Abstract
Keloids are a type of abnormal scar tissue. MicroRNAs (miRNAs) exhibit a pivotal role in the regulation of cell proliferation and metastasis of keloids. miRNA microarray revealed that miR-637 was one of the most frequently altered miRNAs in keloids. Furthermore, upregulation of miR-637 inhibited cell proliferation and metastasis by targeting mothers against decapentaplegic homolog (Smad)3, one of the important proteins that affects the formation of keloids. Further studies demonstrated that miR-637 regulated the proliferation and metastasis of human keloid fibroblast (HKF) cells by mediating the Smad3 signaling pathway. Overall, the present findings suggest that miR-637 may be a promising therapeutic target in keloids.
Keywords: smad3, keloid, miR-637, proliferation, metastasis
Introduction
Keloid is a type of abnormal scar tissue, which is caused by excessive proliferation of fibroblasts, massive deposition of collagen and extracellular matrix, and deposition of core protein (1). However, keloid is difficult to completely cure, therefore it is necessary to find a more efficient strategy prevent and treat keloid.
The current study demonstrated that ~2/3 of the human protein encoding genome could be regulated by microRNAs (miRNAs). It has been reported that in keloid, miRNAs affect the proliferation, metastasis and differentiation of fibroblasts, and may also adjust the deposition of the extracellular matrix, which may affect the development of keloid (2).
The mothers against decapentaplegic homolog (Smad)3 mediated signaling pathway is closely associated with wound healing and scar formation (3). Previous studies have shown that Smad3 is highly expressed in pathological scar tissues, which induces the transition of fibroblasts to myofibroblasts (4). Inhibiting the expression of Smad3 can inhibit the formation of keloid (5).
In the current research, we aimed to investigate the differential expression of miRNAs in the keloid epidermis compared with that in the normal skin epidermis. For the selected, significantly lower expressed miR-637, the effects on the proliferation and metastasis of human keloid fibroblast cells were evaluated. The examinations were carried out to confirm the roles of miR-637 in the regulation of proliferation and metastasis of human keloid fibroblast cells. The involvement of miR-637 in the regulation of Smad3 signaling pathway in keloid fibroblast cells was additionally investigated.
Materials and methods
Tissue samples and cell lines
We enrolled keloid patients at the General Hospital of Shenyang Military Region (Shenyang, China) between January 2012 and January 2015. We obtained 30 paired samples of keloid and the excess normal tissue from the partial skin flap (control). All patients (14 female and 16 male, average age 31.25±7.39) had not received topical and systemic therapy for at least 2 months prior to undergoing a skin biopsy. We also retained three normal skin tissues from trauma patients as normal controls (normal). Written consent was gathered from all participants before the study was performed. The protocols were approved by the Ethics Committee of General Hospital of Shenyang Military Command.
Human keloid fibroblasts (HKF) were purchased from Bioleaf Corporation (Shanghai, China), and human embryonic skin fibroblasts obtained from Beijing Union Medical College cell bank. Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), at 37°C in an environment containing 5% CO2.
RNA isolation
Tissues were stored in liquid nitrogen. Total RNA was extracted from tissues and cells using mirVana® miRNA isolation kit (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer's instructions. The quantity and quality of the extracted RNA were measured with a spectrophotometer (Thermo Fisher Scientific, Inc.). Finally, the sample was stored at −80°C.
MicroRNA microarray analysis
Agilent's Human miRNA Microarray Release 18.0 (Shanghai Biochip Co., Ltd., Shanghai, China) was used to detect the RNA from 3 keloids and 3 adjacent tissues. The slides were scanned by an Agilent Microarray Scanner with Feature Extraction Software 10.7, and raw data were normalized by Quantile algorithm, Gene Spring Software 11.0 (all by Agilent Technologies, Inc., Santa Clara, CA, USA). Differentially expressed microRNAs were identified by significance analysis of microarrays (SAM, http://www-stat.stanford.edu/tibs/SAM/index.html).
Quantitative polymerase chain reaction (qPCR)
qPCR was conducted with qPCR Universal Reagent and the MX3000P qPCR instrument following the protocols. U6 small nuclear RNA was used as an internal control. The expression of miR-637 was detected using Stem-Loop RT-PCR assay, as previously described (6,7). Primer sequences were synthesized as follows (Table I). All the reactions were carried out as described previously (8).
Table I.
Primers of RNAs.
| Name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| miR-637 | ACACTCACTGGGGGCTTT | GCAGAGCCCGTTGAGAGTACA |
| miR-487 | ACACTCAATCATACAGGGA | AACTGGATGTTGAGAGTACAT |
| miR-154 | ACACTCAATCATACACGGT | AATAGGTCTTGAGAGTACAT |
| miR-582 | ACACTCTAACTGGTTGAAC | GGTTCAGTTTTGAGAGTACAT |
| miR-194 | ACACTCCCAGTGGGGCTG | CAGATAACAGTTGAGAGTACAT |
| miR-21 | ACACTCCAACACCAGTCG | ACAGCCCATTTGAGAGTACAT |
| miR-503 | CACTCGGGGTATTGTTTCC | CCTGGCAGCTTGAGAGTACAT |
| miR-550 | ACACTCAGTGCCTGAGGGA | CTCTTACTTGAGAGTACAT |
| U6 | CTCGCTTCGGCAGCACA | ACGCTTCACGAATTTGCGT |
| Smad3 | GGGCTTTGAGGCTGTCTAC | GTCCACGCTGGCATCTTCTG |
| Cyclin D1 | CCAACCTCCTCAACGACC | TGGCACAGAGGGCAACGAAG |
| MMP2 | GATCTTGACCAGAATACCAT | GGCTTGCGAGGGAAGAAGTT |
| GAPDH | CATCCCTTCTCCCCACAC | GTCCCAGGGCTTTGATTTG |
miRNA, microRNA; MMP, matrix metalloproteinase.
Western blot analyses
Cells and tissues were lysed and the protein was extracted using radioimmunoprecipitation assay buffer. Samples were resolved by 10% SDS-PAGE and transferred onto polyvinylidene difluoride PVDF membranes. Target proteins were probed with specific antibodies. Relative expression of relevant proteins were quantified and normalized to GAPDH. Antibodies used for western blotting were: anti-Smad3 (sc-101154, 1:500), anti-Cyclin D1 (sc-70899, 1:300), anti-matrix metallopeptidase (MMP)2 (sc-13594, 1:300), anti-GAPDH (sc-51631, 1:1,000) and horseradish peroxidase-conjugated anti-mouse (sc-2005, 1:5,000) secondary antibodies (Santa Cruz, USA).
Dual luciferase reporter assay
Dual luciferase activity assays were performed as previously described (9). The Smad3 3′-untranslated region (UTR) was PCR amplified and cloned into the pMIR-REPORT™ vector (Ambion; Thermo Fisher Scientific, Inc.). The primers were: Smad3-WT, F: 5′-AGGGCTTTGAGGCTGTCTACC-3′, R: 5′-GTCCACGCTGGCATCTTCTG-3′, Smad3-mut (the site of miR-637 binding was mutated), F: 5′-AAACCAGGCGGCTAAACAAGTG-3′, R: 5′-GCAACAGCAGCAGTGAAGGTG-3′. HKF cells were seeded and co-transfected with the above constructs and miR-637 mimic (Guangzhou Ribobio Co., Ltd, Guangzhou, China), miR-637 antisense (AS) (Guangzhou Ribobio Co., Ltd) or control (Guangzhou Ribobio Co., Ltd). Luciferase activity was determined with the dual luciferase reporter assay system after 36 h transfection; the luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA).
MTT assays
Cell proliferation activity was detected using an MTT assay (Solarbio, China). Cells were seeded at a density of 1×103 cells per well of 96-well plates, and transfected with miR-637 mimic, miR-637 inhibitor (Guangzhou Ribobio Co., Ltd) or negative control; then the cell proliferation was evaluated by MTT assay. A microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to measure the optical densities at a wavelength of 490 nm.
Transwell assay
A total of 12 h following transfection with miR-637 mimic, miR-637 inhibitor or negative control, 1×105 cells in serum-free media were seeded in transwell chambers with or without Matrigel coating, and then transwell assays were conducted. After 8 h cells were fixed by 4% paraformaldehyde, and stained with 0.4% trypan blue. Numbers of cells in different groups were determined using Image-Pro Plus 6.0 software (Nikon Corporation, Tokyo, Japan).
Construction of stable siRNA-expressing cell lines
To stably silence Smad3, cells were transfected with the pRS-si-Smad3 plasmid (Shanghai GeneChem Company) and were selected with G418 (400 µg/ml). After 3 weeks, stable cells were selected, cultured and amplified.
Statistical analysis
All data (showed as mean ± standard deviation) were analyzed with SPSS software, version 17.0 (SPSS Inc., Chicago, IL, USA). Statistical significance between two groups of data was evaluated by Student's t-test (two-tailed) and one-way analysis of variance following Fisher's Least Significant Difference post hoc test. Statistical significance was defined as P<0.05. All experiments were repeated three times.
Results
Differential expression profile of miRNAs in keloid tissues
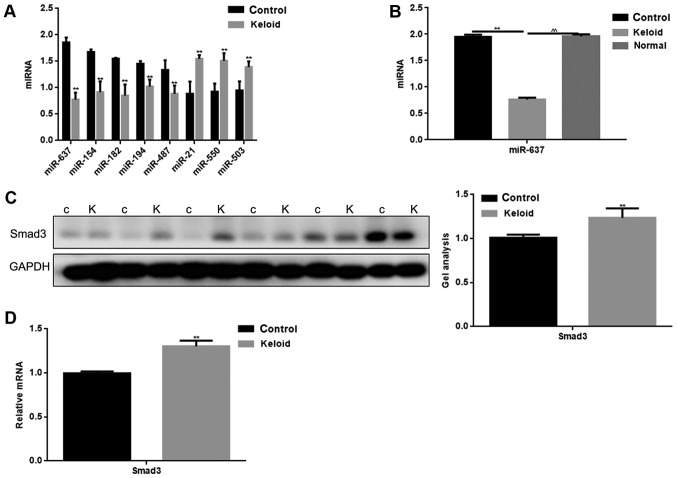
The miRNA microarray revealed 15 downregulated (P<0.05) and 3 upregulated miRNAs (P<0.05) in keloid tissues compared with adjacent tissues (Table II). There were 8 miRNAs tat exhibited a fold change >x30. Among these miRNAs, we found that the change in miR-637 was the greatest. The expression levels of these miRNAs in keloid and adjacent tissues were detected by qPCR (Fig. 1A). The expression of miR-637 was verified to be significantly lower in keloid tissues. We compared the expression of miR-637 in keloid, normal tissues from keloid patients and normal skin tissues from trauma patients (Fig. 1B). It was found that miR-637 expression was low in keloid. It can be seen that miR-637 may play an important role in keloid.
Table II.
Differential miRNAs in keloids.
| miRNA | P-values | Fold change (keloid/adjacent tissues) | Trend |
|---|---|---|---|
| Hsa-miR-34a | 0.001 | 27.265 | Down |
| Hsa-miR-499 | 0.031 | 2.905 | Down |
| Hsa-miR-582 | 7.69×10-05 | 54.608 | Down |
| Hsa-miR-154 | 9.53×10-06 | 65.257 | Down |
| Hsa-miR-126 | 0.023 | 4.167 | Down |
| Hsa-miR-194 | 1.04×10-04 | 50.137 | Down |
| Hsa-miR-101 | 0.011 | 6.072 | Down |
| Hsa-miR-98 | 0.001 | 29.150 | Down |
| Hsa-miR-940 | 0.001 | 29.152 | Down |
| Hsa-miR-200c | 0.009 | 7.689 | Down |
| Hsa-miR-101 | 0.006 | 9.871 | Down |
| Hsa-miR-646 | 0.003 | 13.509 | Down |
| Hsa-miR-106 | 0.004 | 11.097 | Down |
| Hsa-miR-487 | 2.76×10-05 | 31.825 | Down |
| Hsa-miR-637 | 4.05×10-06 | 69.303 | Down |
| Hsa-miR-21 | 6.61×10-06 | 67.042 | Up |
| Hsa-miR-503 | 0.001 | 39.842 | Up |
| Hsa-miR-550 | 9.75×10-06 | 54.187 | Up |
miRNA, micro RNA.
Figure 1.
Expression levels of miRNAs and Smad3 in keloid tissues. (A) Expression levels of miRNAs in keloid tissues and control (adjacent tissues) were detected by qPCR. **P<0.01. (B) The level of miR-637 in keloid tissues, adjacent tissues and normal tissues were detected via qPCR. The expression of miR-637 was lower in keloid tissues. Data are presented as mean ± SEM. **P<0.01, ^^P<0.01 as indicated. (C) The protein level of Smad3 in keloid tissues and adjacent tissues were detected via western blotting. The expression of Smad3 was higher in keloid tissues. Data are presented as mean ± SEM. **P<0.01 vs. control (adjacent tissues) group. (D) mRNA expression of Smad3 in keloid tissues and adjacent tissues were detected respectively by qPCR. The expression of Smad3 was higher in the keloid cancer tissues. Data are presented as mean ± SEM. **P<0.01 vs. control (adjacent tissues) group. Smad, mothers against decapentaplegic homolog; miRNA, microRNA; qPCR, quantitative polymerase chain reaction; SEM, standard error of the mean; C, control (adjacent tissues); K, keloid tissues.
The expression of Smad3 in keloid tissues
By using miRDB software, it was demonstrated that miR-637 targeted multiple proteins. Smad3 is not only an important regulatory protein in keloid, however also contains a binding site to miR-637. The expression of Smad3 in keloid and adjacent tissues was detected by western blotting and qPCR (Fig. 1C and D). The adjacent tissues were used as a control. Results showed that there was a higher expression of Smad3 in keloid tissues, compared with adjacent tissues (P<0.05).
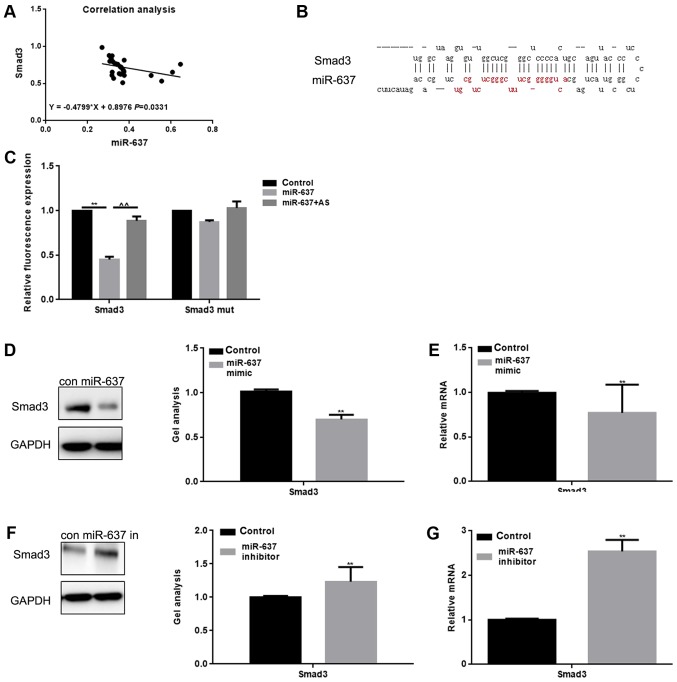
Relationship between miR-637 and Smad3
We analyzed the correlation between the expression levels of miR-637 and Smad3 in keloid, and found that the expression in keloid was negatively correlated (Fig. 2A). Through use of the miRDB tool, we found that miR-637 was able to bind with the 3′-UTR of Smad3 (Fig. 2B). The Luciferase reporter assay showed that miR-637 reduced the expression of Smad3 at the transcriptional level (Fig. 2C). Results showed that when cells were co-transfected with Smad3 and miR-637, a decreased luciferase activity was observed (P<0.05), however, the cells co-transfected with Smad3 mut or miR-637 AS (Antisense) did not result in a significant variation. miR-637 mimic or miR-637 inhibitor were transfected into HKF cells, and then western blot and qPCR were conducted in order to detect the expression of Smad3 (Fig. 2D-G). Results showed that the expression of Smad3 could be downregulated by miR-637 (P<0.05).
Figure 2.
Relationship between miR-637 and Smad3. (A) Correlation between the expression of miR-637 and Smad3 in keloid. (B) miRDB predicted that miR-637 specifically combined with Smad3. (C) The interaction between miR-637 and the Smad3 3′-UTR was tested by luciferase reporter assays. Data are presented as mean ± SEM. **P<0.01, ^^P<0.01 as indicated. (D and E) Western blotting and qPCR showed that when miR-637 was overexpressed, the expression of Smad3 was downregulated. Data are shown as mean ± SEM. **P<0.01 vs. control group (F and G) Western blotting and qPCR showed that when miR-637 was repressed, expression of Smad3 was upregulated. Data are presented as mean ± SEM. **P<0.01 vs. control group. Smad, mothers against decapentaplegic homolog; miRNA, microRNA; qPCR, quantitative polymerase chain reaction; SEM, standard error of the mean.
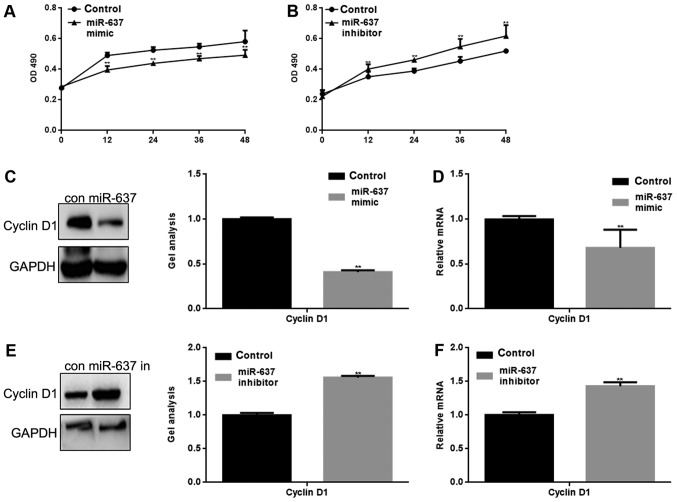
miR-637 inhibits the proliferation of HKF cells by suppressing Cyclin D1
miR-637 mimic or inhibitor was transfected into HKF cells. The proliferation of HKF cells was measured by MTT assay. The result showed that miR-637 mimic significantly inhibited HKF cell proliferation and conversely, miR-637 inhibitor significantly promoted the proliferation of HKF cells (P<0.05) (Fig. 3A and B). Cyclin D1 is an important protein in the regulation of the cell cycle, and is one of the proteins involved in the Smad3 signaling pathway (10). We suspect that the inhibitory effect on HKF cell proliferation may be mediated by regulation of Cyclin D1. Western blotting and qPCR were used to detect the effect of miR-637 on Cyclin D1. The results showed that miR-637 significantly inhibited the expression of Cyclin D1 (P<0.05) (Fig. 3C-F).
Figure 3.
miR-637 inhibits the proliferation of HKF cells by suppressing Cyclin D1. (A) MTT assay showed that following transfection with miR-637 mimic, HKF cell proliferation was repressed. Data are presented as mean ± SEM. **P<0.01 vs. control group. (B) MTT assay of HKF cells following downregulation of miR-637. Data are presented as mean ± SEM. **P<0.01 vs. control group. (C and D) Following transfection with miR-637 mimic in HKF cells, the expression of Cyclin D1 was detected by western blotting and qPCR. Data are presented as mean ± SEM. **P<0.01 vs. control. (E and F) Following downregulation of miR-637, the expression of Cyclin D1 was detected by western blotting and qPCR. Data are presented as mean ± SEM. **P<0.01 vs. control. miRNA, microRNA; qPCR, quantitative polymerase chain reaction; SEM, standard error of the mean; HKF, human keloid fibroblast.
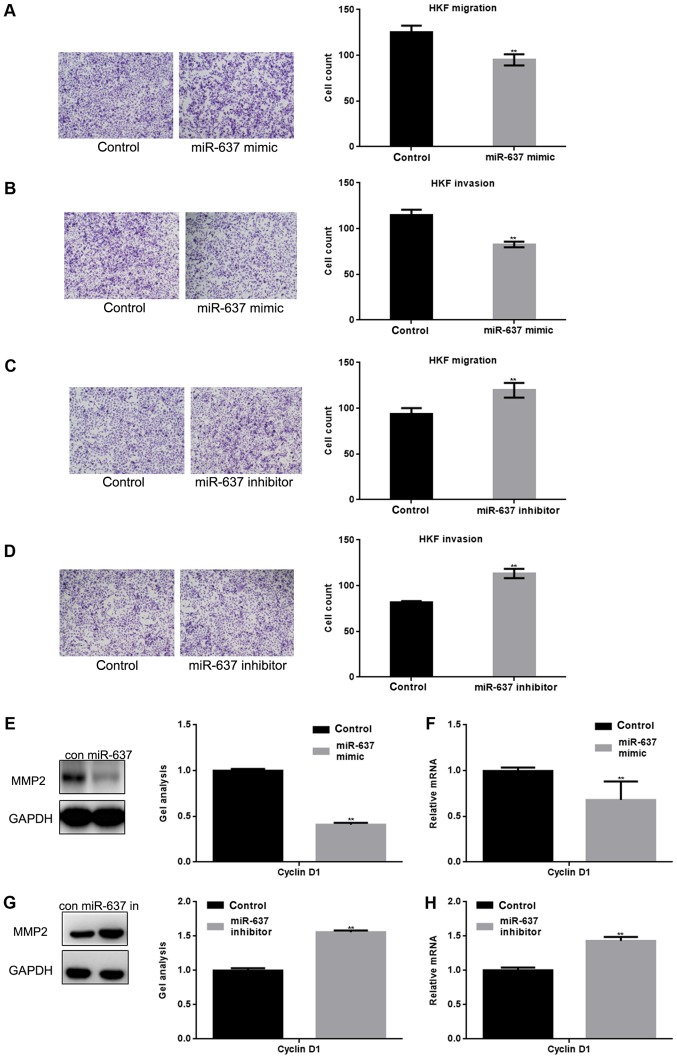
miR-637 inhibits the metastasis of HKF cells by suppressing MMP2
Smad3 can affect the metastasis of cells in many ways, for example MMP2 is one of the downstream proteins of the Smad3 pathway (11). Transwell assays (with or without matrigel) were used to study whether miR-637 is involved in metastasis of HKF cells (Fig. 4A-D). The present study demonstrated that the migration and invasion of HKF cells was significantly inhibited by miR-637 (P<0.05). In addition, western blotting and qPCR showed that MMP2 was significantly downregulated by miR-637 (P<0.05) (Fig. 4E-H).
Figure 4.
miR-637 inhibits the metastasis of HKF cells by suppressing MMP2. (A and B) After overexpression of miR-637 in HKF cells, transwell assays with or without matrigel were performed. Cells were counted and results represent the mean ± standard deviation of three experiments (magnification, ×200). **P<0.01 vs. control. (C and D) After downregulation of miR-637 in HKF cells, transwell assays with or without matrigel were performed. Cells were counted and results represent the mean ± SD of three experiments. **P<0.01 vs. control. (E and F) After transfection with miR-637 mimic in HKF cells, the expression of MMP2 was detected by western blotting and qPCR. Data are presented as mean ± SEM. **P<0.01 vs. control. (G and H) After downregulation of miR-637, the expression of MMP2 was detected by western blotting and qPCR. Data are presented as mean ± SEM. **P<0.01 vs. control. miRNA, microRNA; qPCR, quantitative polymerase chain reaction; SEM, standard error of the mean; SD, standard deviation; HKF, human keloid fibroblast; MMP, matrix metallopeptidase.
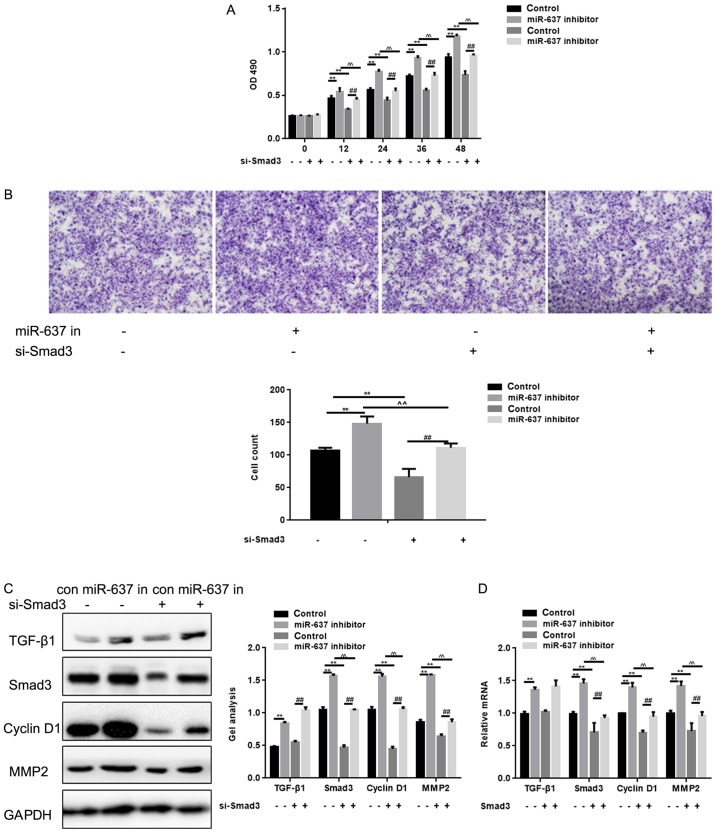
Smad3 is crucial to cell proliferation and metastasis of HKF cells
Smad3 small interfering (si)RNA significantly suppressed cell proliferation and metastasis ability of cells (P<0.05) (Fig. 5A and B). To explore the potential signaling pathways involved in the actions of miR-637 and Smad3 in HKF cells, Cyclin D1 and MMP2 expression levels were detected. Co-treatment with Smad3 siRNA and miR-637 inhibitor in cells significantly downregulated the level of Smad3 to a greater extent compared with miR-637 inhibitor alone (P<0.05), and the same trend was observed in the expression levels of Cyclin D1 and MMP2 (P<0.05) (Fig. 5C and D). The expression of tumor growth factor-β1 was downregulated by miR-637 inhibitor (P<0.05), however was not affected by Smad3 siRNA (Fig. 5C and D). These results suggested that miR-637 may regulate the proliferation and metastasis of HKF cells via Smad3 signaling pathway.
Figure 5.
Smad3 is crucial to cell proliferation and metastasis of HKF cells. (A) Effects of miR-637 siRNA and Smad3 siRNA on cell proliferation ability using MTT. Data are presented as mean ± SEM. **P<0.01, ^^P<0.01 ##P<0.01 as indicated. (B) Effects of miR-637 siRNA and Smad3 siRNA on cell proliferation ability using transwell assay. Cells were counted and results represent the mean ± SD of three experiments (magnification, ×200). **P<0.01, ^^P<0.01 ##P<0.01 as indicated. (C and D) Effects of miR-637 siRNA and Smad3 siRNA on Smad3, Cyclin D1 and MMP2 were detected by western blotting and qPCR. Data are shown as mean ± SEM. HKF, human keloid fibroblast; MMP, Matrix Metallopeptidase; qPCR, quantitative polymerase chain reaction; Smad, mothers against decapentaplegic homolog; SEM, standard error of the mean. **P<0.01, ^^P<0.01 ##P<0.01 as indicated.
Discussion
A variety of miRNAs may be involved in the proliferation and migration of fibroblasts, and many miRNAs have been shown to regulate the formation of keloids (12). It has previously been demonstrated that p63 is affected by a high expression of miR-205 in the skin, and miR-205 influences cell differentiation of skin stem cells, and regulates the proliferation and apoptosis of fibroblasts (13). miR-222 stimulates the growth factor signaling pathway to promote cell cycle progression, thereby regulating the formation of keloids (14). It has also been revealed that miR-29b regulates collagen I in skin fibroblast cells, thereby affecting the formation of keloid (15). In a mouse model, abnormal expression of miR-31 and miR-21-5p has been revealed to be involved in the formation of keloid (16). The migration of fibroblasts may be affected by the regulation of miR-21 on MMPs, and this regulation is crucial in the formation of keloids (13,14,16,17). These findings demonstrated that miRNAs exhibit a key role in the healing of skin wounds, and predicted that they may play an important role in scar formation and skin remodeling. Researchers have reported that miR-637 inhibits the metastasis and growth of glioma cells by targeting the AKT pathway (18). miR-637 has also been shown to inhibit the formation of inflammation by regulating C-Reactive Protein (19–21). However, the effect of miR-637 on keloid has not been studied yet.
Overexpression of Smad3 in keloid significantly upregulates pro-collagen gene expression, promotes deposition of extracellular matrix, proliferation and metastasis of fibroblasts (3,5,22). Therefore, Smad3 is one of the potential targets for keloid treatment (23).
Microarray analysis was used to screen abnormal expression of miRNAs in keloid. The results showed that miR-637, miR-487, miR-154, miR-582 and miR-194 were significantly downregulated in keloid, and the expression levels of miR-21, miR-503 and miR-550 were upregulated. Furthermore, we found that these miRNAs may play an important role in keloid. miR-637 was selected as the primary factor for investigation in the present study. It was proved that miR-637 directly affected the activity of Smad3 through the targeting of Smad3.
A keloid scar is an overgrowth of dense fibrous tissue that develops around a wound. These scars are raised scars that spread beyond the margins of the original wound to normal skin by invasion (24). The downregulation of miR-637 may be responsible for the enhanced capacity of keloid to proliferate and metastasise (25). miR-637 may affect the proliferation and migration of cells by inhibiting the expression of Cyclin D1 and MMP2. These two proteins can be regulated by Smad3 (26,27), leading to the hypothesis that regulation of miR-637 on proliferation and migration of fibroblasts is partly mediated by regulation of Smad3. The present study additionally demonstrated that miR-637 can influence the expression of TGF-β1 to some extent, as TGF-β1 has been proved to play an important role in keloid (28), and can regulate Smad3 activity (18,22,28). miR-637 has been shown to inhibit AKT pathway activity (18), and the inhibitory effect of miR-637 on Smad3 may be through its effects on TGF- β1 and targeting Smad3. This experiment has some limitations, and further experimentation is required in vivo to verify the results.
The results of the present study suggested that miR-637 participated in the regulation of keloid development by inhibition of Smad3, and may therefore act as a future potential target for keloid treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YZ conceived of the present study and performed the molecular studies. BG also performed the molecular studies. QH participated in the design of the study and performed the statistical analysis. WL participated in its design and coordination of the study and helped to draft the manuscript. PC assisted in the conception of the study. KT contributed to analysis and interpretation of data.
Ethics approval and consent to participate
Written consent was gathered from all participants before the study was performed. The protocols were approved by the Ethics Committee of General Hospital of Shenyang Military Command (R201208).
Consent for publication
Patients have provided written informed consent for the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yao X, Cui X, Wu X, Xu P, Zhu W, Chen X, Zhao T. Tumor suppressive role of miR-1224-5p in keloid proliferation, apoptosis and invasion via the TGF-β1/Smad3 signaling pathway. Biochem Biophys Res Commun. 2018;495:713–720. doi: 10.1016/j.bbrc.2017.10.070. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Yang D, Xiao Z, Zhang M. miRNA expression profiles in keloid tissue and corresponding normal skin tissue. Aesthetic Plast Surg. 2012;36:193–201. doi: 10.1007/s00266-011-9773-1. [DOI] [PubMed] [Google Scholar]
- 3.Jin SE, Kim CK, Kim YB. Cellular delivery of cationic lipid nanoparticle-based SMAD3 antisense oligonucleotides for the inhibition of collagen production in keloid fibroblasts. Eur J Pharm Biopharm. 2012;82:19–26. doi: 10.1016/j.ejpb.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Zhao B, Guan H, Liu JQ, Zheng Z, Zhou Q, Zhang J, Su LL, Hu DH. Hypoxia drives the transition of human dermal fibroblasts to a myofibroblast-like phenotype via the TGF-β1/Smad3 pathway. Int J Mol Med. 2017;39:153–159. doi: 10.3892/ijmm.2016.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phan TT, Lim IJ, Aalami O, Lorget F, Khoo A, Tan EK, Mukhopadhyay A, Longaker MT. Smad3 signalling plays an important role in keloid pathogenesis via epithelial-mesenchymal interactions. J Pathol. 2005;207:232–242. doi: 10.1002/path.1826. [DOI] [PubMed] [Google Scholar]
- 6.Feng J, Wang K, Liu X, Chen S, Chen J. The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene. 2009;437:14–21. doi: 10.1016/j.gene.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013;13:68. doi: 10.1186/1475-2867-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zelivianski S, Cooley A, Kall R, Jeruss JS. Cyclin-dependent kinase 4-mediated phosphorylation inhibits Smad3 activity in cyclin D-overexpressing breast cancer cells. Mol Cancer Res. 2010;8:1375–1387. doi: 10.1158/1541-7786.MCR-09-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Clutter S, Benincosa J, Fortney J, Gibson LF. Activation of transforming growth factor-beta1/p38/Smad3 signaling in stromal cells requires reactive oxygen species-mediated MMP-2 activity during bone marrow damage. Stem Cells. 2005;23:1122–1134. doi: 10.1634/stemcells.2004-0354. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Xu D, Li N, Li Y, He Y, Hu X, Lyu L, He L. Downregulation of microRNA-31 inhibits proliferation and induces apoptosis by targeting HIF1AN in human keloid. Oncotarget. 2017;8:74623–74634. doi: 10.18632/oncotarget.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An G, Liang S, Sheng C, Liu Y, Yao W. Upregulation of microRNA-205 suppresses vascular endothelial growth factor expression-mediated PI3K/Akt signaling transduction in human keloid fibroblasts. Exp Biol Med (Maywood) 2017;242:275–285. doi: 10.1177/1535370216669839. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Li C, Bai Y, Liu H, Zuo X, Yao H, Xu Y, Cao M. Comparative study of microRNA profiling in keloid fibroblast and annotation of differential expressed microRNAs. Acta Biochim Biophys Sin (Shanghai) 2013;45:692–699. doi: 10.1093/abbs/gmt057. [DOI] [PubMed] [Google Scholar]
- 15.Zhang GY, Wu LC, Liao T, Chen GC, Chen YH, Zhao YX, Chen SY, Wang AY, Lin K, Lin DM, et al. A novel regulatory function for miR-29a in keloid fibrogenesis. Clin Exp Dermatol. 2016;41:341–345. doi: 10.1111/ced.12734. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Wang X, Yang D, Xiao Z, Chen X. MicroRNA-21 affects proliferation and apoptosis by regulating expression of PTEN in human keloid fibroblasts. Plast Reconstr Surg. 2014;134:561e–573e. doi: 10.1097/PRS.0000000000000577. [DOI] [PubMed] [Google Scholar]
- 17.Wu ZY, Lu L, Liang J, Guo XR, Zhang PH, Luo SJ. Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genet Mol Res. 2014;13:2727–2738. doi: 10.4238/2014.April.14.2. [DOI] [PubMed] [Google Scholar]
- 18.Que T, Song Y, Liu Z, Zheng S, Long H, Li Z, Liu Y, Wang G, Liu Y, Zhou J, et al. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34:4952–4963. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 19.Yi W, Li D, Guo Y, Zhang Y, Huang B, Li X. Sevoflurane inhibits the migration and invasion of glioma cells by upregulating microRNA-637. Int J Mol Med. 2016;38:1857–1863. doi: 10.3892/ijmm.2016.2797. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Hooten Noren N, Dluzen DF, Martindale JL, Gorospe M, Evans MK. Posttranscriptional regulation of the inflammatory marker C-reactive protein by the RNA-binding protein HuR and MicroRNA 637. Mol Cell Biol. 2015;35:4212–4221. doi: 10.1128/MCB.00645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JF, He ML, Fu WM, Wang H, Chen LZ, Zhu X, Chen Y, Xie D, Lai P, Chen G, et al. Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54:2137–2148. doi: 10.1002/hep.24595. [DOI] [PubMed] [Google Scholar]
- 22.Liang CJ, Yen YH, Hung LY, Wang SH, Pu CM, Chien HF, Tsai JS, Lee CW, Yen FL, Chen YL. Thalidomide inhibits fibronectin production in TGF-β1-treated normal and keloid fibroblasts via inhibition of the p38/Smad3 pathway. Biochem Pharmacol. 2013;85:1594–1602. doi: 10.1016/j.bcp.2013.02.038. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Gao Z, Shi Y, Sun Y, Lin Z, Jiang H, Hou T, Wang Q, Yuan X, Zhu X, et al. Inhibition of Smad3 expression decreases collagen synthesis in keloid disease fibroblasts. J Plast Reconstr Aesthet Surg. 2007;60:1193–1199. doi: 10.1016/j.bjps.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Ma X, Chen J, Xu B, Long X, Qin H, Zhao RC, Wang X. Keloid-derived keratinocytes acquire a fibroblast-like appearance and an enhanced invasive capacity in a hypoxic microenvironment in vitro. Int J Mol Med. 2015;35:1246–1256. doi: 10.3892/ijmm.2015.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao H, Dong P, Yan L, Yang Z, Lv X, Li Q, Zong X, Fan J, Fu X, Liu X, Xiao R. TGF-β1 induces polypyrimidine tract-binding protein to alter fibroblasts proliferation and fibronectin deposition in keloid. Sci Rep. 2016;6:38033. doi: 10.1038/srep38033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia W, Lo CM, Poon RYC, Cheung TT, Chan ACY, Chen L, Yang S, Tsao GSW, Wang XQ. Smad inhibitor induces CSC differentiation for effective chemosensitization in cyclin D1- and TGF-β/Smad-regulated liver cancer stem cell-like cells. Oncotarget. 2017;8:38811–38824. doi: 10.18632/oncotarget.16402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu C, Lu G, Li Q, Zhang J, Huang Z, Gao X. Selenium modulates MMP2 expression through the TGFβ1/Smad signalling pathway in human umbilical vein endothelial cells and rabbits following lipid disturbance. J Trace Elem Med Biol. 2017;42:59–67. doi: 10.1016/j.jtemb.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Yao X, Cui X, Wu X, Xu P, Zhu W, Chen X, Zhao T. Tumor suppressive role of miR-1224-5p in keloid proliferation, apoptosis and invasion via the TGF-β1/Smad3 signaling pathway. Biochem Biophys Res Commun. 2018;495:713–720. doi: 10.1016/j.bbrc.2017.10.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.