Abstract
Background:
Numerous studies have indicated the estrogenic effects of polybrominated diphenyl ethers (PBDEs) and hydroxylated PBDEs (OH-PBDEs). However, the previous mechanistic studies focused on their estrogenic effects through genomic transcriptional activation of estrogen receptors.
Objective:
The present study aimed to investigate the estrogenic effects of PBDEs and OH-PBDEs via nongenomic G protein–coupled estrogen receptor (GPER) pathways.
Methods:
The binding affinities of 12 PBDEs and 18 OH-PBDEs with GPER were determined by a fluorescence competitive binding assay in a human breast cancer cell line (SKBR3). Molecular docking was performed to simulate the interactions. Their activities on GPER pathways were investigated by detecting calcium mobilization and cyclic adenosine monophosphate (cAMP) accumulation in SKBR3 cells. The effects on SKBR3 cell migration were investigated using Boyden chamber and wound-healing assays.
Results:
Our results showed that 11 of the OH-PBDEs but none of the PBDEs bound to GPER directly. Relative binding affinities ranged from 1.3% to 20.0% compared to . Docking results suggested that the hydroxyl group played an essential role in the binding of OH-PBDEs to GPER by forming hydrogen bond interactions. Most of the OH-PBDEs activated subsequent GPER signaling pathways. Among them, 4ʹ-OH-BDE-049, 5ʹ-OH-BDE-099, and 3ʹ-OH-BDE-154 displayed the highest activity with lowest effective concentrations (LOECs) of . These three OH-PBDEs also promoted SKBR3 cell migration via GPER pathways with LOECs of .
Conclusion:
OH-PBDEs could bind to GPER, activate the subsequent signaling pathways, and promote SKBR3 cell migration via GPER pathways. OH-PBDEs might exert estrogenic effects by a novel nongenomic mechanism involving the activation of GPER at nanomolar concentrations. https://doi.org/10.1289/EHP2387
Introduction
Polybrominated diphenyl ethers (PBDEs) have been and are currently used as flame retardant additives in a variety of industrial and consumer products such as plastic materials and textile fabrics (Li et al. 2017). As a result of such widespread use, PBDEs are thought to be ubiquitously present in human biological samples; in support of this theory, numerous studies from Europe, Asia, Australia, the United States, and Canada have detected measurable levels of various PBDEs in whole blood, plasma, and serum samples (Fromme et al. 2016). In addition, hydroxylated PBDEs (OH-PBDEs), which are metabolically biotransformed from PBDEs or of natural origin, were also found in the blood serum of children residing in Nicaragua (Athanasiadou et al. 2008) and in the blood of pregnant women in the U.S. state of Indiana (Qiu et al. 2009). There is evidence that the toxicologic effects reported for PBDEs on the thyroid hormone receptor (Li et al. 2010) and the estrogen receptor (Mercado-Feliciano and Bigsby 2008a) might be enhanced by metabolism of the parent compounds to OH-PBDEs.
Specific PBDEs have been shown to disrupt thyroid function in various in vivo animal models, including birds, fish, and rodents (Jugan et al. 2010; Legler 2008) and were associated with changes in the thyroid hormone thyroxine in U.S. adult male sportfish consumers (Turyk et al. 2008). Moreover, PBDEs were shown to increase the migration and invasion of human colorectal carcinoma cells (Wang et al. 2015) and the viability and proliferation of human breast, ovarian, and cervical cancer cells (Li et al. 2012), supporting potential carcinogenic activity. Finally, PBDEs have been associated with neurodevelopmental deficits in rats and humans (Herbstman and Mall 2014). In recent years, there has been growing concern about the estrogen-disrupting activities of PBDEs. For example, in vivo studies showed that exposure of pregnant rats to PBDE-99 () resulted in changes in the regulation of estrogen target genes in the uterus of female offspring (Ceccatelli et al. 2006), decreases in sex steroid levels and disruption of sexual development in male and female offspring (Lilienthal et al. 2006), and exhibition of sexually dimorphic behavior in male offspring (Lilienthal et al. 2006). Using the ovariectomized (OVX) mouse as a model, Mercado-Feliciano and Bigsby (2008b) found that exposure to PBDE mixture DE-71 () led to changes in uterine weight, uterine epithelial height, and vaginal epithelial thickness. In addition, in vitro studies showed that some PBDEs and OH-PBDEs exerted estrogenic effects in Chinese hamster ovary (Kojima et al. 2009) and human breast cancer (Meerts et al. 2001) cell lines.
The mechanisms by which exogenous chemicals are thought to disrupt the estrogen system are numerous and complex (Shanle and Xu 2011). An exogenous chemical may exert estrogenic effects by both classical nuclear estrogen receptors (ERs) and nongenomic G protein–coupled estrogen receptor (GPER) pathways (Shanle and Xu 2011). Earlier mechanistic studies of the exogenous chemicals focused on their estrogenic effects through ER-mediated pathways (Li et al. 2013; Meerts et al. 2001; Mercado-Feliciano and Bigsby 2008a). In recent years, a growing body of evidence has shown that some exogenous chemicals, such as bisphenol A (BPA), atrazine, nonylphenol, kepone, and genistein, might exert their estrogenic effects by activating GPER-mediated pathways (Albanito et al. 2015; Pupo et al. 2012; Thomas and Dong 2006). Understanding how estrogenic compounds activate GPER pathways is essential to thoroughly evaluating potential estrogenic effects. It has been shown that PBDEs and OH-PBDEs had weak activities towards ERs [with activities - to -fold less potent than ()] (Kojima et al. 2009; Li et al. 2013; Meerts et al. 2001; Mercado-Feliciano and Bigsby 2008a). Some OH-PBDEs bind to ERs directly with relatively low binding affinities (with binding potencies of 0.001% to 0.24% compared to that of ) (Li et al. 2013; Mercado-Feliciano and Bigsby 2008a). However, studies on the effects of PBDEs and OH-PBDEs on GPER are very limited. To the best of our knowledge, there has only been one report, which showed that 2, 2ʹ, 4, 4ʹ-tetrabromodiphenyl ether (BDE-047) stimulated the migration of human neuroblastoma SH-SY5Y cells and that the activity was inhibited by a GPER antagonist (G15), suggesting the involvement of GPER (Tian et al. 2016).
In the present study, we quantified the GPER binding potencies of 12 PBDEs (with bromination numbers from 1 to 8) and 18 OH-PBDEs (with hydroxyl positions of ortho-, meta- and para-) using a fluorescence competitive binding assay in a human breast cancer cell line (SKBR3). We used molecular docking to simulate the interactions of these compounds with GPER to determine the structural basis of the observed binding interactions. We used calcium mobilization and cyclic adenosine monophosphate (cAMP) production assays to investigate the activities of these compounds in SKBR3 cells. Finally, we further studied the effects of the compounds on GPER-mediated SKBR3 cell migration using Boyden chamber migration and wound-healing assays.
Methods
Reagents
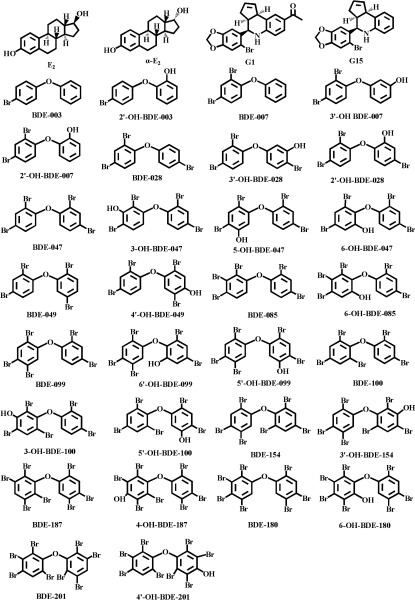
The 12 PBDEs and 18 OH-PBDEs used in this study are listed in Table 1 (see also Supplemental Material, “The full name of the 12 PBDEs and 18 OH-PBDEs”), and the structures are shown in Figure 1. All PBDEs and OH-PBDEs (solid; purity ) were purchased from AccuStandard and were dissolved in dimethyl sulfoxide (DMSO) to make stock solutions. (purity = 98%), (; purity = 98%), and (; purity = 98%) were purchased from Sigma-Aldrich. GPER agonist G1 (purity = 98%) and GPER antagonist G15 (purity = 98%) were purchased from Cayman Chemical Company. Calcium indicator fura-2-acetoxymethyl ester (fura-2 AM) and cAMP-Screen® System were purchased from Invitrogen. All solvents were obtained from Sigma-Aldrich and were used without further purification.
Table 1.
Results of the fluorescence competitive binding assays and molecular docking analysis of the investigated compounds with G protein–coupled estrogen eceptor 1 (GPER).
| Compounds | () | RBA (%) | OH position | Hydrogen bond | Hydrogen bond residue position |
|---|---|---|---|---|---|
| 0.3 | 100.0 | — | Asn276 | TM6 | |
| NA | — | — | — | — | |
| G1 | 0.6 | 51.7 | — | Asn276 | TM6 |
| G15 | 2.1 | 13.8 | — | — | — |
| BDE-003 | NA | — | — | — | — |
| 2ʹ-OH-BDE-003 | 20.0 | 1.3 | ortho | Gln138, Ser134 | TM3 |
| BDE-007 | NA | — | — | — | — |
| 2ʹ-OH-BDE-007 | NA | — | ortho | — | — |
| 3ʹ-OH-BDE-007 | 2.6 | 10.0 | meta | Gln138, Glu218 | TM3, TM5 |
| BDE-028 | NA | — | — | — | — |
| 2ʹ-OH-BDE-028 | NA | — | ortho | — | — |
| 3ʹ-OH-BDE-028 | 3.0 | 8.7 | meta | Glu218 | TM5 |
| BDE-047 | NA | — | — | — | — |
| 3-OH-BDE-047 | 12.0 | 2.2 | meta | Ser134 | TM3 |
| 5-OH-BDE-047 | NA | — | meta | — | — |
| 6-OH-BDE-047 | NA | — | ortho | — | — |
| BDE-049 | NA | — | — | — | — |
| 4ʹ-OH-BDE-049 | 2.8 | 9.3 | para | His282 | TM6 |
| BDE-085 | NA | — | — | — | — |
| 6-OH-BDE-085 | NA | — | ortho | — | — |
| BDE-099 | NA | — | — | — | — |
| 6ʹ-OH-BDE-099 | NA | — | ortho | — | — |
| 5ʹ-OH-BDE-099 | 1.3 | 20.0 | meta | Glu275 | TM6 |
| BDE-100 | NA | — | — | — | — |
| 5ʹ-OH-BDE-100 | NA | — | meta | — | — |
| 3-OH-BDE-100 | 3.6 | 7.2 | meta | — | — |
| BDE-154 | NA | — | — | — | — |
| 3ʹ-OH-BDE-154 | 1.3 | 20.0 | meta | Ser134 | TM3 |
| BDE-180 | NA | — | — | — | — |
| 6-OH-BDE-180 | 4.8 | 5.4 | ortho | His307, Phe208 | TM7, EL2 |
| BDE-187 | NA | — | — | — | — |
| 4-OH-BDE-187 | 6.2 | 4.2 | para | Phe208 | EL2 |
| BDE-201 | NA | — | — | — | — |
| 4ʹ-OH-BDE-201 | 5.3 | 4.9 | para | — | — |
Note: —, no information was collected at that particular examination point; , ; Asn, asparagine; BDE, bromodiphenyl ether; , ; EL, extracellular loop; G1, GPER agonist; G15, GPER antagonist; Gln, glutamine; Glu, glutamic acid; His, histidine; , concentration of a ligand required to displace half of the probes from the receptors; NA, not achieved; OH-BDE, hydroxylated bromodiphenyl ether; Phe, phenylalanine; RBA (relative binding affinity): ratio between of and compounds; Ser, serine; TM, transmembrane helix.
Figure 1.
Structures of the compounds tested in the present study.
Cell Culture Conditions
Human breast cancer SKBR3 cells and human embryonal kidney HEK293 cells were purchased from the American Type Culture Collection. The SKBR3 cells were cultured in phenol red–free Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone Inc.), penicillin (Invitrogen), and streptomycin (Invitrogen) at 37°C in a humidified 5% carbon dioxide () atmosphere. The HEK293 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) with 10% FBS, penicillin, and streptomycin at 37°C in a humidified 5% atmosphere.
Synthesis of Fluorescent Probe
The fluorescent probe () was synthesized by conjugating a fluorescein group to in three steps (see Figure S1): namely, generation of , generation of , and conjugation of with fluorescein. Thin-layer chromatography analysis using Kieselgel 60 F254 plates (Merck) was used to check the progress of the three reactions (Step 1–Step 3). The crude product of each reaction was purified using column chromatography performed on Kieselgel 60 silica gel (300–400 mesh, Merck) columns.
Step 1: Generation of .
was produced as described previously with slight modification (Revankar et al. 2007). Briefly, a solution of palladium(II) acetate [; ] and triphenylphosphine (;) in diethylamine () was stirred under argon (Ar) for 10 min at room temperature (rt, 25°C). Then, copper(I) iodide (CuI; ) and tert-butyl 4-bromobenzylcarbamate () were added and stirred for 5 min at rt. () was added and stirred for 6 h at 60°C. Finally, the solvent was removed by rotary evaporation, and the residue was purified by silica gel column chromatography using polyethylene/ethyl acetate (PE/EtOAc; 2:1) as the eluent to yield () as a light yellow solid.
Step 2: Generation of .
was also produced as described previously with slight modification (Revankar et al. 2007). () was dissolved in ethanol (;), and then 6 M hydrochloric acid (HCl; ) was added dropwise to the solution at 0°C. The resulting solution was stirred for 12 h at rt. The solvent was removed by rotary evaporation, and the residue was diluted with (). Then, the pH was adjusted to 11 with sodium bicarbonate (). The mixture was extracted with EtOAc (), and the solvent was removed by rotary evaporation. Finally, the residue was purified by silica gel column chromatography using methylene chloride/methanol (; 10:1) as the eluent to yield () as a white solid.
Step 3: Conjugation of with fluorescein.
The estrogen derivative () was conjugated to 5-carboxyfluorescein succinimidyl ester (NHS-Fluorescein, CAT number C2210, Invitrogen, ) in DMSO for 16 h at rt in the dark. Then, the solution was diluted with () and extracted with EtOAc () three times. The organic phases from the three extraction processes were combined, dried, concentrated, and then purified by preparative high-performance liquid chromatography (HPLC). The purification was performed using a Hewlett-Packard 1100 series HPLC instrument (Hewlett-Packard) equipped with a Gilson pumping system (Gilson), a Gilson 215 autosampler (Gilson), a Gemini C18 column (Phenomenex), and a photodiode array detector (Hewlett-Packard). The mobile phase [] was set to a linear gradient from 90/10 to 10/90 at a flow rate of , and the elution composition was detected at . After purification, a yellow solid product, (), was obtained. Stock solutions of () were prepared in DMSO and were stored at until use.
Characterization of Probe
Purity determination by HPLC.
The purity of was analyzed by HPLC on an Agilent HPLC instrument (Agilent Technologies) equipped with an XBridge C18 column (Waters). The mobile phase { [0.05% trifluoroacetic acid (TFA)]/ (0.05% TFA)} was set to a linear gradient from 90/10 to 0/100 at a flow rate of , and the elution composition was detected at .
Characterization by proton nuclear magnetic resonance.
Proton nuclear magnetic resonance () spectra were recorded on a Bruker UltraShield 300 MHz NMR spectrometer (Bruker). Briefly, of the product was dissolved in deuterated DMSO, and the solution was then subjected to detection. Chemical shifts (f1) were given in parts per million (ppm) with tetramethylsilane as an internal standard.
Characterization by mass spectrometry.
Mass spectrometry (MS) spectra were recorded on an HP-1100 liquid chromatography–mass spectrometry (LC-MS) instrument (Agilent Technologies) equipped with a Capcell Pak UG 120 ODS column (Shiseido Co., Ltd.). The mobile phase [ (0.05% TFA)/ (0.05% TFA)] was set to a linear gradient from 90/10 to 5/95 at a flow rate of , and the elution composition was detected by MS. All MS experiments were performed using electrospray ionization in positive ion mode.
Western Blotting Assay for the Expression of GPER
SKBR3 and HEK293 cells were seeded in dishes and incubated for 24 h. The cells were lysed and centrifuged at for 10 min at 4°C. The supernatant was collected, and the protein content was quantified using a BCA protein assay kit (Beyotime Biotechnology). Protein samples () were resolved on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Millipore). The membranes were blocked with 5% nonfat milk and then incubated with rabbit antibody against GPER (1:500; Abcam) and mouse antibody against (1:1,000; Cell Signaling Technology) overnight at 4°C. Then, the blots were incubated with appropriate secondary antibodies conjugated to IRDye700 or IRDye800 fluorescence dye, and the protein bands were detected using an Odyssey Infrared Imaging system (Li-COR Biosciences).
SKBR3 Cell–Based Fluorescence Competitive Binding Assay
Binding affinities of the 30 PBDEs/OH-PBDEs to GPER were measured using a SKBR3 cell–based fluorescence competitive binding assay. SKBR3 cells, a type of human breast cancer cell that expresses high levels of GPER but negligible levels of ERs, have previously been used in GPER binding assays (Lappano et al. 2010, 2012). Therefore, we selected this cell line for use in our study. The binding of to GPER was detected in SKBR3 cells by flow cytometry. HEK293 (GPER–) cells were used as negative controls, and fluorescein was used as a negative probe. and two GPER-specific ligands (G1 and G15) were used as positive controls, and inactive compound was used as a negative control to compete with binding of to SKBR3 cells. The cells were seeded in dishes (Corning) at a density of cells/dish. After serum starvation for 24 h, cells were harvested by trypsin-ethylenediaminetetraacetic acid (EDTA; Invitrogen), washed twice with ice-cold phosphate-buffered saline (PBS) buffer [ sodium chloride (NaCl), potassium chloride (KCl), disodium phosphate (), and monopotassium phosphate (), pH 7.4], and placed on ice in phenol red–free RPMI 1640 at a density of cells/mL. In the competitive binding assay, of harvested SKBR3 cells (approximately cells), , and a test compound (, G1, G15, , or one of the 30 PBDEs/OH-PBDEs) were mixed in PBS buffer for a total volume of and incubated for 10 min at 37°C. Cell samples were analyzed in a NovoCyte FCM flow cytometer (ACEA Biosciences). At least 10,000 events were analyzed per sample using forward scatter versus side scatter dot-plot gating to resolve the primary population of cells. The fluorescence intensity of the cells in the fluorescein isothiocyanate (FITC) channel for each sample was recorded in log mode. The maximum specific binding to was calculated by subtracting nonspecific binding (binding of in the presence of 500-fold excess ) from total binding (binding of alone). The displacement activities of the competitors (, G1, G15, at the highest tested concentrations of , , , and , respectively, and 30 PBDEs/OH-PBDEs at the highest tested concentration of or ) with binding were evaluated with competitive binding assays. The displacement of binding by a competitor was expressed as a percentage of the maximum specific binding to obtain the competition curve. The competition curves were fitted with a sigmoidal model using SigmaPlot 12.0 (Systat Software Inc.) to derive the value (the concentration of a ligand required to displace half of the probes from the receptors).
Calcium Mobilization Assay
Calcium mobilization assays were performed as described previously with slight modification (Revankar et al. 2005). The SKBR3 cells were seeded in dishes at a density of cells/dish. After serum starvation for 24 h, cells were incubated with fura-2 AM and 0.05% pluronic acid at 37°C for 30 min. Cells were washed 3 times with phenol red–free RPMI 1640 and incubated for another 30 min. Then, the cells were harvested with trypsin-EDTA, washed once with phenol red–free RPMI 1640, and placed on ice in phenol red–free RPMI 1640 at a density of cells/mL. Calcium mobilization was determined ratiometrically by dual excitation at and measuring fluorescence emission at using a Horiba Fluoromax-4 spectrofluorometer. Twenty microliters of harvested SKBR3 cells (approximately cells) was mixed in of Hank’s Balanced Salt Solution (HBSS) buffer (CAT number 14025134, Invitrogen), and the background fluorescence was detected five times every 20 s. Then, a tested chemical (, G1, or one of 11 OH-PBDEs) was added, and the fluorescence was detected immediately and every 20 s. The relative ratio, which was used to denote the calcium mobilization in SKBR3 cells, was plotted as a function of time. For the G15 inhibitory experiments, the cells were pretreated with G15 for 2 h, and vehicle (0.1% DMSO for 2 h) was used for those without G15 pretreatment.
cAMP Assay
SKBR3 cells ( cells) were seeded in 96-well microplates which were precoated with a capture antibody for the cAMP-antibody complex (Invitrogen). After incubation for 24 h, cells were changed into phenol red–free RPMI 1640 medium without FBS and incubated for another 24 h. Next, cells were treated with 3-isobutyl-1-methylxanthone in phenol red–free RPMI 1640 for 30 min and were then treated with a test chemical (, G1, or one of 11 OH-PBDEs) for 30 min. The final intracellular cAMP was quantified using the competitive immunoassay kit (Invitrogen) according to the manufacturer’s instructions. For the G15 inhibitory experiments, the cells were pretreated with G15 for 2 h, and vehicle (0.1% DMSO for 2 h) was used for those without G15 pretreatment.
Cell Migration Assay
SKBR3 cell migration was evaluated using Boyden chamber and wound-healing assays as described previously with slight modification (Cao et al. 2017). For the Boyden chamber assay, 24-well plates containing cell culture inserts with pore size (Corning) were used. The cells ( cells, ) were seeded in the upper chamber, and the lower chamber was supplied with phenol red–free RPMI 1640 with 10% FBS (Invitrogen). After 12 h, the cells were incubated with phenol red–free RPMI 1640 without FBS for another 12 h. Then, the cells were treated with a test chemical (, G1, or one of three OH-PBDEs) in phenol red–free RPMI 1640 without FBS, and the lower chamber was changed to phenol red–free RPMI 1640 medium with 10% charcoal-stripped FBS (CS-FBS; Invitrogen). After incubation for 48 h, the cells on the upper side of the insert membrane were removed with cotton swabs, and the cells on the lower surface of the membrane were fixed with methanol for 10 min and stained with 0.1% crystal violet (Sigma-Aldrich). Then, the cells were counted under an inverted microscope (ROCTEC) coupled to a camera (Canon), and six fields were randomly chosen per chamber. The relative cell migration was determined by comparing the average number of cells in the six fields of the treated chamber to those in the control chamber. For the G15 inhibitory experiments, the cells were pretreated with G15 for 2 h, and vehicle (0.1% DMSO for 2 h) was used for those without G15 pretreatment.
For the wound-healing assays, SKBR3 cells were seeded in 12-well microplates at a density of cells/well, incubated first in phenol red–free RPMI 1640 with 10% FBS for 12 h and then in phenol red–free RPMI 1640 without FBS for another 12 h. The cell layer was wounded with a sterile pipet tip and washed 3 times with phenol red–free RPMI 1640. The cells were then treated with a test chemical (, G1, or one of three OH-PBDEs) in phenol red–free RPMI 1640 with 10% CS-FBS for 48 h. The closure of the scratch was photographed using an inverted microscope (ROCTEC) coupled to a camera (Canon). The change in the scratch area was evaluated using Image-Pro Plus 6.0 (Media Cybernetics). The relative cell migration was calculated by comparing the change in the wound area in the treated wells with those in the control wells. For the G15 inhibitory experiments, the cells were pretreated with G15 for 2 h, and vehicle (0.1% DMSO for 2 h) was used for those without G15 pretreatment.
Homology Modeling and Molecular Docking Simulation
The structural template for GPER modeling (PDB ID: 3SN6A) was selected by submitting the amino acid sequence of GPER (NCBI Q99527) to the BLAST search engine (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Modeller 9v8 (Šali and Blundell 1993) was used to generate 20 three-dimensional (3D) protein structures using homology modeling with 3SN6A as the template. Then, the 3D structure was validated and evaluated using a Ramachandran plot generated by Discovery Studio Visualizer 4.5, and the best protein model (with 95% of the residues in the favorable regions of the Ramachandran plot) was selected for optimization. The initial structure was built with Membrane Builder on the CHARMM-GUI website (http://www.charmm-gui.org/?doc=input/membrane). Molecular dynamics (MD) simulation and molecular mechanics (MM) minimization were performed using GROMACS 4.5.6 (Van Der Spoel et al. 2005) with an AMBER force field. MD simulations were carried out with periodic boundary conditions. MD simulations were performed for 5 ns in the constant temperature, constant pressure (NPT) ensemble at standard ambient conditions (; ) controlled by Nosé-Hoover thermostat and Hoover barostat with a relaxation time of 0.02 ps. In all simulations, position restraints were applied to optimize the energy rapidly. In addition, Van der Waals forces were treated with a cutoff of , and the particle mesh Ewald method was used to compute electrostatic force with a cutoff. In addition, the LINCS algorithm was used to constrain the lengths of hydrogen bonds, and Cl– ions were used to neutralize the system charge. The final optimized protein structure obtained from the MD simulations was used as the GPER model to perform molecular docking.
For docking simulations, all small-molecule structures were built with Discovery Studio Visualizer 4.5 and optimized with the MOPAC program (Stewart 1990), and the partial atomic charges were computed with PM6 Hamiltonian36, included in MOPAC. The optimized small molecules were then docked with the GPER model using Autodock Vina v1.1.2 (the Scripps Research Institute); AutoDockTools was used to keep polar hydrogens and to add partial charges to the protein using Kollman united charges. The ligand–protein complex obtained from Autodock Vina was further analyzed by MD simulation using the GROMACS program (Van Der Spoel et al. 2005) coupled to molecular mechanics-Poisson Boltzmann surface area (MM-PBSA) calculation (Kumari et al. 2014) to obtain the interaction free energies of all of the complexes.
Statistical Analysis
All experimental results are expressed as the , and the experiments were repeated at least three times. Student’s t-test was used in the statistical analysis, and a p-value was considered statistically significant.
Results
Synthesis and Characterization of
The results from the characterization of are shown in Figures S2–S4. The purity of was determined by HPLC to be (see Figure S2). MS detection showed that the synthetic product had the correct molecular weight of 760.2 (see Figure S3). results were consistent with the correct structure of (see Figure S4). (400 MHz, DMSO): 10.167 (s, 1H), 9.448–9.248 (m, 2H), 9.002 (s, 1H), 8.517 (s, 1H), 8.299–8.091 (m, 2H), 7.723 (s, 1H), 7.410–7.335 (m, 3H), 7.263–7.242 (m, 1H), 7.081–7.060 (m, 1H), 6.695 (s, 2H), 6.611–6.502 (m, 4H), 6.439 (s, 1H), 4.547–4.409 (m, 2H), 2.740–2.705 (m, 2H), 2.551–2.512 (m, 1H), 2.339–1.667 (m, 10H), 1.342–1.244 (m, 3H), 0.819–0.805 (m, 3H). Abbreviations are as follows: br, broad; d, doublet; dd, doublet of doublet; m, multiplet; q, quartet; s, singlet; t, triplet.
Binding Affinities of PBDEs and OH-PBDEs to GPER
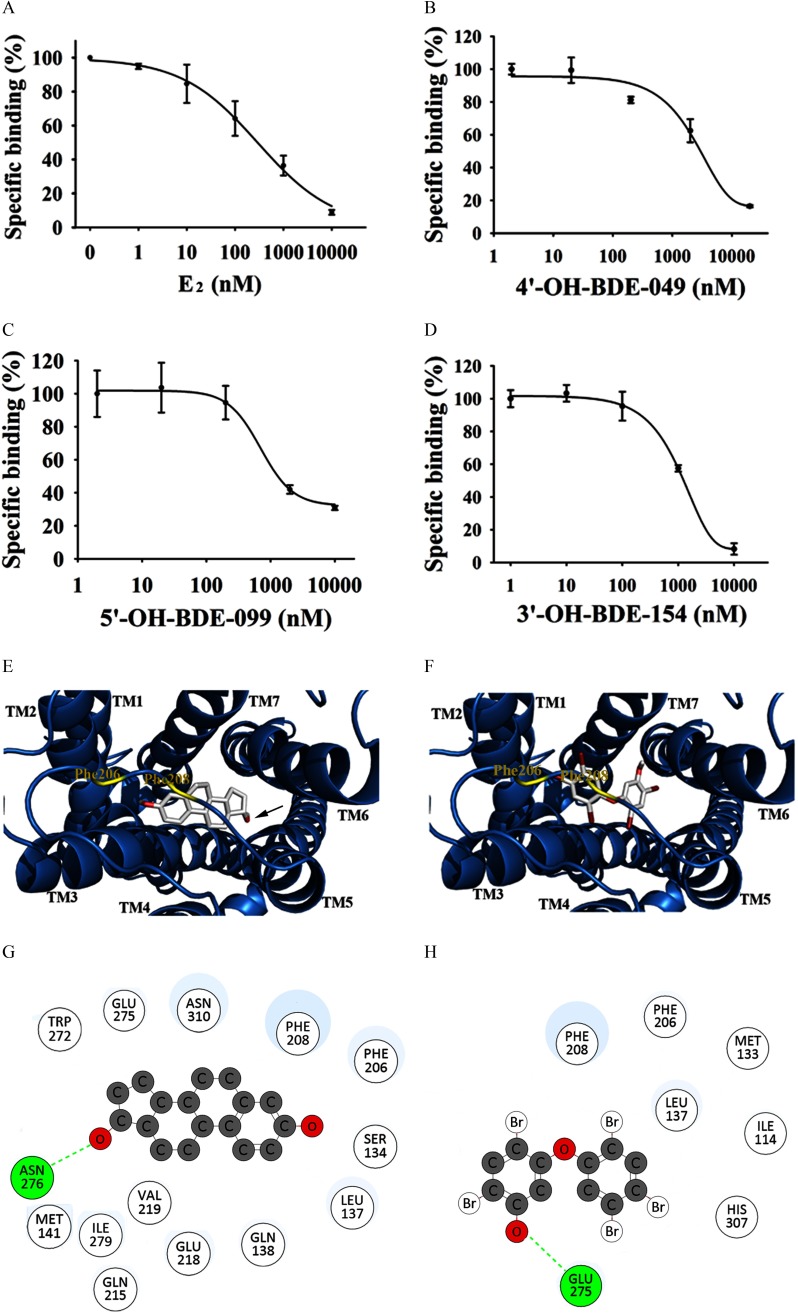
In the present study, we measured the binding affinities of PBDEs and OH-PBDEs with GPER by a novel SKBR3 cell–based fluorescence competitive binding assay. The expression of GPER in the SKBR3 and HEK293 cells used in our experiments were verified by Western blotting assays. SKBR3 cells expressed a high level of GPER protein, whereas GPER protein was undetectable in HEK293 cells (see Figure S5). To demonstrate the specific binding of to GPER, we used HEK293 (GPER–) cells as a negative control and used fluorescein as a negative probe. As shown in Figure S6A, the binding of to SKBR3 cells was higher than that to HEK293 cells, whereas the binding of fluorescein alone to SKBR3 cells was similar to the binding of fluorescein alone to HEK293 cells. Both and fluorescein alone similarly bound to HEK293 cells in a dose-dependent manner (see Figure S6A, B). By contrast, bound to SKBR3 cells more strongly than fluorescein alone (see Figure S6A, B). Because the highest specific binding of to SKBR3 cells (calculated from the ratio or , approximately 2-fold) occurred at (see Figure S6C, D), we used this concentration in the following competitive binding assays. To further confirm the specific binding of , and two GPER-specific ligands (G1 and G15) were used as positive controls, and the inactive compound was used as a negative control to compete with binding of to SKBR3 cells. As shown in Figure 2A (see also Figure S6E), , G1, and G15 inhibited the binding of to SKBR3 cells in a dose-dependent manner with values of , , and , respectively, whereas could not compete with even at the highest concentration () (Table 1). In addition, , G1, and G15 did not significantly affect the binding of to HEK293 cells (see Figure S6F, G) or the nonspecific binding of fluorescein to SKBR3 cells (see Figure S6H).
Figure 2.
Determination of the binding affinities of compounds with G protein–coupled estrogen receptor (GPER) and molecular docking analysis of the interactions between the compounds and GPER. (A–D) Competition curves of () and 3 representative hydroxylated polybrominated diphenyl ethers (OH-PBDEs) determined by SKBR3 cell–based fluorescence competitive binding assay. The concentration of probe is . (E–F) The binding pose of (E) and 5ʹ-OH-BDE-099 (F) in the binding pocket of GPER. Compounds are shown in sticks [carbon (C), oxygen (O), and bromine (Br)], and GPER is shown as blue ribbons produced by PyMOL software (v. 1.5.0.3, Schrodinger, LLC). The helixes are labeled from transmembrane helix 1 (TM1) to TM7. Two residues, phenylalanine (Phe) 206 and Phe208 are labeled with yellow lines to illustrate the binding pocket position. (G–H) Interactions of (G) and 5ʹ-OH-BDE-099 (H) with GPER. The hydrogen bond is indicated by a green dotted line. The binding poses and interactions of and 5ʹ-OH-BDE-099 with GPER were obtained with Autodock Vina v.1.1.2. Note: Asn, asparagine; Gln, glutamine; Glu, glutamic acid; His, histidine; Ile, isoleucine; Leu, leucine; Met, methionine; Trp, tryptophan; Val, valine.
The binding affinities of 12 PBDEs and 18 OH-PBDEs with GPER were then measured. The results are summarized in Table 1. We did not detect obvious binding of any of the 12 PBDEs to GPER, even at the highest concentration tested ( or ) (Table 1; see also Figure S7). Of the 18 OH-PBDEs, we detected binding to GPER by 11 (Figure 2B–D; see also Figure S7), with relative binding affinities (RBAs) ranging from 1.3% to 20.0% compared to (Table 1).
Docking Simulation for the Interactions of PBDEs and OH-PBDEs with GPER
We used molecular docking to simulate interactions between the 30 PBDEs and OH-PBDEs and GPER. Because the crystallographic structure of GPER is not yet available, we built its structure by homology modeling. In our model, the ligand binding pocket was located in a deep cleft formed by residues from transmembrane helix 2 (TM2), TM3, TM5, TM6, and TM7 (Figure 2E, F). First, three compounds with known binding affinity to GPER (, G1, and G15) and one inactive compound () were docked into GPER to validate the model and served as the basis for comparison to the PBDEs/OH-PBDEs of interest. As shown in Figure 2E and 2G, docked into GPER with its hydroxyl group (labeled with an arrow in Figure 2E) extending toward TM6, forming a hydrogen bond interaction with asparagine (Asn) 276. Like , G1 (a GPER agonist) also interacted with Asn276 through hydrogen bonding by the acetyl group oxygen atom (Table 1; see also Figure S8). G15 (a GPER antagonist) and (an inactive compound) did not form hydrogen bonds with any residues (Table 1; see also Figure S8).
We then docked PBDEs and OH-PBDEs into the ligand binding pocket of GPER. Of these compounds, nine OH-PBDEs formed hydrogen bonds with GPER through their hydroxyl groups (Figure 2F, H; see also Figure S8). Unlike , which formed a hydrogen bond with Asn276, the OH-PBDEs formed hydrogen bonds with different residues including serine (Ser) 134, glutamine (Gln) 138, phenylalanine (Phe) 208, glutamic acid (Glu) 218, Glu275, histidine (His) 282, and His307, depending on the OH-PBDE (Table 1).
Agonistic Activities of OH-PBDEs on GPER-Mediated Signaling Pathways
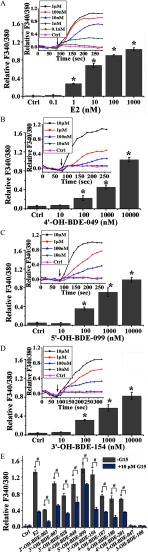
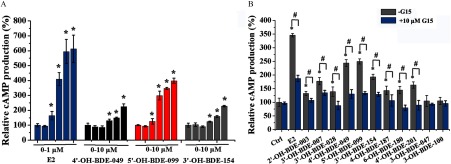
We further investigated the effects of the 11 OH-PBDEs that bound GPER on GPER-mediated signaling pathways. We used SKBR3 cells to examine the effects of the OH-PBDEs on intracellular calcium mobilization and cAMP production; these assays have been widely used to characterize the ligand activation of GPER in cells (Bologa et al. 2006; Revankar et al. 2005; Thomas and Dong 2006). Treatment of the cells with the positive control, , resulted in dose-dependent increases in both calcium mobilization (Figure 3A) and cAMP production (Figure 4A). The lowest effective concentration (LOEC) for was in both of the assays (Figures 3A and 4A). When cells were pretreated with G15, a specific antagonist of GPER, calcium mobilization and cAMP production were significantly lower than in cells pretreated with vehicle. Similar to , treatment of the cells with G1, a GPER-specific agonist, resulted in dose-dependent increases in calcium mobilization (see Figure S9A) and cAMP production (see Figure S9B) with an LOEC of for both assays. The cells were then treated with each of the 11 OH-PBDEs, treatment with nine of which (with the exceptions of 3-OH-BDE-047 and 3-OH-BDE-100) resulted in significant dose-dependent increases in calcium mobilization (Figure 3B–D; see also Figure S10) and cAMP production (Figure 4A; see also Figure S11) compared to control, with LOECs of and in the range of , respectively. To evaluate if the calcium mobilization and cAMP production measured after treatment with OH-PBDEs was mediated by the GPER pathway, we investigated the ability of G15 to reduce the effects of one micromolar of each of the OH-PBDEs. Pretreatment of SKBR3 cells with G15 resulted in less calcium mobilization (Figure 3E) and cAMP production (Figure 4B) than in cells treated with each OH-PBDE alone.
Figure 3.
Effects of () and 11 hydroxylated polybrominated diphenyl ethers (OH-PBDEs) on calcium mobilization in SKBR3 cells and the inhibitory effects of G15. (A–D) Calcium mobilization induced by different concentrations of , 4ʹ-OH-BDE-049, 5ʹ-OH-BDE-099, and 3ʹ-OH-BDE-154. The insets denote the change of F340/380 (the ratio of fluorescence intensity at and of calcium indicator fura-2-acetoxymethyl ester) with time, and the arrows indicate the time at which the compounds were added. (E) Calcium mobilization induced by and OH-PBDEs in the absence or presence of G15, a G protein–coupled estrogen receptor antagonist. The relative value of F340/380 after (the average of the last three data points in the time course curve as shown in the insets of A–D) and before adding the compounds is used to characterize the calcium mobilization. * compared with the control group (Ctrl, 0.1% dimethyl sulfoxide). # compared with the groups treated with compounds in the absence of G15.
Figure 4.
Effects of () and 11 hydroxylated polybrominated diphenyl ethers (OH-PBDEs) on cyclic adenosine monophosphate (cAMP) production in SKBR3 cells and the inhibitory effects of G15. (A) cAMP production induced by different concentrations of (0, , , , , and ) and OH-PBDEs (0, , , , , and ). (B) cAMP production induced by and OH-PBDEs in the absence or presence of G15, a G protein–coupled estrogen receptor antagonist. * compared with the control group (Ctrl, 0.1% dimethyl sulfoxide). # compared with the groups treated with compounds in the absence of G15.
Effect of OH-PBDEs on SKBR3 Cell Migration
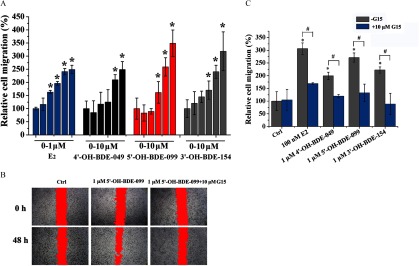
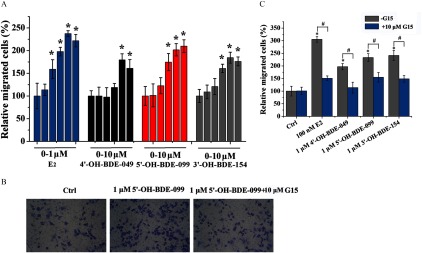
We used Boyden chamber and wound-healing assays to study the effects of OH-PBDEs on the migration of SKBR3 breast cancer cells, which has been shown to be regulated by GPER pathways (Pandey et al. 2009; Marjon et al. 2014). We chose to use 4ʹ-OH-BDE-049, 5ʹ-OH-BDE-099, and 3ʹ-OH-BDE-154 in these assays because they exhibited the highest GPER binding affinities (9.3–20.0% compared to ) and the strongest GPER agonistic activities among the tested OH-PBDEs. As a positive control, promoted the migration of SKBR3 cells in a concentration-dependent manner with an LOEC of (Figures 5A and 6A). Similarly, G1 also promoted SKBR3 cell migration with an LOEC of in both the Boyden chamber and wound-healing assays (see Figure S9C, D). In the wound-healing assay, 4ʹ-OH-BDE-049, 5ʹ-OH-BDE-099, and 3ʹ-OH-BDE-154 promoted SKBR3 cell migration in a concentration-dependent manner with LOECs of , , and , respectively (Figure 5A). The Boyden chamber migration assay showed very similar results for the 3 OH-PBDEs (Figure 6A), with the same LOECs as for the wound-healing assay. When cells were pretreated with G15, we detected significantly less cell migration than when cells were treated with each OH-PBDE alone as measured using the wound-healing (Figure 5B, C) and Boyden chamber (Figure 6B, C) assays.
Figure 5.
Effects of () and three hydroxylated polybrominated diphenyl ethers (OH-PBDEs) on SKBR3 cell migration determined by wound-healing assay and the inhibitory effects of G15. (A) SKBR3 cell migration induced by different concentrations of (0, , , , , and ) and OH-PBDEs (0, , , , , and ). (B) A typical wound-healing assay result for 5ʹ-OH-BDE-099 in the absence or presence of G15, a G protein–coupled estrogen receptor antagonist. The red filling denotes the unhealed scratches. (C) SKBR3 cell migration induced by and OH-PBDEs in the absence or presence of G15. The relative cell migration is quantified by setting the wound area change of the control group (Ctrl) as 100%. * compared with the control group (Ctrl, 0.1% dimethyl sulfoxide). # compared with the groups treated with compounds in the absence of G15.
Figure 6.
Effects of () and three hydroxylated polybrominated diphenyl ethers (OH-PBDEs) on SKBR3 cell migration detected by Boyden chamber assay and the inhibitory effects of G15. (A) SKBR3 cell migration induced by different concentrations of (0, , , , , and ) and OH-PBDEs (0, , , , , and ). (B) A typical Boyden chamber assay result for 5ʹ-OH-BDE-099 in the absence or presence of G15, a G protein–coupled estrogen receptor antagonist. (C) SKBR3 cell migration induced by and OH-PBDEs in the absence or presence of G15. The relative migration of cells is calculated by setting the count of migrated cells of the control group (Ctrl) as 100%. * compared with the control group (Ctrl, 0.1% dimethyl sulfoxide). # compared with the groups treated with compounds in absence of G15.
Discussion
PBDEs and OH-PBDEs have been reported to exert estrogenic effects in numerous in vivo (Ceccatelli et al. 2006; Yu et al. 2014) and in vitro studies (Li et al. 2013; Meerts et al. 2001; Mercado-Feliciano and Bigsby 2008b). However, the previous mechanistic studies focused on their genomic actions mediated by ER pathways (Li et al. 2013; Meerts et al. 2001; Mercado-Feliciano and Bigsby 2008a). Their nongenomic effects mediated by GPER pathways remain unclear. In the present study, by combining receptor binding assays, molecular simulation, receptor signaling pathway evaluation, and cell function detection, we investigated the effects of PBDEs and OH-PBDEs on the GPER pathways.
To evaluate the binding affinity of PBDEs/OH-PBDEs to GPER, we used a SKBR3 cell–based fluorescence competitive binding assay. The binding of probe to SKBR3 cells (GPER+) was higher than that to control HEK293 cells (GPER–), and bound to SKBR3 cells more strongly than did the negative probe fluorescein (see Figure S6A, B), indicating the specific binding of to SKBR3 cells by the part. Three known GPER ligands (, G1, and G15) inhibited the binding of to SKBR3 cells, whereas the inactive compound did not (Figure 2A; see also Figure S6E), further suggesting the specific binding of to GPER in SKBR3 cells. However, , G1, and G15 did not inhibit the nonspecific binding of to control HEK293 cells or the nonspecific binding of the negative probe fluorescein to SKBR3 cells (see Figure S6F–H), suggesting that the competition of the three ligands with to SKBR3 cells was due to specific binding to GPER. Moreover, the binding affinity of the three positive controls (, G1, and G15) and one negative control () to GPER is consistent with previous results determined by a radioisotope method (Lappano et al. 2010, 2012; Thomas et al. 2005), indicating that the established competitive binding assay method is feasible.
We used the established SKBR3 cell–based fluorescence competitive binding assay method to determine the binding affinities of 12 PBDEs and 18 OH-PBDEs to GPER. To the best of our knowledge, we found for the first time that OH-PBDEs but not PBDEs competed with the binding of probe to GPER, suggesting the direct binding of OH-PBDEs to GPER. In previous studies, some OH-PBDEs were demonstrated to bind to ERs directly with RBAs of 0.001–0.24% compared to (Li et al. 2013; Mercado-Feliciano and Bigsby 2008b). Here, we found that some OH-PBDEs possessed binding potency to GPER with RBAs of 1.3–20.0% compared to , two to three orders of magnitude higher than those with ERs. Therefore, we speculated that these OH-PBDEs might be able to trigger rapid nongenomic actions via GPER at much lower concentrations than required for genomic transduction activation of ERs.
Among the 18 OH-PBDEs tested in our study, 5ʹ-OH-BDE-099 and 3ʹ-OH-BDE-154 showed very high affinity for GPER with an RBA of 20.0% compared to . In previous studies, some other environmental chemicals were reported to bind to GPER directly. For example, using a tritiated () displacement assay based on plasma membranes prepared from HEK293 cells transfected with human GPER, BPA was demonstrated to bind to human GPER with an RBA of 2.8% compared to (Thomas and Dong 2006). In addition, Thomas and Dong (2006) found that 2,2-bis(p-chlorophenyl)-1,1,1-trichloroethane, 4-hydroxy-2,2ʹ,5ʹ-trichloro-biphenyl, and 2-(2-chlorophenyl)-2-(4-chlorophenyl)-1,1-dichloroethane bound to human GPER with RBAs of 0.2–1.3%. Some other environmental estrogens, such as genistein, nonylphenol, and kepone, were also found to bind to human GPER with RBAs of 1.3–13.4% (Thomas and Dong 2006). Using a displacement assay based on plasma membranes prepared from HEK293 cells transfected with zebrafish GPER, Fitzgerald et al. (2015) found that BPA bound to zebrafish GPER with an RBA of 15.8% and that tetrabromobisphenol A and tetrachlorobisphenol A showed effective binding at a single tested concentration (). Compared with the environmental chemicals investigated previously, 5ʹ-OH-BDE-099 and 3ʹ-OH-BDE-154 showed much stronger binding affinity to GPER.
We then analyzed the structural characteristics of the binding between GPER and the 12 PBDEs and 18 OH-PBDEs. In our study, only OH-PBDE metabolites but not parent PBDEs bound to GPER, suggesting that the hydroxyl group is an essential factor for GPER binding. Moreover, the hydroxyl position also affected the binding of OH-PBDEs with GPER. In the 18 OH-PBDEs tested, some OH-PBDE isomers are metabolites of the same parent PBDEs but with the hydroxyl group at a different position. However, they showed remarkably different binding potencies for GPER. For example, for the three OH-PBDE metabolites of BDE-047 (3-OH-BDE-047, 5-OH-BDE-047, and 6-OH-BDE-047), only 3-OH-BDE-047 bound with GPER (Table 1). Similar results were also found for the isomers of OH-PBDE metabolites of BDE-007, BDE-028, BDE-099, and BDE-100 (Table 1). However, we did not find a general rule for the position of the hydroxyl group (ortho-, meta- or para-) that could lead to GPER binding because the position of the hydroxyl on the 11 OH-PBDEs that bound to GPER was not confined to a specific location (Table 1).
Molecular docking analysis was performed to provide a structural basis for the differences in binding affinities observed in the competitive experiments. Similar to previous homology modeling structures of GPER (Chimento et al. 2014; Lappano et al. 2010; Méndez-Luna et al. 2015; Moreno-Ulloa et al. 2015), our results (Figure 2E, F) also showed that the ligand binding pocket is located in a deep cleft formed by residues from TM2, TM3, TM5, TM6, and TM7. The docking results for known ligands (, G1, and G15) were in line with the previously published results obtained from homology modeling and molecular docking (Arnatt and Zhang 2013), indicating the reliability of our simulation method. Of the 11 OH-PBDEs that bound to GPER, all formed hydrogen bond interactions with GPER through their hydroxyl groups except for 3-OH-BDE-100 and 4ʹ-OH-BDE-201. However, their corresponding parent PBDEs, without any hydroxyl groups, formed no hydrogen bonds. Furthermore, by comparing different OH-PBDE isomers of the same parent PBDEs (BDE-007, BDE-028, BDE-047, BDE-099, and BDE-100), we found that not all OH-PBDE isomers could form hydrogen bond interactions with GPER through their hydroxyl groups (Table 1). Only the isomers that bound to GPER in the competition experiment displayed hydrogen bond interactions with GPER in the docking simulations (Table 1). Because the OH-PBDE isomers of the same parent PBDEs showed different hydrogen bond interactions, we speculate that the position of the hydroxyl group on the OH-PBDEs also has a significant impact on their capacity to form hydrogen bond interactions with GPER. This finding is understandable considering the results showing distinct binding affinities for and , which differ only in the spatial conformation of the hydroxyl group (Table 1). , with its hydroxyl group in , could not form a hydrogen bond interaction with GPER (Table 1). Overall, our results indicate that having a hydroxyl group in the proper position is highly important in the binding of OH-PBDEs with GPER involving hydrongen bond interactions.
We further investigated the effects of the 11 OH-PBDEs that bound with GPER on the subsequent signaling pathways mediated by GPER. The results of both the calcium mobilization and the cAMP accumulation assays suggested that most of these OH-PBDEs activated GPER and that this activation did not depend on the degree of bromination or on the position of the hydroxyl group (Figures 3E and 4B). In general, the OH-PBDEs with higher binding potency exhibited higher agonistic activity, such as 5ʹ-OH-BDE-099, 3ʹ-OH-BDE-154, and 4ʹ-OH-BDE-049. However, no close correlation was present between the binding affinity and the responses (calcium mobilization and cAMP production) of the OH-PBDEs. This lack of correlation may be partly because the activity of a ligand depends not only on its affinity to a receptor but also on its binding configuration. As demonstrated in our molecular docking analysis (Table 1), these OH-PBDEs do show some difference in their binding interactions with GPER. We also investigated whether G15 could inhibit the ability of each OH-PBDE to induce calcium mobilization and cAMP production. Pretreatment with G15 resulted in significantly lower calcium and cAMP concentration in SKBR3 cells than treatment with OH-PBDEs alone, suggesting that the GPER pathway was involved in the biological responses. However, the possibility of OH-PBDEs leading to the responses in SKBR3 cells through some pathway other than GPER cannot be ruled out. This possibility likely explains why the inhibition efficiency of G15 is different for different OH-PBDEs. In addition, the inhibition efficiency of G15 for some OH-PBDEs was limited when compared to . These results also indicate that the OH-PBDE responses might be mediated by other pathways that could not be inhibited by G15.
The activities of OH-PBDEs with have been studied extensively, and the general trend seems to be that low-brominated OH-PBDEs tend to act as agonists, whereas high-brominated congeners are antagonists (Li et al. 2013). In contrast to the findings for , our study showed that both low-brominated and high-brominated OH-PBDEs exerted agonistic activities to GPER. For example, 6-OH-BDE-085, 5ʹ-OH-BDE-099, 3-OH-BDE-100, and 3ʹ-OH-BDE-154 were antagonists (Li et al. 2013), but these compounds displayed GPER agonistic activities in our studies. The combined results suggest that the high-brominated OH-PBDEs might show opposite activity toward GPER and . Therefore, the estrogen disruption effects of OH-PBDEs might be complex, and their effects on ERs and GPER pathways need to be considered together.
We further investigated the effects of OH-PBDEs on the cell function regulated by GPER. The activation of GPER pathways has been demonstrated to modulate the progression of various cancers by natural estrogen and exogenous chemicals (Liu et al. 2015; Pandey et al. 2009; Prossnitz and Barton 2014; Pupo et al. 2012). In both Boyden chamber and wound-healing assays, we observed increased cell migration after treatment of SKBR3 cells with one of three OH-PBDEs (4ʹ-OH-BDE-049, 5ʹ-OH-BDE-099, and 3ʹ-OH-BDE-154), and we found that pretreatment with G15 resulted in significant reduction of this cell migration, suggesting that it was mediated through activation of the GPER pathway. Therefore, OH-PBDEs might exert estrogenic effects directly via GPER pathways.
According to the docking results from both a previous study (Arnatt and Zhang 2013) and our own, for known GPER agonists such as and G1, the hydrogen bond with Asn276 on TM6 likely plays a key role in the activation of GPER. However, we found that the OH-PBDEs did not form any hydrogen bonds with Asn276. Alternatively, they formed hydrogen bonds with other GPER residues located on TM3, TM6, and TM7, as well as on extracellular loop 2 (EL2). It has been proposed that there might be multiple activation mechanisms for most G protein–coupled receptors, involving different “molecular switches” such as a TM3–TM6 ionic lock switch, a TM3–TM7 lock switch, a TM6 transmission switch, and a TM7 tyrosine toggle switch (Trzaskowski et al. 2012). We speculate that the OH-PBDEs might activate GPER by mechanisms different from those of and G1. However, the exact activation mechanism of the OH-PBDEs requires further investigation.
Based on the abovementioned results, we suggest that OH-PBDEs exert estrogenic effects via GPER pathways. Note that treatment of SKBR3 cells with nanomolar concentrations of either one of two OH-PBDEs (5ʹ-OH-BDE-099 and 3ʹ-OH-BDE-154) appeared to trigger GPER signaling pathways and to promote GPER-mediated cell migration. Human biomonitoring studies have shown the presence of 5ʹ-OH-BDE-099 at lipid ( when the blood lipid concentration was considered to be ) in fetal blood and lipid () in maternal blood (Qiu et al. 2009). The present human exposure concentration of 5ʹ-OH-BDE-099 may be quite low by comparison with the effective concentration for the activation of GPER pathways observed in our study; however, the total OH-PBDE concentration in human blood is much higher, in the range of tens to hundreds ng/g lipid (Athanasiadou et al. 2008; Qiu et al. 2009; Ren et al. 2011). These OH-PBDEs may act together to exert estrogenic effects. Moreover, the persistence and bioaccumulation of OH-PBDEs in humans and the continuous metabolism of PBDEs to OH-PBDEs might lead to increased concentrations in human blood. Therefore, more attention should be paid to the estrogenic effects of OH-PBDEs via GPER pathways, and the synergistic effects of more than one PBDE need further study in the future.
According to the results of previous studies (Li et al., 2013) and our present study, GPER and ER are two potential target molecules of OH-PBDEs in the human body. Previous studies have shown that OH-PBDEs also had other potential target molecules. For example, Ren et al. (2012; 2013) reported that OH-PBDEs could bind to some important proteins related to the thyroid hormone system, including thyroxine-binding globulin (TBG), transthyretin (TTR), and thyroid hormone receptor (TR). Therefore, OH-PBDEs might target both the estrogen system and the thyroid hormone system in organisms. By comparing the RBAs of OH-PBDEs to these proteins [GPER (1.3–20%) and ER (0.001–0.24%) compared with (Li et al. 2013); TBG (0.7–154%) and TTR (6–236%) compared with thyroxine (Ren and Guo 2012); TR (0.6–51%) compared with triiodothyronine (Ren et al. 2013)], it could be interpreted that OH-PBDEs have a higher RBA to the proteins related to the thyroid hormone system than to those related to the estrogen system. However, the specificity of OH-PBDEs depends not only on the RBA to the proteins but also on the expression level of these proteins in the tissues or cells. The specificity of OH-PBDEs toward different targets in organisms needs to be studied more comprehensively.
Conclusion
Although it is likely that OH-PBDEs may exert their estrogenic effects via multiple mechanisms, the combined data from our study strongly suggest that one of those mechanisms includes direct binding to GPER and subsequent activation of downstream signaling pathways. Our study enhances understanding of the mode of action and the molecular initiating events of OH-PBDE interference on estrogen functions.
Supplemental Material
Acknowledgments
This work was supported by the Chinese Academy of Sciences (grant nos. XDB14040100, QYZDJ-SSW-DQC020) and by the National Natural Science Foundation of China (grant nos. 21407168, 21621064, 91543203, 21577163, and 21477146).
References
- Albanito L, Lappano R, Madeo A, Chimento A, Prossnitz ER, Cappello AR, et al. 2015. Effects of atrazine on estrogen receptor α- and G protein-coupled receptor 30-mediated signaling and proliferation in cancer cells and cancer-associated fibroblasts. Environ Health Perspect 123(5):493–499, PMID: , 10.1289/ehp.1408586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnatt CK, Zhang Y. 2013. G protein-coupled estrogen receptor (GPER) agonist dual binding mode analyses toward understanding of its activation mechanism: a comparative homology modeling approach. Mol Inf 32(7):647–658, PMID: , 10.1002/minf.201200136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadou M, Cuadra SN, Marsh G, Bergman A, Jakobsson K. 2008. Polybrominated diphenyl ethers (PBDEs) and bioaccumulative hydroxylated PBDE metabolites in young humans from Managua, Nicaragua. Environ Health Perspect 116(3):400–408, PMID: , 10.1289/ehp.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologa CG, Revankar CM, Young SM, Edwards BS, Arterburn JB, Kiselyov AS, et al. 2006. Virtual and biomolecular screening converge on a selective agonist for GPR30. Nat Chem Biol 2(4):207–212, PMID: , 10.1038/nchembio775. [DOI] [PubMed] [Google Scholar]
- Cao LY, Ren XM, Li CH, Zhang J, Qin WP, Yang Y, et al. 2017. Bisphenol AF and bisphenol B exert higher estrogenic effects than bisphenol A via G protein-coupled estrogen receptor pathway. Environ Sci Technol 51(19):11423–11430, PMID: , 10.1021/acs.est.7b03336. [DOI] [PubMed] [Google Scholar]
- Ceccatelli R, Faass O, Schlumpf M, Lichtensteiger W. 2006. Gene expression and estrogen sensitivity in rat uterus after developmental exposure to the polybrominated diphenylether PBDE 99 and PCB. Toxicology 220(2–3):104–116, PMID: , 10.1016/j.tox.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Chimento A, Casaburi I, Rosano C, Avena P, De Luca A, Campana C, et al. 2014. Oleuropein and hydroxytyrosol activate GPER/GPR30-dependent pathways leading to apoptosis of ER-negative SKBR3 breast cancer cells. Mol Nutr Food Res 58(3):478–489, PMID: , 10.1002/mnfr.201300323. [DOI] [PubMed] [Google Scholar]
- Fitzgerald AC, Peyton C, Dong J, Thomas P. 2015. Bisphenol A and related alkylphenols exert nongenomic estrogenic actions through a G protein-coupled estrogen receptor 1 (Gper)/epidermal growth factor receptor (Egfr) pathway to inhibit meiotic maturation of zebrafish oocytes. Biol Reprod 93(6):135, PMID: , 10.1095/biolreprod.115.132316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme H, Becher G, Hilger B, Völkel W. 2016. Brominated flame retardants – exposure and risk assessment for the general population. Int J Hyg Environ Health 219(1):1–23, PMID: , 10.1016/j.ijheh.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Herbstman JB, Mall JK. 2014. Developmental exposure to polybrominated diphenyl ethers and neurodevelopment. Curr Environ Health Rep 1(2):101–112, PMID: , 10.1007/s40572-014-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugan ML, Levi Y, Blondeau JP. 2010. Endocrine disruptors and thyroid hormone physiology. Biochem Pharmacol 79(7):939–947, PMID: , 10.1016/j.bcp.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Kojima H, Takeuchi S, Uramaru N, Sugihara K, Yoshida T, Kitamura S. 2009. Nuclear hormone receptor activity of polybrominated diphenyl ethers and their hydroxylated and methoxylated metabolites in transactivation assays using Chinese hamster ovary cells. Environ Health Perspect 117(8):1210–1218, PMID: , 10.1289/ehp.0900753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R, Kumar R, Open Source Drug Discovery Consortium, Lynn A. 2014. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962, PMID: , 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Lappano R, Rosano C, De Marco P, De Francesco EM, Pezzi V, Maggiolini M. 2010. Estriol acts as a GPR30 antagonist in estrogen receptor-negative breast cancer cells. Mol Cell Endocrinol 320(1–2):162–170, PMID: , 10.1016/j.mce.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Lappano R, Rosano C, Santolla MF, Pupo M, De Francesco EM, De Marco P, et al. 2012. Two novel GPER agonists induce gene expression changes and growth effects in cancer cells. Curr Cancer Drug Targets 12(5):531–542, PMID: , 10.2174/156800912800673284. [DOI] [PubMed] [Google Scholar]
- Legler J. 2008. New insights into the endocrine disrupting effects of brominated flame retardants. Chemosphere 73(2):216–222, PMID: , 10.1016/j.chemosphere.2008.04.081. [DOI] [PubMed] [Google Scholar]
- Li F, Xie Q, Li X, Li N, Chi P, Chen J, et al. 2010. Hormone activity of hydroxylated polybrominated diphenyl ethers on human thyroid receptor-beta: in vitro and in silico investigations. Environ Health Perspect 118(5):602–606, PMID: , 10.1289/ehp.0901457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Y, Xiao W. 2017. Polybrominated diphenyl ethers in articles: a review of its applications and legislation. Environ Sci Pollut Res 24(5):4312–4321, PMID: , 10.1007/s11356-015-4515-6. [DOI] [PubMed] [Google Scholar]
- Li X, Gao Y, Guo L-H, Jiang G. 2013. Structure-dependent activities of hydroxylated polybrominated diphenyl ethers on human estrogen receptor. Toxicology 309:15–22, PMID: , 10.1016/j.tox.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Li ZH, Liu XY, Wang N, Chen JS, Chen YH, Huang JT, et al. 2012. Effects of decabrominated diphenyl ether (PBDE-209) in regulation of growth and apoptosis of breast, ovarian, and cervical cancer cells. Environ Health Perspect 120(4):541–546, PMID: , 10.1289/ehp.1104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilienthal H, Hack A, Roth-Härer A, Grande SW, Talsness CE. 2006. Effects of developmental exposure to 2,2ʹ,4,4ʹ,5-pentabromodiphenyl ether (PBDE-99) on sex steroids, sexual development, and sexually dimorphic behavior in rats. Environ Health Perspect 114(2):194–201, PMID: , 10.1289/ehp.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liao Y, Fan S, Tang H, Jiang Z, Zhou B, et al. 2015. G protein-coupled estrogen receptor (GPER) mediates NSCLC progression induced by 17β-estradiol (E-2) and selective agonist G1. Med Oncol 32(4):104, PMID: , 10.1007/s12032-015-0558-2. [DOI] [PubMed] [Google Scholar]
- Marjon NA, Hu C, Hathaway HJ, Prossnitz ER. 2014. G protein-coupled estrogen receptor regulates mammary tumorigenesis and metastasis. Mol Cancer Res 12(11):1644–1654, PMID: , 10.1158/1541-7786.MCR-14-0128-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerts IA, Letcher RJ, Hoving S, Marsh G, Bergman A, Lemmen JG, et al. 2001. In vitro estrogenicity of polybrominated diphenyl ethers, hydroxylated PBDEs, and polybrominated bisphenol A compounds. Environ Health Perspect 109(4):399–407, PMID: , 10.1289/ehp.01109399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Luna D, Martínez-Archundia M, Maroun RC, Ceballos-Reyes G, Fragoso-Vázquez MJ, González-Juárez DE, et al. 2015. Deciphering the GPER/GPR30-agonist and antagonists interactions using molecular modeling studies, molecular dynamics, and docking simulations. J Biomol Struct Dyn 33(10):2161–2172, PMID: , 10.1080/07391102.2014.994102. [DOI] [PubMed] [Google Scholar]
- Mercado-Feliciano M, Bigsby RM. 2008a. Hydroxylated metabolites of the polybrominated diphenyl ether mixture DE-71 are weak estrogen receptor-alpha ligands. Environ Health Perspect 116(10):1315–1321, PMID: , 10.1289/ehp.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Feliciano M, Bigsby RM. 2008b. The polybrominated diphenyl ether mixture DE-71 is mildly estrogenic. Environ Health Perspect 116(5):605–611, PMID: , 10.1289/ehp.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Ulloa A, Mendez-Luna D, Beltran-Partida E, Castillo C, Guevara G, Ramirez-Sanchez I, et al. 2015. The effects of (–)-epicatechin on endothelial cells involve the G protein-coupled estrogen receptor (GPER). Pharmacol Res 100:309–320, PMID: , 10.1016/j.phrs.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey DP, Lappano R, Albanito L, Madeo A, Maggiolini M, Picard D. 2009. Estrogenic GPR30 signalling induces proliferation and migration of breast cancer cells through CTGF. Embo J 28(5):523–532, PMID: , 10.1038/emboj.2008.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossnitz ER, Barton M. 2014. Estrogen biology: new insights into GPER function and clinical opportunities. Mol Cell Endocrinol 389(1–2):71–83, PMID: , 10.1016/j.mce.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo M, Pisano A, Lappano R, Santolla MF, De Francesco EM, Abonante S, et al. 2012. Bisphenol A induces gene expression changes and proliferative effects through GPER in breast cancer cells and cancer-associated fibroblasts. Environ Health Perspect 120(8):1177–1182, PMID: , 10.1289/ehp.1104526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Bigsby RM, Hites RA. 2009. Hydroxylated metabolites of polybrominated diphenyl ethers in human blood samples from the United States. Environ Health Perspect 117(1):93–98, PMID: , 10.1289/ehp.11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XM, Guo LH. 2012. Assessment of the binding of hydroxylated polybrominated diphenyl ethers to thyroid hormone transport proteins using a site-specific fluorescence probe. Environ Sci Technol 46(8):4633–4640, PMID: , 10.1021/es2046074. [DOI] [PubMed] [Google Scholar]
- Ren G, Yu Z, Ma S, Zheng K, Wang Y, Wu M, et al. 2011. Determination of polybrominated diphenyl ethers and their methoxylated and hydroxylated metabolites in human serum from electronic waste dismantling workers. Anal Methods 3(2):408–413, 10.1039/C0AY00571A. [DOI] [PubMed] [Google Scholar]
- Ren XM, Guo LH, Gao Y, Zhang BT, Wan B. 2013. Hydroxylated polybrominated diphenyl ethers exhibit different activities on thyroid hormone receptors depending on their degree of bromination. Toxicol Appl Pharmacol 268(3):256–263, PMID: , 10.1016/j.taap.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. 2005. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science 307(5715):1625–1630, PMID: , 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- Revankar CM, Mitchell HD, Field AS, Burai R, Corona C, Ramesh C, et al. 2007. Synthetic estrogen derivatives demonstrate the functionality of intracellular GPR30. ACS Chem Biol 2(8):536–544, PMID: , 10.1021/cb700072n. [DOI] [PubMed] [Google Scholar]
- Šali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815, PMID: , 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Shanle EK, Xu W. 2011. Endocrine disrupting chemicals targeting estrogen receptor signaling: identification and mechanisms of action. Chem Res Toxicol 24(1):6–19, PMID: , 10.1021/tx100231n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JJ. 1990. MOPAC: a semiempirical molecular orbital program. J Comput Aided Mol Des 4(1):1–103, PMID: , 10.1007/BF00128336. [DOI] [PubMed] [Google Scholar]
- Thomas P, Dong J. 2006. Binding and activation of the seven-transmembrane estrogen receptor GPR30 by environmental estrogens: a potential novel mechanism of endocrine disruption. J Steroid Biochem Mol Biol 102(1-5):175–179, PMID: , 10.1016/j.jsbmb.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Thomas P, Pang Y, Filardo EJ, Dong J. 2005. Identity of an estrogen membrane receptor coupled to a G protein in human breast cancer cells. Endocrinology 146(2):624–632, PMID: , 10.1210/en.2004-1064. [DOI] [PubMed] [Google Scholar]
- Tian PC, Wang HL, Chen GH, Luo Q, Chen Z, Wang Y, et al. 2016. 2,2ʹ,4,4ʹ-Tetrabromodiphenyl ether promotes human neuroblastoma SH-SY5Y cells migration via the GPER/PI3K/Akt signal pathway. Hum Exp Toxicol 35(2):124–134, PMID: , 10.1177/0960327115578974. [DOI] [PubMed] [Google Scholar]
- Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. 2012. Action of molecular switches in GPCRs – theoretical and experimental studies. Curr Med Chem 19(8):1090–1109, PMID: , 10.2174/092986712799320556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turyk ME, Persky VW, Imm P, Knobeloch L, Chatterton R Jr, Anderson HA. 2008. Hormone disruption by PBDEs in adult male sport fish consumers. Environ Health Perspect 116(12):1635–1641, PMID: , 10.1289/ehp.11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. 2005. GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718, PMID: , 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Wang F, Ruan XJ, Zhang HY. 2015. BDE-99 (2,2ʹ,4,4ʹ,5-pentabromodiphenyl ether) triggers epithelial-mesenchymal transition in colorectal cancer cells via PI3K/Akt/snail signaling pathway. Tumori 101(2):238–245, PMID: , 10.5301/tj.5000229. [DOI] [PubMed] [Google Scholar]
- Yu L, Liu C, Chen Q, Zhou B. 2014. Endocrine disruption and reproduction impairment in zebrafish after long-term exposure to DE-71. Environ Toxicol Chem 33(6):1354–1362, PMID: , 10.1002/etc.2562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.