Abstract
Acinetobacter baumannii has been recognized as one of the most challeging pathogens in clinical settings worldwide. Outer membrane porins play a significant role in Acinetobacter antibiotic resistance and virulence. A. baumannii carbapenem resistance and virulence factor porin Omp33-36 was the subject of this study. We investigated the omp33-36 gene transcriptional response in the growth phase, its response to carbapenems, and the effect of contact with host cells. Additionally, the cytotoxic effect of A. baumannii towards keratinocytes was assessed, as well as correlation between omp33-36 gene transcription and cytotoxicity. Further, Acinetobacter spp. Omp33-36 was classified and its characteristics relevant for vaccine candidature were determined. The level of the omp33-36 gene transcription varied between growth phases, but a common pattern could not be established among different strains. Treatment with subinhibitory concentrations of carbapenems decreased, while contact with keratinocytes increased omp33-36 expression in the analysed A. baumannii strains. Variations in omp33-36 mRNA levels did not correlate with cytotoxicity levels. Decrease of omp33-36 mRNA during treatment with subinhibitory concentrations of carbapenems, indicated the importance of transcriptional changes in reversible resistance to carbapenems due to the absence of Omp33-36. The transcription of omp33-36 increased after contact with keratinocytes, indicating the important role of de novo transcription during the initial phase of A. baumannii infection. Primary structural analysis of Acinetobacter spp. Omp33-36 revealed three distinct groups (among four A. baumannii variants). Although we have shown that Omp33-36 was highly polymorphic, we propose a potential antigen (PLAEAAFL motif) for vaccine development. According to PROVEAN analysis, the highly polymorphic structure of Omp33-36 porin should not influence its function significantly.
Introduction
Acinetobacter spp. are causative agents of infections that are predominantly nosocomial in origin, although community-acquired infections are increasingly reported as well [1]. Although species of the genus Acinetobacter are predominantly non-pathogenic, pathogenic members have been recognized as one of the most challenging in clinical settings. Acinetobacter baumannii causes the vast majority of Acinetobacter infections, followed by Acinetobacter pittii and Acinetobacter nosocomialis [2].
The pathogenic success of Acinetobacter species has been enabled by intrinsic resistance to a number of antibiotics, disinfectants and desiccation, as well as by its ability to easily acquire antibiotic resistance and virulence via horizontal gene transfer [1]. The increasing resistance to last resort antibiotics effective against Acinetobacter, such as carbapenems, has significantly limited therapeutic options [3]. Furthermore, reported pandrug-resistant A. baumannii strains make the situation more alarming [4]. Due to these findings, the World Health Organization (WHO) has positioned carbapenem-resistant A. baumannii on the top of the list compiled of pathogens for which new antibiotics are urgently needed [5]. In addition to new antibiotics, active and passive immunization, among other approaches, have been proposed for eradication of Acinetobacter infections [1].
The porins involved in the carbapenem resistance of A. baumannii are carbapenem resistance-associated outer membrane protein (CarO) and Omp33-36 [6,7]. CarO porin forms a channel with a binding site specific for imipenem, while specific binding sites for carbapenems in Omp33-36 porin have not been described so far [7]. It has been established that Omp33-36 porin participates in A. baumannii virulence and fitness [8]. The Omp33-36 porin facilitates adherence, invasion and cytotoxicity of A. baumannii, enabling survival in different organs and progression of infection within the host [8]. Additionally, Omp33-36 ability to modulate autophagy of human cells in order to survive within the host represents another aspect of its virulence [9]. Recently, several studies have assessed various outer membrane-associated vaccine candidates against A. baumannii, with promising results [10–16]. According to these studies, the Omp33-36 porin could be taken into account as a convenient target candidate for a preventive or therapeutic vaccine effective against Acinetobacter infections. It has been shown that cell density, treatment with subinhibitory concentrations of antibiotics and interaction with eukaryotic cells affect A. baumannii pathogenicity [17–21]. Although of significance, the role of Omp33-36 in those processes was not sufficiently studied.
In our previous study we revised the CarO porin classification of genus Acinetobacter [22]. Accordingly, the aim of this study was to investigate the prevalence and conservation of Omp33-36 porin among the genus Acinetobacter in order to establish whether it could be considered a good candidate for vaccine development. Furthermore, the dependence of omp33-36 transcription on cell density, the effects of carbapenem treatment, and contact with keratinocytes were studied in carbapenem-susceptible and carbapenem-resistant A. baumannii in order to assess the contribution of Omp33-36 to the overall pathogenic potential of A. baumannii.
Materials and methods
Bacterial strains and species identification
One carbapenem-susceptible (813) and two carbapenem-resistant A. baumannii clinical strains (1995/12 –resistance due to oxacillinases production and 4031 –resistance due to oxacillinase production and inactivated carO gene) were isolated at the Institute for Mother and Child Health Care "Dr. Vukan Čupić", a tertiary care paediatric hospital in Belgrade, Serbia. The carbapenem-resistant strains were identified by a Vitek 2 automated system (BioMérieux, Marcy l’Étoile, France) and amplification of genes for 16S rRNA [23] in our previous study [22], while identification of carbapenem-susceptible strain 813 was done in this study by the same methods.
The carO and the omp33-36 gene analysis
The amplification and analysis of the carO and omp33-36 genes were carried out as previously described [22,24]. The PCR products were purified with a GeneJET PCR Purification Kit (Thermo Scientific, Lithuania) and sequenced by the Macrogen service (Macrogen Inc., Netherlands).
Carbapenem susceptibility testing
Carbapenem susceptibility of the strains used in this study was analyzed by the microdilution method, according to the European Committee on Antimicrobial Susceptibility Testing recommendations (http://www.eucast.org) [25].
Subinhibitory carbapenem treatment
Bacterial cells were grown in Mueller-Hinton (MH) broth at 37°C with shaking overnight. Strains 813 and 1995/12 were grown in the presence of a subinhibitory concentration (1/10 of minimal inhibitory concentration—MIC- value) of meropenem or imipenem, while strain 4031 was incubated only with 1/10 MIC of imipenem. The selection of antibiotics was based on the role of CarO porin in imipenem resistance [7]. The bacterial cell density at 600nm (OD600) was monitored and stopped in different growth phases (early, middle, late exponential and stationary phase). The collected cells were centrifuged at 4500 x g for 5 min and used for RNA isolation. As controls, bacterial cells were cultivated in the absence of carbapenems. Experiments were done in triplicate.
HaCaT keratinocyte infection with A. baumannii
A HaCaT keratinocyte cell line was cultured in high glucose DMEM supplemented with 2mM L-glutamine, 10% fetal bovine serum (FBS), 100U/ml penicillin and 100μg/ml streptomycin (Gibco, Life Technologies). For treatment, 1x106 of HaCaT cells were plated in T25 cell culture flasks (Sarstedt, Germany) and incubated at 37°C with 5% CO2 overnight. Before infection the cells were washed in phosphate-buffered saline (PBS) and suspended in the aforementioned medium without antibiotics.
Bacterial cultures were grown in MH broth at 37°C with shaking overnight. The cells (1x108 CFU/ml) were harvested and washed in PBS. After centrifugation at 4500 x g for 5 min, the pellets were suspended in high glucose DMEM containing 2mM L-glutamine and 10% FBS.
Infection of HaCaT keratinocytes with A. baumannii was performed by addition of the bacterial cell suspension to a flask with HaCaT cells (multiplicity of infection was 100). The HaCaT and A. baumannii cells were coincubated at 37°C with 5% CO2 for 21h. After treatment, the cell culture medium with bacterial cells was centrifuged at 405 x g for 5 min to pellet residual non-adherent HaCaT cells. For harvesting bacterial cells, supernatant was centrifuged at 4500 x g for 5 min and used for RNA isolation. Controls were incubated and purified as described above, but in the absence of HaCaT cells. Experiments were done in triplicate.
Transcriptional analysis by real-time quantitative PCR (RT-qPCR)
The total RNA from A. baumannii cells was isolated with an RNeasy Mini Kit (Qiagen, Germany), with a modified lysis step [26]. DNase I treatment was performed by an Ambion DNA-free™ Kit (Thermo Fisher Scientific, MA, USA). Reverse transcription was done with a RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific, MA, USA) according to the manufacturer′s protocol. RT-qPCR was used for determination of omp33-36 expression level. Primers and conditions applied for RT-qPCR are listed in Table 1 [27]. RT-qPCR was performed with a KAPA SYBR Fast qPCR Kit (KAPA Biosystems, MA, USA) in a 7500 Real Time PCR System thermocycler (Applied Biosystems, Thermo Fischer Scientific, MA, USA). Normalization was done against the rpoB gene using the ΔΔCT method (relative) [28]. The obtained values were then normalized against those from the early exponential phase in the absence of carbapenem. Also, the results from each growth phase were compared to those reached in the same phase without carbapenem treatment. Additionally, dependence of omp33-36 transcription on contact with HaCaT cells was determined by normalization against values acquired from incubation without HaCaT cells. RT-qPCR experiments were done in triplicate.
Table 1. List of primers and conditions used in RT-qPCR.
| Primer name | Sequence (5'-3') | Cycling condition | Reference |
|---|---|---|---|
| Omp33-RT_F | GCTTATCAATTTGAAGTTCAAGGTC | 95°C/3 min | This study |
| Omp33-RT_R | GCTTGGTTTAAGAAAGCTGC | 95°C/15 s- | |
| RpoB_For | TCCGCACGTAAAGTAGGAAC | 60°C/60s | [27] |
| RpoB_Rev | ATGCCGCCTGAAAAAGTAAC | [40 cycles] |
Cytotoxicity Assay
The cytotoxicity levels after infection of HaCaT keratinocytes with A. baumannii cells were measured with a lactate dehydrogenase (LDH) Cytotoxicity Assay Kit (Thermo Scientific, Lithuania). Quantitative determination of LDH released from dead cells was measured in the collected supernatants according to the manufacture′s protocol. The absorbance was measured at 490 nm in a microplate reader (Tecan Austria, Gmbh, Austria). Experiments were done in triplicate.
Database search and data analysis
To build the Omp33-36 protein profile the PSI-BLAST [29] homology searches were restricted to either Moraxellaceae or A. baumannii with an e-value cutoff of 1e-4. All hits that exhibited at least 27% identity and over 71% of the query fragment length were retained for further analysis. As an Omp33-36 prototype, Omp33-36 CAH40841.2 was used. Proteins with sequence sizes significantly different from the average were removed. Amino acid sequences of Omp33-36 from the NCBI database (S1 File) and sequences from Serbian strains were aligned using Clustal W version 1.7 [30].
Phylogenetic analysis
Phylogenetic trees were constructed using MEGA version 7.0 [31]. The Clustal W multiple sequence alignments were performed using default parameters. The maximum-likelihood (ML) method, using a Jones-Taylor-Thornton (JTT) distance matrix, was used for the construction of an Omp33-36 porin phylogenetic tree. The A. baumannii strains from this study were also involved in analysis. A bootstrap consensus tree inferred from 1000 replicates was taken to deduce confidence levels for the ML trees. An online tool (iTOL, v3) for the display and annotation of the tree was used [32].
Topology prediction and analysis of Omp33-36 porin isoforms
Four A. baumannii Omp33-36 porin variants were included as representatives of Acinetobacter spp. Omp33-36 groups. Transmembrane topology predictions for the Omp33-36 isoforms were performed by PRED-TMBB (http://bioinformatics.biol.uoa.gr/PRED-TMBB) [33].
The Shannon entropy [34] of each amino acid position in Omp33-36, multiple alignments of the three main Omp33-36 groups among Moraxellaceae, was calculated using the following equation:
where the Pi is the frequency of amino acid residue i in that site and M is the number of amino acid types. Shannon entropy (H) ranges from 0 (only one residue is represented in a given position) to 4.32 (all 20 residues are equally represented in a given position). The amino acid changes observed in the different Omp33-36 variants relative to the canonical Omp33-36 variant 1 were analyzed using PROVEAN software (http://provean.jcvi.org/), which predicts whether an amino acid substitution, insertion or deletion has any impact on the biological function of a protein.
Statistical analysis
All results are presented as mean values ± standard deviations. One-way ANOVA, followed by Tukey’s post hoc test, was used to compare multiple treatments. In experiments with HaCaT cells, a Student′s t-test was performed to compare differences between the control and experimental groups. Values at p0.05 or less were considered to be statistically significant. All data were analyzed using SPSS 20.0 v.
Results
The strains used in this study were identified as A. baumannii. The selection of strains was based on carbapenem resistance phenotype.
The carO and omp33-36 gene analyses
The nucleotide alignment of the carO gene (S2 File) in carbapenem-susceptible strain 813, compared to the BLAST database, revealed 99% identity to the corresponding gene of the clinical strain A. baumannii C71 from China (KX517489.1), while the nucleotide sequence of omp33-36 was 100% identical to an outer membrane protein gene of clinical strain A. baumannii IOMTU433 from Nepal (AP014649.1, from 219 644 to 218 747 bp). The carO and omp33-36 nucleotide sequences of carbapenem-resistant strains 1995/12 and 4031 were determined in our previous study [22]. Nucleotide sequences of omp33-36 genes from strains used in this study are reported in S3 File.
Carbapenem susceptibility
The carbapenem susceptibility testing was performed for all strains, with the exception of 4031, which was not tested against meropenem due to its lack of functional CarO porin (Table 2).
Table 2. MIC values determined by microdilution method.
| Strain | Imipenem MIC (μg/ml) | Meropenem MIC (μg/ml) |
|---|---|---|
| 813 | 0.1 (S) | 0.1 (S) |
| 1995/12 | 16 (R) | 32 (R) |
| 4031 | 32 (R) | NT |
S-susceptible; R-resistant; NT-not tested.
Effects of growth phase and carbapenems on omp33-36 gene transcription
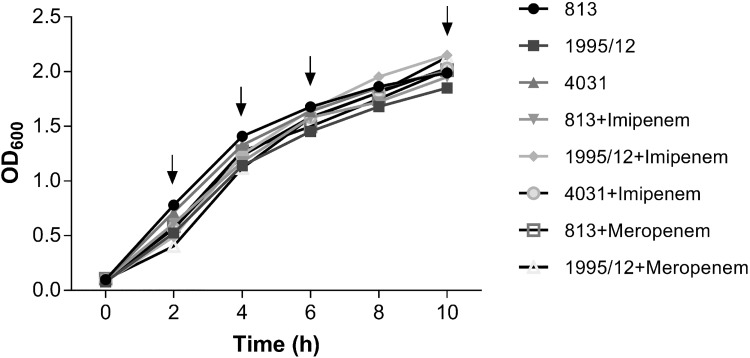
The growth curves of carbapenem-treated and untreated strains are presented in Fig 1. Time points corresponding to early (2h), middle (4h), late exponential (6h) and stationary growth phase (10h) were chosen according to measured OD600 values. Changes in the omp33-36 mRNA levels during growth phases and exposure to carbapenems, investigated by RT-qPCR, are presented in Fig 2.
Fig 1. The growth curves of A. baumannii strains, untreated or treated with subinhibitory carbapenems (1/10 of MIC value).
A. baumannii strains were incubated in the presence of a subinhibitory concentration of imipenem (813, 1995/12 and 4031) or meropenem (813 and 1995/12). The arrows indicate time points (2h, 4h, 6h and 10h) in which aliquots were collected for expression analysis.
Fig 2. Changes of omp33-36 mRNA levels during exposure to carbapenems through growth phases.
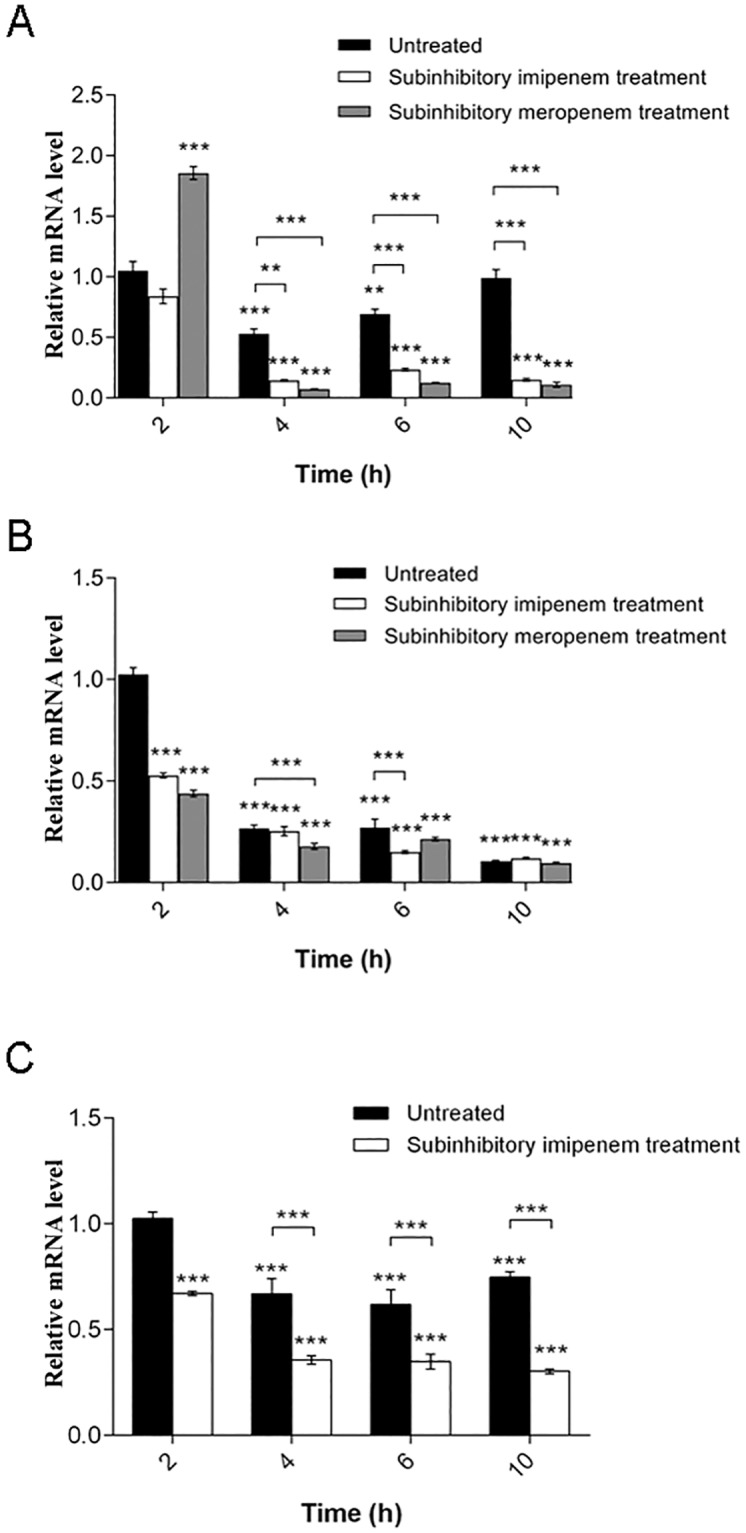
(A) Carbapenem-susceptible strain 813. (B) Carbapenem-resistant strain 1995/12. (C) Carbapenem-resistant strain 4031. All expression results were normalized relative to rpoB by the 2−ΔΔCt method. Values are the means from results obtained in triplicate. Error bars represent the standard deviation of the mean value. One-way ANOVA with Tukey′s post hoc test was used to compare the gene expression results relative to the expression of the omp33-36 gene in the early exponential phase of an untreated strain and to the expression of the untreated strain for each time point (** p< 0.01, *** p< 0.001).
According to the obtained results, transcriptional expression of the omp33-36 gene varied during different growth phases. All strains have shown a trend of omp33-36 mRNA level decrease in transition from early to middle exponential phase (Fig 2). After this decrease, strains 813 and 4031 slowly returned to the omp33-36 mRNA levels starting point (Fig 2A and 2C). In contrast, omp33-36 mRNA levels showed a constant decline in strain 1995/12 (Fig 2B).
Subinhibitory concentrations of carbapenems reduced omp33-36 gene transcription throughout growth phases in all strains (Fig 2). The change was most expressive in carbapenem-susceptible strain 813 (Fig 2A). The exception was meropenem treatment in the early exponential phase compared to the untreated control (Fig 2A).
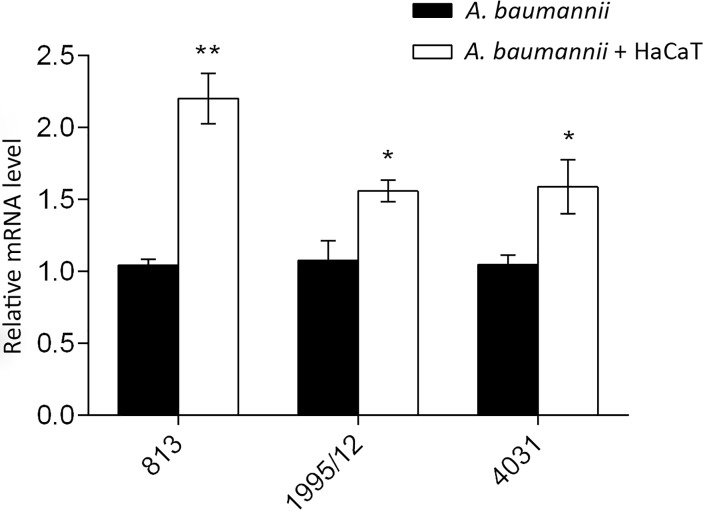
Effect of contact with keratinocytes on the omp33-36 gene transcription
The impact of contact with HaCaT keratinocytes to theomp33-36 gene transcription was investigated by RT-qPCR. All strains significantly increased their omp33-36 mRNA levels after contact with HaCaT keratinocytes (Fig 3). The change was themost prominent in carbapenem-susceptible strain 813, while moderate in carbapenem-resistant strains 1995/12 and 4031 (Fig 3).
Fig 3. Inducibility of the omp33-36 gene expression after contact with HaCaT keratinocytes.
All expression results were normalized relative to rpoB by the 2−ΔΔCt method. Values are the means of results obtained in triplicate. Error bars represent the standard deviation of the mean value. A Student’s t-test was used to compare the experimental group to the control (* p< 0.05, ** p< 0.01).
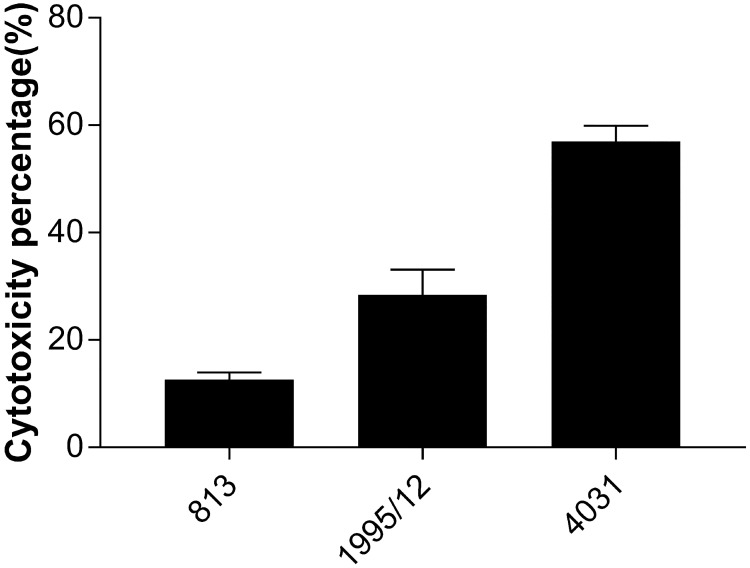
Cytotoxic effect of A. baumannii strains on HaCaT keratinocytes
The results obtained from an LDH assay demonstrated that the most prominent cytotoxic potential was observed for carbapenem-resistant strain 4031 (56,97 ± 2,9%) followed by carbapenem-resistant strain 1995/12 (28,41 ± 4,7%). Carbapenem-susceptible strain 813 showed the weakest cytotoxicity on HaCaT cells (12,6 ± 1,34%) (Fig 4).
Fig 4. Cytotoxic effect of carbapenem-susceptible and carbapenem-resistant A. baumannii strains on HaCaT keratinocyte cells.
Cytotoxic levels caused by carbapenem-susceptible strain 813 and carbapenem-resistant strains 1995/12 and 4031 on HaCaT cells were measured by LDH assay. Values are the means from results obtained in triplicate. Error bars represent the standard deviation of the mean value.
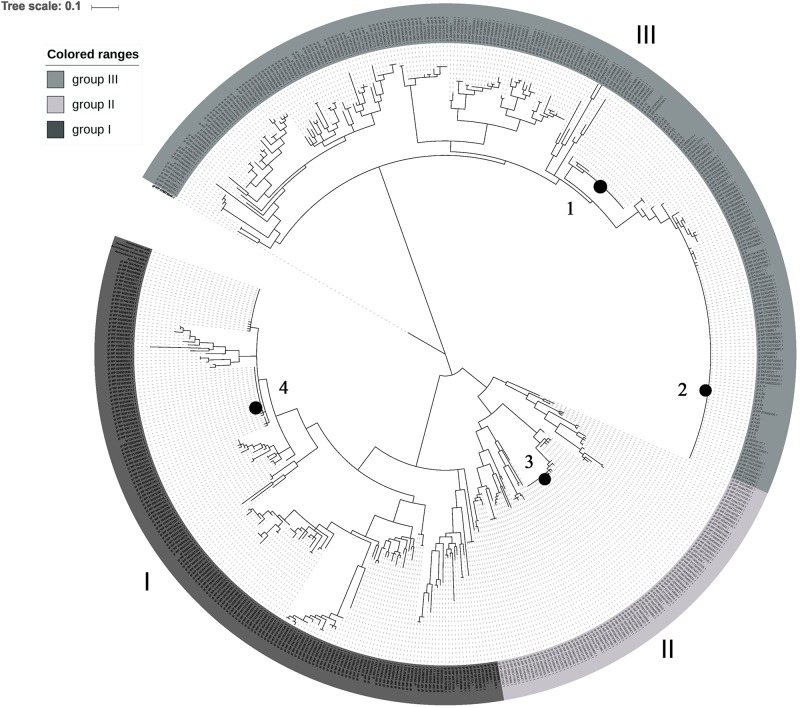
Phylogenetic analysis and polymorphism determination of Omp33-36 porin
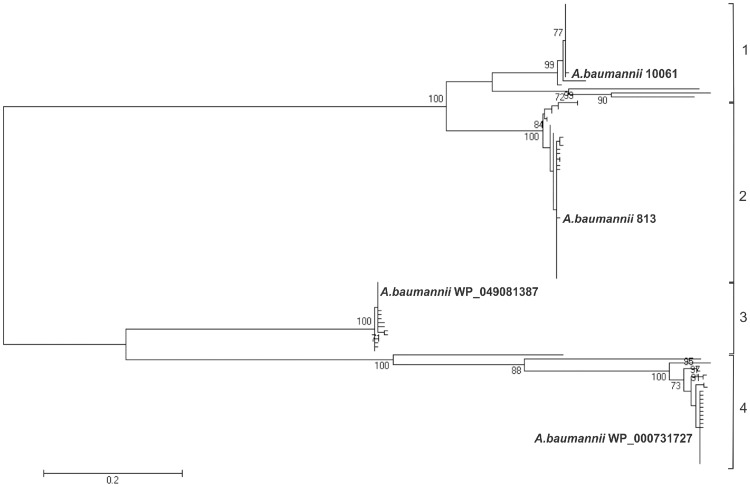
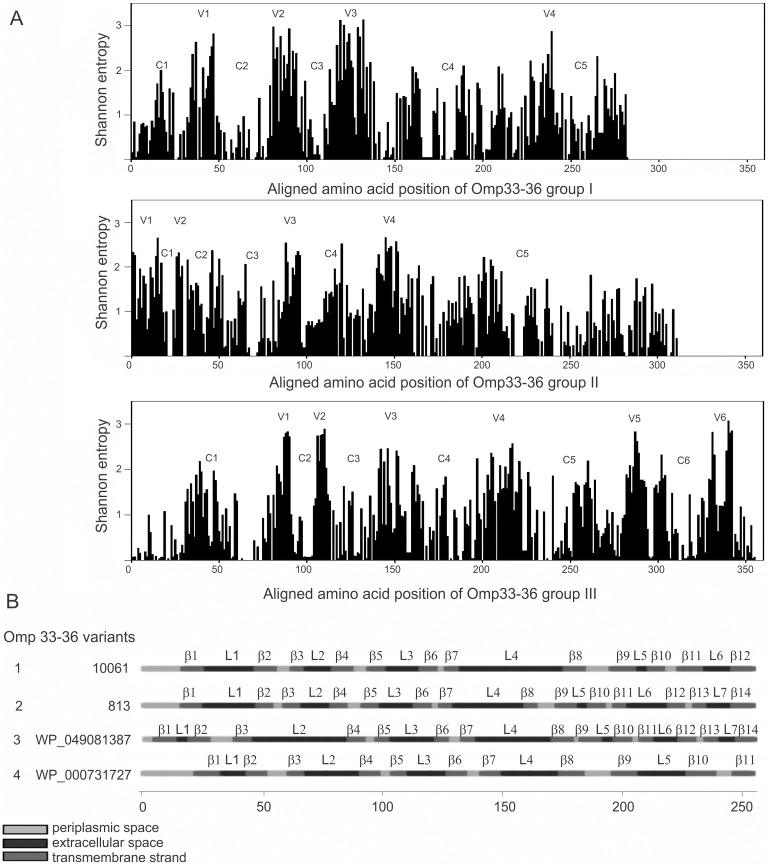
The maximum likelihood (ML) phylogenetic analysis of Omp33-36 porins from the Acinetobacter genus separated Omp33-36 into three groups (I-III) (Fig 5). Furthermore, ML analysis of only A. baumannii species separated Omp33-36 into four clearly distinct isoform groups, referred to as A. baumannii Omp33-36 variants 1 to 4 (Fig 6). The most comprehensive, but least conserved, group III, was divided into two A. baumannii variants (1 and 2) (Fig 5). The Omp33-36 porin of carbapenem-susceptible strain 813 and carbapenem-resistant strain 1995/12 was classified as variant 2, while the Omp33-36 of carbapenem-resistant strain 4031 belonged to variant 1. The smallest, group II, includes Omp33-36 variant 3, while group I includes variant 4 (Fig 5). Multiple sequence alignment comparisons revealed that 86% of the amino acid positions of Omp33-36 porins were polymorphic. The polymorphic regions, as well as conserved regions, were well defined in all three Omp33-36 groups (Fig 7A). There were 87% polymorphic amino acid positions of Omp33-36 in group I (245 out of 284 amino acid positions), 83% in group II (260 out of 311 amino acid positions), and 87% in group III (310 out of 355 amino acid positions). Omp33-36 group I and II had four variable (V1-V4) and five conserved regions (C1-C5), while group III had six variable (V1-V6) and six conserved regions (C1-C6). Among variable regions, the amino acids of group III and V2 and V3 of group I were the most polymorphic, with H mainly higher than 2.5. Regarding the conserved regions, the C4 of group I, C5 of groups II and III at the C terminus, as well as the C1 of group III at the N terminus were the most conserved regions (Fig 7A). Also, the Omp33-36 variants showed the presence of PLAEAAFL motif at their N terminal region, which was conserved among all the analyzed Acinetobacter Omp33-36 porins (S4 File).
Fig 5. Phylogenetic inferences of Omp33-36 porin among Acinetobacter spp.
A phylogenetic tree of Omp33-36 porins was constructed with the maximum likelihood (ML) method using a Jones-Taylor-Thornton (JTT) model distance matrix. The confidence levels were calculated from 1000 bootstrap resamples of alignment used for phylogenetic inferences by ML method. The gray shades represent the three dinstinct groups (I-III) consisting of Acinetobacter spp. from the database. A. baumannii variants are indicated by black circles. In group I is variant 4 in group II is variant 3 and in group III are variants 1 and 2.
Fig 6. Phylogenetic inferences of Omp33-36 porins among A. baumannii.
An ML phylogenetic tree was constructed from the Omp33-36 amino acid sequences available in the NCBI database. The corresponding Omp33-36 variant type (1–4) is indicated and given in bold. The representatives of variants 1–4 were 10061 (Omp33-36 from a Serbian A. baumannii strain [22]), 813 (Omp33-36 from Serbian A. baumannii strain used in this study), WP 049081387 (a putative A. baumannii porin from the NCBI database) and WP_000731727 (a putative A. baumannii porin from the NCBI database), respectively. The percentage of 1000 bootstrap resamples supporting the different clusters are indicated at the bifurcations (only bootstrap results above 70% are reported).
Fig 7. Distinct structural Omp33-36 porins among the Acinetobacter spp. population.
(A) Shannon variability of Omp33-36 sequences within groups I, II and III. The height of each bar indicates the entropy of an amino acid residue at a given position. The conserved (C1-C6) regions and variable (V1-V6) regions are shown. (B) Predicted transmembrane topology of the different A. baumannii Omp33-36 variants (1–4) representing Omp33-36- groups. External loops (L1-L7), transmembrane spanning regions (β1-β14) and periplasmic regions are depicted.
Transmembrane topology of Omp33-36 porin
Transmembrane topology predictions for Omp33-36 variants 2 and 3 showed the same topology scores, 14 β-strand-spanning regions (β1-β14) and seven external loops (L1-L7). However, for the Omp33-36 variants 1 and 4, 12 and 11 β-strand-spanning regions, as well as 6 and 5 external loops were predicted, respectively (Fig 7B).
PROVEAN analysis
In order to correlate Omp33-36 porin with carbapenem resistance, besides expression level, the Omp33-36 primary sequence also has to be considered. PROVEAN analysis was applied to all four A. baumannii Omp33-36 variants to reveal whether the change in the primary structure could have an impact on carbapenem resistance. A deleterious effect on variant 1’s protein function resulted from 11 distinct amino acid substitutions and 2 deletions. Omp33-36 variant 2 had 22% deleterious amino acid exchanges, 13 amino acid substitutions, 2 deletions and 1 insertion. The PROVEAN analysis of variant 3 revealed that 28.8% of the mutations had a deleterious effect, 13 amino acid substitutions and 8 deletions. Results for members of variant 4 showed that between 21% and 29.5% of the changes in Omp33-36’s primary structure were deleterious, and the changes were quite diverse (61 distinct mutations). There were 49 amino acid substitutions, 9 deletions and 3 insertions (S1 Table).
Discussion
Outer membrane porins have been recognized as important factors contributing to both the antimicrobial resistance and virulence of Acinetobacter spp. [3,6–9]. Although the role of Omp33-36 porin in A. baumannii pathogenic potential has been tackled previously [3,6,8,9], data about its gene expression during infection are limited [26,35] and the potential of this porin as a vaccine candidate has been completely neglected.
We investigated the omp33-36 gene’s transcriptional expression in order to establish its role in reversible resistance to carbapenems caused by inappropriate antibiotic dosing (subinhibitory concentrations of imipenem or meropenem) during different growth phases. Additionally, Omp33-36 porin has been shown to be a virulence factor [8,9], thus we examined the inducibility of the omp33-36 gene expression during contact with HaCaT keratinocytes. Further, we wanted to determine if there were differences in the transcriptional response of omp33-36 in one carbapenem-susceptible strain and two carbapenem-resistant strains (with different mechanisms underlying resistance–only production of oxacillinases or in combination with inactivation of the carO gene) [22] under the same experimental conditions.
The growth phase dependence of omp33-36 transcription was undoubtedly demonstrated, since it has been decreased in middle and late exponential phases in all strains. However, the analyzed strains showed different transcriptional responses in the stationary phase. In general, data regarding porin gene expression in Acinetobacter spp. are limited and previously it was only observed that the carO gene showed significant increase of transcription in the middle exponential phase and subsequent decrease in the following phases, as well as a general trend of decrease of oprD gene expression after the early exponential phase [18]. The results obtained in our study indicate that growth phase dependence of the omp33-36 expression, although showing similar trends, is probably strain-specific, and that correlation with transcription of either the carO or the oprD genes cannot be taken out. Given that our study analyzed expression at the transcriptional level, the possible variations in the presence of Omp33-36 porin in the outer membrane due to translational or posttranslational regulation should be considered.
Our results suggest the importance of Omp33-36 porin during adaptation to the presence of carbapenems. The most dramatic change in omp33-36 expression after treatment with a subinhibitory concentration of carbapenems was detected for the carbapenem-susceptible strain 813. We can assume that this decrease in transcription could further contribute to reduction of Omp33-36 molecules’ number in the outer membrane, limiting entrance of carbapenems and allowing adaptation of the bacteria to new conditions. This indicates how inappropriate carbapenem dosing could lead to reversible resistance, and depicts the role of Omp33-36 during this process. These findings on the hormesis effect of antibiotics, which leads to altered gene expression of resistome genes, are in line with previous studies which have shown that subinhibitory concentrations of antibiotics could lead to amended expression of genetic determinants included in resistance as well as intracellular survival of clinically relevant bacteria [36,37]. Furthermore, the only data that connect the presence of subinhibitory concentrations of antibiotics and the presence of Omp33-36 porin in the membrane are those for tetracycline [35]. Considering that inadequate antibiotic treatment could stimulate the pathogenic properties of clinically relevant bacteria such as A. baumannii, controlled and proper antibiotic use is imperative for every single infection treatment [38]. The results obtained in this study support previous conclusions, since carbapenem-susceptible strains can become resistant, and MIC values for carbapenem-resistant bacteria can be increased as a result of exposure of bacteria to subinhibitory concentrations of carbapenems.
Since Acinetobacter spp. have been recognized as prominent infective agents which cause wound infections [39], their response to contact with keratinocytes is of great importance. The strains we analyzed showed significant increases of omp33-36 transcription in the presence of HaCaT keratinocytes. Similarly, the increased production of virulence factor OmpA after incubation of A. baumannii in host airway-like conditions was observed in a previous study [20]. Further, the role of Omp33-36 porin in adhesion and invasion of human lung epithelial cells was established previously [8]. Accordingly, our study revealed that interaction with keratinocytes stimulated the transcription of the omp33-36 gene encoding virulence factor Omp33-36 in A. baumannii, indicating that de novo synthesis of Omp33-36 porin is important during the initial phases of infection. However, we could not correlate omp33-36 transcription level and cytotoxicity on HaCaT cells, and we hypothesize that the effect of other virulence factors is more prominent in this interaction than Omp33-36 porin. However, it should be taken into account that the posttranscriptional regulation of omp33-36 expression, as well as the varying extent of its release in outer membrane vesicles could affect this process [10].
Considering the established importance of Omp33-36 porin in the pathogenicity of Acinetobacter spp. [3,6,8,9], the second aim of our study was to classify these outer membrane porins among the genus Acinetobacter and species A. baumannii. Information about Omp33-36 distribution and conservation are needed for assessment of its potential as a vaccine candidate. This analysis was performed according to our previous study in which we revised the classification of another porin, CarO, included in carbapenem resistance[22]. Since the detected average of amino acid polymorphism was over 80%, Omp33-36 could be considered a polymorphic porin. Accordingly, the organization of conserved regions, as well as transmembrane topology, varied significantly among the three different groups. Unlike previously described conserved outer membrane proteins, which showed efficient vaccine characteristics [10–16], only one conserved motif of eight amino acids (PLAEAAFL) was present at the N-terminal region in all three Acinetobacter spp. Omp33-36 groups. Since this motif was part of the outside loop L2 of Omp33-36 group II it could be a potential antigen for induction of antibody production against Acinetobacter spp. belonging to this group. The investigation on the immunogenicity of this motif and its role in the possible protection of hosts from Acinetobacter spp. need to be elucidated in further studies. Although Omp33-36 showed noticeable polymorphism, the majority of these alterations have no influence on its biological function, according to PROVEAN analysis.
Conclusions
Treatment with subinhibtory concentrations of carbapenems decreased the mRNA levels of omp33-36, meaning that inadequate dosing of carbapenem therapy could lead to the development of reversible resistance due to changes in Omp33-36 levels. Contact with keratinocytes increased the expression of omp33-36, indicating that the de novo synthesis of Omp33-36 is of importance during the initial phases of infection. Bioinformatics analysis revealed the polymorphic structure of Omp33-36 in Acinetobacter spp., however, according to PROVEAN analysis, the majority of polymorphism probably does not affect its functionality. We determined a potential antigen region (PLAEAAFL motif) within Omp33-36 from Acinetobacter spp. that could be used for vaccine development.
Supporting information
(TXT)
(DOC)
(DOCX)
(DOCX)
(DOC)
Acknowledgments
We would like to thank Dr Zorica Vasiljević Institute for Mother and Child Health Care "Dr. Vukan Čupić", Belgrade, Serbia for supplying bacterial strains.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Ministry of Education, Science and Technological Development, Grant No. 173019 to MK, http://www.mpn.gov.rs/. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017;30:409–447. 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al Atrouni A, Joly-Guillou ML, Hamze M, Kempf M. Reservoirs of Non-baumannii Acinetobacter Species. Front Microbiol. 2016;7:49 10.3389/fmicb.2016.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbott I, Cerqueira GM, Bhuiyan S, Peleg AY. Carbapenem resistance in Acinetobacter baumannii: Laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev Anti Infect Ther. 2013;11:395–409. 10.1586/eri.13.21 [DOI] [PubMed] [Google Scholar]
- 4.Qureshi ZA, Hittle LE, O’Hara JA, Rivera JI, Syed A, Shields RK, et al. Colistin-resistant Acinetobacter baumannii: Beyond carbapenem resistance. Clin Infect Dis. 2015;60:1295–1303. 10.1093/cid/civ048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO publishes list of bacteria for which new antibiotics are urgently needed. 2017. http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf
- 6.Del Mar Tomás M, Beceiro A, Pérez A, Velasco D, Moure R, Villanueva R, et al. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49:5172–5175. 10.1128/AAC.49.12.5172-5175.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catel-Ferreira M, Coadou G, Molle V, Mugnier P, Nordmann P, Siroy A, et al. Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J Antimicrob Chemother. 2011;66:2053–2056. 10.1093/jac/dkr267 [DOI] [PubMed] [Google Scholar]
- 8.Smani Y, Dominguez-Herrera J, Pachón J. Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii. J Infect Dis. 2013;208: 1561–1570. 10.1093/infdis/jit386 [DOI] [PubMed] [Google Scholar]
- 9.Rumbo C, Tomás M, Moreira EF, Soares NC, Carvajal M, Santillana E, et al. The Acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infect Immun. 2014;82:4666–4680. 10.1128/IAI.02034-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McConnell MJ, Rumbo C, Bou G, Pachón J. Outer membrane vesicles as an acellular vaccine against Acinetobacter baumannii. Vaccine. 2011;29:5705–5710. 10.1016/j.vaccine.2011.06.001 [DOI] [PubMed] [Google Scholar]
- 11.Luo G, Lin L, Ibrahim AS, Baquir B, Pantapalangkoor P, Bonomo RA, et al. Active and passive immunization protects against lethal, extreme drug resistant-Acinetobacter baumannii infection. PLoS ONE. 2012;7:e29446 10.1371/journal.pone.0029446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badmasti F, Ajdary S, Bouzari S, Fooladi AAI, Shahcheraghi F, Siadat SD. Immunological evaluation of OMV [PagL]+Bap [1-487aa] and AbOmpA [8-346aa]+Bap [1-487aa] as vaccine candidates against Acinetobacter baumannii sepsis infection. Mol Immunol. 2015;67:552–558. 10.1016/j.molimm.2015.07.031 [DOI] [PubMed] [Google Scholar]
- 13.Huang W, Wang S, Yao Y, Xia Y, Yang X, Long Q, et al. OmpW is a potential target for eliciting protective immunity against Acinetobacter baumannii infections. Vaccine. 2015;33:4479–4485. 10.1016/j.vaccine.2015.07.031 [DOI] [PubMed] [Google Scholar]
- 14.Garg N, Singh R, Shukla G, Capalash N, Sharma P. Immunoprotective potential of in silico predicted Acinetobacter baumannii outer membrane nuclease, NucAb. Int J Med Microbiol. 2016;306:1–9. 10.1016/j.ijmm.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 15.Huang W, Yao Y, Wang S, Xia Y, Yang X, Long Q, et al. Immunization with a 22-kDa outer membrane protein elicits protective immunity to multidrug-resistant Acinetobacter baumannii. Sci Rep. 2016;6:20724 10.1038/srep20724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R, Garg N, Shukla G, Capalash N, Sharma P. Immunoprotective efficacy of Acinetobacter baumannii outer membrane protein, FilF, predicted in silico as a potential vaccine candidate. Front Microbiol. 2016;7:158 10.3389/fmicb.2016.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nucleo E, Steffanoni L, Fugazza G, Migliavacca R, Giacobone E, Navarra A, et al. Growth in glucose-based medium and exposure to subinhibitory concentrations of imipenem induce biofilm formation in a multidrug-resistant clinical isolate of Acinetobacter baumannii. BMC Microbiol. 2009;9:270 10.1186/1471-2180-9-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernando D, Kumar A. Growth phase-dependent expression of RND efflux pump-and outer membrane porin-encoding genes in Acinetobacter baumannii ATCC 19606. J Antimicrob Chemother. 2011;67:569–572. 10.1093/jac/dkr519 [DOI] [PubMed] [Google Scholar]
- 19.Giannouli M, Antunes LC, Marchetti V, Triassi M, Visca P, Zarrilli R. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis. 2013;13:282 10.1186/1471-2334-13-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Méndez JA, Mateos J, Beceiro A, Lopez M, Tomás M, Poza M, et al. Quantitative proteomic analysis of host-pathogen interactions: a study of Acinetobacter baumannii responses to host airways. BMC Genom. 2015;16:422 10.1186/s12864-015-1608-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhabaan GN, AbuBakar S, Cerqueira GM, Al-Haroni M, Pang SP, Hassan H. Imipenem treatment induces expression of important genes and phenotypes in a resistant Acinetobacter baumannii isolate. Antimicrob Agents Chemother. 2016;60:1370–1376. 10.1128/AAC.01696-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novovic K, Mihajlovic S, Vasiljevic Z, Filipic B, Begovic J, Jovcic B. Carbapenem-resistant Acinetobacter baumannii from Serbia: Revision of CarO classification. PLoS ONE. 2015;10:e0122793 10.1371/journal.pone.0122793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jovcic B, Begovic J, Lozo J, Topisirovic L, Kojic M. Dynamics of sodium dodecyl sulfate utilization and antibiotic susceptibility of strain Pseudomonas sp. ATCC19151. Arch Biol Sci. 2009;61:159–164. [Google Scholar]
- 24.Mussi MA, Limansky AS, Relling V, Ravasi P, Arakaki A, Actis LA, et al. Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel. J Bact. 2011;193:4736–4748. 10.1128/JB.01533-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.0, 2017.http://www.eucast.org.
- 26.Hood MI, Jacobs AC, Sayood K, Dunman PM, Skaar EP. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob Agents Chemother. 2010;54:1029–1041. 10.1128/AAC.00963-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Perichon B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54:4389–4393. 10.1128/AAC.00155-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letunic I, Bork P. Interactive tree of life [iTOL] v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. A Hidden Markov Model method, capable of predicting and discriminating β-barrel outer membrane proteins. BMC Bioinformatics. 2004;5:29 10.1186/1471-2105-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423, 623–656. [Google Scholar]
- 35.Yun SH, Choi CW, Park SH, Lee JC, Leem SH, Choi JS, et al. Proteomic analysis of outer membrane proteins from Acinetobacter baumannii DU202 in tetracycline stress condition. J Microbiol. 2008;46:720–727. 10.1007/s12275-008-0202-3 [DOI] [PubMed] [Google Scholar]
- 36.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. 10.1016/j.mib.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 37.Hua X, Chen Q, Li X, Yu Y. Global transcriptional response of Acinetobacter baumannii to a subinhibitory concentration of tigecycline. Int J Antimicrob Agents. 2014;44:337–344. 10.1016/j.ijantimicag.2014.06.015 [DOI] [PubMed] [Google Scholar]
- 38.Lee CR, Cho IH, Jeong BC, Lee SH. Strategies to minimize antibiotic resistance. Int J Environ Res Public Health. 2013;10:4274–4305. 10.3390/ijerph10094274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mihu MR, Martinez LR. Novel therapies for treatment of multi-drug resistant Acinetobacter baumannii skin infections. Virulence. 2011;2:97–102. 10.4161/viru.2.2.15061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TXT)
(DOC)
(DOCX)
(DOCX)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.