Abstract
Cerebral ischemia/reperfusion (I/R) injury results in detrimental complications. However, little is known about the underlying molecular mechanisms involved in the reperfusion stage. The aim of the present study was to identify a gene expression profile associated with cerebral ischemia/reperfusion injury. The GSE23160 dataset, which comprised data from sham control samples and post-I/R injury brain tissues that were obtained using a middle cerebral artery occlusion (MCAO) model at 2, 8 and 24 h post-reperfusion, was downloaded from the Gene Expression Omnibus database. The differentially expressed genes (DEGs) in the MCAO samples compared with controls were screened using the GEO2R web tool. Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for DEGs was performed using the online tool DAVID. Furthermore, a protein-protein interaction (PPI) network was constructed using the STRING database and Cytoscape software. In total, 32 DEGs at 2 h post-reperfusion, 39 DEGs at 8 h post-reperfusion and 91 DEGs at 24 h post-reperfusion were identified, while 15 DEGs were common among all three groups. GO analysis revealed that the DEGs at all three time-points were enriched in ‘chemotaxis’ and ‘inflammatory response’ terms, while KEGG pathway analysis demonstrated that DEGs were significantly enriched in the ‘chemokine signaling pathway’. Furthermore, following PPI network construction, Cxcl1 was identified as the only hub gene that was common among all three time-points. In conclusion, the present study has demonstrated a global view of the potential molecular differences following cerebral I/R injury and may contribute to an improved understanding of the reperfusion stage, which may ultimately aid in the development of future clinical strategies.
Keywords: differentially expressed genes, gene ontology, cerebral ischemia/reperfusion injury, inflammatory response, chemokines
Introduction
Globally, stroke is the second most frequent cause of mortality and the primary cause of serious long-term disability worldwide (1). Of all strokes, 87% are ischemic (2). Various mechanisms underlying ischemic stroke are driven by cell-cell interactions within brain, including excitotoxicity, calcium dysregulation, oxidative and nitrosative Stress, cortical spreading depolarizations, inflammation, necrosis, necroptosis and autophagy (3). In addition to a narrow therapeutic time window (4), ischemic stroke remains difficult to manage.
Although reperfusion has been proven to be beneficial for ischemic stroke (5), reperfusion may result in detrimental secondary damage, which is termed ischemia/reperfusion (I/R) injury. Early reperfusion of ischemic brain tissue has been associated with various negative consequences, including blood-brain barrier breakdown, which may result in cerebral edema and/or brain hemorrhage, neurovascular damage and neuronal death (6). Angiogenesis and vasculogenesis have also been detected following reperfusion (7). In addition, inflammation is induced by reperfusion injury and contributes negatively to long-term disease prognosis (8). The inflammatory response may result in subsequent oxidative injury, excitotoxicity and neuronal cell death (9). Chemokines, produced by resident microglial cells and other immune cells in the brain, contribute to the recruitment of circulating leukocytes and exaggerate the inflammatory response. Chemokines have been demonstrated to have both deleterious and beneficial roles in ischemia/reperfusion injury (10).
Microarray analysis has been previously employed to identify molecular variations in cerebral I/R injury (11,12). However, gene expression profiles at different reperfusion periods have not been investigated extensively. Therefore, the present study employed a microarray dataset from the Gene Expression Omnibus (GEO) database and screened for differentially expressed genes (DEGs) between control samples and cerebral I/R samples at 2, 8 and 24 h post-reperfusion, and subsequently analyzed the functions and interactions of these DEGs. The results of the current study may aid in improving the understanding of the molecular mechanisms underlying cerebral I/R injury.
Materials and methods
Microarray data
Microarray gene expression profiles from GSE23160 (12) were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/), which is based on the platform of GPL6885 using Illumina MousRef-8 v2.0 Expression BeadChip (Illumina, Inc., San Diego, CA, USA). All of the samples were taken from male C57BL/6J mice (8–10 weeks). Following 2 h suture-induced middle cerebral artery occlusion (MCAO), the animals underwent reperfusion for 2, 8 or 24 h. Tissue extractions at 2, 8 and 24 h post-reperfusion and sham controls (n=4 per group) were included in this dataset.
Identification of DEGs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), an R-based web application (13), was employed to analyze DEGs between MCAO samples and sham samples. P<0.05 and |logFC|≥1.2 were set as the threshold criteria to identify genes that were differentially expressed in MCAO models. Subsequently, the DEGs at 2, 8 and 24 h post-reperfusion were screened for subsequent analyses. A Venn diagram was produced to indicate the intersection among DEGs in the various MCAO groups using FunRich software (version 2.1.1; www.funrich.org) (14).
Functional enrichment analysis of DEGs
To identify the biological processes, cellular components, molecular functions and biological pathways that the DEGs were significantly enriched in, Gene Ontology (GO) enrichment (15) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (16) were performed using the online tool Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/). P<0.05 was considered to indicate a significantly enriched term or pathway, which was calculated using a hypergeometric test. Heat map illustration of DEGs was performed with heat map illustrator software (version 1.0.3.7; http://hemi.biocuckoo.org) (17).
Construction of the PPI network
To further investigate the underlying molecular mechanisms of cerebral I/R injury, protein-protein interaction (PPI) networks for the DEGs were constructed using the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://www.string-db.org/) (18). A combined score of >0.4 was selected to construct the PPI networks. The obtained PPI networks at 24 h post-reperfusion were subsequently visualized using Cytoscape software (version 3.5.1) (19). Finally, the topological properties of the networks at 2, 8 and 24 h post-reperfusion were analyzed and the degree of each node was calculated; genes with a degree >10 were defined as hub genes.
Results
Identification of DEGs
As demonstrated in Fig. 1, 32 DEGs at 2 h post-reperfusion, 39 DEGs at 8 h post-reperfusion and 91 DEGs at 24 h post-reperfusion were identified in the MCAO samples compared with the controls. Among them, 15 DEGs were common to all three injury samples, including C-C motif chemokine ligand (CCL)7, suppressor of cytokine signaling 3, CCL4, activating transcription factor 3, lipocalin 2, hemoglobin α adult chain 1, gap junction protein β2, CD14 antigen, CCL3, heat shock protein 1A, S100 calcium-binding protein A8 (calgranulin A), C-X-C motif chemokine ligand 1 (CXCL1), epithelial membrane protein 1, tissue inhibitor of metalloproteinase 1 and zinc finger protein 36, all of which were upregulated in the MCAO samples (Table I). Heat map and PPI network analysis at 2 and 8 h post-reperfusion are not presented due to the small number of identified DEGs.
Figure 1.
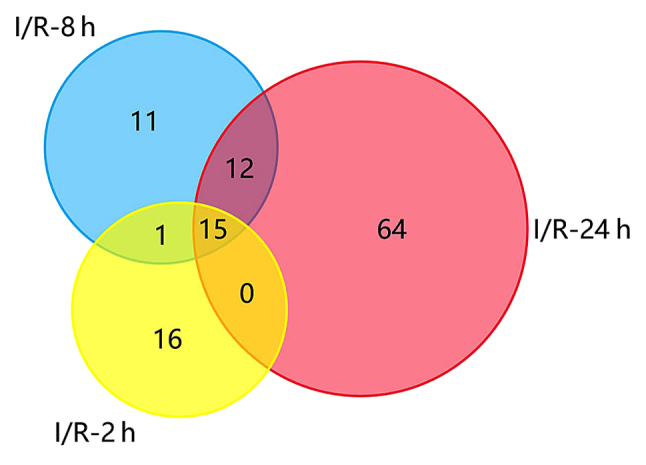
Venn diagram presenting the DEGs between cerebral I/R and sham control samples at 2, 8 and 24 h post-reperfusion. A total of 15 DEGs were common among all three post-reperfusion time-points. DEGs, differentially expressed genes; I/R, ischemia/reperfusion.
Table I.
DEGs between cerebral I/R and sham control samples that were common among 2, 8 and 24 h post-reperfusion time-points.
| Post-reperfusion time-point | |||||||
|---|---|---|---|---|---|---|---|
| 2 h | 8 h | 24 h | |||||
| Gene symbol | Gene name | P-value | Log FC | P-value | Log FC | P-value | Log FC |
| ATF3 | Activating transcription factor 3 | 5.01×10-6 | 2.026964 | 3.56×10-5 | 1.692171 | 7.49×10-3 | 1.591308 |
| CCL3 | C-C motif chemokine ligand 3 | 5.13×10-10 | 3.259239 | 7.05×10-6 | 3.291652 | 4.81×10-4 | 2.300334 |
| CCL4 | C-C motif chemokine ligand 4 | 1.32×10-10 | 4.429954 | 1.29×10-6 | 4.47245 | 5.83×10-5 | 3.679271 |
| CCL7 | C-C motif chemokine ligand 7 | 2.18×10-5 | 1.220192 | 7.63×10-6 | 1.426973 | 4.00×10-3 | 1.707457 |
| CD14 | CD14 antigen | 1.38×10-7 | 1.757282 | 5.92×10-7 | 2.578573 | 2.56×10-3 | 2.353608 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 3.29×10-6 | 1.806643 | 4.81×10-8 | 2.878735 | 1.43×10-3 | 2.596958 |
| EMP1 | Epithelial membrane protein 1 | 1.68×10-6 | 1.688657 | 6.78×10-7 | 1.383669 | 1.89×10-3 | 2.161470 |
| GJB2 | Gap junction protein β2 | 3.16×10-8 | 1.999569 | 1.15×10-7 | 1.717101 | 9.13×10-5 | 1.822626 |
| HBA-A1 | Hemoglobin α, adult chain 1 | 5.38×10-7 | 2.436023 | 2.15×10-8 | 3.757753 | 4.48×10-4 | 3.807134 |
| HSPA1A | Heat shock protein 1A | 1.69×10-6 | 3.054788 | 3.80×10-5 | 2.545466 | 1.59×10-2 | 1.299110 |
| LCN2 | Lipocalin 2 | 4.06×10-9 | 1.312245 | 4.47×10-15 | 3.250233 | 1.72×10-5 | 3.408973 |
| S100A8 | S100 calcium-binding protein A8 (calgranulin A) | 2.29×10-6 | 1.240885 | 1.66×10-6 | 1.655962 | 1.90×10-3 | 3.780381 |
| SOCS3 | Suppressor of cytokine signaling 3 | 3.54×10-8 | 2.009771 | 1.74×10-8 | 2.004998 | 5.48×10-4 | 2.361302 |
| TIMP1 | Tissue inhibitor of metalloproteinase 1 | 1.02×10-8 | 2.376190 | 3.61×10-18 | 3.403190 | 1.52×10-5 | 3.874533 |
| ZFP36 | Zinc finger protein 36 | 2.01×10-5 | 1.595428 | 6.26×10-7 | 1.404682 | 1.17×10-2 | 1.330419 |
Bioinformatics analyses of DEGs
To further the understanding of the screened DEGs and determine their potential roles following I/R injury, GO functional and KEGG pathway enrichment analyses were performed.
A total of 51 GO enriched terms for biological processes at 2, 8 and 24 h post-reperfusion were obtained. The 10 most enriched GO terms according to the P-value for 2, 8 and 24 h post-reperfusion groups are presented in Table II. Furthermore, ‘chemotaxis’ (GO:0006935), ‘inflammatory response’ (GO:0006954), ‘immune response’ (GO:0006955), ‘G-protein coupled receptor signaling pathway’ (GO:0007186), ‘response to toxic substance’ (GO:0009636), ‘neutrophil chemotaxis’ (GO:0030593), ‘positive regulation of tumor necrosis factor production’ (GO:0032760), ‘positive regulation of GTPase activity’ (GO:0043547), ‘lymphocyte chemotaxis’ (GO:0048247), ‘positive regulation of inflammatory response’ (GO:0050729), ‘cell chemotaxis’ (GO:0060326), ‘chemokine-mediated signaling pathway’ (GO:0070098), ‘positive regulation of ERK1 and ERK2 cascade’ (GO:0070374), ‘cellular response to interferon-gamma’ (GO:0071346), ‘cellular response to interleukin-1’ (GO:0071347), ‘monocyte chemotaxis’ (GO:0002548) and ‘cellular response to tumor necrosis factor’ (GO:0071356) were significantly enriched at all three post-reperfusion time-points (2, 8 and 24 h).
Table II.
Top 10 enriched GO biological process terms for DEGs between cerebral ischemia/reperfusion and sham control samples.
| A, Top 10 enriched GO biological process terms for DEGs at 2 h post-reperfusion | |||||
|---|---|---|---|---|---|
| GO ID | GO term | Count | % | P-value | Genes |
| GO:0030593 | Neutrophil chemotaxis | 5 | 15.63 | 5.65×10-6 | CXCL1, CCL3, S100A8, CCL4, CCL7 |
| GO:0032570 | Response to progesterone | 4 | 12.50 | 1.80×10-5 | FOS, OXT, FOSB, GJB2 |
| GO:0006954 | Inflammatory response | 7 | 21.88 | 2.24×10-5 | CXCL1, CCL3, S100A8, CCL4, CCL7, CD14, IL1A |
| GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 10 | 31.25 | 2.99×10-5 | FOS, CCL3, EGR2, ATF3, EGR4, FOSB, NPAS4, JUNB, IL1A, CYR61 |
| GO:0071356 | Cellular response to tumor necrosis factor | 5 | 15.63 | 3.59×10-5 | LCN2, ZFP36, CCL3, CCL4, CCL7 |
| GO:2000503 | Positive regulation of natural killer cell chemotaxis | 3 | 9.38 | 4.25×10-5 | CCL3, CCL4, CCL7 |
| GO:0006935 | Chemotaxis | 5 | 15.63 | 4.73×10-5 | CCL3, S100A8, CCL4, CCL7, CYR61 |
| GO:0051591 | Response to cAMP | 4 | 12.50 | 8.99×10-5 | FOS, OXT, FOSB, JUNB |
| GO:0070098 | Chemokine-mediated signaling pathway | 4 | 12.50 | 1.13×10-4 | CXCL1, CCL3, CCL4, CCL7 |
| GO:0050729 | Positive regulation of inflammatory response | 4 | 12.50 | 1.69×10-4 | CCL3, S100A8, CCL4, CCL7 |
| B, Top 10 enriched GO biological process terms for DEGs at 8 h post-reperfusion | |||||
| GO ID | GO term | Count | % | P-value | Genes |
| GO:0030593 | Neutrophil chemotaxis | 10 | 25.64 | 8.69×10-15 | CXCL1, CCL12, CCL3, S100A8, LGALS3, CCL9, CCL4, CCL7, FCGR3, CCL17 |
| GO:0002548 | Monocyte chemotaxis | 8 | 20.51 | 1.19×10-12 | CCL12, CCL3, FLT1, LGALS3, CCL9, CCL4, CCL7, CCL17 |
| GO:0070098 | Chemokine-mediated signaling pathway | 7 | 17.95 | 1.09×10-9 | CXCL1, CCL12, CCL3, CCL9, CCL4, CCL7, CCL17 |
| GO:0071356 | Cellular response to tumor necrosis factor | 8 | 20.51 | 1.83×10-9 | LCN2, ZFP36, CCL12, CCL3, CCL9, CCL4, CCL7, CCL17 |
| GO:0050729 | Positive regulation of inflammatory response | 7 | 17.95 | 2.52×10-9 | CCL12, CCL3, S100A8, CCL9, TLR2, CCL4, CCL7 |
| GO:0006935 | Chemotaxis | 8 | 20.51 | 3.01×10-9 | CCL12, CCL3, FLT1, S100A8, CCL9, CCL4, CCL7, CCL17 |
| GO:0071346 | Cellular response to interferon-gamma | 7 | 17.95 | 4.02×10-9 | CCL12, CCL3, CCL9, CCL4, GBP2, CCL7, CCL17 |
| GO:0048247 | Lymphocyte chemotaxis | 6 | 15.38 | 5.34×10-9 | CCL12, CCL3, CCL9, CCL4, CCL7, CCL17 |
| GO:0071347 | Cellular response to interleukin-1 | 7 | 17.95 | 1.09×10-8 | LCN2, CCL12, CCL3, CCL9, CCL4, CCL7, CCL17 |
| GO:0006954 | Inflammatory response | 10 | 25.64 | 1.76×10-8 | CXCL1, CCL12, CCL3, S100A8, CCL9, TLR2, CCL4, CCL7, CD14, CCL17 |
| C, Top 10 enriched GO biological process terms for DEGs at 24 h post-reperfusion | |||||
| GO ID | GO term | Count | % | P-value | Genes |
| GO:0006954 | Inflammatory response | 21 | 22.83 | 1.99×10-16 | CXCL1, CCL3, CCL2, S100A8, CCL21C, S100A9, CCL9, TLR2, CCL21A, PF4, FPR2, IDO1, CCL4, CCL7, CXCL10, SLC11A1, CYBA, CCL12, CHIL3, CD14, SPP1 |
| GO:0006955 | Neutrophil chemotaxis | 13 | 14.13 | 5.60×10-16 | CXCL1, CCL3, CCL2, S100A8, LGALS3, CCL21C, S100A9, CCL21A, CCL9, CCL4, CCL7, CCL12, SPP1 |
| GO:0006956 | Chemokine-mediated signaling pathway | 11 | 11.96 | 1.08×10-13 | CXCL1, CCL12, CCL3, CCL2, CCL21C, CCL9, CCL21A, PF4, CCL4, CCL7, CXCL10 |
| GO:0006957 | Monocyte chemotaxis | 9 | 9.78 | 1.54×10-11 | CCL12, CCL3, CCL2, LGALS3, CCL21C, CCL9, CCL21A, CCL4, CCL7 |
| GO:0006958 | Positive regulation of inflammatory response | 10 | 10.87 | 1.92×10-11 | CCL12, CCL3, CCL2, S100A8, S100A9, CCL9, TGM2, TLR2, CCL4, CCL7 |
| GO:0006959 | Cellular response to tumor necrosis factor | 11 | 11.96 | 1.39×10-10 | LCN2, ZFP36, CCL12, CYBA, CCL3, CCL2, CCL21C, CCL9, CCL21A, CCL4, CCL7 |
| GO:0006960 | Cellular response to interleukin-1 | 10 | 10.87 | 1.76×10-10 | LCN2, CCL12, CCL3, CCL2, CCL21C, CCL9, SAA3, CCL21A, CCL4, CCL7 |
| GO:0006961 | Lymphocyte chemotaxis | 8 | 8.70 | 1.96×10-10 | CCL12, CCL3, CCL2, CCL21C, CCL9, CCL21A, CCL4, CCL7 |
| GO:0006962 | Chemotaxis | 11 | 11.96 | 2.80×10-10 | CCL12, CCL3, CCL2, S100A8, S100A9, CCL9, PF4, FPR2, CCL4, CCL7, CXCL10 |
| GO:0006963 | Cellular response to interferon-gamma | 9 | 9.78 | 1.33×10-9 | CCL12, CCL3, CCL2, CCL21C, CCL9, CCL21A, CCL4, GBP2, CCL7 |
GO, Gene Ontology; DEGs, differentially expressed genes.
Additionally, DEGs were enriched in various GO cellular component terms; at 2 and 8 h post-reperfusion, DEGs were enriched in ‘extracellular region’ (GO:0005576), while ‘membrane’ (GO:0016020) was significantly enriched at both 8 and 24 h post-reperfusion (Table III). Furthermore, Table IV indicates that DEGs were significantly enriched in ‘cytokine activity’ (GO:0005125) and ‘chemokine activity’ (GO:0008009) GO molecular function terms at 2, 8 and 24 h post-reperfusion.
Table III.
GO cellular component terms for DEGs between cerebral ischemia/reperfusion and sham control samples.
| A, Enriched GO cellular component terms for DEGs at 2 h post-reperfusion | |||||
|---|---|---|---|---|---|
| GO ID | GO term | Count | % | P-value | Genes |
| GO:0005576 | Extracellular region | 14 | 43.75 | 9.56×10-7 | CXCL1, CCL3, AVP, S100A8, PMCH, OXT, CCL4, CCL7, TIMP1, LCN2, NPTX2, IL1A, CD14, CYR61 |
| GO:0005615 | Extracellular space | 12 | 37.50 | 1.02×10-5 | LCN2, CXCL1, AVP, CCL3, S100A8, PMCH, OXT, CCL4, CCL7, CD14, IL1A, TIMP1 |
| B, Enriched GO cellular component terms for DEGs at 8 h post-reperfusion | |||||
| GO ID | GO term | Count | % | P-value | Genes |
| GO:0005615 | Extracellular space | 16 | 41.03 | 4.81×10-8 | CXCL1, CCL3, FLT1, S100A8, LGALS3, PMCH, CCL9, CCL4, CCL7, TIMP1, CCL17, LCN2, CCL12, SERPINA3N, DMKN, CD14 |
| GO:0005576 | Extracellular region | 14 | 35.90 | 1.39×10-5 | CXCL1, CCL3, S100A8, LGALS3, PMCH, CCL9, CCL4, CCL7, TIMP1, LCN2, CCL12, SERPINA3N, DMKN, CD14 |
| GO:0009897 | External side of plasma membrane | 5 | 12.82 | 3.17×10-3 | LGALS3, OSMR, TLR2, CD14, FCGR3 |
| GO:0016020 | Membrane | 20 | 51.28 | 4.79×10-2 | GPR84, FLT1, S100A8, LGALS3, OSMR, FKBP5, MS4A6D, TLR2, SLC10A6, GJB2, FCGR3, HBA-A1, CH25H, PLIN4, HMOX1, ITGAD, SLC15A3, GBP2, EMP1, CD14 |
| C, Enriched GO cellular component terms for DEGs at 24 h post-reperfusion | |||||
| GO ID | GO term | Count | % | P-value | Genes |
| GO:0009897 | External side of plasma membrane | 8 | 8.70 | 6.31×10-4 | FCGR2B, LGALS3, PDPN, CCL21C, FCGR4, TLR2, CD14, CXCL10 |
| GO:0048237 | Rough endoplasmic reticulum lumen | 3 | 3.26 | 4.29×10-4 | LYZ2, LYZ1, CHIL3 |
| GO:0009986 | Cell surface | 10 | 10.87 | 2.33×10-3 | SLC11A1, THBD, FCGR2B, LGALS3, TNFRSF12A, IFITM3, FCGR4, TLR2, CD14, ANXA2 |
| GO:0005886 | Plasma membrane | 34 | 36.96 | 8.00×10-3 | GPR182, GPR84, S100A8, IFITM2, TNFRSF12A, IFITM3, VIM, S100A9, TLR2, CD52, FPR2, SLC11A1, P2RY6, DAB2, PLIN2, HMOX1, TGM2, STRA6, CLEC4D, ANGPT2, ACTB, PDPN, LILRB4A, GJB2, ANXA2, CYBA, THBD, FCGR2B, HSPB1, SCN4B, RGS9, EMP3, CD14, EMP1 |
| GO:0016020 | Membrane | 44 | 47.83 | 1.14×10-2 | GPR182, GPR84, GFAP, TSPO, S100A8, IFITM2, TNFRSF12A, IFITM3, S100A9, TLR2, CD52, FPR2, FXYD5, GLIPR2, SLC11A1, P2RY6, DAB2, PLIN2, HMOX1, CH25H, TGM1, TGM2, STRA6, CLEC4D, ACTB, LGALS3, PDPN, MS4A6D, LILRB4A, GJB2, ANXA2, HBA-A1, CYBA, RAB32, THBD, FCGR2B, SCN4B, RGS9, EMP3, SLC15A3, GBP2, CD14, EMP1, MVP |
GO, Gene Ontology; DEGs, differentially expressed genes.
Table IV.
Enriched GO molecular function terms for DEGs between cerebral ischemia/reperfusion and sham control samples.
| A, Enriched GO molecular function terms for DEGs at 2 h post-reperfusion | |||||
| GO ID | GO term | Count | % | P-value | Genes |
|---|---|---|---|---|---|
| GO:0005125 | Cytokine activity | 6 | 18.75 | 2.48×10-5 | CXCL1, FOS, CCL3, CCL4, CD14, IL1A |
| GO:0008009 | Chemokine activity | 4 | 12.50 | 6.79×10-5 | FOS, CCL3, CCL4, CD14 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 6 | 18.75 | 2.84×10-4 | FOS, EGR2, ATF3, FOSB, NPAS4, JUNB |
| GO:0001077 | Transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | 5 | 15.63 | 9.82×10-4 | FOS, EGR2, FOSB, NPAS4, JUNB |
| GO:0003690 | Double-stranded DNA binding | 3 | 9.38 | 2.13×10-2 | FOS, FOSB, JUNB |
| GO:0003677 | DNA binding | 8 | 25.00 | 2.85×10-2 | ZFP36, FOS, EGR2, ATF3, EGR4, FOSB, NPAS4, JUNB |
| B, Enriched GO molecular function terms for DEGs at 8 h post-reperfusion | |||||
| GO ID | GO term | Count | % | P-value | Genes |
| GO:0008009 | Chemokine activity | 7 | 17.95 | 3.98×10-10 | CXCL1, CCL12, CCL3, CCL9, CCL4, CCL7, CCL17 |
| GO:0005125 | Cytokine activity | 8 | 20.51 | 1.54×10-7 | CXCL1, CCL12, CCL3, CCL9, CCL4, CCL7, CCL17, TIMP1 |
| GO:0048020 | CCR chemokine receptor binding | 4 | 10.26 | 2.14×10-5 | CCL3, CCL9, CCL4, CCL17 |
| C, Enriched GO molecular function terms for DEGs at 24 h post-reperfusion | |||||
| GO ID | GO term | Count | % | P-value | Genes |
| GO:0008009 | Chemokine activity | 11 | 11.96 | 1.69×10-14 | CXCL1, CCL12, CCL3, CCL2, CCL21C, CCL9, CCL21A, PF4, CCL4, CCL7, CXCL10 |
| GO:0005125 | Cytokine activity | 12 | 13.04 | 4.78×10-9 | CXCL1, CCL12, CCL3, CCL2, CCL9, PF4, CCL4, CCL7, SPP1, TIMP1, CXCL10, IL11 |
| GO:0048020 | CCR chemokine receptor binding | 5 | 5.44 | 8.52×10-6 | CCL3, CCL21C, CCL9, CCL21A, CCL4 |
| GO:0031727 | CCR2 chemokine receptor binding | 3 | 3.26 | 1.30×10-4 | CCL12, CCL2, CCL7 |
| GO:0008201 | Heparin binding | 4 | 4.35 | 3.43×10-2 | CCL2, PF4, CCL7, CXCL10 |
| GO:0020037 | Heme binding | 4 | 4.35 | 4.94×10-2 | HBA-A1, CYBA, HMOX1, IDO1 |
GO, Gene Ontology; DEGs, differentially expressed genes.
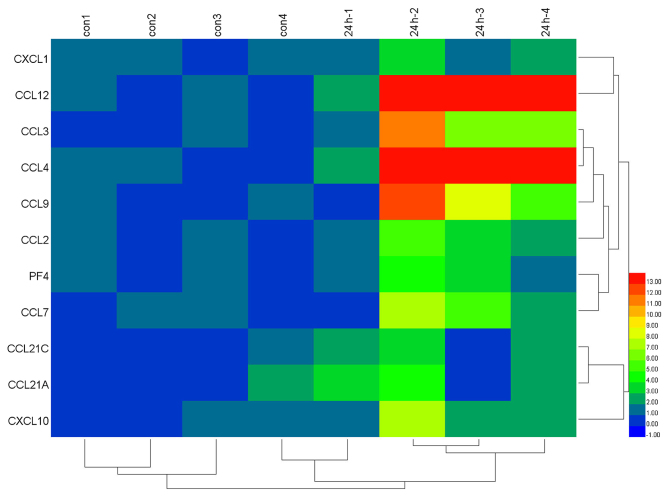
Table V presents the KEGG pathways that were significantly enriched in DEGs. KEGG analysis indicated that ‘chemokine signaling pathway’, ‘cytokine-cytokine receptor interaction’ and ‘toll-like receptor signaling pathway’ were significantly enriched in DEGs at 2, 8 and 24 h post-reperfusion. Furthermore, as demonstrated in Fig. 2, a total of 11 chemokine signaling pathway-associated genes were overexpressed in 24 h post-reperfusion injury samples compared with the sham control samples.
Table V.
Enriched KEGG pathways for DEGs between cerebral ischemia/reperfusion and sham control samples.
| A, Enriched KEGG pathways for DEGs at 2 h post-reperfusion | |||||
|---|---|---|---|---|---|
| KEGG entry | Pathway name | Count | % | P-value | Genes |
| mmu05132 | Salmonella infection | 6 | 18.75 | 1.27×10-6 | CXCL1, FOS, CCL3, CCL4, CD14, IL1A |
| mmu04380 | Osteoclast differentiation | 5 | 15.63 | 2.68×10-4 | FOS, SOCS3, FOSB, JUNB, IL1A |
| mmu04620 | Toll-like receptor signaling pathway | 4 | 12.50 | 2.11×10-3 | FOS, CCL3, CCL4, CD14 |
| mmu04062 | Chemokine signaling pathway | 4 | 12.50 | 1.34×10-2 | CXCL1, CCL3, CCL4, CCL7 |
| mmu04060 | Cytokine-cytokine receptor interaction | 4 | 12.50 | 2.41×10-2 | CCL3, CCL4, CCL7, IL1A |
| mmu05166 | HTLV-I infection | 4 | 12.50 | 3.35×10-2 | ZFP36, FOS, EGR2, ATF3 |
| B, Enriched KEGG pathways for DEGs at 8 h post-reperfusion | |||||
| KEGG entry | Pathway name | Count | % | P-value | Genes |
| mmu04062 | Chemokine signaling pathway | 7 | 17.95 | 2.28×10-5 | CXCL1, CCL12, CCL3, CCL9, CCL4, CCL7, CCL17 |
| mmu04060 | Cytokine-cytokine receptor interaction | 6 | 15.38 | 8.01×10-4 | CCL12, CCL3, FLT1, OSMR, CCL4, CCL7 |
| mmu05132 | Salmonella infection | 4 | 10.26 | 1.72×10-3 | CXCL1, CCL3, CCL4, CD14 |
| mmu04620 | Toll-like receptor signaling pathway | 4 | 10.26 | 3.60×10-3 | CCL3, TLR2, CCL4, CD14 |
| mmu04145 | Phagosome | 4 | 10.26 | 1.61×10-2 | TLR2, TUBB6, CD14, FCGR3 |
| mmu05142 | Chagas disease (American trypanosomiasis) | 3 | 7.69 | 4.02×10-2 | CCL12, CCL3, TLR2 |
| C, Enriched KEGG pathways for DEGs at 24 h post-reperfusion | |||||
| KEGG entry | Pathway name | Count | % | P-value | Genes |
| mmu04062 | Chemokine signaling pathway | 11 | 11.96 | 2.50×10-7 | CXCL1, CCL12, CCL3, CCL2, CCL21C, CCL9, CCL21A, PF4, CCL4, CCL7, CXCL10 |
| mmu04060 | Cytokine-cytokine receptor interaction | 11 | 11.96 | 1.95×10-6 | CCL12, CCL3, CCL2, TNFRSF12A, CCL21C, CCL21A, PF4, CCL4, CCL7, CXCL10, IL11 |
| mmu05323 | Rheumatoid arthritis | 6 | 6.52 | 1.43×10-4 | CCL12, CCL3, CCL2, TLR2, MMP3, IL11 |
| mmu04620 | Toll-like receptor signaling pathway | 6 | 6.52 | 3.80×10-4 | CCL3, TLR2, CCL4, CD14, SPP1, CXCL10 |
| mmu04668 | TNF signaling pathway | 6 | 6.52 | 5.41×10-4 | CXCL1, CCL12, CCL2, SOCS3, MMP3, CXCL10 |
| mmu04145 | Phagosome | 7 | 7.61 | 6.73×10-4 | ACTB, CYBA, FCGR2B, FCGR4, TLR2, TUBB6, CD14 |
| mmu05144 | Malaria | 4 | 4.35 | 3.20×10-3 | HBA-A1, CCL12, CCL2, TLR2 |
| mmu05164 | Influenza A | 6 | 6.52 | 3.99×10-3 | ACTB, CCL12, CCL2, SOCS3, HSPA1A, CXCL10 |
| mmu05142 | Chagas disease (American trypanosomiasis) | 4 | 4.35 | 2.58×10-2 | CCL12, CCL3, CCL2, TLR2 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
Figure 2.
Heat map illustration of chemokine signaling pathway-associated genes in 24 h post-reperfusion and sham samples. A total of 11 chemokine signaling pathway-associated genes were included. The color code depicts the value of each gene following median normalization, with blue indicating the lowest and red as the highest value.
PPI network analysis
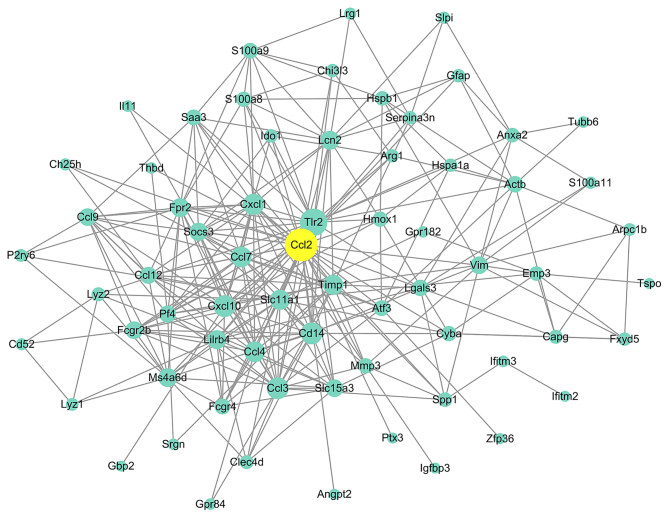
Genes with an interaction degree >10 in the PPI network analysis of DEGs at 2, 8 and 24 h post-reperfusion were defined as hub genes, which are listed in Table VI. CXCL1 was the only gene that was considered to be a hub gene at 2, 8 and 24 h post-reperfusion (Table VI). The constructed PPI network of 24 h post-reperfusion samples is presented in Fig. 3, which contains 67 nodes and 281 edges. Each node represents a DEG and each edge represents a PPI between two DEGs. At 24 h post-reperfusion, 23 genes served as hub genes, and of these hub genes, CCL2 exhibited the highest degree (Fig. 3).
Table VI.
Hub genes identified in PPI networks.
| A, Hub genes in the PPI network at 2 h post-reperfusion | |
|---|---|
| Gene | Degree |
| FOS | 16 |
| CXCL1 | 11 |
| ATF3 | 11 |
| B, Hub genes in the PPI network at 8 h post-reperfusion | |
| Gene | Degree |
| TLR2 | 14 |
| CXCL1 | 13 |
| CD14 | 12 |
| CCL4 | 12 |
| CCL3 | 12 |
| CCL7 | 12 |
| TIMP1 | 10 |
| C, Hub genes in the PPI network at 24 h post-reperfusion | |
| Gene | Degree |
| CCL2 | 37 |
| TLR2 | 28 |
| CCL3 | 19 |
| CD14 | 19 |
| CXCL10 | 19 |
| CXCL1 | 18 |
| CCL7 | 17 |
| CCL4 | 17 |
| SLC11A1 | 16 |
| TIMP1 | 16 |
| MS4A6D | 14 |
| LCN2 | 14 |
| LILRB4 | 14 |
| FPR2 | 14 |
| SOCS3 | 13 |
| SLC15A3 | 12 |
| VIM | 12 |
| FCGR2B | 12 |
| CCL9 | 12 |
| CCL12 | 11 |
| PF4 | 11 |
| LGALS3 | 10 |
| ACTB | 10 |
PPI, protein-protein interaction.
Figure 3.
Protein-protein interaction network for DEGs between cerebral ischemia/reperfusion and sham control samples at 24 h post-reperfusion. Circles represent nodes and lines between nodes represent edges, which indicate DEGs and interactions between two DEGs, respectively. C-C motif chemokine ligand 2 was identified as the hub gene with the highest interaction degree and is indicated in yellow. DEGs, differentially expressed genes.
Discussion
In the present study, 32 DEGs at 2 h, 39 DEGs at 8 h and 91 DEGs at 24 h post-reperfusion injury were identified between cerebral I/R and sham control samples. Previous studies have performed bioinformatics analysis to identify DEGs between MCAO models and controls (11,20–23). However, to the best of our knowledge, the present study is the first to perform global gene expression profiling at three time-points following reperfusion, and the findings may lead to improvements in the understanding of the pathophysiological process of cerebral I/R injury. DEGs associated with inflammation have previously been associated with cerebral I/R injury (20,22), and the results of the present study were consistent with these previous reports, indicating a persistent inflammatory response in cerebral I/R injury.
In the current study, enrichment analysis revealed that ‘chemotaxis’, ‘chemokine activity’ and ‘chemokine signaling pathway’ terms were significantly enriched for the obtained DEGs. Furthermore, members of the chemokine family were the most abundant among the upregulated genes that were common among 2, 8 and 24 h post-reperfusion time-points, including CCL3, CCL4, CCL7 and CXCL1. Chemokines have been reported to have complex and essential roles in I/R injury, which involves extensive leukocyte and neutrophil infiltration, subsequently exaggerating the ischemic area (24). Following ischemic stroke, chemokines are primarily produced by resident microglial cells in the brain and infiltrating immune cells, which leads to further leukocyte recruitment and activation (25). In the present study, CCL2 exhibited the highest degree in the PPI network at 24 h post-reperfusion, which is consistent with previous studies (26,27). Accordingly, CCL2 mRNA expression was initially increased at 6 h post-reperfusion, peaking 2 days later. Additionally, CCL3 was previously described to be upregulated post-I/R injury via the induction of monocyte accumulation in the ischemic brain (28,29), and the expression of CCL3 post-reperfusion has been reported to be time-dependent (30). CCL7, as a mast cell-derived product, has been reported to be involved in the recruitment of inflammatory cells into the ischemic sites (31), subsequently contributing to stroke pathology (32). CXCL1, identified as a hub gene in the PPI networks at 2, 8 and 24 h post-reperfusion in the present study, was reported to be increased in the cerebrospinal fluid of patients that have suffered from a stroke (33). However, both neurotoxic and neuroprotective effects have been demonstrated for chemokines in post-stroke inflammation (28).
It is established that inflammation is a major contributor to stroke pathophysiology, and the immune system has been implicated in all stages of the ischemic cascade, from the acute damaging events to the progression of tissue repair (34). Microglia cells, which are closely associated with inflammation, were reported to become rapidly activated following ischemia (35). Furthermore, various pro-inflammatory factors, including interleukin (IL)-1β, IL-6, tumor necrosis factor-α, reactive oxygen species, nitric oxide and prostaglandin E2, were reported to be produced by activated microglia and contribute to neuronal death in cerebral ischemia (36). In the present study, specific cytokines were not measured. Further studies are required to investigate the association between the chemokine family and pro-inflammatory cytokines, which further elucidate the pathophysiological process following cerebral I/R injury.
The results of the present study revealed that the toll-like receptor signaling pathway was significantly enriched at 2 h post-reperfusion, suggesting early transcriptional activation. TLR2, a vital factor in the inflammatory response and tissue damage, has been reported to be implicated in cerebral ischemic damage (37). Microglia cells produce cytokines and chemokines following the stimulation of TLR2 (38). Furthermore, leukocyte and microglial infiltration, and neuronal death, were reported to be attenuated by TLR2 inhibition (39), indicating a potential novel therapeutic strategy.
In conclusion, the current study identified a set of DEGs that were altered between cerebral I/R injury samples and sham control samples. The findings may provide novel insight into the potential mechanisms underlying the development of cerebral I/R injury. Our future studies will be aimed at unveiling the potential diagnostic and prognostic value of these hub genes, which may ultimately aid the translation of these targets into clinical practice.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Projects of Medical and Health Technology Program in Zhejiang Province (grant no. 201482575) and the Projects of Technology Development Program in Hangzhou City (grant no. 20140633B66).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XS and WB were responsible for study design. XH, HJ and XS were responsible for data acquisition, analysis, and interpretation. WB and ZY drafted the manuscript. ZY interpreted the results. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Donnan GA, Fisher M, Macleod M, Davis SM. Stroke. Lancet. 2008;371:1612–1623. doi: 10.1016/S0140-6736(08)60694-7. [DOI] [PubMed] [Google Scholar]
- 2.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, et al. Heart disease and stroke statistics-2014 update: A report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moskowitz MA, Lo EH, Iadecola C. The science of stroke: Mechanisms in search of treatments. Neuron. 2010;67:181–198. doi: 10.1016/j.neuron.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hacke W, Kaste M, Bluhmki E, Brozman M, Dávalos A, Guidetti D, Larrue V, Lees KR, Medeghri Z, Machnig T, et al. Thrombolysis with alteplase 3 to 4.5 h after acute ischemic stroke. N Engl J Med. 2008;359:1317–1329. doi: 10.1056/NEJMoa0804656. [DOI] [PubMed] [Google Scholar]
- 5.Lansberg MG, Bluhmki E, Thijs VN. Efficacy and safety of tissue plasminogen activator 3 to 4.5 h after acute ischemic stroke: A metaanalysis. Stroke. 2009;40:2438–2441. doi: 10.1161/STROKEAHA.109.552547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung JE, Kim GS, Chen H, Maier CM, Narasimhan P, Song YS, Niizuma K, Katsu M, Okami N, Yoshioka H, et al. Reperfusion and neurovascular dysfunction in stroke: From basic mechanisms to potential strategies for neuroprotection. Mol Neurobiol. 2010;41:172–179. doi: 10.1007/s12035-010-8102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding G, Jiang Q, Li L, Zhang L, Zhang ZG, Ledbetter KA, Gollapalli L, Panda S, Li Q, Ewing JR, Chopp M. Angiogenesis detected after embolic stroke in rat brain using magnetic resonance T2*WI. Stroke. 2008;39:1563–1568. doi: 10.1161/STROKEAHA.107.502146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McColl BW, Allan SM, Rothwell NJ. Systemic infection, inflammation and acute ischemic stroke. Neuroscience. 2009;158:1049–1061. doi: 10.1016/j.neuroscience.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 9.Chamorro Á, Dirnagl U, Urra X, Planas AM. Neuroprotection in acute stroke: Targeting excitotoxicity, oxidative and nitrosative stress, and inflammation. Lancet Neurol. 2016;15:869–881. doi: 10.1016/S1474-4422(16)00114-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Chu SF, Liu DD, Zhang Z, Kong LL, Zhou X, Chen NH. Chemokines play complex roles in cerebral ischemia. Neurochem Int. 2018;112:146–158. doi: 10.1016/j.neuint.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Zhao L, Han S, Li J, Li D. Identification and functional analysis of MicroRNAs in mice following focal cerebral ischemia injury. Int J Mol Sci. 2015;16:24302–24318. doi: 10.3390/ijms161024302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MJ, Wong CH, Peng ZF, Manikandan J, Melendez AJ, Tan TM, Crack PJ, Cheung NS. A global transcriptomic view of the multifaceted role of glutathione peroxidase-1 in cerebral ischemic-reperfusion injury. Free Radic Biol Med. 2011;50:736–748. doi: 10.1016/j.freeradbiomed.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 13.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altermann E, Klaenhammer TR. PathwayVoyager: Pathway mapping using the kyoto encyclopedia of genes and genomes (KEGG) database. BMC Genomics. 2005;6:60. doi: 10.1186/1471-2164-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: A toolkit for illustrating heatmaps. PLoS One. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Liu M, Pan Y, Bai B, Chen J. Global gene expression profile of cerebral ischemia-reperfusion injury in rat MCAO model. Oncotarget. 2017;8:74607–74622. doi: 10.18632/oncotarget.22995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Yu Y, Yang J, Zhao X, Li Z. Dissecting Xuesaitong's mechanisms on preventing stroke based on the microarray and connectivity map. Mol Biosyst. 2015;11:3033–3039. doi: 10.1039/C5MB00379B. [DOI] [PubMed] [Google Scholar]
- 22.White RE, Palm C, Xu L, Ling E, Ginsburg M, Daigle BJ, Han R, Patterson A, Altman RB, Giffard RG. Mice lacking the β2 adrenergic receptor have a unique genetic profile before and after focal brain ischaemia. ASN Neuro. 2012;4 doi: 10.1042/AN20110020. pii: e00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Jiang Y, Zhu J, Wen Z, Xu X, Xu X, Xie Y, Yang L, Xu L, Lan W, et al. Orosomucoid1: Involved in vascular endothelial growth factor-induced blood-brain barrier leakage after ischemic stroke in mouse. Brain Res Bull. 2014;109:88–98. doi: 10.1016/j.brainresbull.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Chu SF, Liu DD, Zhang Z, Kong LL, Zhou X, Chen NH. Chemokines play complex roles in cerebral ischemia. Neurochem Int. 2018;112:146–158. doi: 10.1016/j.neuint.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Goazigo Réaux-Le A, Van Steenwinckel J, Rostène W, Parsadaniantz Mélik S. Current status of chemokines in the adult CNS. Prog Neurobiol. 2013;104:67–92. doi: 10.1016/j.pneurobio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Yue TL, Barone FC, Feuerstein GZ. Monocyte chemoattractant protein-1 messenger RNA expression in rat ischemic cortex. Stroke. 1995;26:661–666. doi: 10.1161/01.STR.26.4.661. [DOI] [PubMed] [Google Scholar]
- 27.Yamagami S, Tamura M, Hayashi M, Endo N, Tanabe H, Katsuura Y, Komoriya K. Differential production of MCP-1 and cytokine-induced neutrophil chemoattractant in the ischemic brain after transient focal ischemia in rats. J Leukoc Biol. 1999;65:744–749. doi: 10.1002/jlb.65.6.744. [DOI] [PubMed] [Google Scholar]
- 28.Mirabelli-Badenier M, Braunersreuther V, Viviani GL, Dallegri F, Quercioli A, Veneselli E, Mach F, Montecucco F. CC and CXC chemokines are pivotal mediators of cerebral injury in ischaemic stroke. Thromb Haemost. 2011;105:409–420. doi: 10.1160/TH10-10-0662. [DOI] [PubMed] [Google Scholar]
- 29.Cowell RM, Xu H, Galasso JM, Silverstein FS. Hypoxic-ischemic injury induces macrophage inflammatory protein-1alpha expression in immature rat brain. Stroke. 2002;33:795–801. doi: 10.1161/hs0302.103740. [DOI] [PubMed] [Google Scholar]
- 30.Boddeke EW, Meigel I, Frentzel S, Gourmala NG, Harrison JK, Buttini M, Spleiss O, Gebicke-Härter P. Cultured rat microglia express functional beta-chemokine receptors. J Neuroimmunol. 1999;98:176–184. doi: 10.1016/S0165-5728(99)00096-X. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Li X, Yaish-Ohad S, Sarau HM, Barone FC, Feuerstein GZ. Molecular cloning and expression of the rat monocyte chemotactic protein-3 gene: A possible role in stroke. Brain Res Mol Brain Res. 1999;71:304–312. doi: 10.1016/S0169-328X(99)00203-X. [DOI] [PubMed] [Google Scholar]
- 32.Arac A, Grimbaldeston MA, Nepomuceno AR, Olayiwola O, Pereira MP, Nishiyama Y, Tsykin A, Goodall GJ, Schlecht U, Vogel H, et al. Evidence that meningeal mast cells can worsen stroke pathology in mice. Am J Pathol. 2014;184:2493–2504. doi: 10.1016/j.ajpath.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Losy J, Zaremba J, Skrobanski P. CXCL1 (GRO-alpha) chemokine in acute ischaemic stroke patients. Folia Neuropathol. 2005;43:97–102. [PubMed] [Google Scholar]
- 34.Iadecola C, Anrather J. The immunology of stroke: From mechanisms to translation. Nat Med. 2011;17:796–808. doi: 10.1038/nm.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schilling M, Besselmann M, Leonhard C, Mueller M, Ringelstein EB, Kiefer R. Microglial activation precedes and predominates over macrophage infiltration in transient focal cerebral ischemia: A study in green fluorescent protein transgenic bone marrow chimeric mice. Exp Neurol. 2003;183:25–33. doi: 10.1016/S0014-4886(03)00082-7. [DOI] [PubMed] [Google Scholar]
- 36.Jin R, Yang G, Li G. Inflammatory mechanisms in ischemic stroke: Role of inflammatory cells. J Leukoc Biol. 2010;87:779–789. doi: 10.1189/jlb.1109766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abe T, Shimamura M, Jackman K, Kurinami H, Anrather J, Zhou P, Iadecola C. Key role of CD36 in Toll-like receptor 2 signaling in cerebral ischemia. Stroke. 2010;41:898–904. doi: 10.1161/STROKEAHA.109.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marsh BJ, Williams-Karnesky RL, Stenzel-Poore MP. Toll-like receptor signaling in endogenous neuroprotection and stroke. Neuroscience. 2009;158:1007–1020. doi: 10.1016/j.neuroscience.2008.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziegler G, Freyer D, Harhausen D, Khojasteh U, Nietfeld W, Trendelenburg G. Blocking TLR2 in vivo protects against accumulation of inflammatory cells and neuronal injury in experimental stroke. J Cereb Blood Flow Metab. 2011;31:757–766. doi: 10.1038/jcbfm.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.