Abstract
The present study aimed to identify novel intervertebral disc degeneration (IDD)-associated long noncoding (lnc)RNAs and genes. The lncRNA and mRNA microarray dataset GSE56081 was downloaded from the Gene Expression Omnibus database and included 5 samples from patients with degenerative lumbar nucleus pulposus and 5 normal controls. Differentially expressed lncRNAs or differentially expressed genes (DEGs) were identified and co-expression network analysis was performed followed by functional analysis for genes in the network. Additionally, a microRNA (miRNA)-lncRNA-mRNA competing endogenous RNA (ceRNA) regulatory network was constructed based on DEGs and lncRNAs in the co-expression network. Furthermore, a literature search was performed to identify specific miRNAs that had been previously associated with IDD and a specific miRNA-associated ceRNA network was extracted from the co-expression network. A total of 967 genes and 137 lncRNAs were differentially expressed between IDD samples and controls. A co-expression network was constructed and contained 39 differentially expressed lncRNAs and 209 DEGs, which were primarily involved in ‘skeletal system development’, ‘response to mechanical stimulus’ and ‘bone development’. Furthermore, a ceRNA network was established, including 79 miRNAs, 9 downregulated lncRNAs and 148 DEGs. The identified miRNAs included a previously reported disease-associated miRNA, hsa-miR-140. The present study demonstrated that hsa-miR-140 was regulated by three lncRNAs in the hsa-miR-140-associated ceRNA network, including KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1), OIP5 antisense RNA 1 (OIP5-AS1) and UGDH antisense RNA 1 (UGDH-AS1). KCNQ1OT1 was co-expressed with neurochondrin (NCDN) and lon peptidase 2, peroxisomal. In addition, the lncRNAs OIP5-AS1 and UGDH-AS1 targeted several overlapping co-expressed genes, including forkhead box F1 (FOXF1) and polycystin 1, transient receptor potential channel interacting (PKD1). Therefore, KCNQ1OT1 may regulate the expression of NCDN, and OIP5-AS1 and UGDH-AS1 may affect the expression of FOXF1 and PKD1 in IDD. Further experiments are required to validate the results of the present study, which may provide valuable insights into the identification of novel biomarkers associated with IDD.
Keywords: intervertebral disc degeneration, differentially expressed long noncoding RNAs, co-expression network, competing endogenous RNA
Introduction
Intervertebral disc degeneration (IDD) is among the causes of lower back pain, which leads to disability and societal burden, and has a negative socioeconomic impact worldwide (1). It is estimated that worldwide, lower back pain affects ≤84% of people at a certain point in their lifetime, with 10% becoming chronically disabled (2). Currently, therapies for IDD primarily aim to relieve pain symptoms, offering only temporary benefits rather than a permanent cure (3). Therefore, it is necessary to elucidate the underlying mechanisms of IDD and lower back pain in order to develop novel effective treatment methods for these conditions.
The intervertebral disc is a complex structure that consists of three anatomical sub-structures, including the nucleus pulposus (NP), annulus fibrosus and cartilaginous endplates, which are essential for the maintenance of normal function of the disc (4). Early IDD typically occurs in the central NP and is characterized by a decrease in the number of active cells and a loss of extracellular matrix within the NP region (5). Previous studies have identified certain molecules associated with IDD (6,7). For instance, one study reported that early IDD is associated with the downregulation of caveolin-1 expression and inhibition of canonical Wnt signaling (6). Furthermore, a recent study indicated that long non-coding RNAs (lncRNAs), a subset of non-protein-coding transcripts that are >200 nucleotides in length, are involved in a series of biological processes (8). Additionally, competing endogenous RNAs (ceRNAs) participate in microRNA (miRNA)-dependent crosstalk and contribute to the complexity of gene regulation, and may aid in the understanding of disease pathobiology (9). lncRNAs regulate the expression of neighboring coding genes through various mechanisms and may function as ceRNAs to modulate the derepression of miRNA targets (10). In recent years, the roles of lncRNAs in several diseases and cancers have been investigated (11,12). However, potential roles of lncRNAs in IDD are yet to be elucidated.
Wan et al (13) performed microarray data profiling of NP samples from patients with IDD and identified numerous differentially expressed lncRNAs and mRNAs in degenerative discs. Reanalysis of previous datasets using different, updated methods may lead to the identification of novel molecular markers associated with the analyzed dataset (14,15). In the present study, lncRNA and mRNA microarray data from patients with IDD, including 5 samples with degenerative lumbar NP and 5 normal controls, were downloaded and reanalyzed. Differentially expressed lncRNAs or differentially expressed genes (DEGs) were identified and co-expression network analysis was performed, followed by functional analysis of genes in the network. Additionally, a ceRNA regulatory network was constructed based on DEGs and lncRNAs in the co-expression network. Furthermore, a literature search was performed to identify specific miRNAs associated with IDD and a specific miRNA-associated ceRNA network was extracted from the co-expression network. The present study aimed to identify novel IDD-associated lncRNAs and genes, which may guide further investigations and contribute to the development of novel therapeutic strategies for the treatment of IDD.
Materials and methods
Collection of microarray data
The lncRNA and mRNA microarray of IDD was downloaded from the Gene Expression Omnibus database (16) (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE56081 (13), which is a subseries of the SuperSeries GSE67567 (13,17,18). The Arraystar Human lncRNA microarray (version 2.0; Agilent_033010 Probe Name version; Arraystar Inc., Rockville, MD, USA) platform was used. A total of 5 degenerative lumbar NP samples derived from patients with IDD and 5 non-degenerative specimens from cadaveric donors were included in this dataset. As described in the original study by Wan et al (13), all specimens were collected within 3 h following disc excision (for discs from patients with IDD) or mortality (for discs from cadaveric donors).
Data preprocessing and DEG screening
Raw data was initially normalized using the robust multi-array average method (19). Subsequently, limma package version 3.28.21 (20) of Bioconductor 3.5 (http://www.bioconductor.org/packages/3.5/bioc/html/limma.html) was adopted to identify DEGs between the IDD samples and normal controls. False discovery rate (FDR) using Benjamini-Hochberg method (21) in multi-test package version 2.2.0 of Bioconductor (22) (http://www.bioconductor.org/packages/2.5/bioc/html/multtest.html) was used to correct multiple testing to avoid false positives. Only genes and lncRNAs with |log2fold change (FC)| >1 and FDR <0.05 were selected as significantly differentially expressed. Sequences and probe IDs were provided by the platform but not gene and lncRNA names or symbols, therefore, the sequences were aligned with hg19 genome annotation (Ensembl) (23) to identify genes corresponding with each ID. Furthermore, heatmaps were constructed to present expression profiles of differentially expressed lncRNAs and DEGs using hierarchical clustering, which was performed using pheatmap package (http://cran.r-project.org/web/packages/pheatmap/index.html) in R based on Euclidean distances.
Co-expression network analysis of differentially expressed lncRNAs and DEGs
To determine the functions of differentially expressed lncRNAs, Pearson correlation analysis of the DEGs and differentially expressed lncRNAs was performed using the cor() function in R. Pearson correlation coefficients between-1 and 0 or 0 and 1 were considered to represent positive and negative correlation, respectively. In the present study, co-expressed lncRNA-mRNA pairs were identified when absolute Pearson correlation coefficient was >0.6. In addition, lncRNA-mRNA co-expression network was visualized using Cytoscape version 2.8.0 (24).
Functional analysis of genes in the network
An ontology-based tool, clusterProfiler, offers three methods for biological term classification and enrichment analyses of genes, including group Gene Ontology (GO), enrichGO and enrich Kyoto Encyclopedia of Genes and Genomes (KEGG) (25). In the present study, in order to elucidate the biological function of genes enriched in the co-expression network, GO biological process (BP) terms and KEGG pathway analysis of genes in the network were performed using enrichGO and enrichKEGG functions in R package clusterProfiler with P<0.05 as a cutoff value (25).
Construction of ceRNA regulatory network
To elucidate the roles of lncRNAs and mRNAs in the co-expression network, data pertaining to lncRNAs and mRNAs was combined with miRNA data in order to construct a miRNA-lncRNA-mRNA ceRNA regulatory network. Initially, miRNAs that had regulatory associations with lncRNAs in the co-expression network were searched for in two databases; miRcode (26) (http://www.mircode.org/) and starBase (27) (http://starbase.sysu.edu.cn/). Subsequently, the targets of the identified miRNAs were determined using the miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw), which includes experimentally validated microRNA-target interactions (28). The overlapping genes between targets of the identified miRNAs and mRNAs contained in the co-expression network were retained for ceRNA construction. ceRNA and disease-specific ceRNA networks were subsequently constructed. Based on the identified miRNAs, miRNA targets in the co-expression network and differentially expressed lncRNAs, a ceRNA regulatory network was constructed and visualized using Cytoscape version 2.8.0 (24). Furthermore, specific miRNAs associated with IDD were identified in previously reported articles by combining search terms ‘intervertebral disc degeneration’ AND ‘human’ AND ‘miRNA’ in PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), and a IDD-specific ceRNA network containing specific miRNAs was extracted from the co-expression ceRNA-regulated network and visualized using Cytoscape version 2.8.0 (24).
Results
Identification of DEGs and differentially expressed lncRNAs
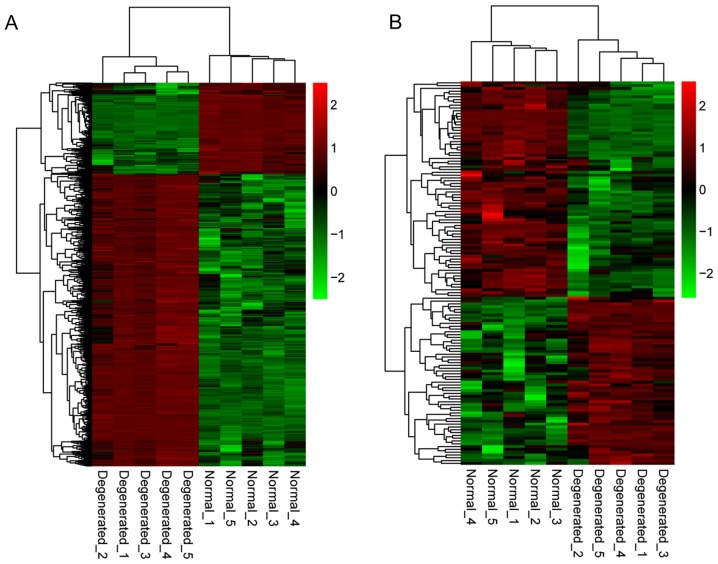
Following data normalization, differential expression analysis of genes and lncRNAs was performed using the dataset that included 5 IDD samples and 5 controls. A total of 967 genes and 137 lncRNAs were significantly differentially expressed within the FDR <0.05 and |log2FC| >1 criteria. Among them, 737 genes were upregulated and 230 genes were downregulated; 60 lncRNAs were upregulated and 77 lncRNAs were downregulated. Additionally, hierarchical clustering of gene expression levels and lncRNA expression levels indicated clustering of normal groups and IDD groups (Fig. 1).
Figure 1.
Heatmaps of DEGs and differentially expressed lncRNAs. (A) Heatmap of DEGs between IDD and non-degenerative control samples. (B) Heatmap of differentially expressed lncRNAs between IDD and non-degenerative samples. Red color indicates upregulation and green color indicates downregulation. Normal and degenerated refer to samples obtained from non-degenerative controls and patients with IDD, respectively. DEGs, differentially expressed genes; lncRNA, long noncoding RNA; IDD, intervertebral disc degeneration.
Differential lncRNA-mRNA co-expression network analysis
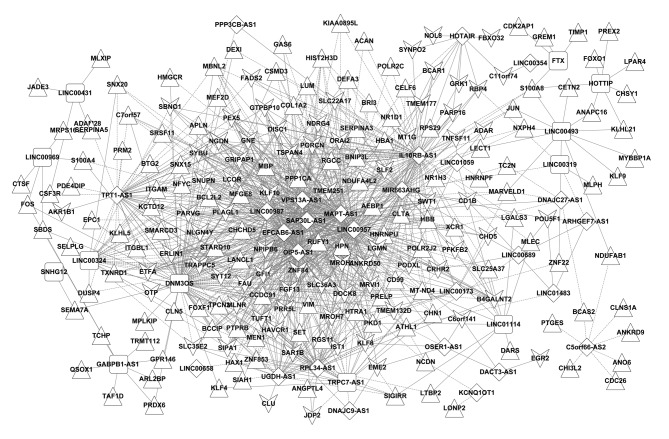
Differentially expressed lncRNAs and DEGs with Pearson correlation coefficients >0.6 were selected. The lncRNA-mRNA co-expression network, containing 248 nodes and 1,082 edges (448 negative and 634 positive connections), was constructed (Fig. 2). A total of 39 differentially expressed lncRNAs (27 downregulated and 12 upregulated) and 209 DEGs (75 downregulated and 134 upregulated) were included in this co-expression network.
Figure 2.
lncRNA-mRNA co-expression network. Triangular and vee-shaped nodes represent upregulated and downregulated genes, respectively. Diamond-shaped and rectangular nodes indicate upregulated and downregulated lncRNAs, respectively. lncRNA, long noncoding RNA. Continuous line represents the connections between lncRNA and miRNA; dotted lines represent connections between miRNA and gene or lncRNA and gene.
Functional analysis of genes and lncRNAs in the co-expression network
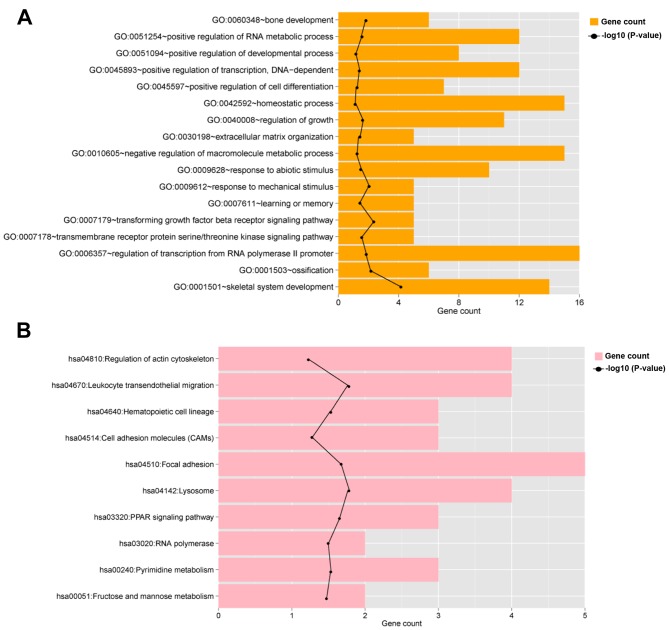
To determine the potential functions of the identified DEGs and lncRNAs in the co-expression network, GO and KEGG pathway enrichment analyses were performed in the co-expression network. The analyses revealed the significant enrichment of 17 GO BP terms and 10 KEGG pathways (Table I; Fig. 3). BP and KEGG terms that were significantly associated with members of the co-expression network included ‘skeletal system development’, ‘response to mechanical stimulus’, ‘bone development’ and ‘focal adhesion’ (Table I; Fig. 3).
Table I.
GO and KEGG pathway enrichment analysis for genes in the lncRNA-mRNA co-expression network.
| A, GO terms enriched in the lncRNA-mRNA co-expression network | |||
|---|---|---|---|
| Description (ID, name) | Count | FDR | Genes |
| GO:0001501, skeletal system development | 14 | 0.000067700 | RBP4, AEBP1, LGALS3, TUFT1, KLF10, PRELP, MEN1, MEF2D, SBDS, LECT1, TNFSF11, COL1A2, ACAN, PKD1 |
| GO:0009612, response to mechanical stimulus | 5 | 0.003660942 | FOS, BTG2, JUN, PKD1, GFI1 |
| GO:0007179, transforming growth factor beta receptor signaling pathway | 5 | 0.004694322 | FOS, LTBP2, KLF10, JUN, COL1A2 |
| GO:0040008, regulation of growth | 11 | 0.005411270 | MEN1, EPC1, RBP4, HTRA1, LGMN, BCAR1, NOL8, SIPA1, PEX5, GREM1, GAS6 |
| GO:0001503, ossification | 6 | 0.009830659 | MEN1, MEF2D, SBDS, TNFSF11, TUFT1, KLF10 |
| GO:0060348, bone development | 6 | 0.012887071 | MEN1, MEF2D, SBDS, TNFSF11, TUFT1, KLF10 |
| GO:0006357, regulation of transcription from RNA polymerase II promoter | 16 | 0.017700242 | JDP2, EGR2, KLF9, KLF10, FOXO1, NFYC, MEN1, PLAGL1, FOS, EPC1, SMARCD3, JUN, GFI1, LCOR, KLF4, NR1H3 |
| GO:0045893, positive regulation of transcription, DNA-dependent | 12 | 0.019627483 | PLAGL1, MEN1, FOS, EPC1, EGR2, SMARCD3, JUN, FOXF1, NFYC, FOXO1, KLF4, NR1H3 |
| GO:0051254, positive regulation of RNA metabolic process | 12 | 0.020731482 | PLAGL1, MEN1, FOS, EPC1, EGR2, SMARCD3, JUN, FOXF1, NFYC, FOXO1, KLF4, NR1H3 |
| GO:0009628, response to abiotic stimulus | 10 | 0.024048392 | MEN1, FOS, RBP4, TNFSF11, BTG2, HMGCR, JUN, PKD1, GFI1, GRK1 |
| GO:0007178, transmembrane receptor protein serine/threonine kinase signaling pathway | 5 | 0.029381231 | FOS, LTBP2, KLF10, JUN, COL1A2 |
| GO:0030198, extracellular matrix organization | 5 | 0.030296507 | LGALS3, LUM, FOXF1, COL1A2, ACAN |
| GO:0007611, learning or memory | 5 | 0.037173540 | FOS, SBDS, EGR2, HMGCR, JUN |
| GO:0010605, negative regulation of macromolecule metabolic process | 15 | 0.038521124 | JDP2, KLF10, CDC26, SIGIRR, TIMP1, MEN1, EPC1, SET, JUN, SERPINA5, BNIP3L, GFI1, LCOR, KLF4, ADAR |
| GO:0051094, positive regulation of developmental process | 8 | 0.038680685 | MEN1, TNFSF11, KLF10, JUN, CLU, GFI1, OTP, ANGPTL4 |
| GO:0042592, homeostatic process | 15 | 0.045211148 | CLNS1A, RBP4, EGR2, NCDN, MBP, TIMP1, MEN1, TNFSF11, PRDX6, JUN, SERPINA3, TXNRD1, XCR1, QSOX1, CLN5 |
| GO:0045597, positive regulation of cell differentiation | 7 | 0.045820821 | MEN1, TNFSF11, KLF10, JUN, CLU, GFI1, OTP |
| B, KEGG pathways enriched in the lncRNA-mRNA co-expression network | |||
| Description (entry, name) | Count | FDR | Genes |
| hsa04142, Lysosome | 4 | 0.014272775 | CLTA, LGMN, CLN5, CTSF |
| hsa04670, Leukocyte transendothelial migration | 4 | 0.014534463 | BCAR1, SIPA1, CD99, ITGAM |
| hsa03320, PPAR signaling pathway | 3 | 0.018064527 | FADS2, ANGPTL4, NR1H3 |
| hsa04510, Focal adhesion | 5 | 0.018720786 | PARVG, PPP1CA, JUN, BCAR1, COL1A2 |
| hsa04640, Hematopoietic cell lineage | 3 | 0.025076738 | CD1B, CSF3R, ITGAM |
| hsa03020, RNA polymerase | 2 | 0.027129221 | POLR2C, POLR2J2 |
| hsa00240, Pyrimidine metabolism | 3 | 0.028840787 | TXNRD1, POLR2C, POLR2J2 |
| hsa00051, Fructose and mannose metabolism | 2 | 0.031922905 | PFKFB2, AKR1B1 |
| hsa04810, Regulation of actin cytoskeleton | 4 | 0.043576076 | PPP1CA, BCAR1, FGF13, ITGAM |
| hsa04514, Cell adhesion molecules (CAMs) | 3 | 0.043833628 | CD99, SELPLG, ITGAM |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, long noncoding RNA; count, number of genes enriched in the GO terms or pathways; FDR, false discovery rate.
Figure 3.
Functional analysis of members of the long noncoding RNA-mRNA co-expression network. Bar plots presenting enriched (A) GO biological process terms and (B) Kyoto Encyclopedia of Genes and Genomes pathways in the co-expression network. The black line indicates-log10 (P-value). GO, Gene Ontology.
Construction of ceRNA regulatory network and disease-specific ceRNA network
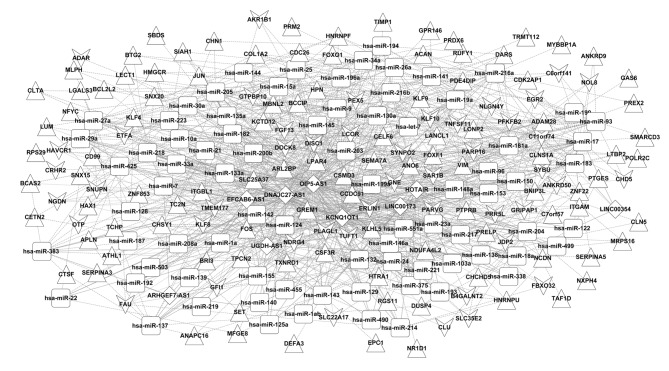
A total of 79 associated miRNAs, which were targeted by 9 lncRNAs in the co-expression network, were screened by integrating information from miRcode and starBase databases to determine regulatory associations between miRNAs and lncRNAs. A total of 647 pairs of miRNA-gene interactions were obtained by identifying targets of these miRNAs from genes in the co-expression network using the miRTarBase database. The ceRNA regulatory network was therefore constructed based on combining the identified miRNA-gene and lncRNA-miRNA associations (Fig. 4). The ceRNA network was composed of 236 nodes [79 miRNAs, 9 downregulated lncRNAs, 148 DEGs (including 46 downregulated and 102 upregulated DEGs)] and 1,065 edges (647 miRNA-gene connections, 142 lncRNA-gene connections and 276 lncRNA-miRNAs connections).
Figure 4.
Construction of ceRNA network. Triangular and vee-shaped nodes denote upregulated and downregulated genes, respectively. Diamond-shaped nodes indicate downregulated long noncoding RNAs and rectangular nodes indicate miRNAs. ceRNA, competing endogenous RNA; miRNA/miR, microRNA.
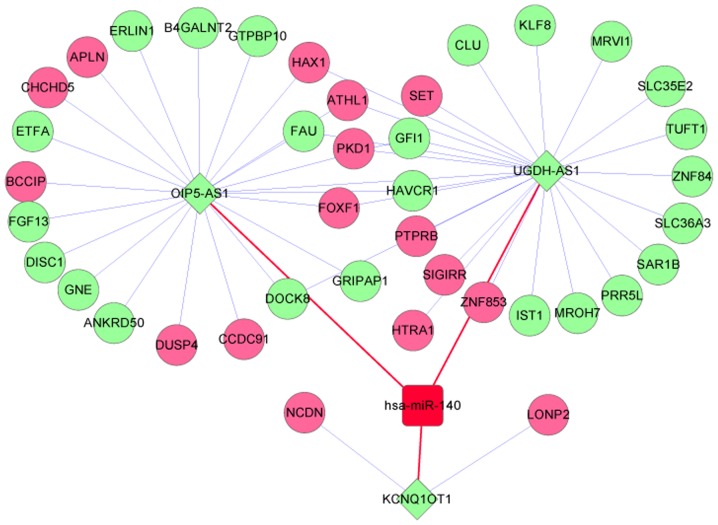
Furthermore, IDD-associated miRNAs from previous studies were searched and the collected data indicated that hsa-miR-140 may be implicated in human IDD (29,30). Subsequently, connections between hsa-miR-140 and other DEGs, lncRNAs and miRNAs were extracted from the co-expression ceRNA-regulated network and a hsa-miR-140-associated ceRNA network was constructed (Fig. 5). The disease-specific ceRNA network included 47 nodes (1 miRNA, 3 lncRNAs, 27 downregulated DEGs, and 16 upregulated DEGs) and 55 edges. hsa-miR-140 was demonstrated to be regulated by three lncRNAs in the hsa-miR-140-associated ceRNA network, including KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1), OIP5 antisense RNA 1 (OIP5-AS1) and UGDH antisense RNA 1 (UGDH-AS1). The results of the present study indicated that the aforementioned lncRNAs may be associated with IDD and the genes that are co-expressed with these lncRNAs may also be associated with IDD. KCNQ1OT1 was co-expressed with neurochondrin (NCDN) and lon peptidase 2, peroxisomal (LONP2). lncRNAs OIP5-AS1 and UGDH-AS1 were co-expressed with several overlapping genes, including forkhead box F1 (FOXF1) and polycystin 1, transient receptor potential channel interacting (PKD1).
Figure 5.
Disease-specific competing endogenous RNA network. Pink and green circular nodes denote upregulated and downregulated genes, respectively. Green diamond-shaped nodes denote downregulated lncRNAs and red square nodes indicate intervertebral disc degeneration-associated miRNAs. Blue lines represent co-expression associations between lncRNAs and genes. Red lines indicate lncRNA-miRNA regulation associations. lncRNA, long noncoding RNA; miRNA, microRNA.
Discussion
In the present study, 967 genes (737 upregulated and 230 downregulated) and 137 lncRNAs (60 upregulated and 77 downregulated) were differentially expressed between the IDD samples and controls. A co-expression network was constructed containing 39 differentially expressed lncRNAs and 209 DEGs that were primarily involved in ‘skeletal system development’, ‘response to mechanical stimulus’, ‘bone development’ and ‘focal adhesion’. Furthermore, a ceRNA network was established, which included 79 miRNAs, 9 downregulated lncRNAs and 148 DEGs. Among the miRNAs, hsa-miR-140 was a previously reported disease-associated miRNA and therefore, a hsa-miR-140-associated ceRNA network was extracted. It was determined that hsa-miR-140 was regulated by three lncRNAs, including KCNQ1OT1, OIP5-AS1 and UGDH-AS1. KCNQ1OT1 was co-expressed with NCDN and LONP2. In addition, lncRNAs OIP5-AS1 and UGDH-AS1 targeted several overlapping co-expressed genes, including FOXF1 and PKD1.
Aberrant expression of miR-140 was reported to contribute to osteoarthritis, a degenerative disease characterized by pathological alterations similar to those identified in patients with IDD (31,32). miRNA-140 has also been demonstrated to serve roles in cartilage development and homeostasis by regulating the expression of matrix metalloprotease-13, insulin-like growth factor binding protein-5, Sp1 transcription factor, interleukin-1 and Smad family member 3 (31,33,34). The results of the present study support the hypothesis that miR-140 serves a role in the pathogenesis of degenerative disorders such as IDD. The present study identified three lncRNAs, KCNQ1OT1, OIP5-AS1 and UGDH-AS1, which were associated with disease-specific miRNAs, including miR-140.
KCNQ1OT1 has been demonstrated to mediate transcriptional silencing (35). In the miR-140-associated ceRNA network, KCNQ1OT1 was co-expressed with NCDN, which was enriched in the GO term ‘positive regulation of developmental process’. A recent study reported that NCDN served as a neuronal target antigen in patients with autoimmune cerebellar degeneration (36). NCDN, a cytoplasmic leucine-rich protein that functions in neurite outgrowth and chondrocyte differentiation, was demonstrated to be NP-specific and age-associated, indicating its potential functional implication in the development of NP (37). Therefore, we hypothesize that the downregulation of KCNQ1OT1 may be associated with the degeneration of the intervertebral disc through the regulation of NCDN expression.
Previous studies have demonstrated that lncRNA OIP5-AS1 acts as a sponge to prevent RNA-binding protein HuR from binding to its target mRNA and reduce cell proliferation (38). Stimulation of disc cell proliferation and accumulation of extracellular matrix may slow or even reverse the degenerative alterations resulting from IDD (7). To the best of our knowledge, the role of UGDH-AS1 has not been previously investigated. In the present study, OIP5-AS1 and UGDH-AS1 shared several overlapping co-expressed genes, including FOXF1 and PKD1. FOXF1 is a member of the forkhead family of transcription factors, which are characterized by a distinct forkhead domain (39). A previous study indicated that FOXF1 promoted the production of extracellular matrix (40). Importantly, IDD is associated with an increased breakdown of extracellular matrix (41). Boulter et al (42) reported that PKD1 may serve a role in skeletal development. Based on the results of the aforementioned studies and data analyzed in the present study, it may be hypothesized that the lncRNAs OIP5-AS1 and UGDH-AS1 are involved in the molecular mechanisms underlying IDD by regulating the expression of FOXF1 and PKD1. The results of the present study require further investigation to clarify and confirm the roles of the identified molecules in IDD.
The present study had several limitations, including the small sample size of the dataset employed; further analyses with an increased sample size are necessary to confirm the results. Furthermore, non-degenerative specimens from cadaveric donors were used as controls in the dataset that was analyzed in the present study. Patients with normal discs or with mild disc degeneration are not typically subjected to surgical intervention. The use of normal discs from cadaveric donors reflected limited access to less degenerated tissues from surgical patients. However, cadaveric tissues were collected within 3 h following mortality to ensure that the tissues remained intact (13). Additionally, the results of the analysis in the present study were not experimentally verified. IDD is a complex and multi-factorial process that is influenced by genetic predispositions, aging, biomechanical loading and physical activities, lifestyle and other health-associated factors, including diabetes (43). Numerous clinical investigations have indicated that cell-based biological therapies, including disc cell augmentation, application of growth factors/cytokines and gene therapy, may be employed for the treatment of IDD (44,45). The development of biomarkers for IDD is valuable in the early stages of disc degeneration (45). lncRNAs have been implicated in multiple pathological processes of IDD (46), and lncRNAs and DEGs in patients with disc degeneration may have potential as diagnostic or prognostic biomarkers (47). In order to determine the potential for the clinical application of lncRNAs and DEGs identified in the present study, an in vivo murine IDD model may be used to confirm the roles of these molecules in IDD in future studies.
In conclusion, the present study identified several potential genes and lncRNAs, including KCNQ1OT1, OIP5-AS1, UGDH-AS1, NCDN, FOXF1 and PKD1, that were associated with IDD. KCNQ1OT1 may be involved in the degeneration of the intervertebral disc by regulating the expression of NCDN, while the lncRNAs OIP5-AS1 and UGDH-AS1 may be implicated in the molecular mechanisms of IDD by affecting the expression of FOXF1 and PKD1. The results of the present study may contribute to elucidating the molecular pathogenesis of IDD and aid in the identification of novel biomarkers associated with IDD.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZGQ, ZQ and YL provided the concept and design of the research. QZ, ZYW and QS acquired the data. XZ, CF and ZW analyzed and interpreted the data. FX, YDL and YW performed the statistical analysis. ZGQ and ZQ drafted the manuscript. YL and ZW revised the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Vadalà G, Russo F, Ambrosio L, Papalia R, Denaro V. Mesenchymal stem cells for intervertebral disc regeneration. J Biol Regul Homeost Agents. 2016;30(4 Suppl 1):S173–S179. [PubMed] [Google Scholar]
- 2.Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat Rev Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gantenbein B, Illien-Jünger S, Chan SC, Walser J, Haglund L, Ferguson SJ, Iatridis C J, Grad S. Organ culture bioreactors-platforms to study human intervertebral disc degeneration and regenerative therapy. Curr Stem Cell Res Ther. 2015;10:339–352. doi: 10.2174/1574888X10666150312102948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombier P, Clouet J, Hamel O, Lescaudron L, Guicheux J. The lumbar intervertebral disc: From embryonic development to degeneration. Joint Bone Spine. 2014;81:125–129. doi: 10.1016/j.jbspin.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Yang W, Yu XH, Wang C, He WS, Zhang SJ, Yan YG, Zhang J, Xiang YX, Wang WJ. Interleukin-1β in intervertebral disk degeneration. Clin Chim Acta. 2015;450:262–272. doi: 10.1016/j.cca.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 6.Smolders LA, Meij BP, Onis D, Riemers FM, Bergknut N, Wubbolts R, Grinwis GC, Houweling M, Koerkamp MJ, van Leenen D, et al. Gene expression profiling of early intervertebral disc degeneration reveals a down-regulation of canonical Wnt signaling and caveolin-1 expression: implications for development of regenerative strategies. Arthritis Res Ther. 2013;15:R23. doi: 10.1186/ar4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang SZ, Rui YF, Tan Q, Wang C. Enhancing intervertebral disc repair and regeneration through biology: Platelet-rich plasma as an alternative strategy. Arthritis Res Ther. 2013;15:220. doi: 10.1186/ar4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen K, Sarkar A, Maji RK, Ghosh Z, Gupta S, Ghosh TC. Deciphering the cross-talking of human competitive endogenous RNAs in K562 chronic myelogenous leukemia cell line. Mol Biosyst. 2016;12:3633–3642. doi: 10.1039/C6MB00568C. [DOI] [PubMed] [Google Scholar]
- 11.Chen EG, Zhang JS, Xu S, Zhu XJ, Hu HH. Long non-coding RNA DGCR5 is involved in the regulation of proliferation, migration and invasion of lung cancer by targeting miR-1180. Am J Cancer Res. 2017;7:1463–1475. [PMC free article] [PubMed] [Google Scholar]
- 12.Fayda M, Isin M, Tambas M, Guveli M, Meral R, Altun M, Sahin D, Ozkan G, Sanli Y, Isin H, et al. Do circulating long non-coding RNAs (lncRNAs) (LincRNA-p21, GAS 5, HOTAIR) predict the treatment response in patients with head and neck cancer treated with chemoradiotherapy? Tumour Biol. 2016;37:3969–3978. doi: 10.1007/s13277-015-4189-1. [DOI] [PubMed] [Google Scholar]
- 13.Wan ZY, Song F, Sun Z, Chen YF, Zhang WL, Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: A microarray related study. Arthritis Res Ther. 2014;16:465. doi: 10.1186/s13075-014-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C, Nie F, Qin Z, Li B, Zhao X. A snapshot of gene expression signatures generated using microarray datasets associated with excessive scarring. Am J Dermatopathol. 2013;35:64–73. doi: 10.1097/DAD.0b013e31825ba13f. [DOI] [PubMed] [Google Scholar]
- 15.Minsky N, Roeder RG. Inhibition of adhesion molecule gene expression and cell adhesion by the metabolic regulator PGC-1α. PLoS One. 2016;11:e0165598. doi: 10.1371/journal.pone.0165598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Che L, Xie YK, Hu QJ, Ma CJ, Pei YJ, Wu ZG, Liu ZH, Fan LY, Wang HQ. Noncoding RNAs in human intervertebral disc degeneration: An integrated microarray study. Genom Data. 2015;5:80–81. doi: 10.1016/j.gdata.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan PH, Liu ZH, Pei YJ, Wu ZG, Yu Y, Yang YF, Liu X, Che L, Ma CJ, Xie YK, et al. Landscape of RNAs in human lumbar disc degeneration. Oncotarget. 2016;7:63166–63176. doi: 10.18632/oncotarget.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: From disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1869. [DOI] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 22.Pollard KS, Dudoit S, Van Der Laan MJ. Multiple testing procedures: The multtest package and applications to genomics, in Bioinformatics and computational biology solutions using R and bioconductor. Springer; 2005. pp. 249–271. [Google Scholar]
- 23.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Banet Fernandez J, Billis K, Giron Garcia C, Hourlier T, et al. The Ensembl gene annotation system. Database (Oxford) 2016;2016:pii:baw093. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2010;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeggari A, Marks DS, Larsson E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JH, Liu S, Zhou H, Qu LH, Yang JH. Starbase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou CH, Chang NW, Sirjana S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swingler TE, Wheeler G, Carmont V, Elliott HR, Barter MJ, Abuelmagd M, Donell ST, Boothandford RP, Hajihosseini MK, Münsterberg A, et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64:1909–1919. doi: 10.1002/art.34314. [DOI] [PubMed] [Google Scholar]
- 30.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 31.Tardif G, Hum D, Pelletier JP, Duval N, Martel-Pelletier J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord. 2009;10:148. doi: 10.1186/1471-2474-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tardif G, Pelletier JP, Fahmi H, Hum D, Zhang Y, Kapoor M, Martel-Pelletier J. NFAT3 and TGF-β/SMAD3 regulate the expression of miR-140 in osteoarthritis. Arthritis Res Ther. 2013;15:R197. doi: 10.1186/ar4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Qin S, Yi C, Ma G, Zhu H, Zhou W, Xiong Y, Zhu X, Wang Y, He L, Guo X. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 2011;585:2992–2997. doi: 10.1016/j.febslet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK, Asahara H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P, Kanduri C. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miske R, Gross CC, Scharf M, Golombeck KS, Hartwig M, Bhatia U, Schulte-Mecklenbeck A, Bönte K, Strippel C, Schöls L, et al. Neurochondrin is a neuronal target antigen in autoimmune cerebellar degeneration. Neurol Neuroimmunol Neuroinflamm. 2017;4:e307. doi: 10.1212/NXI.0000000000000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang X, Jing L, Chen J. Changes in the molecular phenotype of nucleus pulposus cells with intervertebral disc aging. PLoS One. 2012;7:e52020. doi: 10.1371/journal.pone.0052020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Abdelmohsen K, Yang X, De S, Grammatikakis I, Noh JH, Gorospe M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016;44:2378–2392. doi: 10.1093/nar/gkw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahlapuu M, Enerbäck S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development. 2001;128:2397–2406. doi: 10.1242/dev.128.12.2397. [DOI] [PubMed] [Google Scholar]
- 40.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- 41.Mccann MR, Séguin CA. Notochord cells in intervertebral disc development and degeneration. J Dev Biol. 2016;4:1–18. doi: 10.3390/jdb4010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulter C, Mulroy S, Webb S, Fleming S, Brindle K, Sandford R. Cardiovascular, skeletal, and renal defects in mice with a targeted disruption of the Pkd1 gene. Proc Natl Acad Sci USA. 2001;98:12174–12179. doi: 10.1073/pnas.211191098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13:318–330. doi: 10.1016/j.spinee.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Gruber HE, Leslie K, Ingram J, Norton HJ, Hanley EN. Cell-based tissue engineering for the intervertebral disc: In vitro studies of human disc cell gene expression and matrix production within selected cell carriers. Spine J. 2004;4:44–55. doi: 10.1016/S1529-9430(03)00425-X. [DOI] [PubMed] [Google Scholar]
- 45.Gruber HE, Hanley EN. Do we need biomarkers for disc degeneration? Biomark Insights. 2007;1:131–133. [PMC free article] [PubMed] [Google Scholar]
- 46.Chen WK, Yu XH, Yang W, Wang C, He WS, Yan YG, Zhang J, Wang WJ. lncRNAs: Novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50 doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.