much larger proportion of the CTV predictions are within a factor of 10 of the gold standard regulatory toxicity, whereas more than half of HTS-based predictions have an absolute deviation greater than a factor of 10. Moreover, based on values, CTV predictions explain much more of the variation in regulatory toxicity values than HTS-based predictions. Overall, when compared with the gold standard of using regulatory toxicity values, CTV gives more precise and more accurate toxicity value predictions than those derived from currently available HTS assays and IVIVE approaches.
Table 3.
Summary of conditional toxicity value versus high-throughput screening toxicity value comparisons.
| “Gold standard” toxicity value | In silico/in vitro predicted toxicity value | Linear model slope (log10-transformed) (p-value) | Median absolute deviationa | Adjusted |
|---|---|---|---|---|
| NOAEL () | CTV NOAEL | 1.25 () | 0.66 | 0.47 |
| 0.52 (0.02) | 1.11 | 0.12 | ||
| BMDL () | CTV BMDL | 1.17 () | 0.58 | 0.59 |
| 0.35 (0.20) | 1.06 | 0.06 | ||
| RfD () | CTV RfD | 0.99 () | 0.72 | 0.36 |
| 0.32 (0.02) | 2.68 | 0.09 |
Note: BMDL, benchmark dose lower confidence limit; CTV, conditional toxicity value; , high-throughput screening–based oral equivalent dose lower 5% confidence limit; NOAEL, no observed adverse effect level; RfD, reference dose.
The median of absolute residuals between predicted and “gold standard” toxicity values.
Figure 7.
Comparison of conditional toxicity value (CTV)-based (A, C, E) or high-throughput screening (HTS) assay-based (B, D, F) toxicity value predictions with “gold standard” regulatory toxicity values. In each panel, the x-axis is the toxicity value predicted from CTV (left panels) or based on HTS assays (right panels), which is compared with regulatory toxicity values on the y-axis (all values are in units of ). Comparisons are made for regulatory NOAELs (panels A and B), BMDLs (panels C and D), or RfDs (panels E and F). In all cases, the predictions from CTV are based on cross-validation (panels A, C, and E). Panels A–E also include lines indicating equality and a factor of 10 greater or less than equality (solid and dotted lines); for panel F, the line is offset by a factor of to account for the fact that the HTS-based oral equivalent dose lower 5% confidence limit () is a point of departure and is not meant to be equivalent to an RfD. The offset is approximately equivalent to treating as a surrogate for the RfD. The value of this offset was determined by the intercept of the linear regression. Each panel also includes linear regression lines (dashed lines), along with the number of compounds (n) and the adjusted . Note: BMDL, benchmark dose lower confidence limit; HTS OED05, high-throughput screening–based oral equivalent dose lower 5% confidence limit; NOAEL, no observed adverse effect level; RfD, reference dose.
Fully understanding the implications for risk assessment of these alternative approaches would require calculating margins of exposure (MoEs), hazard quotients (HQs) or both. Because of the additional uncertainties and assumptions involved in such calculations, they are described in the Supplemental Material (see Figure S5) as an illustrative example.
Discussion
One of the gaps addressed by this study and the accompanying web tool is that QSAR methodologies can be applied to chemicals of interest, regardless of the data available. Thus, this study’s output provides a means to derive a “conditional” toxicity value for a chemical when one does not already exist. Additionally, experimental data and regression methodologies can be used to supplement QSAR-based predictions when available.
CTV fills a critical gap not currently covered by existing chemical structure–based approaches. For example, most available QSAR tools, such as TOPKAT, ToxTree, OECD QSAR toolbox, RepDose, and others (Bhatia et al. 2015), are designed to identify hazards by placing compounds in categories (e.g., nontoxic vs. toxic) and do not provide quantitative outputs (e.g., PODs) that can be used for risk characterization. One exception is the threshold of toxicological concern (TTC), which assigns chemicals to structural classes, each of which is associated with a TTC representing a “conservative” POD (Patlewicz et al. 2014, 2015). Another approach that can provide PODs is read-across, in which chemicals with PODs are used to fill data gaps for chemicals without PODs (Cote et al. 2016; Judson et al. 2011). CTV can be thought of as a hybrid between the TTC approach and read-across because molecular descriptors are used to generate a “custom category” for each chemical via machine learning, and toxicity values are derived accordingly. Similar to how a TTC is commonly defined as the lower fifth percentile of the values within the structural class, the lower confidence bound for the CTV-derived toxicity value can then be thought of as a chemical-specific TTC.
The use of in vitro data such as HTS assays combined with IVIVE using reverse toxicokinetics represents another major effort to address the lack of toxicity values for the majority of chemicals to which humans are exposed in the environment (Wetmore 2015). However, the chemical space to which HTS assays can be applied is limited (e.g., restrictions related to molecular weight, solubility, volatility). Moreover, conversion of in vitro activity to exposure estimates requires toxicokinetic modeling, further limiting the applicability to the few chemicals for which relevant data on toxicokinetics are available or can be generated (Holman et al. 2017a, b). Thus, CTV, given its broad domain of applicability, can provide a complementary approach to HTS-based methods. Additionally, our direct comparison between CTV- and HTS assay–based toxicity values suggests that, given the present state of HTS-based risk assessment, CTV might currently provide more accurate and precise estimates for use in risk assessment when benchmarked against peer-reviewed toxicity values developed by federal or state agencies.
It is also worth noting that there is a limit to how accurately a toxicity value can be predicted given the extensive scientific judgment involved in their development. Indeed, risk estimates can vary widely across regulatory settings even for the same chemical and based on the same underlying observational data (NRC 2009). For example, for the same chemical, differences across agencies in the RfDs from our database are typically approximately 0.6 units. This value is only slightly smaller than the typical absolute error estimate for CTV’s RfD predictions of units, suggesting that our models are close to the accuracy/precision limit imposed by inherent heterogeneity in the underlying toxicity values. Part of this heterogeneity is likely because the RfDs are not precisely defined and, as described by the NRC (2009), do not take into account the probability of harm. Probabilistic approaches to calculating toxicity values that provide quantitative risk estimates for noncancer effects are available (Chiu and Slob 2015; WHO/IPCS 2014) and have recently been applied to recalculate RfDs for approximately 600 chemicals and replace them with “” values that reflect human dose (HD) estimates for a specific magnitude of effect M at a specific population incidence level I (Chiu et al. In Press). Thus, development of future QSAR models based on probabilistic toxicity values may be beneficial.
An additional limitation is that there were inadequate data to develop models for organ-specific toxicity values. Organ-specific information was not available for many chemicals (e.g., a NOAEL does not always specify a particular target organ), so developing QSAR-based organ-specific toxicity value predictions would reduce the size of the data set to preclude confident modeling (Tropsha 2010). However, there are many reasons why end point–specific values would be beneficial, such as addressing cumulative risk. Moreover, it would be interesting to ascertain whether QSAR models perform better for certain organ systems or end points. Therefore, as health assessment programs such as the U.S. EPA’s IRIS move toward developing “end point–specific” reference values, and as HTS assays, perhaps through adverse outcome pathways, become more closely associated with target organ toxicities, future QSARs may be able to address this limitation.
We anticipate a number of refinements to CTV in the future. First, additional models can be developed for specific needs, such as for additional state- or agency-specific toxicity values or for probabilistic toxicity values as they become available. At the same time, as new toxicity values are developed by federal and state agencies, it will be necessary to update the CTV databases and models periodically. Additionally, we plan to solicit user feedback to improve the presentation and visualization of our web portal to better communicate the modeling results and their uncertainties. Furthermore, “ensemble” models based on combining multiple molecular descriptor sets and multiple machine learning approaches can be evaluated regarding their value for increasing predictive accuracy and precision. Finally, it may be possible to incorporate biological data, such as HTS assay results, into the predictive models. However, a key question will be the extent to which adding such data improves model performance, which can be evaluated in a value-of-information approach.
The need for a publicly accessible computational tool, such as CTV, to predict quantitative toxicity values within an order of magnitude of traditionally derived toxicity values has been emphasized in multiple scientific and risk assessment venues. In 2009, the National Academies explicitly suggested that the U.S. EPA “perform quantitative structure activity relationship (QSAR) analyses … for developing distributions of toxicity parameter values derived from data on representative data-rich chemicals” (NRC 2009). Creation of a companion web-based portal to enable wide accessibility to the predictive quantitative QSAR models, based on transparently reported data and methods, enables stakeholders to make predictions on chemicals of interest and directly meets the challenges faced by decision makers at all levels of government.
Specifically, the predicted values derived from CTV can be applied in a variety of risk assessment settings, including in ranking and prioritization of compounds for additional study and evaluation, as well as when decisions about chemical safety and risk management are needed. Accordingly, we anticipate that groups across state and federal governments who are mandated to make decisions about chemical safety and the need for remediation efforts, such as CalEPA or the U.S. EPA Office of Land and Emergency Management, would be able to use CTV to inform those risk assessment decisions when no other data are available. Additionally, as shown by our example with 4-methylcyclohexanemethanol, CTV could be very useful in deriving screening levels for application in an emergency situation such as a chemical spill or a natural disaster, where rapid decisions are necessary to ensure protection of public health and the environment. Although rigorous health assessments of chemicals that are of concern to state and federal governments remain important and will continue, this project will facilitate the work of health assessors at multiple levels when decisions are needed quickly, or when a chemical of concern has not yet been tested or reviewed. Thus, the outputs of this study fill a gap in the current risk assessment paradigm that requires extensive experimental/human data, time-consuming systematic review, and rigorous peer review (Mansouri et al. 2016).
Conclusion
We have developed an in silico tool that can predict toxicity values such as a point of departure, a reference dose or concentration, a cancer oral slope factor, or an inhalation unit risk, with typical absolute error of greater than a factor of 10. The resulting conditional toxicity value, or CTV, can be used with exposure information to provide quantitative indications of risk for the vast majority of environmental chemicals for which toxicity data or human health assessments are unavailable, thereby filling a critical gap in the current risk assessment/management paradigm.
Supplemental Material
Supplemental Material
Acknowledgments
The authors acknowledge L. Zeise (California EPA) and M. Martin [formerly U.S. Environmental Protection Agency (EPA), present affiliation: Pfizer] for valuable suggestions and comments. Additionally, the authors wish to thank D. Liu for technical assistance in developing the toxvalue.org web portal.
This publication was made possible, in part, by U.S. EPA grant number STAR RD83516602, and National Institutes of Health (NIH) grant P42 ES027704. Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the U.S. EPA or NIH. Further, the U.S. EPA and NIH do not endorse the purchase of any commercial products or services mentioned in the publication.
References
- Anderson E, Veith G, Weininger D. 1987. “SMILES: A Line Notation and Computerized Interpreter for Chemical Structures.” EPA/600/M-87/021 (NTIS PB88130034). Washington, DC:U.S. Environmental Protection Agency; https://cfpub.epa.gov/si/si_public_record_report.cfm?dirEntryId=33186&keyword=syntax&actType=&TIMSType=+&TIMSSubTypeID=&DEID=&epaNumber=&ntisID=&archiveStatus=Both&ombCat=Any&dateBeginCreated=&dateEndCreated=&dateBeginPublishedPresented=&dateEndPublishedPresented=&dateBeginUpdated=&dateEndUpdated=&dateBeginCompleted=&dateEndCompleted=&personID=&role=Any&journalID=&publisherID=&sortBy=revisionDate&count=50&CFID=61591074&CFTOKEN=96057771 [accessed 8 May 2018]. [Google Scholar]
- Bhatia S, Schultz T, Roberts D, Shen J, Kromidas L, Marie Api A. 2015. Comparison of Cramer classification between Toxtree, the OECD QSAR Toolbox and expert judgment. Regul Toxicol Pharmacol 71(1):52–62, PMID: 25460032, 10.1016/j.yrtph.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Bhhatarai B, Wilson DM, Parks AK, Carney EW, Spencer PJ. 2016. Evaluation of TOPKAT, Toxtree, and Derek Nexus in silico models for ocular irritation and development of a knowledge-based framework to improve the prediction of severe irritation. Chem Res Toxicol 29(5):810–822, PMID: 27018716, 10.1021/acs.chemrestox.5b00531. [DOI] [PubMed] [Google Scholar]
- CDC (Centers for Disease Control and Prevention). 2014. “Summary Report of Short-Term Screening Level Calculation and Analysis of Available Animal Studies for MCHM.” Atlanta, GA:Centers for Disease Control and Prevention. [Google Scholar]
- Chiu WA, Axelrad DA, Dalaijamts C, Dockins C, Shao K, Shapiro AJ, Paoli G. In Press. Beyond the RfD: broad application of a probabilistic approach to improve chemical dose-response assessment for non-cancer effects. Environ Health Perspect 10.1289/EHP3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WA, Slob W. 2015. A unified probabilistic framework for dose–response assessment of human health effects. Environ Health Perspect 123(12):1241–1254, PMID: 26006063, 10.1289/ehp.1409385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote I, Andersen ME, Ankley GT, Barone S, Birnbaum LS, Boekelheide K, et al. 2016. The next generation of risk assessment multi-year study-highlights of findings, applications to risk assessment, and future directions. Environ Health Perspect 124(11):1671–1682, PMID: 27091369, 10.1289/EHP233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov SD, Diderich R, Sobanski T, Pavlov TS, Chankov GV, Chapkanov AS, et al. 2016. QSAR toolbox - workflow and major functionalities. SAR QSAR Environ Res 27(16):203–219, PMID: 26892800, 10.1080/1062936X.2015.1136680. [DOI] [PubMed] [Google Scholar]
- Fernández-Delgado M, Cernadas E, Barro S, Amorim D. 2014. Do we need hundreds of classifiers to solve real world classification problems? J Mach Learn Res 15(1):3133–3181. [Google Scholar]
- Fourches D, Muratov E, Tropsha A. 2010. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model 50(7):1189–1204, PMID: 20572635, 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbraikh A, Tropsha A. 2002. Beware of q2!. J Mol Graph Model 20(4):269–276, PMID: 11858635, 10.1016/S1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- Guha R. 2007. Chemical informatics functionality in R. J Stat Softw 18(5):1–16, 10.18637/jss.v018.i05. [DOI] [Google Scholar]
- Guyton KZ, Kyle AD, Aubrecht J, Cogliano VJ, Eastmond DA, Jackson M, et al. 2009. Improving prediction of chemical carcinogenicity by considering multiple mechanisms and applying toxicogenomic approaches. Mutat Res 681(2–3):230–240, PMID: 19010444, 10.1016/j.mrrev.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Holman E, Francis R, Gray G. 2017a. Part II: Quantitative evaluation of choices used in setting noncancer chronic human health reference values across organizations. Risk Anal 37(5):879–892, PMID: 27654007, 10.1111/risa.12699. [DOI] [PubMed] [Google Scholar]
- Holman E, Francis R, Gray G. 2017b. Part I––comparing noncancer chronic human health reference values: an analysis of science policy choices. Risk Anal 37(5):861–878, PMID: 27663864, 10.1111/risa.12700. [DOI] [PubMed] [Google Scholar]
- IPCS/IOMC(International Programme on Chemical Safety/Inter-Organization Programme for the Sound Management of Chemicals). 2014. Guidance Document on Evaluating and Expressing Uncertainty in Hazard Characterization. Geneva, Switzerland:World Health Organization; http://www.who.int/ipcs/methods/harmonization/areas/hazard_assessment/en/ [accessed 8 May 2018]. [Google Scholar]
- Jolly R, Ahmed KB, Zwickl C, Watson I, Gombar V. 2015. An evaluation of in-house and off-the-shelf in silico models: implications on guidance for mutagenicity assessment. Regul Toxicol Pharmacol 71(3):388–397, PMID: 25656493, 10.1016/j.yrtph.2015.01.010. [DOI] [PubMed] [Google Scholar]
- Judson RS, Kavlock RJ, Setzer RW, Cohen Hubal EA, Martin MT, Knudsen TB, et al. 2011. Estimating toxicity-related biological pathway altering doses for high-throughput chemical risk assessment. Chem Res Toxicol 24(4):451–462, PMID: 21384849, 10.1021/tx100428e. [DOI] [PubMed] [Google Scholar]
- Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, et al. 2015. Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol Sci 148(1):137–154, PMID: 26272952, 10.1093/toxsci/kfv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, et al. 2009. The toxicity data landscape for environmental chemicals. Environ Health Perspect 117(5):685–695, PMID: 19479008, 10.1289/ehp.0800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, et al. 2012. Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol 25(7):1287–1302, PMID: 22519603, 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Krewski D, Westphal M, Andersen ME, Paoli GM, Chiu WA, Al-Zoughool M, et al. 2014. A framework for the next generation of risk science. Environ Health Perspect 122(8):796–805, PMID: 24727499, 10.1289/ehp.1307260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. 2002. Classification and Regression by randomForest. R News 2(3):18–22. [Google Scholar]
- Low YS, Sedykh AY, Rusyn I, Tropsha A. 2014. Integrative approaches for predicting in vivo effects of chemicals from their structural descriptors and the results of short-term biological assays. Curr Top Med Chem 14(11):1356–1364, PMID: 24805064, 10.2174/1568026614666140506121116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltarollo VG, Gertrudes JC, Oliveira PR, Honorio KM. 2015. Applying machine learning techniques for ADME-Tox prediction: a review. Expert Opin Drug Metab Toxicol 11(2):259–271, PMID: 25440524, 10.1517/17425255.2015.980814. [DOI] [PubMed] [Google Scholar]
- Mansouri K, Abdelaziz A, Rybacka A, Roncaglioni A, Tropsha A, Varnek A, et al. 2016. CERAPP: Collaborative Estrogen Receptor Activity Prediction Project. Environ Health Perspect 124(7):1023–1033, PMID: 26908244, 10.1289/ehp.1510267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri A, Consonni V, Pavan M, Todeschini R. 2006. DRAGON software: an easy approach to molecular descriptor calculations. MATCH Commun Math Comput Chem 56:237–248. [Google Scholar]
- National Academies of Science, Engineering, and Medicine. 2017. Using 21st Century Science to Improve Risk-Related Evaluations. Washington, DC:The National Academies Press; 10.17226/24635 [accessed 8 May 2018]. [DOI] [PubMed] [Google Scholar]
- NRC (National Research Council). 2009. Science and Decisions: Advancing Risk Assessment. Washington, DC:National Academies Press; 10.17226/12209 [accessed 8 May 2018]. [DOI] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co-operation and Development). 2007. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationships [(Q)SAR] Models. Paris, France:Organisation for Economic Co-operation and Development. [Google Scholar]
- Patlewicz G, Ball N, Becker RA, Booth ED, Cronin MT, Kroese D, et al. 2014. Read-across approaches–misconceptions, promises and challenges ahead. ALTEX 31(4):387–396, PMID: 25368965. [DOI] [PubMed] [Google Scholar]
- Patlewicz G, Ball N, Boogaard PJ, Becker RA, Hubesch B. 2015. Building scientific confidence in the development and evaluation of read-across. Regul Toxicol Pharmacol 72(1):117–133, PMID: 25857293, 10.1016/j.yrtph.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Patlewicz G, Worth AP, Ball N. 2016. Validation of computational methods. Adv Exp Med Biol 856:165–187, PMID: 27671722, 10.1007/978-3-319-33826-2_6. [DOI] [PubMed] [Google Scholar]
- Pinto CL, Mansouri K, Judson R, Browne P. 2016. Prediction of estrogenic bioactivity of environmental chemical metabolites. Chem Res Toxicol 29(9):1410–1427, PMID: 27509301, 10.1021/acs.chemrestox.6b00079. [DOI] [PubMed] [Google Scholar]
- Ruggiu F, Marcou G, Varnek A, Horvath D. 2010. ISIDA property-labelled fragment descriptors. Mol Inform 29(12):855–868, PMID: 27464350, 10.1002/minf.201000099. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Sedykh A, Low Y, Guyton KZ, Tropsha A. 2012. Predictive modeling of chemical hazard by integrating numerical descriptors of chemical structures and short-term toxicity assay data. Toxicol Sci 127(1):1–9, PMID: 22387746, 10.1093/toxsci/kfs095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedykh A, Zhu H, Tang H, Zhang L, Richard A, Rusyn I, et al. 2011. Use of in vitro HTS-derived concentration–response data as biological descriptors improves the accuracy of QSAR models of in vivo toxicity. Environ Health Perspect 119(3):364–370, PMID: 20980217, 10.1289/ehp.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERA (Toxicology Excellence for Risk Assessment). 2014. “Report of Expert Panel Review of Screening Levels for Exposure to Chemicals from the January 2014 Elk River Spill.” Charleston, WV:Toxicology Excellence for Risk Assessment; https://www.tera.org/Peer/WV/WV%20Expert%20Report%2012%20May%202014.pdf [accessed 8 May 2018]. [Google Scholar]
- Thomas RS, Wesselkamper SC, Wang NC, Zhao QJ, Petersen DD, Lambert JC, et al. 2013. Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol Sci 134(1):180–194, PMID: 23596260, 10.1093/toxsci/kft094. [DOI] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR. 2013. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121(7):756–765, PMID: 23603828, 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis CC, Pack SA, Saulsbury AW, Yambert MW. 1990. Prediction of carcinogenic potency from toxicological data. Mutat Res 241(1):21–36, PMID: 2333083, 10.1016/0165-1218(90)90106-C. [DOI] [PubMed] [Google Scholar]
- Tropsha A. 2010. Best practices for QSAR model development, validation, and exploitation. Mol Inform 29(6–7):476–488, PMID: 27463326, 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, et al. 2013. High-throughput models for exposure-based chemical prioritization in the ExpoCast project. Environ Sci Technol 47(15):8479–8488, PMID: 23758710, 10.1021/es400482g. [DOI] [PubMed] [Google Scholar]
- Wambaugh JF, Wetmore BA, Pearce R, Strope C, Goldsmith R, Sluka JP, et al. 2015. Toxicokinetic triage for environmental chemicals. Toxicol Sci 147(1):55–67, PMID: 26085347, 10.1093/toxsci/kfv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore BA. 2015. Quantitative in vitro-to-in vivo extrapolation in a high-throughput environment. Toxicology 332:94–101, PMID: 24907440, 10.1016/j.tox.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Allen B, Ferguson SS, Sochaski MA, Setzer RW, et al. 2015. Toxicol Sci 148(1):121–36, PMID: 26251325, 10.1093/toxsci/kfv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall JA, Shapiro AJ, Wright FA, Woodruff TJ, Chiu WA, Guyton KZ, et al. 2014. Standardizing benchmark dose calculations to improve science-based decisions in human health assessments. Environ Health Perspect 122(5):499–505, PMID: 24569956, 10.1289/ehp.1307539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeise L, Wilson R, Crouch E. 1984. Use of acute toxicity to estimate carcinogenic risk. Risk Anal 4(3):187–199, 10.1111/j.1539-6924.1984.tb00138.x. [DOI] [Google Scholar]
- Zhu H, Ye L, Richard A, Golbraikh A, Wright FA, Rusyn I, et al. 2009. A novel two-step hierarchical quantitative structure-activity relationship modeling work flow for predicting acute toxicity of chemicals in rodents. Environ Health Perspect 117(8):1257–1264, PMID: 19672406, 10.1289/ehp.0800471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.