Abstract
Drug resistance is a principal contributor to the poor prognosis of ovarian cancer (OC). Therefore, identifying factors that affect drug resistance in OC is critical. In the present study, 51 OC specimens from lab collections were immunohistochemically tested, public data for 489 samples from The Cancer Genome Atlas cohort and 1,656 samples from the Kaplan-Meier Plotter were downloaded, and data were retrieved from Oncomine. It was identified that the mRNA and protein expression of the potassium calcium-activated channel subfamily N member 3 (KCNN3) was markedly lower in OC tissues compared with normal tissues, and in drug-resistant OC tissues compared with sensitive OC tissues. Low KCNN3 expression consistently predicted shorter disease-free and overall survival (OS). Specifically, low KCNN3 expression predicted shorter OS in 395 patients with low expression levels of mucin-16. There was additional evidence that KCNN3 expression is mediated by microRNA-892b. Furthermore, text mining and analyses of protein and gene interactions indicated that KCNN3 affects drug resistance. To the best of the authors' knowledge, this is the first report to associate KCNN3 with poor prognosis and drug resistance in OC. The present findings indicated that KCNN3 is a potential prognostic marker and therapeutic target for OC.
Keywords: potassium calcium-activated channel subfamily N member 3, ovarian cancer, prognosis, drug resistance
Introduction
Ovarian cancer (OC) is a principal cause of cancer mortality in women, with an estimated 14,080 cases of mortality in the USA (1) every year, and 22,500 cases of mortality annually in China (2). Although patients with OC are initially responsive to standard therapy (generally debulking surgery followed by platinum-centered chemotherapy) (3), ~70% of them develop recurrent disease (4). The 5-year overall survival (OS) is very poor as the relapsed disease is frequently incurable (5), with little improvement over the last 30 years (6). The emergence of a drug-resistant disease is thus a primary obstacle in the clinical management of OC. To overcome these unsatisfactory treatment outcomes, understanding the molecular mechanisms that contribute to drug resistance and identifying predictive biomarkers are critical (7).
Accumulating evidence indicates that ion channels serve crucial roles in cancer biology (8,9) and mediate numerous aspects of cancer pathology, including apoptosis, angiogenesis, cell growth, migration, invasion and metastasis (10). Among the ion channels, potassium channels are the most diverse and ubiquitous, and represent easily accessible cancer biomarkers and targets for therapy (11,12). In OC, potassium channels have been demonstrated to be closely associated with cancer progression and outcomes. Potassium two pore domain channel subfamily K member 9 is involved in the oncogenesis of OC, although its prominent expression is paradoxically associated with better survival (13); potassium voltage-gated channel subfamily H (KCNH) member 1 and KCNH member 2 (KCNH2) expression is associated with poor prognosis (14,15), and KCNH2 channel activity is associated with tumor drug resistance (16).
Potassium calcium-activated channel subfamily N member 3 (KCNN3), a potassium channel of the small conductance Ca2+-activated potassium channel family (17), contributes to the development and progression of numerous solid tumors. In melanoma cells, upregulation of KCNN3 enhances cell motility by hyperpolarizing the cell membrane potential (18); in breast cancer cells, KCNN3 is a mediator of cell migration (19), and together with P2X purinoceptor 7, contributes to cysteine cathepsin-dependent cell invasiveness (20); and in colon cancer, KCNN3, together with short transient receptor potential channel 1 (TRPC1) and calcium release-activated calcium channel protein 1 (orai-1), regulates store operated calcium entry (SOCE)-dependent cell migration (21). Furthermore, KCNN3 is upregulated by a 16-fold change in bortezomib-resistant BN myeloma cells (22), which suggests that its expression is associated with drug resistance. However, KCNN3 has not been widely studied with respect to cancer, and research on its role in OC is rare. To the best of the authors' knowledge, the present study, which used public data, bioinformatics and immunohistochemistry analyses, is the first to report the association of KCNN3 with prognosis and drug resistance in OC.
Materials and methods
Data acquisition
Data regarding gene expression determined using Log2 median-centered intensity in OC and normal controls were retrieved from microarrays downloaded from The Cancer Genome Atlas (TCGA) Ovarian cohort, including 586 ovarian serous adenocarcinoma and 8 normal control samples; and the Yoshihara Ovarian cohort, including 43 ovarian serous adenocarcinoma and 10 normal control samples, deposited in Oncomine (https://www.oncomine.org/resource/main.html) (23,24)) (Tables I and II). The mRNA expression values of 489 tissues from a total of 586 patient samples with ovarian serous adenocarcinoma were determined via Agilent microarray analysis (Agilent Technologies, Inc., Santa Clara, CA, USA) (25), and these data were downloaded from the cBioportal (http://www.cbioportal.org/) (26,27). Corresponding data of microRNA (miR), DNA methylation and clinical data of the 489 OC tissues were also downloaded from cBioportal (http://www.cbioportal.org/). Among the 489 tissues, 197 were platinum-sensitive and 90 were resistant tissues. KCNN3 mRNA expression (probe: 205903_s_at) data and survival information of the 1,656 patients with OC [including 395 patients with OC who had low cancer antigen (CA) 125 expression levels] were downloaded from the KM Plotter (http://kmplot.com/), which is a collection of 14 independent microarrays (data set nos. GSE14764, GSE15622, GSE18520, GSE19829, GSE23554, GSE26193, GSE26712, GSE27651, GSE30161, GSE3149, GSE51373, GSE63885, GSE65986 and GSE9891) from the Gene Expression Omnibus (GEO) profiles and TCGA ovarian cohort (28). No alterations were made to any of the aforementioned data used in the analysis.
Table I.
Association between KCNN3 expression and clinical factors of patients with ovarian cancer in TCGA cohort (489 patients) and lab collection (51 patients).
| TCGA cohort, 489 patients | Lab collection, 51 patients | |||||||
|---|---|---|---|---|---|---|---|---|
| KCNN3 mRNA expression | KCNN3 protein expression | |||||||
| Clinical factors | No. of patients (Percentage of total cohort) | High (%) | Low (%) | P-valuea | No. of patients (Percentage of total cohort) | High (%) | Low (%) | P-valuea |
| Drug resistance | 287 | 0.003 | 51 | 0.011 | ||||
| Resistance | 90 (31.4%) | 33 (36.7) | 57 (63.3) | 24 (47.1) | 6 (25.0) | 18 (75.0) | ||
| Sensitive | 197 (68.6) | 110 (55.8) | 87 (44.2) | 27 (52.9) | 17 (63.0) | 10 (37.0) | ||
| Grade | 477 | 1.000 | 51 | 0.002 | ||||
| I–II | 57 (11.9) | 29 (50.9) | 28 (49.1) | 15 (29.4) | 12 (20.0) | 3 (80.0) | ||
| III | 420 (88.1) | 210 (50.0) | 210 (50.0) | 36 (70.6) | 11 (30.6) | 25 (69.4) | ||
| Stage | 484 | 1.000 | 51 | 1.000 | ||||
| I–II | 24 (5.0) | 12 (50.0) | 12 (50.0) | 11 (21.6) | 5 (45.5) | 6 (54.5) | ||
| III–IV | 460 (95.0) | 229 (49.8) | 231 (50.2) | 40 (78.4) | 18 (45.0) | 22 (55.0) | ||
| Primary therapy outcome success | 0.098 | |||||||
| Stable and progressive disease | 62 (15.7) | 24 (38.7) | 38 (61.3) | |||||
| Complete response and partial response | 333 (84.3) | 168 (50.5) | 165 (49.5) | |||||
| Serum CA 125, U/ml | 51 | 0.012 | ||||||
| <400 | 23 (45.1) | 15 (65.2) | 8 (34.8) | |||||
| ≥400 | 28 (54.9) | 8 (28.6) | 20 (71.4) | |||||
Evaluated using Pearson's χ2 test (2-sided). KCNN3, potassium calcium-activated channel subfamily N member 3; TCGA, The Cancer Genome Atlas.
Table II.
On the basis of the microarray data retrieved from Oncomine, KCNN3 is differentially expressed and downregulated in the majority of tumor types.
| Datasetsa | |||
|---|---|---|---|
| Cancer type | All | KCNN3 upregulated | KCNN3 downregulated |
| Bladder | 6 | 2 | |
| Brain and central nervous system | 13 | 3 | 1 |
| Breast | 10 | 1 | |
| Cervical | 6 | – | – |
| Colorectal | 11 | 1 | |
| Esophageal | 7 | – | – |
| Gastric | 6 | 1 | |
| Head and neck cancer | 18 | – | – |
| Kidney | 7 | 1 | |
| Leukemia | 14 | 1 | |
| Liver | 8 | – | – |
| Lung | 13 | – | – |
| Lymphoma | 11 | 1 | |
| Melanoma | 5 | – | – |
| Myeloma | 4 | 1 | |
| Ovarian | 8 | 2 | |
| Pancreatic | 9 | 1 | 2 |
| Prostate | 15 | – | – |
| Sarcoma | 6 | 2 | |
| Other | 14 | 2 | |
| Sum | 191 | 5 | 17 |
Datasets deposited in Oncomine. P<0.05, fold change ≥2.0. ‘−’ indicates that no significant change was detected; KCNN3, potassium calcium-activated channel subfamily N member 3.
Samples
OC specimens were collected from adult patients (aged 18–87 years old) with ovarian serous adenocarcinoma who were treated at The Affiliated Tumor Hospital of Guangxi Medical University (Nanning, China) between April 2005 and December 2012. All patients underwent optimal cytoreductive surgeries (residual <2 cm), and at least six cycles of platinum-paclitaxel chemotherapy following surgery. The classification of response to chemotherapy was defined as sensitive (complete remission and relapse >6 months following stopping chemotherapy; n=27) or resistant (complete remission and relapse <6 months following stopping chemotherapy; n=24) to primary chemotherapy. Specimens were fixed in 10% formalin for 48 h at room temperature and then embedded in paraffin. Paraffin-embedded sections (4 µm) from 51 patients (aged 26–71 years old; median age, 49 years old) were subsequently stained with 0.5% hematoxylin for 8 min at room temperature and 0.5% eosin for 1 min at room temperature (H&E), and the stained sections were evaluated by two independent pathologists. The present study was approved by The Ethics Committee of Guangxi Medical University and was performed in accordance with The Declaration of Helsinki. Informed consent was obtained from all individual participants included in the present study. Furthermore, commercially available adult human normal tissue arrays were purchased from Cybrdi, Inc. (Rockville, MD, USA; cat. no. OV241c), consisting of six OC tissues and six adjacent normal ovarian tissues. H&E staining on these tissues was performed prior to purchase by Cybridi, Inc. In total, six normal ovarian tissues and 57 OC tissues were included in the present study.
Immunohistochemistry
The primary antibody used in the present study was rabbit monoclonal antibody against human KCNN3 (1:1,000; Abcam, Cambridge, UK; cat. no. ab192515), and the secondary antibody was goat anti-rabbit immunoglobulin G heavy and light chains (horseradish peroxidase; 1:2,000; Abcam; cat. no. ab97051). The sections was incubated with 3% peroxidase blocking solution (SPlink Detection kits, OriGene Technologies, Inc., Beijing, China; cat. no. SP-9000) for 15 min at room temperature, and then incubated with 10% goat serum (SPlink Detection kits; OriGene Technologies, Inc.; cat. no. SP-9000) for 15 min at room temperature. Subsequently, the sections were incubated with primary antibody in 0.01 M PBS for 12 h at 4°C, and then incubated with secondary antibody for 1 h at room temperature. The slides were imaged using an EVOS FL Auto Imaging System (Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA; magnification, ×10 and ×40). All slides were evaluated independently by two pathologists. Slide immunostaining was scored based on the percentage and intensity of the stained tumor cells (29). The intensity of immunostaining was graded as following: 0, negative staining; 1+, weak staining; 2+, moderate staining; and 3+, strong staining. The staining percentage was graded as following: 0, stained tumor cells <25%; 1+, stained tumor cells 25–50%; 2+, stained tumor cells 50–75%; and 3+, stained tumor cells >75%. The final immunostaining score was calculated by multiplying the staining intensity score by the staining percentage score. Final values ranged between 0 and 9. Scores <5 were considered ‘low expression’ and scores ≥5 were considered ‘high expression’.
Bioinformatics analysis
Biological process annotation was performed using Coremine Medical (http://www.coremine.com/medical/) (30). A protein-gene interaction network was generated using GeneMania (http://www.genemania.org/) (31,32). miR-mRNA predictions used miRsystem, which has seven prediction tools, including TARGETSCAN, RNA22, PICTAR, DIANA, MIRANDA, MIRBRIDGE and PITA (http://mirsystem.cgm.ntu.edu.tw/) (33).
Statistical analysis
The data were analyzed using SPSS 20.0 software (IBM Corp., Armonk, NY, USA). Gene mRNA expression levels are presented as the mean ± standard deviation. Homogeneity of variance was analyzed using the Student's t-test. Correlations between gene-protein expression and clinicopathological factors were evaluated using the Pearson's χ2 test and Spearman's correlation (2-sided). The probability of survival and significance was calculated using the Kaplan-Meier method. Gene expression values were dichotomized into high and low expression, using the median as a cut-off in all the above analyses (34). Correlations between miRs/DNA methylation and gene expression were analyzed using bivariate correlations. P<0.05 was considered to indicate a statistically significant difference.
Results
KCNN3 expression is decreased in OC tissues and drug-resistant tissues compared with normal tissues
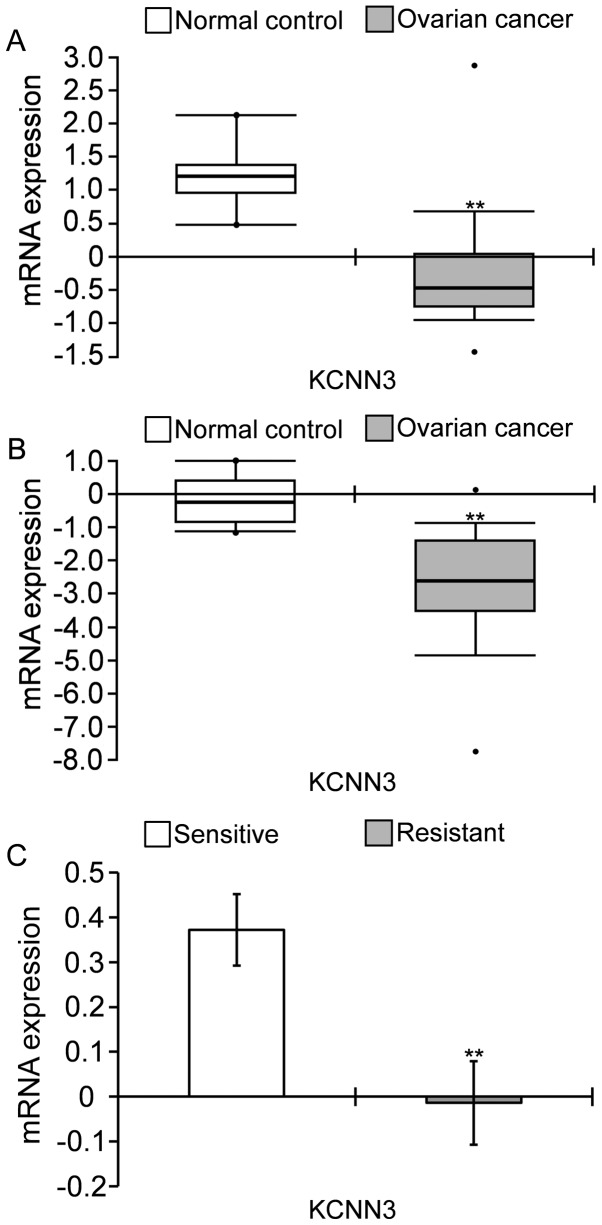
KCNN3 expression was significantly decreased in OC tissues compared with the normal control, according to the TCGA Ovarian cohort and Yoshihara Ovarian cohort deposited in Oncomine. (Fig. 1A and B). KCNN3 mRNA was significantly lower in 586 samples ovarian serous cystadenocarcinomas compared with eight normal ovaries, by 2.770-fold change obtained from the TCGA Ovarian cohort, as determined via Log2 median-centered intensity (Fig. 1A); and was lower in 40 ovarian serous cystadenocarcinomas compared with 10 normal peritoneal samples, by 5.778-fold changes, according to the Yoshihara Ovarian cohort (Fig. 1B). Furthermore, KCNN3 was significantly lower in drug-resistant OC tissues compared with sensitive tissues. KCNN3 mRNA expression in 90 platinum-resistant tissues was significantly lower compared with in 197 platinum-sensitive tissues in TCGA cohort (Fig. 1C; P<0.01; Table I).
Figure 1.
KCNN3 expression is lower in OC tissues compared with normal controls, and in drug-resistant OC tissues compared with sensitive tissues, according to microarray analysis. (A) KCNN3 was significantly decreased in 586 ovarian serous cystadenocarcinomas compared with eight normal ovaries (TCGA ovarian cohort; P=0.0000275). (B) KCNN3 was significantly decreased in 40 ovarian serous cystadenocarcinomas compared with 10 peritoneal samples (Yoshihara ovarian analysis; P=0.00000000656). Gene expression in the analyses was determined using Log2 median-centered intensity in Oncomine. (C) Average KCNN3 expression was lower in 90 platinum-resistant OC tissues compared with 197 sensitive tissues (TCGA ovarian cohort downloaded from cBioportal). **P<0.01 vs. sensitive tissues. KCNN3, potassium calcium-activated channel subfamily N member 3; OC, ovarian cancer; TCGA, The Cancer Genome Atlas.
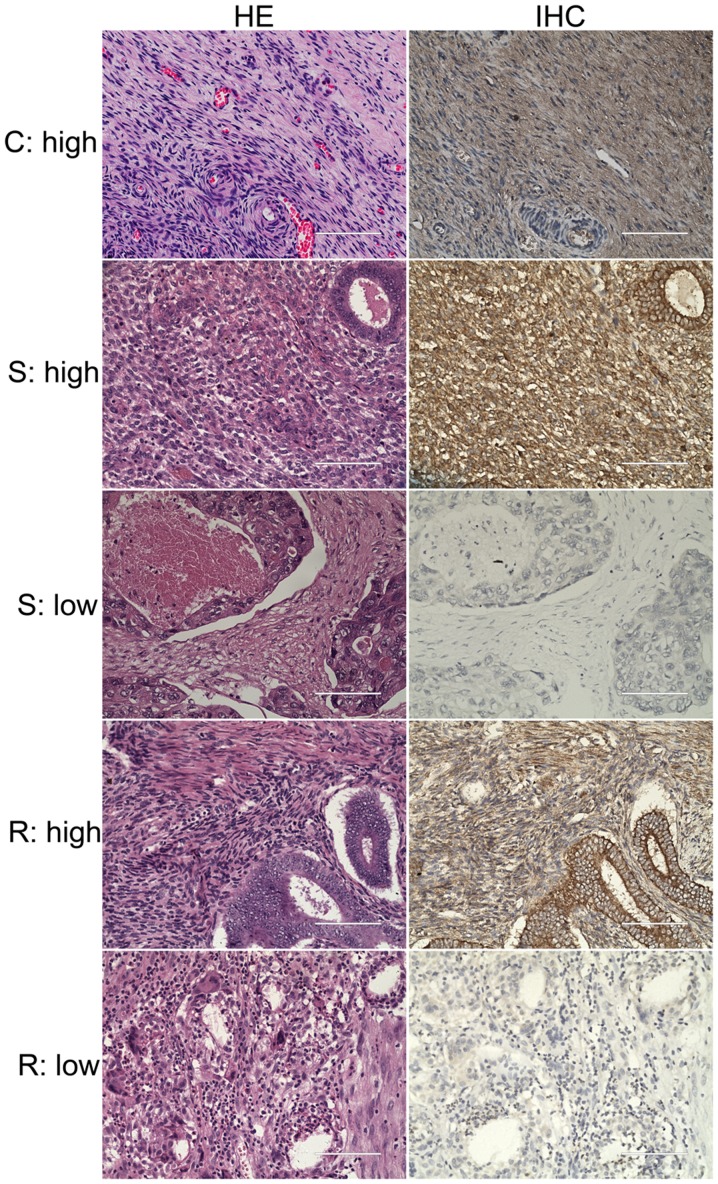
The morphology of 57 OC tissues and 6 normal ovarian tissues were investigated via H&E staining (Fig. 2). The immunohistochemistry results of these tissues demonstrated that the majority of OC tissues revealed low expression levels of KCNN3 (33/57 cases); whereas, all six normal controls revealed high expression levels of the protein (Fig. 2). Statistical analysis using the χ2 test revealed a significant low expression of KCNN3 in OC tissues, as determined via imaging of immunohistochemistry results (P=0.009). Furthermore, among the 51 OC specimens from the lab collection, KCNN3 protein expression was significantly lower in 24 drug-resistant OC tissues compared with 27 sensitive tissues, as determined via imaging of immunohistochemistry results (Fig. 2; Table I; P=0.011). The percentage of drug-resistant tissues with low expression of KCNN3 was 75% (18/24 cases), whereas the low expression of the gene in drug-sensitive tissues was only 37% (10/27 cases; P=0.011; Table I).
Figure 2.
IHC analysis of KCNN3 protein expression in ovarian cancer tissues. A total of two serial sections from the same paraffin-embedded block of patients with ovarian cancer were used for protein detection. Representative staining of KCNN3 high and low expression in normal ovarian tissues; drug-sensitive and resistant tissues are demonstrated. Scale bars, 100 µm. HE, hematoxylin-eosin staining; IHC, immunohistochemistry; C, normal ovarian tissues as control; S, drug-sensitive; R, drug-resistant; KCNN3, potassium calcium-activated channel subfamily N member 3.
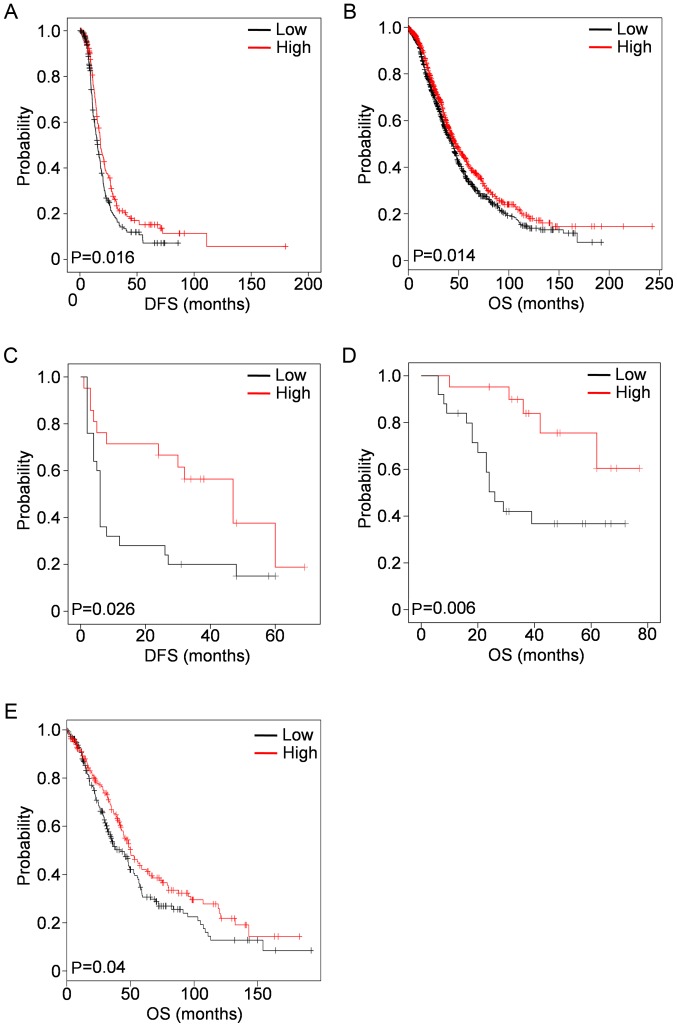
Low KCNN3 expression predicts shorter disease-free survival (DFS) and OS in OC
KCNN3 expression was analyzed against OC clinical factors, and identified to be associated with prognosis. Lower KCNN3 mRNA expression was significantly associated with shorter DFS in 489 patients with OC in TCGA ovarian cohort (low vs. high groups; average values, 22.630±1.720 vs. 35.231±4.684; median values, 15.410±1.049 vs. 18.040±1.253; P=0.016; Fig. 3A), although its association with OS was not significant (low vs. high groups; average values, 49.126±3.051 vs. 59.070±4.489; median values, 43.400±3.858 vs. 43.890±2.449; P=0.153; data not shown). However, lower KCNN3 mRNA expression was significantly associated with poor OS in a large sample of 1,656 patients with OC from the KM Plotter (low vs. high groups; average values, 61.517±2.608 vs. 77.324±4.214; median values, 44.430±2.001 vs. 47.820±2.564; P=0.014; Fig. 3B), which included the above 489 patients in TCGA cohort (28). These results were consistent with observations in specimens from 51 patients with OC, in which lower KCNN3 protein expression was significantly associated with poor DFS (low vs. high groups, average values; 16.920±4.230 vs. 38.559±5.764; median values. 6.000±0.400 vs. 47.000±10.441; P=0.026; Fig. 3C) and OS (low vs. high groups; average values, 38.950±5.388 vs. 68.712±5.025; P=0.006; Fig. 3D). There is no median data for OS as the mortality rate in this subgroup was <50%.
Figure 3.
Low KCNN3 expression is associated with DFS and OS in OC, as determined using KM survival plots. (A) Low KCNN3 mRNA expression was associated with shorter DFS in 489 patients with OC (TCGA ovarian cohort). (B) Low KCNN3 mRNA expression (probe: 205903_s_at) was associated with shorter OS in 1,656 patients (data from KM Plotter). Low KCNN3 protein expression was associated with (C) shorter OS and (D) DFS in 51 OC specimens. (E) Low KCNN3 mRNA expression was associated with shorter OS in 395 patients with OC whose CA 125 expression levels were in the lowest quartile (data from KM Plotter). mRNA expression values were dichotomized into high and low, using the median as a cutoff. Protein expression values were dichotomized into high and low, according to slide immunostaining scores. KCNN3, potassium calcium-activated channel subfamily N member 3; DFS, disease-free survival; OS, overall survival; OC, ovarian cancer; KM, Kaplan-Meier; CA 125, cancer antigen 125.
A low expression level of KCNN3 protein in the subgroup of patients with OC with high expression levels of mucin-16 [cancer antigen (CA) 125] (≥400 µl/ml) was additionally observed, compared with a higher KCNN3 expression level in patients with low expression levels of CA 125 (<400 µl/ml; Table I). Of these 395 patients with OC who had low CA 125 expression levels (in the lowest quartile), low KCNN3 expression was notably associated with shorter OS in the KM Plotter cohort (Fig. 3E). In addition, low KCNN3 expression was significantly associated with higher histological grade (Grade III) in the 51 OC specimens from the lab collection, although no significant association was detected between KCNN3 and tumor stage (Table I).
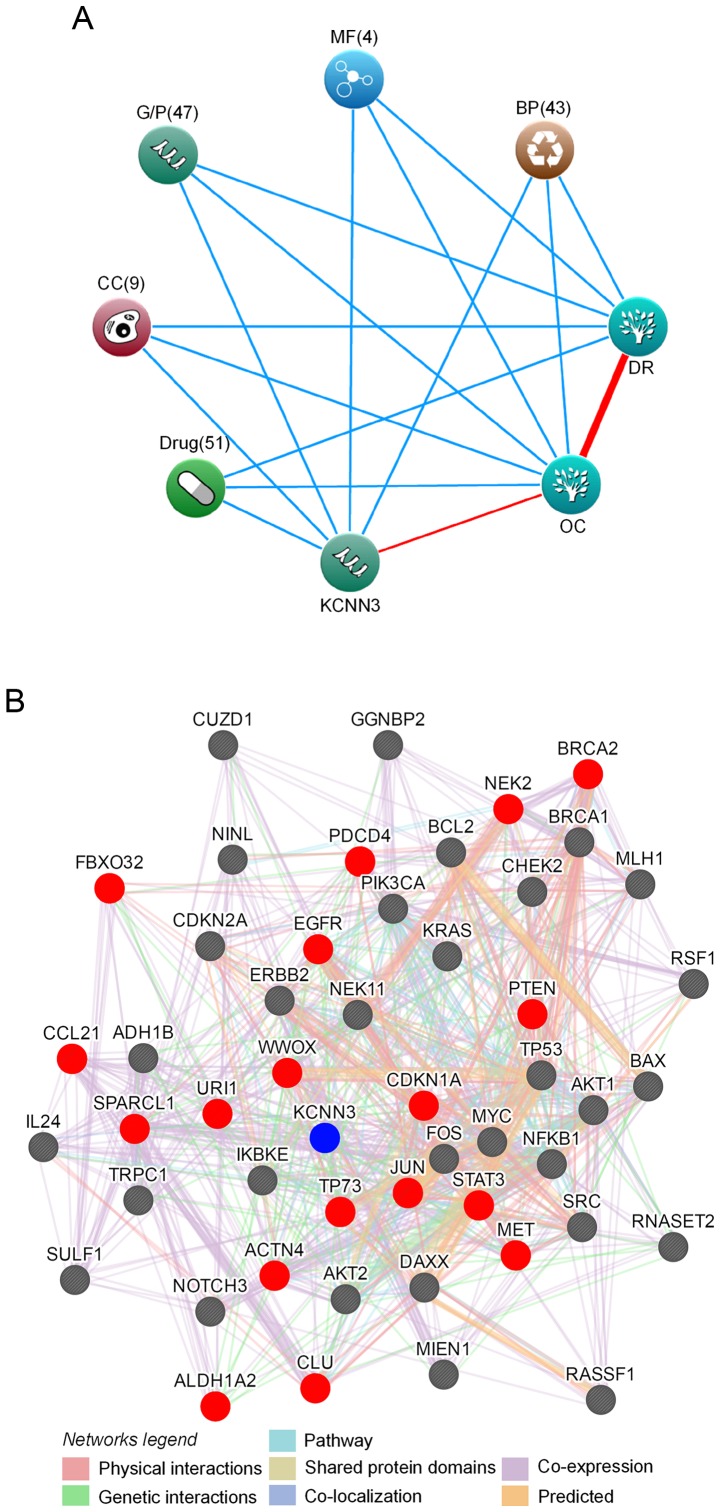
Bioinformatics analyses suggest that KCNN3 mediates drug resistance
Bioinformatics approaches were performed, including protein interaction and text mining, to predict the function of KCNN3 in OC drug resistance. The search terms ‘KCNN3’, ‘OC’ and ‘drug resistance (DR)’ were significantly associated with 51 drugs (e.g. cisplatin, oxaliplatin and doxorubicin), 47 genes and proteins (including, TP53, JUN and CD34), 43 biological processes (including, ‘apoptosis’, ‘DNA repair’ and ‘cell cycle’), nine cellular components and four molecular functions (Fig. 4A). Possible networks of protein or gene interactions with KCNN3 included 49 gene or gene products associated with drug-resistance in OC, which further explained its association with drug resistance. These genes/gene products included 25 oncogenes (35); URI1, STAT3, SRC, RSF1, PIK3CA, NOTCH3, NINL, NFKB1, MYC, MIEN1, MET, KRAS, JUN, IKBKE, FOS, ERBB2, EGFR, DAXX, CUZD1, CLU, BCL2, BAX, AKT2, AKT1 and ACTN4 and 15 tumor suppressors (36) including BRCA1, BRCA2, CHEK2, FBXO32, MLH1, SULF1, IL24, CDKN2A, CDKN1A, TP53, TP73, PDCD4, PTEN, RASSF1 and WWOX, as well as 9 other genes, including CCL21 and SPARCL1 (37), GGNBP2 and RNASET2 (38), NEK2 (39), NEK11 (40), ALDH1A2 and ADH1B (41) and TRPC1 (42). KCNN3 directly interacted with 18 of these genes or gene products, and exhibited indirect interactions with the rest (Fig. 4B). As there were associations between KCNN3 and a wide range of drugs, genes/proteins, biological processes, cellular components and molecular functions with known roles in OC and drug resistance (Fig. 4), it was concluded that KCNN3 expression possibly affects drug resistance in OC.
Figure 4.
Bioinformatics analyses of associations between KCNN3 and drug resistance in OC. (A) KCNN3 associations with drug resistance and OC, as determined by text mining using Coremine Medical. The input terms were ‘KCNN3’, ‘drug resistance’ and ‘ovarian cancer’. (B) A protein interaction network of KCNN3 with 49 drug resistance-associated proteins in OC, as generated using GeneMANIA. Red and grey circles represent target proteins that interact with KCNN3 directly and indirectly, respectively. KCNN3, potassium calcium-activated channel subfamily N member 3; OC, ovarian cancer; DR, drug resistance; BP, biological process; MF, molecular function; G/P, genes and proteins; CC, cellular component.
KCNN3 expression is potentially regulated by miR-892b
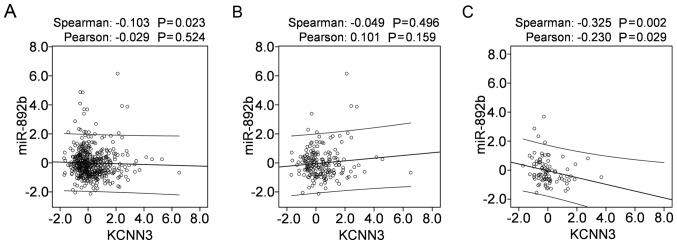
To investigate the possible mechanism that mediates KCNN3 expression in OC, mRNA-miR prediction was conducted using the miRSystem, which predicted 35 miRNAs that potentially target KCNN3. Of these, 24 with expression data available in TCGA were downloaded from cBioportal, from which correlations were analyzed between miRNA expression and KCNN3 mRNA expression in 489 OC tissues. Among the 24 miRNAs, only the expression of miR-892b was negatively correlated with KCNN3 mRNA expression in the 489 OC tissues when determined using Spearman's correlation (Fig. 5A; P<0.05); however, not when determined using Pearson's χ2 test (Fig. 5A; P>0.05). In 197 platinum-sensitive tissues of the 489 OC tissues, miR-892b was not correlated with KCNN3 (Fig. 5B); however, they were significantly and negatively correlated in 90 platinum-resistant tissues of the 489 OC tissues (Fig. 5C; P<0.05). The present results support the possibility that miR-892b targets KCNN3 and contributes to its downregulation in OC, particularly in platinum-resistant tissues.
Figure 5.
Correlations between miR-892b and KCNN3 expression in 489 OC tissues from TCGA cohort downloaded from cBioPortal. Correlations between miR-892b and KCNN3 expression in (A) 489 OC tissues, (B) 197 platinum-sensitive tissues and (C) 90 platinum-resistant tissues. Correlations between miR and gene expression were analyzed using bivariate correlations. miR, microRNA; KCNN3, potassium calcium-activated channel subfamily N member 3; OC, ovarian cancer; TCGA, The Cancer Genome Atlas.
DNA methylation with KCNN3 mRNA expression levels in 489 OC tissues (including 197 sensitive tissues and 90 resistant tissues) was additionally analyzed; however, no correlation was observed, although DNA methylation of KCNN3 was negatively correlated with mRNA expression (data not shown).
Discussion
Novel technologies have led to an increase in the volume and diversity of large-scale public data (43), which are a vital pillar of open science and a key enabler of reproducibility and novel discoveries (44). Reuse of public data may potentially answer questions beyond those originally envisioned (45), and provide a systems-level approach to predicting treatment response and disease progression, and to developing precision therapies (43,46). Computational approaches based on these public datasets may additionally facilitate more rapid annotation of protein function and guide laboratory experiments (47). In the present study, microarrays and associated clinical data retrieved from Oncomine, TCGA and KM Plotter were used to identify genes associated with prognosis and drug resistance in OC.
It was identified that KCNN3 was significantly lower in OC tissues compared with normal controls, in agreement with the two independent microarrays, Yoshihara ovarian statistics and TCGA ovarian cohort. This result was consistent with findings that KCNN3 was significantly downregulated in ≥10 tumor types and upregulated in only three different tumors. Further analyses based on TCGA cohort indicated significantly lower KCNN3 expression in drug-resistant OC tissues, which was supported by experiments conducted with 51 OC specimens. Low KCNN3 expression in OC, particularly in drug-resistant tissues, appears to be regulated by miR-892, which has been demonstrated to affect cancer growth, migration, invasion, metastasis and angiogenesis (48,49).
Significantly lower expression of KCNN3 in OC and drug-resistant OC suggests that KCNN3 mediates cancer progression and drug resistance. This hypothesis is supported by bioinformatics analyses, including text mining and protein interaction analyses, and is consistent with a previous study, in which KCNN3 was predicted to be one of 1,298 genes that contribute to drug resistance in OC (50). A previous study demonstrated that KCNN3 together with the TRPC1 and orai-1 complex regulates SOCE-dependent colon cancer cell migration (21). Specifically, acquisition of drug resistance in multiple myeloma is associated with the suppression of inositol 1,4,5-triphosphate receptor type 1, phospholipase C, transient receptor potential cation channel subfamily M member 7 and TRPC1 expression, and reducing the expression of TRPC1 markedly inhibits drug-induced cell death (51). It was observed that decreased expression of TRPC1 is associated with drug resistance in OC (42), and the protein interacts with KCNN3. Thus, it was concluded that low expression of KCNN3 may contribute to drug resistance via interactions with TRPC1, through inhibition of drug induced cell death.
Downregulation of KCNN3 predicted worse DFS and OS in 51 patients with OC, and consistently predicted worse DFS and OS in 489 and 1,656 patients, respectively, suggesting that it may be a marker for prognosis in OC, in particular for OS. Low KCNN3 expression was notably associated with decreased OS in 395 patients with OC with CA 125 expression levels in the lowest quartile. The downregulation of KCNN3 was associated with higher serum CA 125 (≥400 µl/ml); whereas, KCNN3 upregulation was associated with lower serum CA 125 (<400 µl/ml), thus it was concluded that the gene may be used to predict OS among patients whose serum CA 125 is <400 µl/ml.
In conclusion, the present study reported for the first time, to the best of the authors' knowledge, the associations between KCNN3 and drug resistance and prognosis in OC, which indicate that KCNN3 is a potential therapeutic target and prognostic marker in the treatment of OC. Further in vitro and in vivo studies are required to validate and clarify the present results.
Acknowledgements
The authors would like to thank Mrs Marla Brunker, from Liwen Bianji, Edanz Group China www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
The present study was supported by National Natural Science Foundation of China (grant nos. 81460397, 81660606 and 81302283), China Postdoctoral Science Foundation (grant nos. 2014M552535XB and 2014M552291), and Natural Science Foundation of Guangxi (grant nos. 2015GXNSFAA139151, 2014GXNSFBA118155, 2015GXNSFBA139115 and 2014GXNSFCA118010).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FY designed the study. XL performed bioinformatics analyses and data mining. LW and BZ collected samples and clinical data. XC and CD performed immunohistochemical analysis. XL and FY wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Ethics Committee of Guangxi Medical University (Nanning, China) with the 1964 Helsinki declaration and its later ethical standards. Informed consent was obtained from all individual participants included in the present study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Cooke SL, Brenton JD. Evolution of platinum resistance in high-grade serous ovarian cancer. Lancet Oncol. 2011;12:1169–1174. doi: 10.1016/S1470-2045(11)70123-1. [DOI] [PubMed] [Google Scholar]
- 4.Miller DS, Blessing JA, Krasner CN, Mannel RS, Hanjani P, Pearl ML, Waggoner SE, Boardman CH. Phase II evaluation of pemetrexed in the treatment of recurrent or persistent platinum-resistant ovarian or primary peritoneal carcinoma: A study of the Gynecologic Oncology Group. J Clin Oncol. 2009;27:2686–2691. doi: 10.1200/JCO.2008.19.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfisterer J, Ledermann JA. Management of platinum-sensitive recurrent ovarian cancer. Semin Oncol. 2006;33(2 Suppl):S12–S16. doi: 10.1053/j.seminoncol.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 7.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: An evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 8.Arcangeli A, Crociani O, Lastraioli E, Masi A, Pillozzi S, Becchetti A. Targeting ion channels in cancer: A novel frontier in antineoplastic therapy. Curr Med Chem. 2009;16:66–93. doi: 10.2174/092986709787002835. [DOI] [PubMed] [Google Scholar]
- 9.Cuddapah VA, Sontheimer H. Ion channels and transporters [corrected] in cancer. 2. Ion channels and the control of cancer cell migration. Am J Physiol Cell Physiol. 2011;301:C541–C549. doi: 10.1152/ajpcell.00102.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prevarskaya N, Skryma R, Shuba Y. Ion channels and the hallmarks of cancer. Trends Mol Med. 2010;16:107–121. doi: 10.1016/j.molmed.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 11.D'Amico M, Gasparoli L, Arcangeli A. Potassium channels: Novel emerging biomarkers and targets for therapy in cancer. Recent Pat Anticancer Drug Discov. 2013;8:53–65. doi: 10.2174/1574892811308010053. [DOI] [PubMed] [Google Scholar]
- 12.Pardo LA, Stühmer W. The roles of K(+) channels in cancer. Nat Rev Cancer. 2014;14:39–48. doi: 10.1038/nrc3635. [DOI] [PubMed] [Google Scholar]
- 13.Innamaa A, Jackson L, Asher V, Van Shalkwyk G, Warren A, Hay D, Bali A, Sowter H, Khan R. Expression and prognostic significance of the oncogenic K2P potassium channel KCNK9 (TASK-3) in ovarian carcinoma. Anticancer Res. 2013;33:1401–1408. [PubMed] [Google Scholar]
- 14.Asher V, Khan R, Warren A, Shaw R, Schalkwyk GV, Bali A, Sowter HM. The Eag potassium channel as a new prognostic marker in ovarian cancer. Diagn Pathol. 2010;5:78. doi: 10.1186/1746-1596-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cicek MS, Koestler DC, Fridley BL, Kalli KR, Armasu SM, Larson MC, Wang C, Winham SJ, Vierkant RA, Rider DN, et al. Epigenome-wide ovarian cancer analysis identifies a methylation profile differentiating clear-cell histology with epigenetic silencing of the HERG K+ channel. Hum Mol Genet. 2013;22:3038–3047. doi: 10.1093/hmg/ddt160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pillozzi S, Masselli M, De Lorenzo E, Accordi B, Cilia E, Crociani O, Amedei A, Veltroni M, D'Amico M, Basso G, et al. Chemotherapy resistance in acute lymphoblastic leukemia requires hERG1 channels and is overcome by hERG1 blockers. Blood. 2011;117:902–914. doi: 10.1182/blood-2010-01-262691. [DOI] [PubMed] [Google Scholar]
- 17.Köhler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, Adelman JP. Small-conductance, calcium-activated potassium channels from mammalian brain. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 18.Chantome A, Girault A, Potier M, Collin C, Vaudin P, Pagès JC, Vandier C, Joulin V. KCa2.3 channel-dependent hyperpolarization increases melanoma cell motility. Exp Cell Res. 2009;315:3620–3630. doi: 10.1016/j.yexcr.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Potier M, Joulin V, Roger S, Besson P, Jourdan ML, Leguennec JY, Bougnoux P, Vandier C. Identification of SK3 channel as a new mediator of breast cancer cell migration. Mol Cancer Ther. 2006;5:2946–2953. doi: 10.1158/1535-7163.MCT-06-0194. [DOI] [PubMed] [Google Scholar]
- 20.Jelassi B, Chantome A, Alcaraz-Pérez F, Baroja-Mazo A, Cayuela ML, Pelegrin P, Surprenant A, Roger S. P2X(7) receptor activation enhances SK3 channels- and cystein cathepsin-dependent cancer cells invasiveness. Oncogene. 2011;30:2108–2122. doi: 10.1038/onc.2010.593. [DOI] [PubMed] [Google Scholar]
- 21.Gueguinou M, Harnois T, Crottes D, Uguen A, Deliot N, Gambade A, Chantôme A, Haelters JP, Jaffrès PA, Jourdan ML, et al. SK3/TRPC1/Orai1 complex regulates SOCE-dependent colon cancer cell migration: A novel opportunity to modulate anti-EGFR mAb action by the alkyl-lipid Ohmline. Oncotarget. 2016;7:36168–36184. doi: 10.18632/oncotarget.8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Pennisi A, Zhan F, Sawyer JR, Shaughnessy JD, Yaccoby S. Establishment and exploitation of hyperdiploid and non-hyperdiploid human myeloma cell lines. Br J Haematol. 2007;138:802–811. doi: 10.1111/j.1365-2141.2007.06742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network: Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 29.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 30.Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28:21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- 31.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC, Chuang EY. miRSystem: An integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One. 2012;7:e42390. doi: 10.1371/journal.pone.0042390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedditch EL, Gao B, Russell AJ, Lu Y, Emmanuel C, Beesley J, Johnatty SE, Chen X, Harnett P, George J, et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju149. pii: dju149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Oncogenes associated with drug resistance in ovarian cancer. J Cancer Res Clin Oncol. 2015;141:381–395. doi: 10.1007/s00432-014-1765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin F, Liu X, Li D, Wang Q, Zhang W, Li L. Tumor suppressor genes associated with drug resistance in ovarian cancer (review) Oncol Rep. 2013;30:3–10. doi: 10.3892/or.2013.2446. [DOI] [PubMed] [Google Scholar]
- 37.Yin F, Liu X, Li D, Wang Q, Zhang W, Li L. Bioinformatic analysis of chemokine (C-C motif) ligand 21 and SPARC-like protein 1 revealing their associations with drug resistance in ovarian cancer. Int J Oncol. 2013;42:1305–1316. doi: 10.3892/ijo.2013.1819. [DOI] [PubMed] [Google Scholar]
- 38.Yin F, Liu L, Liu X, Li G, Zheng L, Li D, Wang Q, Zhang W, Li L. Downregulation of tumor suppressor gene ribonuclease T2 and gametogenetin binding protein 2 is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;32:362–372. doi: 10.3892/or.2014.3175. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Upregulation of NEK2 is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;31:745–754. doi: 10.3892/or.2013.2910. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Downregulation of NEK11 is associated with drug resistance in ovarian cancer. Int J Oncol. 2014;45:1266–1274. doi: 10.3892/ijo.2014.2503. [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Gao Y, Zhao B, Li X, Lu Y, Zhang J, Li D, Li L, Yin F. Discovery of microarray-identified genes associated with ovarian cancer progression. Int J Oncol. 2015;46:2467–2478. doi: 10.3892/ijo.2015.2971. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Zou J, Su J, Lu Y, Zhang J, Li L, Yin F. Downregulation of transient receptor potential cation channel, subfamily C, member 1 contributes to drug resistance and high histological grade in ovarian cancer. Int J Oncol. 2016;48:243–252. doi: 10.3892/ijo.2015.3254. [DOI] [PubMed] [Google Scholar]
- 43.Sparks R, Lau WW, Tsang JS. Expanding the immunology toolbox: Embracing public-data reuse and crowdsourcing. Immunity. 2016;45:1191–1204. doi: 10.1016/j.immuni.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Grechkin M, Poon H, Howe B. Wide-Open: Accelerating public data release by automating detection of overdue datasets. PLoS Biol. 2017;15:e2002477. doi: 10.1371/journal.pbio.2002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet. 2013;14:89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- 46.Kannan L, Ramos M, Re A, El-Hachem N, Safikhani Z, Gendoo DM, Davis S, Gomez-Cabrero D, Castelo R, Hansen KD, et al. Public data and open source tools for multi-assay genomic investigation of disease. Brief Bioinform. 2016;17:603–615. doi: 10.1093/bib/bbv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin SS, Park SS, Hwang B, Moon B, Kim WT, Kim WJ, Moon SK. MicroRNA-892b influences proliferation, migration and invasion of bladder cancer cells by mediating the p19ARF/cyclin D1/CDK6 and Sp-1/MMP-9 pathways. Oncol Rep. 2016;36:2313–2320. doi: 10.3892/or.2016.5052. [DOI] [PubMed] [Google Scholar]
- 49.Jiang L, Yu L, Zhang X, Lei F, Wang L, Liu X, Wu S, Zhu J, Wu G, Cao L, et al. miR-892b silencing activates NF-κB and promotes aggressiveness in breast cancer. Cancer Res. 2016;76:1101–1111. doi: 10.1158/0008-5472.CAN-15-1770. [DOI] [PubMed] [Google Scholar]
- 50.Lloyd KL, Cree IA, Savage RS. Prediction of resistance to chemotherapy in ovarian cancer: A systematic review. BMC Cancer. 2015;15:117. doi: 10.1186/s12885-015-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emmons MF, Anreddy N, Cuevas J, Steinberger K, Yang S, McLaughlin M, Silva A, Hazlehurst LA. MTI-101 treatment inducing activation of Stim1 and TRPC1 expression is a determinant of response in multiple myeloma. Sci Rep. 2017;2:7. doi: 10.1038/s41598-017-02713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.