 Biscarbene gold(i) complexes were prepared and evaluated for their effects against cancer cells and pathogenic bacteria.
Biscarbene gold(i) complexes were prepared and evaluated for their effects against cancer cells and pathogenic bacteria.
Abstract
A series of gold(i) complexes with two N-heterocyclic carbene ligands (biscarbene gold complexes) were prepared and evaluated for their effects against cancer cells and pathogenic bacteria. Proliferation inhibition was observed in cancer cells and in Gram-positive bacteria, whereas Gram-negative bacteria were less sensitive towards the compounds. The protein binding and cellular uptake were quantified and the combined results indicated a strong correlation between cellular bioavailability and antiproliferative effects. The biscarbene gold complexes inhibited bacterial and mammalian TrxRs with low to moderate potency. However, based on the obtained structure–activity-relationships and the high cellular accumulation levels, TrxR inhibition can be considered as a relevant contributor to the cellular pharmacology of biscarbene gold(i) complexes.
Introduction
Gold complexes have attracted major attention in inorganic medicinal chemistry and are currently considered as metallodrug candidates for several possible therapeutic applications including cancer or infectious diseases.1–5 The history of gold in medicine traces back thousands of years, and the element had been of high relevance in alchemy.6 In modern science, the probably first report on the antibacterial effects of gold salts was published by the famous bacteriologist Robert Koch,7 and some gold complexes are nowadays used therapeutically in the treatment of rheumatoid arthritis. Moreover, the strong antiproliferative properties of many gold complexes indicate their potential as anticancer agents, which have currently been evaluated in several ongoing clinical trials with auranofin.
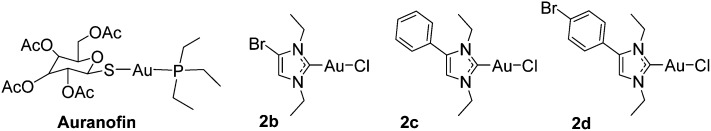
Regarding gold metallodrug design, the element has been most frequently used in the oxidation state +1 with thiolate and phosphane ligands (which are present in the lead compound auranofin, see Fig. 1). However, traditional gold drugs face major challenges regarding their stability, as these ligands are readily replaced under physiological conditions.1,2,8 More recently, organometallic gold complexes have attracted the attention of inorganic medicinal chemists, as they offer options for a more robust coordination of ligands to the gold central atom. Among them, N-heterocyclic carbene ligands are especially promising, because antiproliferative effects, apoptosis induction, inhibition of thioredoxin reductase (TrxR), antibacterial properties and other biological effects have been reported for many derivatives.9–27 Recently, we described a series of chlorido gold(i) NHC complexes with anticancer and antibacterial properties.17 The complexes were effective inhibitors of both mammalian and bacterial TrxRs. Of particular importance were the Gram-positive directed antibacterial effects, which might be the consequence of the high dependence of these bacteria on an intact Trx/TrxR system. Their dual activity against bacterial and mammalian TrxRs, together with the observed cytotoxic and antibacterial effects, provides the rationale to study gold complexes as both cytotoxic and antibacterial agents. In this report, we extend these studies to biscarbene compounds containing two NHC ligands. The cytotoxicity, cellular uptake, antibacterial effects, inhibition of mammalian and bacterial TrxRs, and the resulting structure–activity-relationships are presented.
Fig. 1. Auranofin and examples of previously reported gold(i) NHC complexes.17.
Chemistry
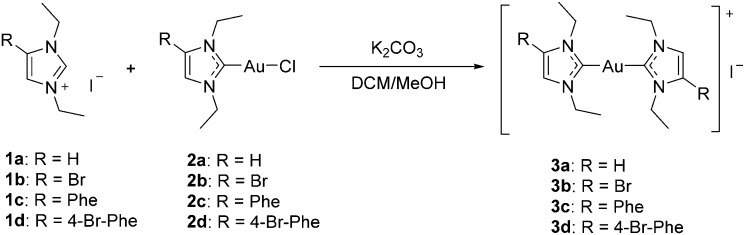
Starting from the imidazolium cations 1a–d, the chlorido gold(i) NHC complexes 2a–d were prepared as recently described.17 The target compounds 3a–d were obtained by reacting 2a–d with the respective imidazolium cations. All complexes were characterized by 1H-, 13C-NMR, and MS spectroscopy, and high purity of all target compounds was noted by elemental analysis. Mass spectrometry confirmed the presence of the respective [M – I]+ and [I]– ions and showed the expected isotope pattern in the case of the bromine containing compounds. In comparison to the mono-NHC complexes (2a–d), no significant changes could be observed in the 1H-NMR spectra of 3a–d. In the 13C-NMR spectra of complexes 3a–d the signal for the carbon at position 2, which is coordinated to gold(i), is observed at significantly higher ppm values (approx. +12–13 ppm) than that in the mono-NHC precursors 2a–d (Scheme 1).17
Scheme 1. Synthesis of NHC–Au–NHC complexes.
Cytotoxicity and bacterial growth inhibition
The cytotoxicity and growth inhibition of 3a–d were determined against three cancer cell lines (HT-29 colon carcinoma cells and MCF-7/MDA-MB-231 breast carcinoma cells), one non-tumorigenic kidney cell line (RC-124), and seven bacterial strains of the ESKAPE panel (A. baumannii, E. coli, K. pneumoniae, P. aeruginosa, E. faecium, MRSA RKI, MRSA DSM) in comparison to auranofin. Regarding their cytotoxicity against cell lines, all complexes were active in the low micromolar to nanomolar range and there was a clear positive effect of the phenyl residues as well as the bromine substituent. Complexes 3c and 3d with the phenylimidazole-based NHC ligands had substantially higher activities than auranofin and reached IC50 values in the range of 0.05–0.18 μM. The bromine containing compounds triggered lower IC50 values than the respective bromine free compounds (comparing the results of the couples 3a/3b and 3c/3d). In no case could a tumor selective toxicity be achieved as the effects against the human kidney cell line RC-124 were comparable with those against the tumor cell lines.
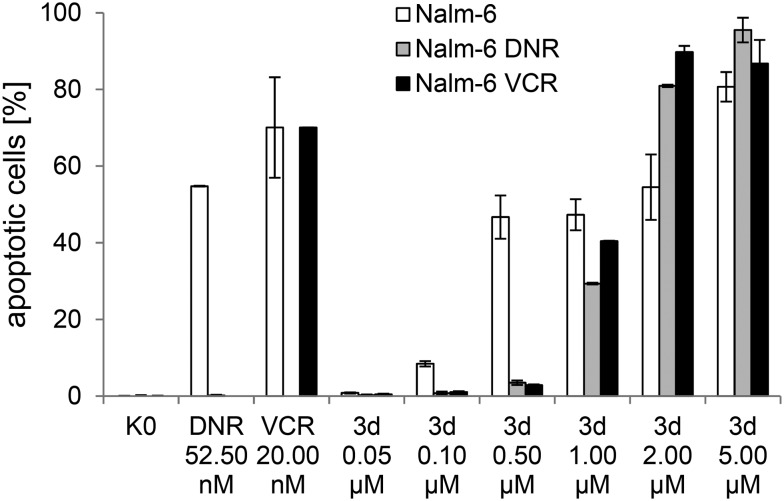
The most cytotoxic compound 3d was subjected to further experiments on apoptosis induction in drug resistant p-glycoprotein overexpressing Nalm cells based on our previous studies with structurally related compounds (e.g.2b).14,17 In good agreement with previous observations, 3d was able to overcome drug resistance in the daunorubicin and vincristine-resistant lines (see Fig. 2). Importantly, its apoptosis induction was 3–5 fold higher than that of 2b in the same assay.17
Fig. 2. DNA fragmentation after 72 h for complex 3d in Nalm-6 and drug resistant Nalm-6 cells. DNR: daunorubicin, VCR: vincristine; Nalm-6: wild-type cells; Nalm-6-DNR: daunorubicin-resistant Nalm-6 cells; Nalm-6-VCR: vincristine-resistant Nalm-6 cells. K0: untreated control; the IC50 values of 3d against the different cell lines are: Nalm-6: 0.9 μM, Nalm-6-DNR: 1.3 μM, Nalm-6-VCR: 1.0 μM.
Evaluation of the inhibition of bacterial growth confirmed the previously reported preference for Gram-positive strains (E. faecium, MRSA1 and MRSA2) of many gold complexes, as only 3c and auranofin triggered moderate activity against Gram-negative bacteria (MIC values of 19.5 μM or higher against A. baumannii, E. coli, K. pneumoniae, P. aeruginosa). Against Gram-positive bacteria, the order of activity for the gold NHC complexes was 3c > 3b ∼ 3d > 3a, indicating the positive effect of the phenyl substituent. The most potent compound 3c was in particular active against MRSA with MIC values of approx. 2 μM (Table 2).
Table 2. Antibacterial activities of 3a–d and auranofin. Minimal inhibitory concentrations (MIC) are given in μM (standard deviations in brackets); MRSA = methicillin-resistant Staphylococcus aureus. As positive control antibiotics, amikacin (P. aeruginosa), linezolid (S. aureus) and ciprofloxacin (all other strains) have been used.
| Compound | MRSA RKI | MRSA DSM | E. faecium | E. coli | P. aeruginosa | A. baumannii | K. pneumoniae |
| 3a | >100 (0) | 79.0 (29.2) | >100 (0) | >100 (0) | >100 (0) | >100 (0) | >100 (0) |
| 3b | 12.7 (3.0) | 12.2 (8.0) | 70.0 (18.7) | >100 (0) | >100 (0) | >100 (0) | >100 (0) |
| 3c | 2.3 (1.2) | 1.7 (0.4) | 8.7 (0.5) | 47.7 (10.7) | 19.5 (6.1) | 48.7 (11.0) | 85.3 (26.9) |
| 3d | 16.6 (7.7) | 9.6 (3.4) | 25.5 (12.6) | >100 (0) | >100 (0) | >100 (0) | >100 (0) |
| Auranofin | 0.6 (0.7) | 0.4 (0.3) | 0.3 (0.3) | 45.7 (5.1) | >100 (44.1) | 55.0 (18.0) | 81.0 (48.0) |
| Antibiotic | 2.4 (1.4) | 4.7 (5.8) | 9.5 (2.0) | 0.1 (0) | 7.3 (1.5) | 0.9 (0.6) | 0.2 (0.1) |
Protein binding and cellular uptake
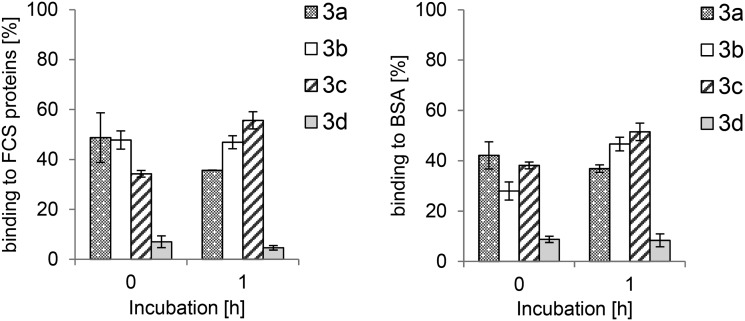
Previous studies on gold NHC complexes had shown correlations between the cytotoxic effects in cells, cellular bioavailability and protein binding.15,17 Binding studies of 3a–d with serum albumin and proteins from fetal calf serum in a precipitation assay showed low to moderate binding efficacy. Complex 3c afforded the highest protein binding (52% with serum albumin and 56% with calf serum after 1 h), complexes 3a and 3b were bound in the rather narrow range of 36–47% after 1 h, while 3d showed the lowest protein binding (8% with serum albumin and 5% with calf serum after 1 h). The levels remained rather stable over time with only small variations (e.g. some increase in the case of 3d, see the ESI†) (Fig. 3).
Fig. 3. Protein binding of 3.0 μM bis-NHC gold(i) complexes 3a–d to fetal calf serum proteins (left) and bovine serum albumin (right).
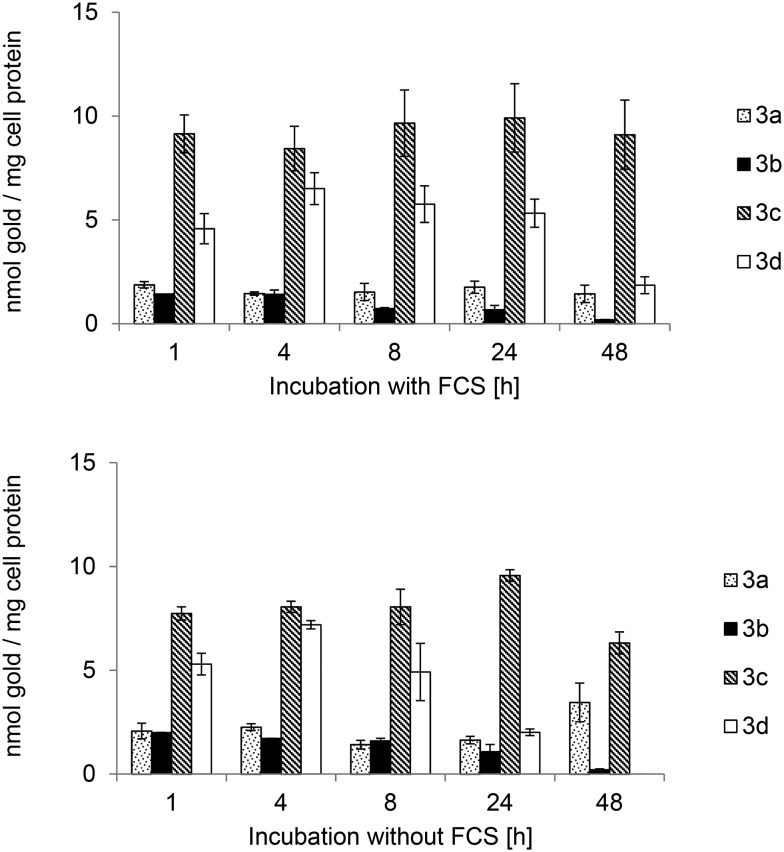
The cellular uptake was quantified in MCF-7 cells, which had been most sensitive towards the complexes (see Table 1). In order to evaluate the influence of protein binding on the bioavailability, the experiments were performed using serum containing and serum free cell culture media (see Fig. 4). However, there was no significant influence of the fetal calf serum proteins on the cellular uptake, as the cellular levels of 3a–d were comparable for both experimental setups. All compounds showed fast cellular uptake and reached high values within the first hour of exposure. The highest cellular concentration was noted for complex 3c (maximum concentration: 9.9 (+FCS)/9.6 (–FCS) nmol gold per mg cell protein) after 24 h. The most striking result was the substantial difference between the imidazole (3a, b) and phenylimidazole (3c, d) based gold NHC complexes as the latter ones reached much higher uptake values (2.5–7.9-fold higher after 24 h). The bromine substituent had a negative impact on the cellular bioavailability as the levels of 3b and 3d were, within 24 h, in most cases up to 4.8-fold lower than those obtained with the respective non-bromine-substituted counterparts 3a and 3c. Moreover, the levels of 3b and 3d decreased over continued incubation (see 48 h values in Fig. 4).
Table 1. Antiproliferative effects of auranofin and complexes 3a–d expressed as IC50 values (μM) with standard errors as superscripts.
| Compound | HT-29 | MCF-7 | MDA-MB-231 | RC-124 |
| Auranofin | 3.79±0.18 | 2.00±0.05 | 1.54±0.12 | 1.44±0.03 |
| 3a | 5.54±0.79 | 4.20±0.72 | 5.08±0.60 | 3.44±0.38 |
| 3b | 1.51±0.10 | 0.72±0.07 | 1.08±0.06 | 0.94±0.10 |
| 3c | 0.15±0.01 | 0.16±0.03 | 0.18±0.02 | 0.09±0.02 |
| 3d | 0.14±0.02 | 0.06±0.00 | 0.18±0.01 | 0.05±0.01 |
Fig. 4. Cellular uptake in MCF-7 breast carcinoma cells of bis-NHC complexes 3a–d with (top) or without (bottom) fetal calf serum incubation concentration of 3.0 μM.
Inhibition of TrxRs
The inhibition of mammalian and bacterial TrxRs was evaluated as a possible mode of drug action using purified rat liver TrxR and TrxR from E. coli (Table 3). Against rat TrxR, complexes 3a–d displayed low efficacy, however, the same structure–activity-relationships as for cytotoxicity were noted (positive effects of the phenyl and bromine substituents). The most active gold NHC complex was 3d with an IC50 value of 16.3 μM. Also against bacterial TrxR from E. coli only low activities were observed for 3a–d. In this case, complex 3c was the most active agent with an IC50 value of 30.0 μM. Notably, 3c had triggered the strongest antibacterial effects among the gold NHC complexes in this study. Auranofin showed much higher activity against both forms of TrxR.
Table 3. IC50 values of rat TrxR and E. coli TrxR inhibition tests. Standard errors are given as superscripts.
| Compound | Rat TrxR IC50 [μM] | E. coli TrxR IC50 [μM] |
| Auranofin | 0.093±0.009 | 0.296±0.069 |
| 3a | 127.8±6.1 | 410.9±42.1 |
| 3b | 71.9±5.4 | 57.2±6.2 |
| 3c | 30.3±4.6 | 30.0±7.1 |
| 3d | 16.3±3.2 | 96.6±13.4 |
Discussion and conclusions
The biscarbene complexes 3a–d displayed high cytotoxic activities in cancer cells (IC50 of 0.05–5.54 μM), which were most pronounced in the case of the phenylimidazole-based and bromine-containing NHC ligands (the most active complex is 3d: IC50 of 0.05–0.18 μM). In comparison to the previously studied monocarbene derivatives (2a–d) substantially stronger activity was noted with similar structure–activity-relationships (e.g. positive effect of the phenylimidazole structure; for comparison, IC50 [μM] values for 2a/3a: 10.58–16.97/3.44–5.54, 2b/3b: 5.49–12.05/0.72–1.51, 2c/3c: 4.46–8.13/0.09–0.18 and 2d/3d: 4.73–7.20/0.05–0.18).17 The effective antiproliferative effects in this study are overall in good agreement with the reported high potencies of various biscarbene NHC complexes.10,15,18,19,28 Cellular uptake studies provided an explanation for the enhanced activities of the phenylimidazole derived compounds, as complexes 3c and 3d exhibited the highest cellular gold levels. Moreover, in the presence of serum the uptake of various biscarbene compounds appears in general higher than those of the respective monocarbene analogues.10,15,17 The higher cellular uptake and higher cytotoxic activity are likely the consequence of the larger lipophilicity of 3c and 3d compared to 3a and 3b.
On the other hand, there was a negative effect of the bromine substituent on the cellular uptake, contrasting with the results of the cytotoxicity study. This negative effect of halide substituents on the cellular uptake had also been observed with the analogous monocarbene complexes recently.17 It can be assumed that the biscarbene complexes of this work are stable in organic solvents, like DMF or DMSO, and water-based matrices, like cell culture medium or buffer. Ongoing stability studies with a related biscarbene gold(i) complex showed that the complex was stable for several days in different solvents (unpublished data). Taken together, this indicates that the intact complexes are taken up into the cells, where they trigger their biological activity.
Complexes 3a–d displayed comparably low protein binding (4–58%), and the cellular accumulation efficiency was largely independent of the presence of serum in the cell culture media. With this property biscarbene complexes differ from analogous monocarbene derivatives, which show high protein binding (80–100%) and in consequence lower cellular concentrations.10,15,17 Our recent studies confirmed the relevance of an intact gold–NHC fragment for cellular accumulation.17 Taken together with the low protein binding of biscarbene complexes and their lipophilic/cationic nature, this provides a possible explanation for the generally enhanced cellular bioavailability of gold(i) biscarbene complexes.
Complexes 3a–d also inhibited the growth of Gram-positive bacteria, although with in most cases lower potency than the monocarbene analogues 2a–d (for comparison, MIC [μM] values for 2a/3a: 2.55–2.97/79.0–100.0, 2b/3b: 0.64–3.12/12.2–70.0, 2c/3c: 4.16–6.45/1.7–8.7 and 2d/3d: 6.25–12.51/9.6–25.5).17 The preference for Gram-positive over Gram-negative bacteria has also been reported for auranofin and is likely related to the strong dependence of these bacteria on an intact Trx/TrxR system.29 Experiments on the inhibition of mammalian and bacterial TrxRs afforded moderate to very low activities (IC50 16.3–410.9 μM), however, the most active complexes 3c and 3d were also the most cytotoxic and most efficient antibacterial agents. It should be noted here that much higher activity against TrxRs could be achieved with various monocarbene species (e.g.2a–d: IC50 0.042–0.446 μM), whereas biscarbene gold(i) NHC complexes are usually weaker TrxR inhibitors.15,17,18 The relevance of TrxR inhibition to the biological activity of 3a–d remains elusive at this stage. On the one hand, the excellent accumulation capability of the complexes led to elevated cellular levels that makes a contribution of TrxR inhibition very likely. For example, taking the characteristics of MCF-7 cells into account30,31 the value obtained with 3c after 24 h (9.91 nmol per mg cellular protein uptake from serum containing medium, Fig. 4 top) corresponds to a molar concentration of 1119 μM and with this to a 373-fold accumulation compared to the exposure concentration (3.0 μM). On the other hand, other mechanisms of action (TrxR related and TrxR unrelated) have been identified for various biscarbene gold NHC derivatives, including the targeting of mitochondria,9,15 binding to DNA G-quadruplexes,12,32 interference with the ASK1-p38-MAPK signaling,33 or antiangiogenic and antivascular effects.10
The antimicrobial effects of auranofin and other gold(i) complexes against various microorganisms have recently attracted increasing attention, and studies on the underlying mode of action are of great interest.26,29,34–40 Regarding the antibacterial effects, the inhibition of bacterial TrxRs by auranofin has been demonstrated recently and appears to be of major relevance.17,26,29,38
In conclusion, biscarbene gold(i) complexes represent strongly cytotoxic and moderately antibacterial agents. The strong cellular uptake was found to be a crucial factor for obtaining highly antiproliferative agents. In particular, high activities were noted for derivatives containing a phenylimidazole-based NHC ligand. Several types of gold complexes have demonstrated both antibacterial and cytotoxic effects. The underlying reasons are not completely understood; however, most likely similar modes of action are important in both species (e.g. inhibition of TrxRs). For future therapeutic applications, it will be important to provide structure–activity-relationship data to separate cytotoxic from antibacterial properties.
Experimental
General
All reagents were obtained from Sigma-Aldrich (Switzerland) or Fluka Analytical. Bovine serum albumin was from Sigma Aldrich, fetal calf serum (FCS) was from Biochrom GmbH Berlin, cell culture media were from Thermo Fisher Scientific Bremen, cell lines and bacteria strains were from CLS Cell Lines Service GmbH Eppelheim (RC-124) or Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures GmbH Braunschweig (bacteria, HT-29, MCF-7, MDA-MB-231), tryptic soy broth was from MP Biomedicals GmbH Eschwege, Bacto yeast extract and Bacto agar were from BD Becton Dickinson Heidelberg, Müller–Hinton broth was from Carl Roth GmbH + Co. KG Karlsruhe and d(+) glucose was from Merck KGaA Darmstadt. Tryptic soy yeast contains tryptic soy broth (30 g L–1) and Bacto yeast extract (3 g L–1). Müller–Hinton-broth plus glucose contains Müller–Hinton-broth (21 g L–1) and d(+) glucose (1%). Bacto agar was used in a concentration of 15 g L–1. The purities of the newly synthesized compounds were proved by elemental analysis (Flash EA 1112, Thermo Quest) and differed by less than 0.5% from the predicted values. 1H NMR spectra and 13C NMR spectra were recorded using a Bruker AV II-400 or a Bruker DRX-400 AS NMR spectrometer. Mass spectra were recorded on an Agilent Technologies 6120 Quadrupole LC/MS (ESI source, positive mode) or an Advion expressionL CMS (ESI source, negative mode). For the absorption measurements in both enzyme assays a Perkin Elmer 2030 Multilabel Reader VICTOR™ X4 was used. The amount of gold was detected using an HR-CS AAS contrAA-700 from Analytik Jena. Compounds 1a–d and 2a–d were prepared as described recently.17
Synthesis
General procedure for synthesis of NHC–Au–NHC+ I– complexes 3a–d
Equivalent amounts of 0.23–0.34 mmol of the respective (phenyl)imidazolium gold(i) chloride and the corresponding imidazolium iodide were dissolved in 20 mL of a 1 : 1 mixture of dichloromethane/methanol. After addition of 1.1 equivalents of potassium carbonate the reaction was stirred over 10 days at room temperature with light protection. To remove excess potassium carbonate, the solvents were removed and the residue was resuspended in dichloromethane and filtered. The complexes were isolated by evaporating the filtrate and drying under vacuum at 50 °C over a period of 72 h.
[Bis(1,3-diethyl-imidazol-2-ylidene)]gold(i) iodide 3a
The synthesis of the imidazolium cation of 3a with different counter ions has been reported;9,20 see General procedure; starting material: chlorido(1,3-diethyl-imidazol-2-ylidene)gold(i) (120.0 mg, 0.34 mmol) and 1,3-diethyl-imidazolium iodide (84.8 mg, 0.34 mmol), yield: 141.7 mg (0.25 mmol, 74%), white powder; 1H-NMR (400 MHz, CDCl3-d1) δ ppm 7.25 (s, 4H, Im-H4 + Im-H5), 4.31 (q, 3JH,H= 7.3 Hz, 8H, CH2), 1.55 (t, 3JH,H = 7.4 Hz, 12H, CH3); 13C-NMR (101 MHz, CDCl3-d1) δ ppm 182.4 (2C, Im-C2quat.), 121.2 (4C, Im-C4H + Im-C5H), 46.6 (4C, CH2), 17.1 (4C, CH3); elemental analysis: C14H24AuIN4 (calc.%/found%) C (29.38/29.57), H (4.23/4.17), N (9.79/9.64); MS (ESI): m/z 445.2 [M – I]+ and m/z 127.0 [I]–.
[Bis(4-bromo-1,3-diethyl-imidazol-2-ylidene)]gold(i) iodide 3b
See General procedure; starting material: chlorido(4-bromo-1,3-diethyl-imidazol-2-ylidene)gold(i) (120.0 mg, 0.28 mmol) and 4-bromo-1,3-diethyl-imidazolium iodide (91.2 mg, 0.28 mmol), yield: 131.5 mg (0.18 mmol, 66%), pale yellow powder; 1H-NMR (400 MHz, CDCl3-d1) δ ppm 7.27 (s, 2H, Im-H5), 4.41 (q, 3JH,H= 7.3 Hz, 4H, CH2), 4.36 (q, 3JH,H= 7.3e Hz, 4H, CH2), 1.56 (t, 3JH,H = 7.4 Hz, 6H, CH3), 1.51 (t, 3JH,H = 7.3 Hz, 6H, CH3); 13C-NMR (101 MHz, CDCl3-d1) δ ppm 184.2 (2C, Im-C2quat.), 121.1 (2C, Im-C5H), 105.5 (2C, Im-C4Brquat.), 47.6 (2C, CH2), 45.7 (2C, CH2), 16.6 (2C, CH3), 16.4 (2C, CH3); elemental analysis: C14H22AuBr2IN4 (calc.%/found%) C (23.03/23.50), H (3.04/2.97), N (7.67/7.67); MS (ESI): m/z 603.0 [M – I]+ and m/z 127.0 [I]–.
[Bis(1,3-diethyl-4-phenyl-imidazol-2-ylidene)]gold(i) iodide 3c
See General procedure; starting material: chlorido(1,3-diethyl-4-phenyl-imidazol-2-ylidene)gold(i) (120.0 mg, 0.28 mmol) and 1,3-diethyl-4-phanyl-imidazolium iodide (91.0 mg, 0.28 mmol), yield: 173.2 mg (0.24 mmol, 86%), white powder; 1H-NMR (400 MHz, CDCl3-d1) δ ppm 7.50 (m, 6H, Ph-Hpara/meta), 7.41 (m, 4H, Ph-Hortho), 7.17 (s, 2H, Im-H5), 4.43 (q, 4H, 3JH,H= 7.3 Hz, CH2), 4.27 (q, 4H, 3JH,H= 7.3 Hz, CH2), 1.62 (t, 6H, 3JH,H = 7.2 Hz, CH3), 1.43 (t, 6H, 3JH,H = 7.2 Hz, CH3); 13C-NMR (101 MHz, CDCl3-d1) δ ppm 183.5 (2C, Im-C2quat.), 135.0 (2C, Im-C4quat.), 129.7 (2C, Ph-C4H), 129.5 (4C, Ph-C3H + Ph-C5H), 129.1 (4C, Ph-C2H + Ph-C6H), 127.4 (2C, Ph-C1quat.), 119.0 (2C, Im-C5H), 47.1 (2C, CH2), 44.0 (2C, CH2), 17.6 (2C, CH3), 17.1 (2C, CH3); elemental analysis: C26H32AuIN4 (calc.%/found%) C (43.11/43.47), H (4.45/4.34), N (7.73/7.59); MS (ESI): m/z 597.2 [M – I]+ and m/z 127.0 [I]–.
[Bis(4-(4-bromophenyl)-1,3-diethyl-imidazol-2-ylidene)]gold(i) iodide 3d
See General procedure; starting material: chlorido(4-(4-bromophenyl)-1,3-diethyl-imidazol-2-ylidene)gold(i) (120.0 mg, 0.23 mmol) and (4-(4-bromophenyl)-1,3-diethyl-imidazolium iodide (95.5 mg, 0.23 mmol), yield: 163.5 mg (0.18 mmol, 79%), white powder; 1H-NMR (400 MHz, CDCl3-d1) δ ppm 7.64 (m, 4H, Ph-H2 + Ph-H6), 7.31 (m, 4H, Ph-H3 + Ph-H5), 7.21 (s, 2H, Im-H5), 4.42 (q, 4H, 3JH,H= 7.3 Hz, CH2), 4.28 (q, 4H, 3JH,H= 7.3 Hz, CH2), 1.61 (t, 6H, 3JH,H = 7.2 Hz, CH3), 1.41 (t, 6H, 3JH,H = 7.2 Hz, CH3); 13C-NMR (101 MHz, CDCl3-d1) δ ppm 183.9 (2C, Im-C2quat.), 133.7 (2C, Im-C4quat.), 132.4 (4C, Ph-C3H + Ph-C5H), 131.0 (4C, Ph-C2H + Ph-C6H), 126.3 (2C, Ph-C1quat.), 124.2 (2C, Ph-C4Brquat.), 119.4 (2C, Im-C5H), 47.1 (2C, CH2), 44.2 (2C, CH2), 17.5 (2C, CH3), 17.0 (2C, CH3); elemental analysis: C26H30AuBr2IN4 (calc.%/found%) C (35.40/35.71), H (3.43/3.45), N (6.35/6.19); MS (ESI): m/z 755.0 [M – I]+ and m/z 127.0 [I]–.
Cell culture
HT-29 colon carcinoma cells, MDA-MB-231 breast cancer cells and MCF-7 breast carcinoma cells were maintained in Dulbecco's modified Eagle medium (4.5 g L–1d-glucose, l-glutamine, pyruvate), which was supplemented with gentamycin (50 mg L–1) and fetal bovine serum superior, standardized (Biochrom GmbH, Berlin) (10% v/v), and were passaged once a week. RC-124 healthy human kidney cells were maintained in McCoy's 5A (modified, with l-glutamine) medium which was supplemented with gentamycin (50 mg L–1) and fetal bovine serum superior, standardized (Biochrom GmbH, Berlin) (10% v/v), and were also passaged once a week. For experiments with RC-124 cells, microtiter plates had been pretreated in the following way: 30 μL of a sterilized gelatine solution (1.5% (m/V)) were added to each well of flat bottom 96-well plates, the plates were covered with their lids and incubated for 1 h at 37 °C, the excess solution was removed, the wells were washed with PBS 7.4 pH, and the new cell-culture medium was added.
Antiproliferative assay in tumorigenic and non-tumorigenic cells
The antiproliferative effects were determined according to a recently used method with minor modifications. In short: a volume of 100 μL of HT-29 cells (2565 cells per mL), MDA-MB-231 cells (4120 cells per mL), MCF-7 cells (4840 cells per mL) or RC-124 cells (1460 cells per mL) was transferred into the wells of 96-well plates (note: for RC-124, pretreated plates were used, see above) and incubated at 37 °C/5% CO2 for 72 h (MCF-7, MDA-MB-231, RC-124) or 48 h (HT-29). Stock solutions of the compounds in dimethylformamide (DMF) were freshly prepared and diluted with the respective cell culture medium to graded concentrations (final concentration of DMF: 0.1% v/v). After 72 h (HT-29) or 96 h (MCF-7, MDA-MB-231, RC-124) of exposure, the cell biomass was determined by crystal violet staining and the IC50 value was determined as the concentration that caused 50% inhibition of cell proliferation compared to the untreated control. Results were calculated as the mean values of three independent experiments.
Apoptosis induction in drug resistant Nalm-6 cells
Measurement of DNA fragmentation in Nalm-6 cells: apoptotic cell death was determined by a modified cell cycle analysis, which detects DNA fragmentation at the single-cell level. For measurement of DNA fragmentation cells were seeded at a density of 1 × 105 cells per mL and treated with different concentrations of 3d. After 72 h of incubation, cells were collected by centrifugation at 300g for 5 min, washed with PBS at 4 °C and fixed in PBS/formaldehyde (2%, v/v) on ice for 30 min. After fixation, cells were incubated with ethanol/PBS (2 : 1, v/v) for 15 min, pelleted, and resuspended in PBS containing RNase A (40 mg mL–1). After incubation for 30 min at 37 °C, cells were pelleted again and finally resuspended in PBS containing propidium iodide (50 mg mL–1). Nuclear DNA fragmentation was then quantified by flow cytometric determination of hypodiploid DNA. Data were collected and analyzed by using a FACScan (Becton Dickinson, Heidelberg, Germany) equipped with CELLQuest software. Data are given in %hypoploidy (subG1), which reflects the number of apoptotic cells.
Antibacterial screening
Overnight cultures of the bacteria were grown aerobically at 37 °C in Müller–Hinton broth with added 1% glucose and pH 7.2 for Gram-negative strains, or with trypticase soy yeast extract medium (TSY – 30 g L–1 trypticase soy broth, 3 g L–1 yeast extract, pH 7.2) for Gram-positive strains. The cultures were adjusted to an OD600nm of 0.001, which resulted in a final starting OD600nm of 0.0005 in the test. 25 μL of test culture was added to 25 μL of a serial dilution of the test compounds in the appropriate medium for the different strains in accordance with standardized procedures in 384 well plates. Test compounds from stock solutions in DMF (complexes 3a–d, auranofin) DMSO (linezolid) or water (ciprofloxacin, amikacin) were used at final concentrations of 100, 50, 25, 12.5, 6.25, 3.125, 1.56, 0.78, 0.39 and 0.2 μM. As positive control compounds, linezolid (both MRSA strains), ciprofloxacin (E. faecium, E. coli, A. baumannii, K. pneumonia) and amikacin (P. aeruginosa) were applied. The highest DMF/DMSO concentration in the assay was 1%, which had no apparent effect on the growth of the bacteria. After an incubation time of 18 h at 37 °C under moist conditions, the optical density at 600 nm was measured with a Fusion Universal Microplate Analyser (Perkin–Elmer, Waltham, USA). The lowest concentration that completely suppressed growth defined the MIC values. The MIC values were determined by curve fitting with SigmaPlot. All values represent averages from at least three independent experiments. The following bacterial strains were used. Gram-negative: Acinetobacter baumannii (DSM 30007), Escherichia coli (DSM 1116), Klebsiella pneumoniae (DSM 11678) and Pseudomonas aeruginosa PA7 (DSM 24068). Gram-positive: Enterococcus faecium (DSM 20477), Staphylococcus aureus MRSA (clinical isolate, RKI 11-02670) and Staphylococcus aureus MRSA (DSM 11822).
Protein binding studies
The precipitation assay, based on the method of Ma et al., was modified and performed with solutions containing bovine serum albumin or with fetal calf serum.17,41 An aliquot (11 mL) of DMEM cell culture medium was supplemented with FCS (1.1 mL; standardized) or with BSA (440 mg). An aliquot (1.0 mL) of the corresponding solution was used for matrix-matched calibration and was treated like the other samples without incubation. Stock solutions of the test compounds were prepared in DMF (concentration: 3.0 mM). An aliquot (10 μL) of each solution was pipetted into cell culture medium (10 mL) containing either FCS or BSA and carefully mixed (final incubation concentration: 3 μM). The reaction mixture was incubated at 37 °C for 48 h under shaking. After the requisite duration (0, 1, 4, 8, 24, 48 h), aliquots (250 μL) of each sample were treated with ice-cooled ethanol (500 μL) and stored at –25 °C for 2 h. Thereafter, the samples were centrifuged (964g at 4 °C for 15 min). An aliquot (350 μL) of the supernatant was separated and stored at –25 °C. The experiment was carried out in duplicate. Aliquots (100 μL) of each sample were treated with 13% nitric acid (10 μL) for stabilization, and the gold contents were quantified by HR-CS AAS (see below). The bonded moiety was calculated as a percentage.
AAS measurements
For the gold and fluorine measurements a contrAA 700 high-resolution continuum-source atomic absorption spectrometer (Analytik Jena AG) was used. Pure samples of the respective complexes were used as standards and calibration was performed in a matrix-matched manner (meaning that all samples and standards were adjusted to the same protein concentration by dilution with distilled water).
Triton-X 100 (1%, 10 μL) as well as ascorbic acid (1%, 10 μL) were added to each standard sample (100 μL). Samples were injected (25 μL) into coated standard graphite tubes (Analytik Jena AG) and thermally processed as previously described in more detail.17 Gold was quantified at a wavelength of 242.79 nm. The mean integrated absorbance of triple injections was used throughout the studies. The final results of gold concentrations were calculated from the data obtained in two independent experiments and are expressed as nmol of metal per mg of cellular protein and as cellular molar concentration [mM]. The procedure for calculating the cellular molar concentration in MCF-7 cell lines is described in the literature.
Cellular uptake studies in MCF-7 cells
The cellular metal uptake was determined according to previously described methods.17,42 In short: MCF-7 breast carcinoma cells were grown until at least 75–80% confluency in 150 cm2 cell-culture flasks. Stock solutions of the compounds in DMF were prepared and diluted with cell-culture medium to a final concentration of 3.0 μM immediately before use (final DMF concentration: 0.1% v/v). The cell culture medium of the flasks was replaced with the medium that contained the metal compound (20 mL) and the flasks were incubated at 37 °C/5% CO2 up to 48 h. After the desired incubation period the uptake was stopped by removing the cell culture medium. The cells were washed with PBS (10 mL), the washing solution was removed, and the cells were isolated after 6 min trypsinization (2.4 mL trypsin solution 0.05%, containing EDTA 0.004%) by centrifugation (5 min, 1096g). The obtained cell pellets were stored at –20 °C for further use. For metal and protein quantification the pellets were resuspended in demineralized water (1.0 mL) and lysed for 30 min by ultra-sonication. The protein content of lysates was determined by the Bradford method and the metal content was determined by AAS as described above.
Inhibition of mammalian TrxR
To determine the inhibition of mammalian TrxR an established microplate reader based assay was performed.14,17 Commercially available rat liver TrxR (from Sigma-Aldrich) was used and diluted with distilled water to achieve a concentration of 3.58 U mL–1. The compounds were freshly dissolved as stock solutions in DMF. 25 μL aliquots of the enzyme solution and 25 μL of either potassium phosphate buffer pH 7.0 containing the compounds in graded concentrations or 25 μL buffer without compounds but DMF (positive control) were added. 50 μL of a blank solution (DMF in buffer) was also prepared (final concentration of DMF: 0.5% v/v). The resulting solutions were incubated with moderate shaking for 75 min at 37 °C in a 96-well plate. To each well, 225 μL of reaction mixture (1 mL reaction mixture consists of 500 μL potassium phosphate buffer pH 7.0, 80 μL EDTA solution (100 mM, pH 7.5), 20 μL BSA solution (0.2%), 100 μL of NADPH solution (20 mM) and 300 μL distilled water) were added and the reaction started immediately by the addition of 25 μL of 20 mM ethanolic DTNB solution. After proper mixing, the formation of 5-TNB was monitored with a microplate reader at 405 nm 10 times in 35 s intervals for about 6 min. The increase in 5-TNB concentration over time followed a linear trend (r2 ≥ 0.990), and the enzymatic activities were calculated as the slopes (increase in absorbance per second) thereof. For each tested compound, its noninterference with the assay components was confirmed by a negative control experiment using an enzyme-free test solution. The IC50 values were calculated as the concentration of compound decreasing the enzymatic activity of the untreated control by 50% and are given as the means and error of three repeated experiments.
TrxR E. coli inhibition assay
The DTNB-coupled thioredoxin reductase inhibition assay for E. coli was partly adopted and modified from Lu et al.17,43 Commercially available E. coli TrxR and its natural substrate E. coli Trx (both from Sigma-Aldrich) were used and diluted with distilled water to achieve a concentration of 35.5 U mL–1 for the enzyme and 0.77 μg mL–1 for the substrate. The compounds were freshly dissolved as stock solutions in DMF. 10 μL aliquots of the enzyme solution, 10 μL substrate solution and 100 μL NADPH (200 mM) in TE buffer were mixed in a well with 20 μL TE buffer pH 7.5 (consists of Tris-HCl 50 mM and EDTA 1 mM in aqueous solution) containing the compounds in graded concentrations or 20 μL buffer solution without compounds (control). Blank solutions: 100 μL NADPH (200 mM) and 40 μL of a DMF/buffer mixture were added and the resulting solutions were incubated with moderate shaking for 75 min at 25 °C in a 96-well plate (final concentrations of DMF: 0.5% v/v). To each well, 100 μL of reaction mixture (containing NADPH 200 μM and DTNB 5 mM in TE buffer solution) were added and the reaction started immediately. After proper mixing, the formation of 5-TNB was monitored with a microplate reader (Perkin-Elmer Victor X4) at 405 nm 10 times in 35 s intervals for about 6 min. The values were corrected using the absorbance of the blank solution. The increase in 5-TNB concentration over time followed a linear trend (r2 ≥ 0.990), and the enzymatic activities were calculated as the slopes (increase in absorbance per second) thereof. For each tested compound, its noninterference with the assay components was confirmed by a negative control experiment, where the highest test compound concentration is used and the enzyme aliquot solution is replaced by the same amount of TE buffer. The IC50 values were calculated as the concentration of compound decreasing the enzymatic activity of the positive control by 50% and are given as the means and error of three repeated experiments.
Conflict of interest
The authors declare no competing interests.
Supplementary Material
Acknowledgments
Financial support from Deutsche Forschungsgemeinschaft (DFG) is gratefully acknowledged (project codes: OT338/12-1 and BR3572/4-1). We thank Dr. Raimo Franke (HZI) for fruitful discussions and his support of this study.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7md00269f
References
- Ott I. Coord. Chem. Rev. 2009;253:1670–1681. [Google Scholar]
- Zou T., Lum C. T., Lok C. N., Zhang J. J., Che C. M. Chem. Soc. Rev. 2015;44:8786–8801. doi: 10.1039/c5cs00132c. [DOI] [PubMed] [Google Scholar]
- Bertrand B., Casini A. Dalton Trans. 2014;43:4209–4219. doi: 10.1039/c3dt52524d. [DOI] [PubMed] [Google Scholar]
- Glisic B. D., Djuran M. I. Dalton Trans. 2014;43:5950–5969. doi: 10.1039/c4dt00022f. [DOI] [PubMed] [Google Scholar]
- Liu W., Gust R. Coord. Chem. Rev. 2016;329:191–213. [Google Scholar]
- Rubbiani R., Wahrig B., Ott I. J. Biol. Inorg. Chem. 2014;19:961–965. doi: 10.1007/s00775-014-1135-4. [DOI] [PubMed] [Google Scholar]
- Koch R., Verhandlungen des X. Internationalen Medizinischen Kongresses, Berlin 1890. 1891 Bd.I. Verlag von August Hirschwald, Berlin, 1890.
- Snyder R. M., Mirabelli C. K., Crooke S. T. Biochem. Pharmacol. 1986;35:923–932. doi: 10.1016/0006-2952(86)90078-x. [DOI] [PubMed] [Google Scholar]
- Hickey J. L., Ruhayel R. A., Barnard P. J., Baker M. V., Berners-Price S. J., Filipovska A. J. Am. Chem. Soc. 2008;130:12570–12571. doi: 10.1021/ja804027j. [DOI] [PubMed] [Google Scholar]
- Muenzner J. K., Biersack B., Albrecht A., Rehm T., Lacher U., Milius W., Casini A., Zhang J. J., Ott I., Brabec V., Stuchlikova O., Andronache I. C., Schuppan D., Kaps L., Schobert R. Chem. – Eur. J. 2016;22:18953–18962. doi: 10.1002/chem.201604246. [DOI] [PubMed] [Google Scholar]
- Bertrand B., Citta A., Franken I. L., Picquet M., Folda A., Scalcon V., Rigobello M. P., Le Gendre P., Casini A., Bodio E. J. Biol. Inorg. Chem. 2015;20:1005–1020. doi: 10.1007/s00775-015-1283-1. [DOI] [PubMed] [Google Scholar]
- Bertrand B., Stefan L., Pirrotta M., Monchaud D., Bodio E., Richard P., Le Gendre P., Warmerdam E., de Jager M. H., Groothuis G. M., Picquet M., Casini A. Inorg. Chem. 2014;53:2296–2303. doi: 10.1021/ic403011h. [DOI] [PubMed] [Google Scholar]
- Rubbiani R., Salassa L., de Almeida A., Casini A., Ott I. ChemMedChem. 2014;9:1205–1210. doi: 10.1002/cmdc.201400056. [DOI] [PubMed] [Google Scholar]
- Rubbiani R., Kitanovic I., Alborzinia H., Can S., Kitanovic A., Onambele L. A., Stefanopoulou M., Geldmacher Y., Sheldrick W. S., Wolber G., Prokop A., Wolfl S., Ott I. J. Med. Chem. 2010;53:8608–8618. doi: 10.1021/jm100801e. [DOI] [PubMed] [Google Scholar]
- Rubbiani R., Can S., Kitanovic I., Alborzinia H., Stefanopoulou M., Kokoschka M., Mönchgesang S., Sheldrick W. S., Wölfl S., Ott I. J. Med. Chem. 2011;54:8646–8657. doi: 10.1021/jm201220n. [DOI] [PubMed] [Google Scholar]
- Rubbiani R., Schuh E., Meyer A., Lemke J., Wimberg J., Metzler-Nolte N., Meyer F., Mohr F., Ott I. Med. Chem. Commun. 2013;4:942–948. [Google Scholar]
- Schmidt C., Karge B., Misgeld R., Prokop A., Franke R., Bronstrup M., Ott I. Chem. – Eur. J. 2017;23:1869–1880. doi: 10.1002/chem.201604512. [DOI] [PubMed] [Google Scholar]
- Liu W., Bensdorf K., Proetto M., Hagenbach A., Abram U., Gust R. J. Med. Chem. 2012;55:3713–3724. doi: 10.1021/jm3000196. [DOI] [PubMed] [Google Scholar]
- Muenzner J. K., Biersack B., Kalie H., Andronache I. C., Kaps L., Schuppan D., Sasse F., Schobert R. ChemMedChem. 2014;9:1195–1204. doi: 10.1002/cmdc.201400049. [DOI] [PubMed] [Google Scholar]
- Baker M., Barnard P. J., Berners-Price S. J., Brayshaw S. K., Hickey J. L., Skelton B. W., White A. H. Dalton Trans. 2006:3708–3715. doi: 10.1039/b602560a. [DOI] [PubMed] [Google Scholar]
- Zou T., Lum C. T., Chui S. S.-Y., Che C.-M. Angew. Chem., Int. Ed. 2013;52:1–5. doi: 10.1002/anie.201209787. [DOI] [PubMed] [Google Scholar]
- Zou T., Lum C. T., Lok C.-N., To W.-P., Low K.-H., Che C.-M. Angew. Chem., Int. Ed. 2014;53:5810–5814. doi: 10.1002/anie.201400142. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gallardo J., Elie B. T., Sanau M., Contel M. Chem. Commun. 2016;52:3155–3158. doi: 10.1039/c5cc09718e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebryanskaya T. V., Zolotarev A. A., Ott I. Med. Chem. Commun. 2015;6:1186–1189. [Google Scholar]
- Maftei C. V., Fodor E., Jones P. G., Freytag M., Franz M. H., Kelter G., Fiebig H. H., Tamm M., Neda I. Eur. J. Med. Chem. 2015;101:431–441. doi: 10.1016/j.ejmech.2015.06.053. [DOI] [PubMed] [Google Scholar]
- Owings J. P., McNair N. N., Mui Y. F., Gustafsson T. N., Holmgren A., Contel M., Goldberg J. B., Mead J. R. FEMS Microbiol. Lett. 2016:363. doi: 10.1093/femsle/fnw148. [DOI] [PubMed] [Google Scholar]
- Kaps L., Biersack B., Muller-Bunz H., Mahal K., Munzner J., Tacke M., Mueller T., Schobert R. J. Inorg. Biochem. 2012;106:52–58. doi: 10.1016/j.jinorgbio.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Arambula J. F., McCall R., Sidoran K. J., Magda D., Mitchell N. A., Bielawski C. W., Lynch V. M., Sessler J. L., Arumugam K. Chem. Sci. 2016;7:1245–1256. doi: 10.1039/c5sc03519h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbut M. B., Vilcheze C., Luo X., Hensler M. E., Guo H., Yang B., Chatterjee A. K., Nizet V., Jacobs Jr. W. R., Schultz P. G., Wang F. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4453–4458. doi: 10.1073/pnas.1504022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust R., Schnurr B., Krauser R., Bernhardt G., Koch M., Schmid B., Hummel E., Schönenberger H. J. Cancer Res. Clin. Oncol. 1998;124:585–597. doi: 10.1007/s004320050220. [DOI] [PubMed] [Google Scholar]
- Reile H., Bernhardt G., Koch M., Schonenberger H., Hollstein M., Lux F. Cancer Chemother. Pharmacol. 1992;30:113–122. doi: 10.1007/BF00686402. [DOI] [PubMed] [Google Scholar]
- Bazzicalupi C., Ferraroni M., Papi F., Massai L., Bertrand B., Messori L., Gratteri P., Casini A. Angew. Chem. Int., Ed. 2016;55:4256–4259. doi: 10.1002/anie.201511999. [DOI] [PubMed] [Google Scholar]
- Cheng X., Holenya P., Can S., Alborzinia H., Rubbiani R., Ott I., Wolfl S. Mol. Cancer. 2014;13:221. doi: 10.1186/1476-4598-13-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelucci F., Sayed A. A., Williams D. L., Boumis G., Brunori M., Dimastrogiovanni D., Miele A. E., Pauly F., Bellelli A. J. Biol. Chem. 2009;284:28977–28985. doi: 10.1074/jbc.M109.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Rosario S., Cowart D., Myers A., Tarrien R., Levine R. L., Scott R. A., Self W. T. J. Biol. Inorg. Chem. 2009;14:507–519. doi: 10.1007/s00775-009-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokai Y., Jurkowicz B., Fernandez-Gallardo J., Zakirkhodjaev N., Sanau M., Muth T. R., Contel M. J. Inorg. Biochem. 2014;138:81–88. doi: 10.1016/j.jinorgbio.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberi T., Fiaschi T., Modesti A., Massai L., Messori L., Balzi M., Magherini F. Int. J. Biochem. Cell Biol. 2015;65:61–71. doi: 10.1016/j.biocel.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Debnath A., Parsonage D., Andrade R. M., He C., Cobo E. R., Hirata K., Chen S., Garcia-Rivera G., Orozco E., Martinez M. B., Gunatilleke S. S., Barrios A. M., Arkin M. R., Poole L. B., McKerrow J. H., Reed S. L. Nat. Med. 2012;18:956–960. doi: 10.1038/nm.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharlow E. R., Leimgruber S., Murray S., Lira A., Sciotti R. J., Hickman M., Hudson T., Leed S., Caridha D., Barrios A. M., Close D., Grogl M., Lazo J. S. ACS Chem. Biol. 2014;9:663–672. doi: 10.1021/cb400800q. [DOI] [PubMed] [Google Scholar]
- Cassetta M. I., Marzo T., Fallani S., Novelli A., Messori L. BioMetals. 2014;27:787–791. doi: 10.1007/s10534-014-9743-6. [DOI] [PubMed] [Google Scholar]
- Ma J., Stoter G., Verweij J., Schellens J. H. Cancer Chemother. Pharmacol. 1996;38:391–394. doi: 10.1007/s002800050501. [DOI] [PubMed] [Google Scholar]
- Scheffler H., You Y., Ott I. Polyhedron. 2010;29:66–69. [Google Scholar]
- Lu J., Vlamis-Gardikas A., Kandasamy K., Zhao R., Gustafsson T. N., Engstrand L., Hoffner S., Engman L., Holmgren A. FASEB J. 2013;27:1394–1403. doi: 10.1096/fj.12-223305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.