Abstract
Cancer cells are known for aberrant methylation patterns leading to altered gene expression and tumor progression. DNA methyltransferases (DNMTs) are responsible for regulating DNA methylation in normal cells. However, many aberrant versions of DNMTs have been identified to date and their role in cancer continues to be elucidated. It has been previously shown that an aberrant version of a de novo methylase, DNMT3B7, is expressed in many cancer cell lines and has a functional role in the progression of breast cancer, neuroblastoma, and lymphoma. It is clear that DNMT3B7 is important to tumor development in vitro and in vivo, but it is unknown if expression of the transcript in all of these cell lines translates to relevant clinical results. In this study, a bioinformatics approach was utilized to test the hypothesis that DNMT3B7 expression corresponds to tumor progression in patient samples across cancer types. Gene expression and clinical data were obtained from the Genomic Data Commons for the 33 cancer types available and analyzed for DNMT3B7 expression with relation to tissue type in matched and unmatched samples, staging of tumors, and patient survival. Here we present the results of this analysis indicating a role for DNMT3B7 in tumor progression of many additional cancer types. Based on these data, future in vitro and in vivo studies can be prioritized to examine DNMT3B7 in cancer and, hopefully, develop novel therapeutics to target this aberrant transcript across multiple tumor types.
Introduction
The American Cancer Society estimates that nearly 1 out of every 3 people will be diagnosed with cancer in their lifetime [1]. While treatments have significantly improved and patient survival has increased in the last decade, cancer continues to be a global health issue and improved targeted therapies are needed. However, due to the heterogeneity of tumors, it has been difficult to identify one gene or protein that could be targeted to improve treatments across multiple cancer types.
It has been well-documented that cancer cells are characterized by abnormal DNA methylation patterns that alter gene expression and function [2, 3]. Tumor suppressor genes are often hypermethylated and transcriptionally inactive while oncogenes are hypomethylated and active. Normal methylation is regulated by three DNA methyltransferases (DNMTs)–DNMT1, DNMT3A, and DNMT3B [4–6]. DNMT1 is a maintenance methylase that is active throughout life while DNMT3A and DNMT3B are de novo methylases that are normally active in early development.
Recently, it has been shown that aberrant versions of DNMT3B are expressed in cancer cells, but not normal cells, and their functional role is still being elucidated [7–11]. Specifically, one of these aberrant transcripts, DNMT3B7, is expressed in 21 out of 25 cancer cell lines tested, including both solid and hematopoietic malignancies, making it a novel target that could potentially be used to treat many cancers at once [7]. DNMT3B7 retains 94bp of intron 10 sequence leading to an early stop codon and truncated protein. Furthermore, this truncated protein retains functional activity as observed by the fact that cell lines stably expressing DNMT3B7 show altered methylation patterns [7]. Shah and colleagues were the first to show that increased DNMT3B7 expression promotes lymphomagenesis in mice and alters methylation patterns in vivo as well as in vitro [9]. Subsequently, our laboratory has shown that expression of DNMT3B7 promotes tumor progression in breast cancer cells leading to hypermethylation of E-cadherin and corresponding changes in cell adhesion, proliferation, and anchorage-independent growth [10]. Interestingly, expression of DNMT3B7 in neuroblastoma showed an opposing effect in that lower levels of the transcript corresponded to tumor progression as measured by increased cell proliferation, angiogenesis, and tumor formation [11]. It is possible that differences in DNMT3B7 function may be related to cell type, such as changes between epithelial and mesenchymal cells, but additional studies are needed.
Because DNMT3B7 is expressed in so many different cancer cell types, and retains an intron sequence not found in other genes, it is an attractive target for novel targeted therapies. However, while we know that DNMT3B7 is expressed in multiple cancer cell lines, it is unknown whether this altered expression is observed in clinical samples. Furthermore, in order to elucidate the role of DNMT3B7 across all cancer types, in vitro and in vivo studies are required. Studies of this size and nature are both time-consuming and costly, therefore our laboratory utilized a bioinformatics approach to test the hypothesis that DNMT3B7 expression promotes tumor progression across cancers as measured by expression in normal versus tumor tissues, staging, and patient survival. The results of this study provide useful information on which cancer types should be further examined in vivo with the ultimate goal of developing novel therapeutics to target this aberrant transcript and potentially treat many different cancer types.
Materials & methods
Collection of data from Genomic Data Commons
RNAseqV2 and clinical data were obtained from the Genomic Data Commons (GDC) Legacy data portal (https://portal.gdc.cancer.gov/legacy-archive/search/f) [12]. Data were organized and processed using a custom C# script and Microsoft Excel (Redmond, Washington) to analyze expression of the retained 94bp sequence of intron 10 that is specific to DNMT3B7. Analyses were conducted on all available patient data for every cancer type available. Clinical staging was measured as stage I (combination of stage I, stage IA, and stage IB), stage II (combination of stage II, stage IIA, stage IIB, and stage IIC), stage III (combination of stage IIIA, stage IIIB, and stage IIIC), or stage IV. In order to determine survival rates, the median of DNMT3B7 expression across all tumor samples at a given site was determined. The samples were then divided in half, based on the median, into “low” and “high” expression groups and compared [13].
Statistical analysis
All statistical analysis was performed using SigmaStat software (Systat, Chicago, IL). Tumor versus normal tissue expression was compared using a Student’s T-test while matched tissues were compared with a Pairwise T-Test. Comparisons among groups for staging were analyzed with a one-way ANOVA with Dunn’s multiple comparisons. Finally, Survival LogRank analysis was utilized to generate a Kaplan-Meier curve and compare survival rates among low and high DNMT3B7 expression groups.
Results
DNMT3B7 expression is up-regulated in a majority of patient tumor samples
In order to determine the role of DNMT3B7 in tumor progression across 33 cancer types, all available data from the GDC were downloaded and analyzed for expression of the 94bp intron sequence specific to DNMT3B7. Table 1 shows the complete results of our analysis, with statistical significance indicated where appropriate.
Table 1. Results of DNMT3B7 expression across all cancer types.
| Cancer Name | GDC Name | Sample Size | Normal vs. Tumor | Matched Normal vs. Tumor |
Stage | Survival |
|---|---|---|---|---|---|---|
| Adrenocortical Carcinoma | ACC | 79 | NA | NA | No | No |
| Bladder Urothelial Carcinoma | BLCA | 430 | Yes (p<0.001) | Yes (p<0.001) | No | No |
| Breast Invasive Carcinoma | BRCA | 1097 | Yes (p<0.001) [10] | Yes (p<0.001) [10] | Yes (p = 0.01) [10] | No (p = 0.053) |
| Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma | CESC | 309 | Yes (p = 0.022) | No | NA | Yes (p = 0.025) |
| Cholangiocarcinoma | CHOL | 45 | No | NA | No | No |
| Colon Adenocarcinoma | COAD | 334 | Yes (p<0.001) | Yes (p<0.001) | No | No |
| Lymphoid Neoplasm Diffuse B cell Lymphoma | DLBC | 48 | NA | NA | NA | No |
| Esophageal Carcinoma | ESCA | 196 | Yes (p<0.001) | Yes (p = 0.001) | Yes (p = 0.012) | No |
| Glioblastoma Multiforme | GBM | 168 | NA (No, primary vs recurrent) | NA (No, primary vs recurrent) | NA | No |
| Head/Neck Squamous Cell Carcinoma | HNSC | 566 | Yes (p<0.001) | Yes (p<0.001) | No | No |
| Kidney Chromophobe | KICH | 91 | Yes (p<0.001)^ | No | No | No |
| Kidney Renal Clear Cell Carcinoma | KIRC | 612 | Yes (p = 0.023) | No | Yes (p = 0.010) | Yes (p = 0.009) |
| Kidney Renal Papillary Cell Carcinoma | KIRP | 323 | Yes (p<0.001) | Yes (p<0.001) | Yes (p = 0.02) | No |
| Acute Myeloid Leukemia | LAML | 173 | NA | NA | Yes (p<0.001) | Yes (p = 0.035) |
| Brain Lower Grade Glioma | LGG | 541 | NA (Yes, primary vs recurrent, p = 0.005) | NA (No, primary vs recurrent) | NA | No (p = 0.054) |
| Liver Hepatocellular Carcinoma | LIHC | 424 | Yes (p<0.001) | Yes (p<0.001) | No | No |
| Lung Adenocarcinoma | LUAD | 576 | Yes (p<0.001) | Yes (p<0.001) | No | No |
| Lung Squamous Cell Carcinoma | LUSC | 553 | Yes (p<0.001) | Yes (p<0.001) | Yes (p = 0.003) | No |
| Mesothelioma | MESO | 87 | NA | NA | No | Yes (p = 0.013) |
| Ovarian Serous Cystadenocarcinoma | OV | 309 | NA (No, primary vs recurrent) | NA (No, primary vs recurrent) | NA | No |
| Pancreatic Adenocarcinoma | PAAD | 183 | No | No | No | No |
| Pheochromocytoma and Paraganglioma | PCPG | 185 | No | No | NA | No |
| Prostate Adenocarcinoma | PRAD | 558 | No | No | NA | No |
| Rectum Adenocarcinoma | READ | 105 | Yes (p<0.001) | NA | No | No |
| Sarcoma | SARC | 266 | No | No | NA | Yes (p = 0.003) |
| Skin Cutaneous Melanoma | SKCM | 473 | NA | NA | No | Yes (p = 0.003) |
| Stomach Adenocarcinoma | STAD | 450 | Yes (p<0.001) | Yes (p<0.001) | No | No |
| Testicular Germ Cell Tumors | TGCT | 154 | NA | NA | Yes (p<0.001) | No |
| Thryoid Cancer | THCA | 572 | Yes (p<0.001)^ | Yes (p<0.001)^ | No | No |
| Thymoma | THYM | 122 | NA | NA | NA | No |
| Uterine Corpus Endometrial Carcinoma | UCEC | 198 | Yes (p<0.001) | Yes (p = 0.014) | NA | No |
| Uterine Carcinosarcoma | UCS | 57 | NA | NA | NA | No |
| Uveal Melanoma | UVM | 80 | NA | NA | No | No |
Normal vs. Tumor includes all patient samples available while Matched Normal vs. Tumor only includes matched samples.
“Yes” indicates data are statistically significant with increased DNMT3B7 expression observed in tumor samples, higher stage, and/or poor survival groups.
“No” indicates data were not statistically significant.
“NA” indicates data were not available so analysis could not be completed.
^ indicates that the pattern was altered and DNMT3B7 expression was higher in normal samples compared to tumor tissues for that sample.
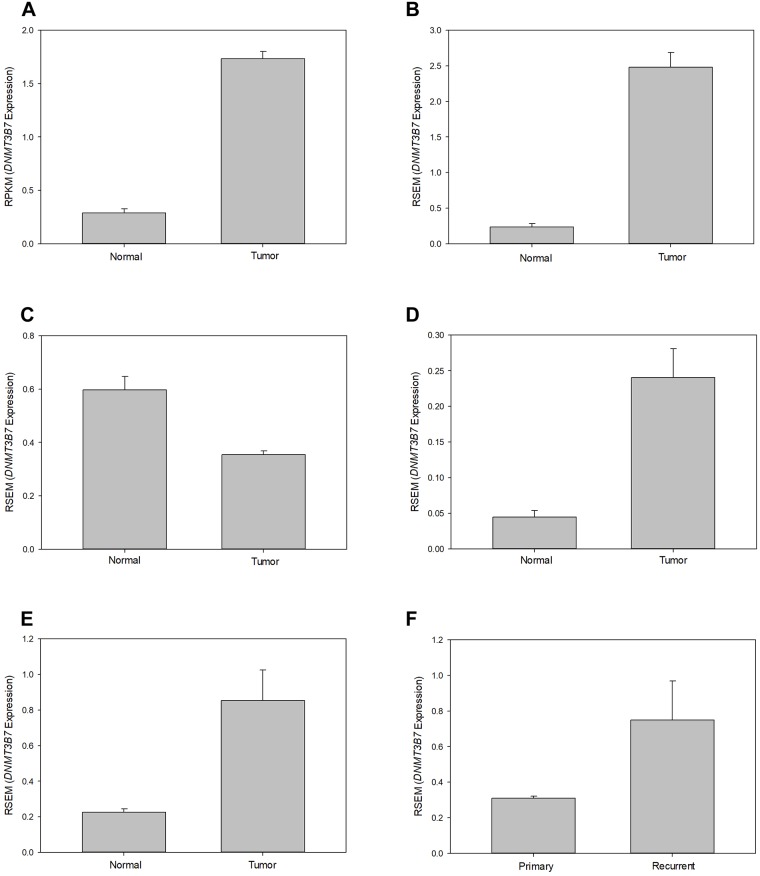
Of the 21 samples in which both normal and tumor tissues were available, 14 (67%) showed increased expression of DNMT3B7 in tumor samples compared to normal tissue, while two—KICH and THCA—showed decreased expression in tumor samples compared to normal (Fig 1A–1C). Of the 16 tumor types that showed significant differences in expression between normal and tumor tissue, 11 had similar patterns in matched patient samples, indicating that these results were not due to outliers in the group but rather genetic changes occurring in patients as their tumor developed and progressed (Fig 1D and 1E). Finally, while 12 of the tumor types did not have normal tissues available for analysis, three of these—GBM, LGG, and OV—did have primary and recurrent tissue samples which were utilized for comparison. Of these three tumor types, LGG showed increased expression in recurrent tumors compared to primary tumors (Fig 1F).
Fig 1. Expression of DNMT3B7 in normal and tumor patient samples.
Representative graphs of 6 different tumor samples showing relative DNMT3B7 expression, as measured by reads per kilobase per million (RPKM) or RNA-Seq by Expectation Maximization (RSEM), in unmatched normal and tumor tissues in (A) HNSC, (B) UCEC, and (C) THCA. Expression of DNMT3B7 in matched patient samples is shown in (D) LIHC and (E) LUAD. DNMT3B7 expression in primary and recurrent tissues in (F) LGG (p = 0.005) was assessed when normal samples were not available. All samples shown here were significant, p < 0.001, unless otherwise stated.
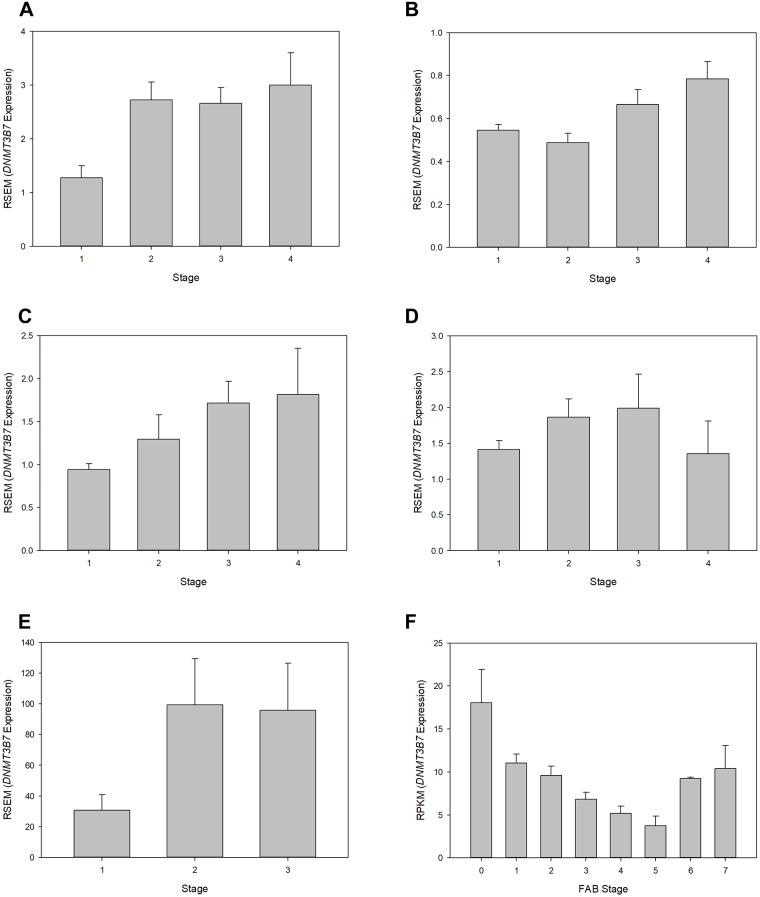
DNMT3B7 expression correlates to increased stage in some cancers
In order to further assess the effects of DNMT3B7 expression on clinical tumor progression, an analysis of expression based on diagnostic staging was completed. Of the 22 tumor samples for which data were available, 7 (32%) showed changes in expression relative to stage. In almost all cases, DNMT3B7 expression increased as clinical stage advanced indicating that DNMT3B7 expression correlates with tumor progression as measured by stage. Fig 2 shows the results of every cancer type with significant results except for BRCA, for which these data were published previously by our laboratory [10].
Fig 2. Relative DNMT3B7 expression correlates to clinical staging.
DNMT3B7 expression was compared to clinical stage and shown to be significantly different in (A) ESCA, p = 0.012; (B) KIRC, p = 0.010; (C) KIRP, p = 0.02; (D) LUSC, p = 0.003; (E) TGCT, p<0.001; and (F) LAML, p<0.001. For (E) TGCT, there were no patient samples with a stage IV diagnosis. (F) LAML staging was measured using the French-American-British (FAB) classifications.
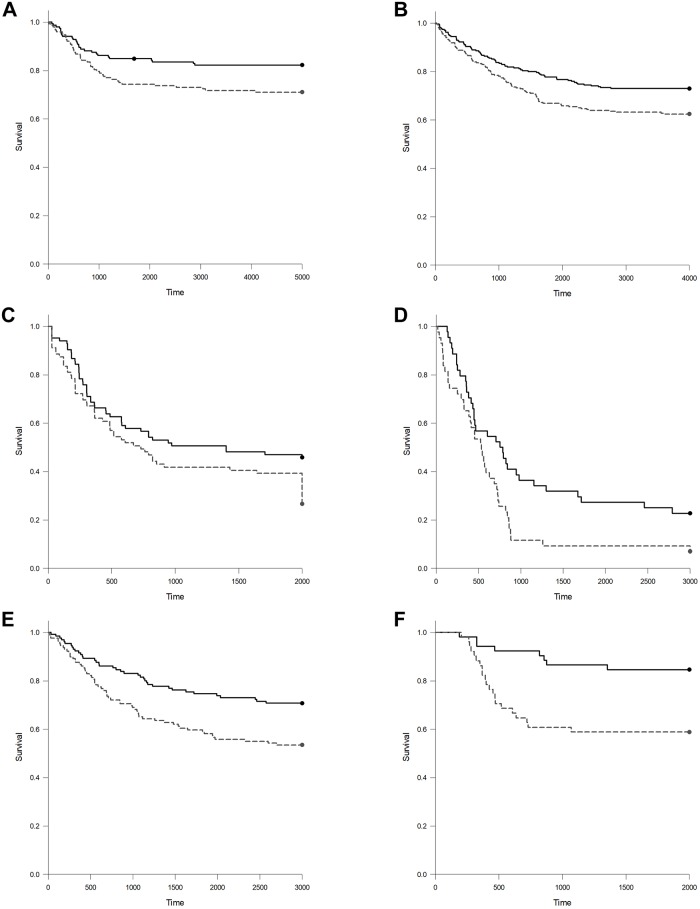
High expression of DNMT3B7 correlates with poorer survival in six tumor types
A final analysis was completed to determine the effect of high DNMT3B7 expression on patient survival. While survival is not a direct measure of tumor progression, due to the availability of therapeutics for specific malignancies, ability to diagnose some tumors at early stages, etc., we thought it was important to determine if there was any relationship between DNMT3B7 and survival as part of this analysis. Therefore, patients in each cancer type were divided into a “high” and “low” expression group based on the median DNMT3B7 expression for that cancer. A Kaplan-Meier analysis was completed and significant results are shown in Fig 3. Of the 33 cancers tested, only 6 (18%) showed a significant change in survival. In all cases, patients with high expression of DNMT3B7 had lower survival rates than those with low expression.
Fig 3. Patients with high levels of DNMT3B7 expression have lower survival rates than those with low expression levels.
The median DNMT3B7 expression for each individual tumor was determined to divide patients with that tumor into “high” (gray, dotted line) and “low” (black, solid line) expression groups. Kaplan-Meier curves were generated and statistical significance was determined for (A) CESC, p = 0.025; (B) KIRC, p = 0.009; (C) LAML, p = 0.035; (D) MESO, p = 0.013; (E) SARC, p = 0.003; and (F) SKCM, p = 0.003.
Discussion
Due to the heterogeneity of tumors, it is rare to find one gene that is mutated across many cancer types that can be targeted by therapeutics. Furthermore, in the rare cases where a gene is mutated in many cancers, (e.g. p53), the mutations are often in different parts of the gene making it impossible to develop one therapeutic that is useful for all patients. However, the results shown here indicate that DNMT3B7 may be a useful target across tumors in the future. Because of the unique sequence it retains of 94bp of intron 10 leading to a premature stop codon, there is a potential for specific targeting of this protein in cells. Our analysis here utilized that unique sequence as a measure of DNMT3B7 expression in all available patient samples on the GDC and showed that this sequence is correlated to tumor progression across all types of cancers—including carcinomas, sarcomas, and leukemias—as measured by tissue type, clinical stage, and survival across many cancer types.
Of the 33 cancers with data available on GDC, 22 (67%) showed significant effects of DNMT3B7 expression with relation to at least one of our measurements presented here. Of the 11 tumor types for which DNMT3B7 expression did not show any effect, five (DLBC, GBM, OV, THYM, and UCS) had no data available to assess anything except survival (Table 1). Therefore, it is possible that DNMT3B7 expression may have an effect in these cancers also, but we are unable to determine that based on the data available at this time. Furthermore, 7 cancers had sample sizes under 100 patients which may have prevented statistical significance from being achieved.
It is always important to determine if the experimental results previously shown in vitro and in vivo match those seen in clinical samples. Overall, we observed that the results shown by Ostler and colleagues [7] in which DNMT3B7 is expressed in virtually all (84%) cancer cell lines tested is confirmed here to a similar degree. Based on the data available on GDC, we were able to show altered expression of DNMT3B7 in 16 out of 21 samples (74%). Furthermore, we see that the previously published in vitro work in breast cancer matches with our clinical analysis (Table 1 and [10]). Shah and colleagues showed that DNMT3B7 expression led to lymphomagenesis [9], however these results could not be recapitulated due to the lack of available data on GDC. Finally, it may have been hypothesized that, based on the results of Ostler and colleagues in neuroblastoma [11], that the brain tumors examined here (GBM and LGG) would have shown a flipped pattern of expression in which higher DNMT3B7 expression correlated with normal samples compared to tumor samples. Unfortunately, normal samples were not available for this analysis, so we are unable to confirm or deny that hypothesis. We did observe that LGG showed increased expression of DNMT3B7 in recurrent tumors compared to primary tumors (Table 1 and Fig 1F) which would oppose the results seen in neuroblastoma. However, this is not overly surprising since neuroblastoma is a pediatric cancer that is caused by problems in early development and neurogenesis while LGG is diagnosed in adult brains and, therefore, is caused by entirely different mechanisms [14].
Our analysis of differences in expression based on clinical stage, while providing some useful data (Fig 2), was not as informative as hoped based on a few factors. First, we were unable to analyze 33% of our total samples due to a lack of available clinical data for our patient samples. It is quite possible that significant changes in DNMT3B7 expression correlate with stage in more cancers than shown here, but we cannot determine that based on the data available. Next, because many of the cancers had relatively small sample sizes to begin with, this was then further exacerbated by the fact that we had to subdivide these samples into smaller groups in order to complete our analysis. Therefore, it is once again possible that DNMT3B7 may play a larger role than observed here, but differences in sample size do not allow us to observe these trends at this time. Additionally, analysis of the effects on clinical stage is dependent on having enough patient samples diagnosed at each stage, which was not always possible. As shown in Fig 2E, some cancers did not have any patients diagnosed at one stage or another, which affected our results. In other cases, such as PAAD, of the 183 patient samples available, 21 were stage I, 151 were stage II, 3 were stage III, and 5 were stage IV. Small sample sizes in all but stage II led to difficulties in achieving any sort of sound statistical analysis. LAML had to be divided based on 8 FAB classifications which led to many more divisions than other tumor types. While our results did obtain statistical significance (Fig 2F), there was no distinguishable pattern except in the most advanced stages (6 and 7).
As stated previously, survival is not a direct measure of tumor progression and it can be affected by many factors including ability to diagnose early and available treatments. However, it is certainly the most important clinical outcome for patients and their families and, for that reason, it was included as part of our analysis (Fig 3). We saw significant differences in survival rates in LAML, which is the tumor type in which DNMT3B7 was originally identified [7]. Conversely, we did not see a significant difference in survival in BRCA (p = 0.053), even with a large sample size and previous in vitro data indicating a role in tumor progression [10]. However, this could be due to the fact that breast cancer is typically diagnosed at an early stage and has many good treatment options available, leading to increased survival rates compared to other cancers.
Our results show that no one cancer had statistically significant results in all three categories tested (normal versus tumor tissue, stage, and survival). However, 7 tumor types (BRCA, CESC, ESCA, KIRC, KIRP, LAML, and LUSC; Table 1) had changes in two different categories, assuming expression in normal and tumor samples in matched and unmatched tissues are considered one category. Because previous in vitro studies in LAML and BRCA have already been completed and confirmed [7, 10], these results suggest that the other 5 cancers listed above should be prioritized for future studies involving DNMT3B7. Specifically, continued in vitro and in vivo work, matched with clinical samples, is imperative to elucidate the functional role of DNMT3B7 in cancer and strengthen the likelihood of therapeutic targeting of this aberrant protein across cancer types in the future. Taken together, the results presented here demonstrate that DNMT3B7 has a role in tumor progression across cancers of all types and is a promising target for future drug development.
Acknowledgments
The authors wish to thank Mark Raimondi for writing all of the code needed to produce our data sets.
Data Availability
RNAseqV2 and clinical data were obtained from the Genomic Data Commons (GDC) Legacy Archives data portal (https://portal.gdc.cancer.gov/legacy-archive/search/f) and all relevant analysis and results are within the paper.
Funding Statement
This work was supported by the Ellen Marks Cancer Foundation (SLR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lifetime Risk of Developing or Dying from Cancer [Internet]. Atlanta: American Cancer Society [cited 29 May 2018]. https://www.cancer.org/cancer/cancer-basics/lifetime-probability-of-developing-or-dying-from-cancer.html
- 2.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005; 6: 597–610. 10.1038/nrg1655 [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007; 16 Spec No: R50–59. [DOI] [PubMed] [Google Scholar]
- 4.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000; 9: 2395–2402. [DOI] [PubMed] [Google Scholar]
- 5.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999; 99: 247–257. [DOI] [PubMed] [Google Scholar]
- 6.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001; 293: 1089–1093. 10.1126/science.1063443 [DOI] [PubMed] [Google Scholar]
- 7.Ostler KR, Davis EM, Payne SL, Gosalia BB, Expósito-Céspedes J, LeBeau MM, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007; 26: 5553–5563. 10.1038/sj.onc.1210351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Wang J, Sun S, Rodriguez M, Yue P, Jang SJ, et al. A novel DNMT3B subfamily, DeltaDNMT3B, is the predominant form of DNMT3B in non-small cell lung cancer. Intl J Onc. 2006; 29: 201–207. [PubMed] [Google Scholar]
- 9.Shah MY, Vasanthakumar A, Barnes NY, Figueroa ME, Kamp A, Hendrick C, et al. DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer Res. 2010; 70: 5840–5850. 10.1158/0008-5472.CAN-10-0847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brambert PR, Kelpsch DJ, Hameed R, Desai CV, Calafiore G, Godley LA, et al. DNMT3B7 expression promotes tumor progression to a more aggressive phenotype in breast cancer cells. PLoS ONE. 2015; 10(1):e0117310 10.1371/journal.pone.0117310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostler KR, Yang Q, Looney TJ, Zhang L, Vasanthakumar A, Tian Y, et al. Truncated DNMT3B isoform DNMT3B7 suppresses growth, induces differentiation, and alters DNA methylation in human neuroblastoma. Cancer Res. 2012; 72: 4714–4723. 10.1158/0008-5472.CAN-12-0886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. New Eng J Med. 2016; 375(12): 1109–1112. 10.1056/NEJMp1607591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H, Leppert JT, Peehl DM. A protective role for androgen receptor in clear cell renal cell carcinoma based on mining TCGA data. PLoS ONE. 2016; 11(1): e0146505 10.1371/journal.pone.0146505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Weille J. On the Genesis of Neuroblastoma and Glioma. Intl J Brain Sci. 2014; 2014 10.1155/2014/217503 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNAseqV2 and clinical data were obtained from the Genomic Data Commons (GDC) Legacy Archives data portal (https://portal.gdc.cancer.gov/legacy-archive/search/f) and all relevant analysis and results are within the paper.