Abstract
Autophagy, part of the innate immune defense mechanisms, is activated during the initial phase of septic insult. Previous studies indicated that micro (mi)RNAs are additionally involved in the host response to sepsis; however, the association between miRNAs and autophagy during this process is not fully understood. To study the role of miRNA (miR)-23a in autophagy initiated by sepsis, macrophages treated with lipopolysaccharides, in addition to blood samples from patients, were evaluated for miR-23a expression levels. Cell viability, inflammatory mediators and autophagic markers were investigated following overexpression or inhibition of miR-23a. The results suggested that miR-23a was suppressed subsequent to septic insult, promoting autophagy and suppressing a hyper inflammatory response, leading to enhanced cell viability. A luciferase assay and western blot analysis confirmed ubiquitin-like protein ATG12 to be the target of miR-23a. The present study revealed that the downregulation of miR-23a regulates an inflammatory response during septic insult via autophagy promotion.
Keywords: autophagy, sepsis, miRNA, innate immune response
Introduction
Sepsis is the third most common disease in the USA, causing ~300,000 cases of mortality annually (1). Despite the availability of various potent antibiotics, the mortality rates of sepsis have failed to reduce (2), suggesting that a more comprehensive approach is required in its management. Genome wide expression studies indicated that pro- and anti-inflammatory cytokines encoding genes were upregulated during the host immune response (3), and the disruption of this balance may lead to hyper-inflammation (4) and immunosuppression (5).
Autophagy is an evolutionary conservative process critical for cell survival under stress (6). A microarray assay demonstrated that autophagy related gene lysosomal associated membrane protein 1 was upregulated in patients with sepsis compared with patients with non-infectious systemic inflammatory response syndrome (SIRS), which is defined as the SIRS associated with previously existing non-infectious diagnosis, while no sign of ongoing infection was observed (7). Polymorphisms of the immunity related GTPase M gene, which is autophagy-associated, was additionally associated with mortality due to sepsis, suggesting a protective role for autophagy (8). To investigate this hypothesis, various factors potentially regulating autophagic activities were studied, including microRNA (miRNA).
miRNAs are a group of short single-stranded RNA, ~21 nucleotides in length. miRNAs cause the degradation of target mRNA by selectively binding to its 3′untranslated region (UTR), fine-tuning gene expression post-transcriptionally (9). Previous studies demonstrated that the expression profile of microRNAs differs significantly between patients with sepsis and patients with non-infectious SIRS (10). A similar alteration was additionally confirmed in microarray studies focused on macrophages stimulated by lipopolysaccharide (LPS) (11). The two aforementioned studies demonstrated miR-23a downregulation in sepsis; however, the underlying mechanism for the involvement of miR-23a in sepsis response remains unclear.
In the present study, microRNA expression profiling data from previous studies were adopted and analyzed. Additionally, the downregulation of miR-23a during septic insult was confirmed with in vivo and in vitro experiments. Further study on the acting mechanism of miR-23a was conducted, aiming to better understand the molecular mechanism for sepsis response.
Materials and methods
Gene expression omnibus (GEO) data analysis
To identify a group of microRNAs that shared a similar expression pattern following LPS stimulation, clustering analysis on a dataset from the GEO database (12) was conducted. The dataset (accession no. GSE55414) contains expression profile from a previous study (11) was adopted in the present study. This dataset (accession no. GSE55414) evaluated miRNA levels at different time points following LPS stimulation. The initial screening of data was performed using Linear Models of Microarray Analysis 3.3 in R 3.4.2 (13), with a false discovery rate and minimal log fold change set as 0.05 and 1, respectively. Clustering analysis of the filtered microarray expression data was conducted with heatmap 1.0.8 (14) utilizing the Euclidean distance algorithm.
Cell culture
To conduct the in vitro study on macrophages, the RAW264.7 macrophage cell line was obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM, Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.) at 37°C. The 3rd passage of cells were used for further experiments.
Cell transfection
Manipulation of the miRNA (miR)-23a expression level was achieved by oligonucleotide transfection. RAW264.7 cells were cultured in six-well plates at a concentration of 2×105/well prior to transfection. miR-23a mimics (100 nM; 5′-AUCACAUUGCCAGGGAUUUCC-3′) and miR-23a inhibitor (100 nM; 5′-GUGGUAAUCCCUGGCAAUGUGAU-3′) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China), and a miRNA negative control was purchased from Exiqon, Inc. (10 nM, www.qiagen.com/us/shop/; cat. no. 479903; Woburn, MA, USA). A Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) system was employed for microRNA transfection following the manufacturer's protocol. Following incubation at 37°C for 1 day, three groups of cells were generated, denoted as the control, miR-23a mimics and miR-23a inhibitor groups, respectively. The expression level of miR-23a was confirmed in all three groups of cells with a reverse transcription-quantitative polymerase chain reaction (RT-qPCR), the detailed protocol was described later in this article.
Immunofluorescence staining
To study the influence of the miR-23a expression level on autophagy activity, immunofluorescence imaging of autophagy markers was employed. RAW264.7 macrophages were fixed with 4% formaldehyde for 15 min at room temperature and incubated in 5% Tris buffered saline with Tween-20 (TBS-T; pH 8.3) diluted non-fat dry milk for 1 h. Immunofluorescence staining was performed using Microtubule-associated protein light chain 3 (LC-3) primary antibody (1:100; Abcam, Cambridge, UK; cat. no. ab62720) incubation overnight at 4°C and subsequent secondary antibody (1:200; Alexa Fluor 488 anti-rabbit IgG; Thermo Fisher Scientific, Inc.; cat. no. A10235) incubation at room temperature for 1 h. Cells were incubated using DAPI (100 ng/ml) for nuclear staining at room temperature for 30 min. Images were obtained from a confocal laser scanning microscope (LSM710; Zeiss, Oberkochen, Germany) at magnification, ×600. LC3 puncta were quantified with Image-J 1.51 (National Institutes of Health, Bethesda, MD, USA) (15).
Blood sampling and miRNA isolation
To further confirm the association between miR-23a levels and the host response to sepsis in the clinical setting, blood samples were collected from 27 patients with sepsis and 22 patients with non-infectious SIRS admitted to the SICU department of The First Affiliated Hospital of Sun Yat-sen University (Guangzhou, China) during the period of June 2016 to February 2017. Patients under the age of 18 or for whom it was impossible to obtain informed consent within 6 h of admission were excluded. Patients were categorized as sepsis or non-infectious SIRS using standard criteria (16), and a blood microbial culture assay result was obtained from all patients as required by the standard criteria (17). Demographic data and Sequential Organ Failure Assessment scores are listed in Table I, revealing no statistically significant difference. miRNA isolation from plasma samples was conducted using a mirVana kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Informed consent was obtained from all individual participants included in the present study. The present study was approved by the Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University.
Table I.
Demographic data and SOFA scores of patients included in the present study.
| Group | Age, years | Statistic | P-value | Male/female ratio | Statistic | P-value | SOFA score | Statistic | P-value |
|---|---|---|---|---|---|---|---|---|---|
| Sepsis, n=27 | 62.3±10.3 | −1.768a | 0.107 | 13:14 | 0.025b | 0.874 | 8.1±2.3 | 1.299a | 0.215 |
| Non-infectious SIRS, | 64.2±11.5 | 12:10 | 7.92±2.6 | ||||||
| n=22 | |||||||||
Independent t-test
χ2test. SOFA, Sequential Organ Failure Assessment; SIRS, systemic inflammatory response syndrome.
ELISA
An ELISA was employed to evaluate the inflammatory response of macrophages subjected to LPS stimulation. RAW264.7 cells were seeded on 24-well plates at 4×104/well. Following LPS (10 µg/ml, Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) stimulation for 4 h in room temperature, the cell culture supernatant was collected. Subsequently, inflammatory cytokine levels of interleukin-6 (IL-6) and tumor necrosis factor (TNF)-α were measured with ELISA kits (cat. nos. 550950 and 560478; BD Biosciences, Franklin Lakes, NJ, USA), following the manufacturer's protocol.
Western blot analysis
Protein expression levels were evaluated with western blot analysis. Cells were seeded on 6 well plates at a density of 1×106 per well. Western blotting was conducted following the protocol reported in a previous study (18). Total protein extracts were obtained with a radioimmunoprecipitation assay and transferred to nitrocellulose membranes. The membranes were incubated with blocking solution overnight, and primary antibodies were subsequently applied and incubated at room temperature for 2 h. Then the membranes were incubated with secondary antibody at room temperature for 1h. Primary antibodies employed in the present study included LC3 (1:1,000; cat. no. ab48394; Abcam), Beclin-1 (1:1,000; cat. no. 3495; Cell Signaling Technology, Inc., Danvers, MA, USA), ATG12 (1:1,000; cat. no. ab155589) and Sequestosome-1 (p62; 1:1,000; cat. no. ab91526; both Abcam), and β-actin (1:1,000; cat. no. sc-130657; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was used as a loading control. HRP-conjugated anti-rabbit (1:5,000; cat. no. ab205718) was used as secondary antibody. Image-J 1.51 was used for densitometry analysis.
RT-qPCR analysis
RNA expression levels were evaluated with RT-qPCR. Total RNA was extracted with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed to cDNA with the QuantiTect RT kit (Qiagen, Inc., Valencia, CA, USA). RT-qPCR was performed with a SYBR Mix (Qiagen, Inc) and the Bio-Rad Real-time PCR System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Following a protocol used in a previous study (19), these cycling parameters were used: initial denaturation at 95°C for 30 sec, followed by 35 cycles: denaturation at 95°C for 10 sec, annealing at 55°C for 10 sec and extension at 72°C for 30 sec. The expression level of mRNAs were first normalized to the GAPDH mRNA level, and the 2−ΔΔCq method (20) was used for quantification. The relative expression level was compared between the three groups of cells. For microRNAs, the U6 level was used as the internal control. AlleleID 6 (Palo Alto, CA, USA, www.premierbiosoft.com/) was used for primer design and sequences are provided in Table II.
Table II.
Primers for reverse transcription-quantitative polymerase chain reaction analysis with the SYBR green system.
| mRNA | Sense (5′-3′) | Anti-sense (5′-3′) |
|---|---|---|
| TNF-α | GTGAGGAGGACGAACATC | GAGCCAGAAGAGGTTGAG |
| IL-6 | TGACCCAACCACAAATGC | TGACCAGAAGAAGGAATGC |
| GAPDH | TCATCCCTGCCTCTACTG | TGCTTCACCACCTTCTTG |
TNF, tumor necrosis factor; IL, i nterleukin.
Luciferase reporter assays
To confirm the targeting association between miR-23a and ATG12 mRNA, luciferase reporter assays were conducted. The PsiCHECK-2 system (Promega Corporation, Madison, WI, USA) was used for the construction of the dual-luciferase assay plasmid. Bioinformatics analysis tool miRanda (microrna.org/) revealed a potential binding sequence for miR-23a in the 3′UTR of ATG12 mRNA. The predicted target sequence was cloned into the PsiCHECK-2 plasmid (Promega), generating an ATG12 wild-type (wt) dual-luciferase reporter plasmid. Subsequently, the mutated target sequence was generated with site-directed mutagenesis and was additionally cloned into the PsiCHECK-2 plasmid system, creating an ATG12 mutant (mut) reporter plasmid. RAW264.7 cells with miR-23a mimics, miR-23a inhibitor and negative controls were first seeded on 96-well plates at a concentration of 1×104/well and transfected with ATG12 wt or ATG12 mut (200 ng/ml) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activities were evaluated with the Modulus™ dual-luciferase reporter assay system (Turner Designs, Sunnyvale, CA, USA) following 48 h incubation, normalized to Renilla activity. and all assays were performed in triplicate.
Cell viability assay
A cell viability assay was performed to study the influence of the miR-23a expression level on cell survival subsequent to LPS stimulation. All three groups of cells were seeded on 96-well plates at 5×103 cells per well. A Cell Counting kit-8 (Engreen Biosystem, Ltd., Auckland, New Zealand) was used for cell viability evaluation, and an absorbance value at 450 nm was used for the quantification of cell viability.
Statistical analysis
All experiments were repeated three times and data are expressed as the mean ± standard deviation. Comparisons between groups were made using one-way analysis of variance, and Fisher's Least Significant Difference test was used for post hoc analysis. Baseline data for patients in sepsis or Non-infectious SIRS groups was compared, independent t-test was used for SOFA score and age comparison, while χ2 test was used for sex ratio comparison. P<0.05 was considered to indicate a statistically significant difference. R studio 1.1.383 (RStudio, Inc., Boston, MA, USA) was used for statistical analysis.
Results
miRNA expression levels in LPS-stimulated macrophages
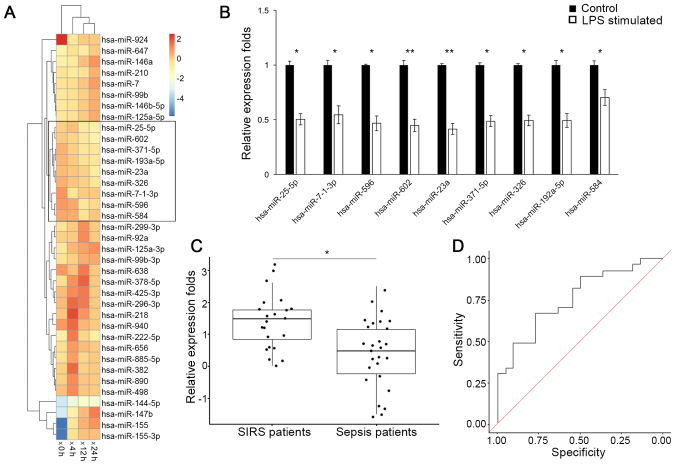
A group of miRNAs that were consistently down regulated following LPS stimulation were identified (Fig. 1A) via clustering analysis on the GEO dataset (GSE55414). Expression levels of these miRNAs were further validated by RT-qPCR analysis in RAW264.7 cells (Fig. 1B), among which the down regulation of miR-23a was the most significant (P<0.01). Consistently, RT-qPCR analysis of circulating miRNAs (Fig. 1C) additionally demonstrated that the miR-23a serum level was decreased in patients with sepsis (0.524; 95% confidence interval (CI), 0.036–1.012), compared with patients with non-infectious SIRS (1.557; 95% CI, 1.191–1.923) with a statistical significance (P<0.05). The receiver operating characteristic curve analysis on the miR-23a level yielded an area under the curve value of 0.758 (Fig. 1D), and at the threshold of 1.01 for miR-23a relative expression level, a sensitivity and specificity of 70.3 and 68.3%, respectively, was achieved for differentiating between patients with sepsis and patients with non-infectious SIRS. These results demonstrated that miR-23a is downregulated in response to sepsis insult.
Figure 1.
Expression analysis of miRNAs during the sepsis response. (A) Heat map for clustering analysis of miRNA expression data; 0, 4, 12 and 24 h are the times elapsed following LPS stimulation. (B) Relative expression fold changes of miRNAs following LPS stimulation. Expression levels prior to LPS administration were used as reference values. (C) Relative expression fold-changes of miR-23a in serum from patients with non-infectious SIRS or sepsis. (D) Receiver operating characteristic curve for the measurement of miR-23a in serum to differentiate between non-infectious SIRS and sepsis. *P<0.05, **P<0.01 vs. respective control. miRNA/miR, microRNA; LPS, lipopolysaccharide; SIRS, systemic inflammatory response syndrome.
Downregulation of miR-23a protects cells by modulating inflammatory mediators
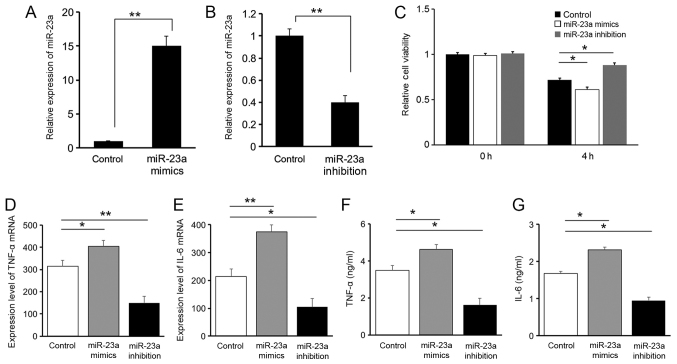
Results of the RT-qPCR analysis on miR-23a expression levels suggested that oligonucleotide transfection was successfully conducted in all three groups of RAW264.7 cells (Fig. 2A and B). The Cell Counting kit-8 result suggested that 4 h following LPS stimulation, the cell viability of RAW264.7 cells decreased in all three groups of cells. Compared with the control group, the cell viability in the miR-23a mimics group was significantly decreased (P<0.05), while inhibition of miR-23a demonstrated a protective effect with an increased cell viability (P<0.05), suggesting that the downregulation of miR-23a is protective for cells following LPS stimulation (Fig. 2C).
Figure 2.
Influence of miR-23a on inflammatory mediators. The expression level of miR-23a in the negative control was compared with (A) the miR-23a mimics and (B) the miR-23a inhibition groups. miR-23a expression in the negative control group was used as reference. (C) Relative cell viability was assessed by Cell Counting kit-8 analysis. The expression levels of (D) TNF-α and (E) IL-6 were measured. ELISA was used to measure the concentrations of (F) TNF-α and (G) IL-6. *P<0.05, **P<0.01. miR, microRNA; TNF, tumor necrosis factor; IL, interleukin.
Expression levels of the pro-inflammatory cytokines IL-6 and TNF-α were evaluated with RT-qPCR (Fig. 2D and E) and ELISA (Fig. 2F and G). A positive association was identified between miR-23a and IL-6 or TNF-α levels, suggesting a pro-inflammatory role for miR-23a. These results indicated that the protective effect of miR-23a downregulation following LPS stimulation is associated with the regulation of inflammatory factors.
Effects of mir-23a on autophagic activity
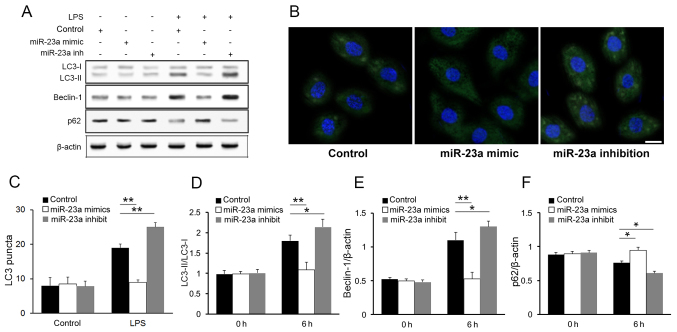
The western blot analysis of the autophagy markers demonstrated an increase in the LC3 conversion (LC3-II/LC3-I ratio) (Fig. 3A) in the miR-23a inhibition group compared with the control group. Immunofluorescence imaging and LC3 puncta quantification further confirmed that miR-23a inhibited autophagic activity in macrophages following LPS stimulation (Fig. 3B and C). The result of western blot analysis also shown that other autophagy markers were influenced by miR-23a level after LPS stimulation (Fig. 3A). Compared with control group, beclin-1 was upregulated while p62 was downregulated in miR-23a inhibited cells. Such negative association between miR-23a level and autophagy markers was further confirmed with densitometry analysis (Fig. 3D-F; P<0.05), while no significant association was found between them in macrophages prior to LPS stimulation. These result is consistent with a previous study (21).
Figure 3.
Modulation of autophagy by miR-23a. (A) Western blotting results for autophagy markers. (B) LC3 immunofluorescence staining results subsequent to LPS stimulation. Scale bar, 20 µm. Densitometry analysis of the autophagy markers (C) LC3-II/LC3-I, (D) beclin-1 and (E) p62. (F) LC3 puncta quantification of immunofluorescence results. Blue staining represents nuclei; green staining represents LC3. *P<0.05, **P<0.01. LPS, lipopolysaccharide; miR, microRNA; LC3-II/LC3-I, microtubule-associated protein light chain 3 conversion ratio.
miR-23a targets ATG12 specifically to modulate autophagic activity
ATG12 serves an integral role in the autophagy pathway by forming a complex with Autophagy protein 5(ATG5) (22). Bioinformatics analysis tools miRanda (microrna.org/) suggested that miR-23a may bind to ATG12 selectively via a specific sequence in the 3′UTR of ATG12 mRNA (Fig. 4A). A dual-luciferase assay was conducted for confirmation and a negative association was identified between luciferase activities and miR-23a levels in cells transfected with the ATG12-wt plasmid; however, no significant difference was observed between groups when ATG12-mut plasmids were applied (P>0.05), suggesting that miR-23a targets ATG12 specifically (Fig. 4B). Downregulation of ATG12 expression by miR-23a was further confirmed with western blot analysis (Fig. 4C).
Figure 4.
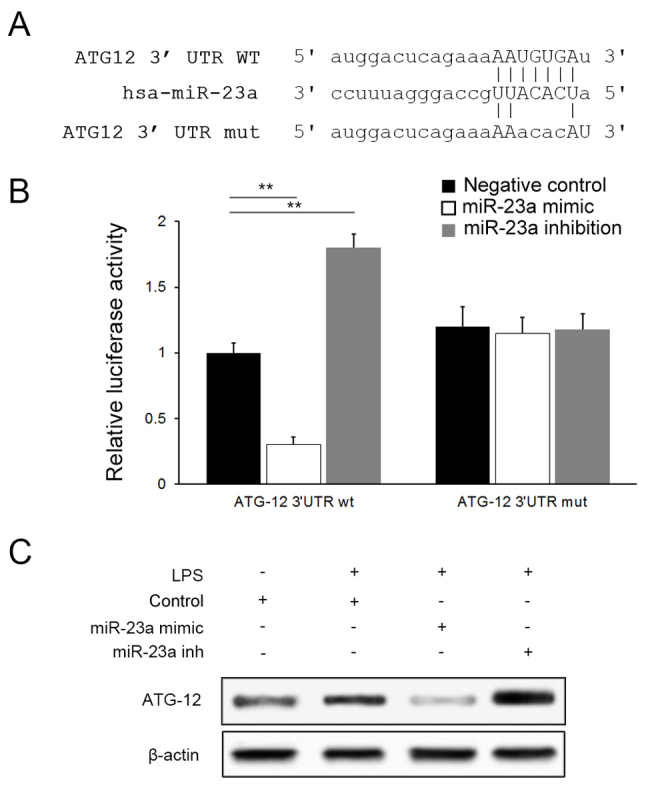
Luciferase and western blot analysis indicated ATG12 to be the target of miR-23a. (A) Predicted binding site for miR-23a in 3′UTR of ATG12 mRNA, and the mutated sequence. (B) Luciferase activity comparison between ATG12-wt and ATG12-mut plasmid. (C) Western blot analysis of ATG12 in the negative control, miR-23a mimics and miR-23a inhibition groups. **P<0.01. ATG12, ubiquitin-like protein ATG12; miR, miRNA; UTR, untranslated region; wt, wild-type; mut, mutant.
Discussion
Sepsis is a principal challenge in the management of critically ill patients (1). The incomplete understanding of mechanisms underlying organ damage and immune defense during septic insult hinder the development of more comprehensive management (23). Accumulating evidence suggests that autophagic activity is increased during the initial phase of sepsis (24,25) and LPS, a key bacterial product, may initiate autophagy through toll-like receptor 2 and toll-like receptor 4 activation (26,27). Two studies indicated a protective role for such a response (24,28); however, the detailed underlying mechanism requires further investigation.
miRNA are a group of small non-coding RNAs that act as post-transcriptional regulators in various processes. The results of the present study demonstrated that multiple microRNAs inhibiting autophagy (29–32) were downregulated subsequent to LPS stimulation. Among these, the downregulation of miR-23a was the most significant. The in vivo study additionally demonstrated that the miR-23a serum level in patients with sepsis differed distinctly compared with patients with non-infectious SIRS, a result consistent with previous studies (33,34). These results collectively demonstrate that miR-23a is involved in the host response to sepsis.
The present results from the western blot analysis and immunofluorescence studies demonstrated for the first time, to the best of the authors' knowledge, that miR-23a is negatively associated with autophagic activities following septic insult. Downregulation of miR-23a mitigates the inhibition of autophagy, leading to the suppression of inflammatory mediators, a role proposed in a previous study for autophagy in sepsis response (35).
The present bioinformatics study and dual luciferase analysis indicated that miR-23a suppressed autophagy via ATG12. The targeting association was demonstrated with the dual-luciferase study, and a negative association was revealed between the miR-23a and ATG12 expression levels. The inhibitory effect that miR-23a exerts on ATG12 was further demonstrated by the LC3-II/LC3-I ratio and immunofluorescence imaging in the present study, since ATG12, along with autophagy protein 5 and autophagy-related protein 16-1 comprise the E3 ubiquitin ligase, which facilitates LC3 family conversion from LC3-I to LC3-II (22).
In conclusion, the present study demonstrated that miR-23a was downregulated during initial septic insult. It was demonstrated for the first time, to the best of the authors' knowledge, that miR-23a downregulation promoted the autophagic activity of macrophages in response to LPS stimulation, and this consequently suppressed inflammatory mediators, preventing an overwhelming inflammatory response. Finally, it was demonstrated that miR-23a may selectively bind to the 3′UTR of ATG12 mRNA, modulating the formation of the E3 ligase and the subsequent facilitation of LC3 conversion.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- DAPI
4′,6-diamidino-2-phenylindole
- GEO
gene expression omnibus
- IL
interleukin
- LPS
lipopolysaccharide
- miRNA/miR
microRNA
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- SIRS
systemic inflammatory response syndrome
- TNF
tumor necrosis factor
- UTR
untranslated region
Funding
The present study was supported by grants from the Fundamental Research Funds for the Central Universities (grant no. 15ykpy14) and Sun Yat-sen University Clinical Research 5010 Program (grant no. 2007015).
Availability of data and materials
The data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XS and DC carried out the statistical analysis and drafted this article. JC and YN were responsible for blood sampling and data analysis. ZJ, M-YC and J-FW carried out western blot and RT-qPCR analyses. X-DG conceived the idea and provided guidance.
Ethics approval and consent to participate
Informed consent was obtained from all individual participants included in the present study. The present study was approved by the Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University [approval no. (2016)025].
Consent for publication
Not applicable.
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.Vincent JL, Marshall JC, Namendys-Silva SA, François B, Martin-Loeches I, Lipman J, Reinhart K, Antonelli M, Pickkers P, Njimi H, et al. Assessment of the worldwide burden of critical illness: the Intensive Care Over Nations (ICON) audit. Lancet Respir Med. 2014;2:380–386. doi: 10.1016/S2213-2600(14)70061-X. [DOI] [PubMed] [Google Scholar]
- 2.Russell JA. Management of sepsis. N Engl J Med. 2006;355:1699–1713. doi: 10.1056/NEJMra043632. [DOI] [PubMed] [Google Scholar]
- 3.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, et al. A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadav H, Cartinceba R. Balance between hyperinflammation and immunosuppression in sepsis. Semin Respir Crit Care Med. 2016;37:42–50. doi: 10.1055/s-0035-1570356. [DOI] [PubMed] [Google Scholar]
- 5.Davis CG, Chang K, Osborne D, Walton AH, Dunne WM, Muenzer JT. Increased susceptibility to Candida infection following cecal ligation and puncture. Biochem Biophys Res Commun. 2011;414:37–43. doi: 10.1016/j.bbrc.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H, Zhang Z, Wang X, Li R, Hou W, Bi W, Zhang X. Inhibition of autophagy induces IL-1β release from ARPE-19 cells via ROS mediated NLRP3 inflammasome activation under high glucose stress. Biochem Biophys Res Commun. 2015;463:1071–1076. doi: 10.1016/j.bbrc.2015.06.060. [DOI] [PubMed] [Google Scholar]
- 7.McHugh L, Seldon TA, Brandon RA, Kirk JT, Rapisarda A, Sutherland AJ, Presneill JJ, Venter DJ, Lipman J, Thomas MR, et al. A molecular host response assay to discriminate between sepsis and infection-negative systemic inflammation in critically ill patients: discovery and validation in independent cohorts. PLoS Med. 2015;12:e1001916. doi: 10.1371/journal.pmed.1001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura T, Watanabe E, Sakamoto T, Takasu O, Ikeda T, Ikeda K, Kotani J, Kitamura N, Sadahiro T, Tateishi Y, et al. Autophagy-related IRGM polymorphism is associated with mortality of patients with severe sepsis. PLoS One. 2014;9:e91522. doi: 10.1371/journal.pone.0091522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, Zhu KM. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394:184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 10.Caserta S, Kern F, Cohen J, Drage S, Newbury SF, Llewelyn MJ. Circulating plasma microRNAs can differentiate human sepsis and systemic inflammatory response syndrome (SIRS) Sci Rep. 2016;6:28006. doi: 10.1038/srep28006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie N, Cui H, Banerjee S, Tan Z, Salomao R, Fu M, Abraham E, Thannickal VJ, Liu G. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J Immunol. 2014;193:327–334. doi: 10.4049/jimmunol.1400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolde R. pheatmap: Pretty Heatmaps. 2015 [Google Scholar]
- 15.Schindelin J, Argandacarreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL, Ramsay G. SCCM/ESICM/ACCP/ATS/SIS: 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 17.Llewelyn MJ, Berger M, Gregory M, Ramaiah R, Taylor AL, Curdt I, Lajaunias F, Graf R, Blincko SJ, Drage S, Cohen J. Sepsis biomarkers in unselected patients on admission to intensive or high-dependency care. Crit care. 2013;17:R60. doi: 10.1186/cc12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S, Liu W, Lei Z. MiR-590-3p regulates osteogenic differentiation of human mesenchymal stem cells by regulating APC gene. Biochem Biophys Res Commun. 2016;478:1582–1587. doi: 10.1016/j.bbrc.2016.08.160. [DOI] [PubMed] [Google Scholar]
- 19.Xiang L, Chen M, He L, Cai B, Du Y, Zhang X, Zhou C, Wang C, Mao JJ, Ling J. Wnt5a regulates dental follicle stem/progenitor cells of the periodontium. Stem Cell Res Ther. 2014;5:135. doi: 10.1186/scrt525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Oh JE, Lee HK. Pattern recognition receptors and autophagy. Front Immunol. 2014;5:300. doi: 10.3389/fimmu.2014.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinatsu O, Zoltan M, Giichi T, Takanori O. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20:59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marik PE. Early management of severe sepsis: Concepts and controversies. Chest. 2014;145:1407–1418. doi: 10.1378/chest.13-2104. [DOI] [PubMed] [Google Scholar]
- 24.Lin CW, Lo S, Perng DS, Wu DB, Lee PH, Chang YF, Kuo PL, Yu ML, Yuan SS, Hsieh YC. Complete activation of autophagic process attenuates liver injury and improves survival in septic mice. Shock. 2014;41:241–249. doi: 10.1097/SHK.0000000000000111. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi W, Watanabe E, Fujimura L, Watanabe-Takano H, Yoshidome H, Swanson PE, Tokuhisa T, Oda S, Hatano M. Kinetics and protective role of autophagy in a mouse cecal ligation and puncture-induced sepsis. Crit Care. 2013;17:R160. doi: 10.1186/cc11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yunanto A, Endharti AT, Widodo A. Neutrophil, TLR2, and TLR4 expression in newborns at risk of sepsis. Paediatrica Indonesiana. 2013;53:132–137. doi: 10.14238/pi53.3.2013.132-7. [DOI] [Google Scholar]
- 27.Wang J, Feng X, Zeng Y, Fan J, Wu J, Li Z, Liu X, Huang R, Huang F, Yu X, Yang X. Lipopolysaccharide (LPS)-induced autophagy is involved in the restriction of Escherichia coli in peritoneal mesothelial cells. Bmc Microbiol. 2013;13:255. doi: 10.1186/1471-2180-13-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin CW, Lo S, Hsu C, Hsieh CH, Chang YF, Hou BS, Kao YH, Lin CC, Yu ML, Yuan SS, Hsieh YC. T-Cell autophagy deficiency increases mortality and suppresses immune responses after sepsis. PLoS One. 2014;9:e102066. doi: 10.1371/journal.pone.0102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie T, Zhang J, Peng C, Lin Y, Chen J. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013–7026. doi: 10.18632/oncotarget.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang YX, Li L. Identification of potential biomarkers of sepsis using bioinformatics analysis. Exp Ther Med. 2017;13:1689–1696. doi: 10.3892/etm.2017.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou W, Wang J, Li Z, Li J, Sang M. MicroRNA-205-5b inhibits HMGB1 expression in LPS-induced sepsis. Int J Mol Med. 2016;38:312–328. doi: 10.3892/ijmm.2016.2613. [DOI] [PubMed] [Google Scholar]
- 32.Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu Y, Yi X, Ma J, Zhao T, Liu L, et al. Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics. 2017;7:2231–2249. doi: 10.7150/thno.18835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge QM, Huang CM, Zhu XY, Bian F, Pan SM. Differentially expressed miRNAs in sepsis-induced acute kidney injury target oxidative stress and mitochondrial dysfunction pathways. PLoS One. 2017;12:e0173292. doi: 10.1371/journal.pone.0173292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Liu C, Wang Z, Huang J, Zeng Q. microRNA-23a-5p acts as a potential biomarker for sepsis-induced acute respiratory distress syndrome in early stage. Cell Mol Biol (Noisy-le-grand) 2016;62:31–37. [PubMed] [Google Scholar]
- 35.Giegerich AK, Kuchler L, Sha LK, Knape T, Heide H, Wittig I, Behrends C, Brüne B, von Knethen A. Autophagy-dependent PELI3 degradation inhibits proinflammatory IL1B expression. Autophagy. 2014;10:1937–1352. doi: 10.4161/auto.32178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.