Abstract
Recent evidence has suggested that competitive endogenous RNAs (ceRNAs) are important regulatory molecules in clear cell kidney carcinoma (KIRC) and their dysregulation may contribute to cancer pathogenesis. However, the critical roles of dysregulated ceRNAs in KIRC remain unknown. In the present study, a KIRC dysregulated ceRNA-ceRNA network (KDCCNet) was constructed based on the ‘ceRNA hypothesis’ by integrating microRNA regulation and expression profiles in cancerous and normal tissues. Two dysregulated patterns of ceRNAs interaction (gain and loss) exist in KDCCNet. The two modules, which are 95% loss interactions and 97% gain interactions, were demonstrated to be able to distinguish normal samples from cancer samples. Two long non-coding (lnc)-RNAs (glucuronidase β pseudogene 11 and LIFR antisense RNA 1) demonstrated significant associations with KIRC prognosis. The present study of the KDCCNet revealed a novel biological mechanism for KIRC and provides novel lncRNAs as candidate prognostic biomarkers.
Keywords: competing endogenous RNAs, network, long noncoding RNAs, clear cell kidney carcinoma
Introduction
Human genome sequence data indicates that >98% of human genome transcripts are non-coding RNAs (ncRNAs), while protein-coding genes account for a small proportion of RNAs (1). Advances in the sensitivity of experimental assays have revealed that ncRNAs are involved in a variety of key biological processes (2). ncRNAs are divided into two groups based on length: Long non-coding RNAs (lncRNAs) and small non-coding RNAs. LncRNAs are a class of pervasively transcribed RNA molecules with lengths of >200 nucleotides. A previous study indicated that lncRNAs serve a critical role in regulating cellular processes (3). MicroRNAs (miRNAs) are small single-stranded RNAs of 18–25 nucleotides in length and are among the most important types of small ncRNAs. miRNAs can repress their targets by binding to miRNA response elements (MREs) in the 3′ untranslated region of mRNAs, causing translational repression or mRNA degradation. Notably, each miRNA regulates numerous RNA targets, including mRNA but also lncRNAs; the majority of RNA molecules harbor several MREs and are repressed by different miRNAs. As a result of this targeted multiplicity, lncRNAs and mRNA may compete for limited pools of miRNAs, therefore acting as competitive endogenous RNAs (ceRNAs) (4).
A number of studies have suggested that lncRNAs serve a critical role in regulating cancer processes through a ceRNA-associated mechanism (5–7). Various studies have investigated the ceRNA mechanism in specific types of cancer by constructing ceRNA networks (8–10). However, ceRNA regulation is required for the functioning of normal and cancerous cells. Therefore, detecting dysregulated ceRNA interactions by comparing ceRNA networks in normal and cancer cells would increase the understanding of the initiation and propagation of cancer.
Clear cell kidney carcinoma (KIRC) is the most common type of renal cell carcinoma, accounting for 70–80% of all renal cell carcinoma cases (11). The incidence and fatality rate of KIRC have increased over the past two decades (12). Although a number of studies have examined KIRC, few biomarkers have been identified (13–15). Therefore, further studies of the key molecular mechanisms involved in KIRC and biomarkers for the early detection of patients with KIRC are urgently required. The dysregulated ceRNA network may be useful for investigating the pathogenesis and identifying novel biomarkers of KIRC.
In the present study, to establish an activated dysregulated ceRNA-ceRNA network, the regulatory association of lncRNAs, miRNAs and mRNAs were integrated based on the ‘ceRNA hypothesis’. Next, the expression patterns of lncRNAs, miRNAs and mRNAs were investigated in KIRC and normal tissues. By comparing ceRNA interactions in KIRC and normal tissues, an activated KIRC dysregulated ceRNA-ceRNA network (KDCCNet) was established. Topological feature analysis of KDCCNet demonstrated that lncRNAs are the key components of the KDCCNet. In the KDCCNet, there are two types of dysregulated patterns of ceRNAs interactions, these being gain and loss interactions. A total of two typical modules were discovered, which could distinguish between normal and cancer samples. The results of the present study provide insight into the underlying mechanisms of dysregulated ceRNA of KIRC and a global view of KDCCNet.
Materials and methods
Data resources
KIRC expression profiles of mRNAs, miRNAs and lncRNAs were obtained from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) database release 1.0 including 517 cancer and 71 adjacent normal samples. All of the expression profiles were log2 transformed.
An independent dataset was downloaded from the GEO database (www.ncbi.nlm.nih.gov/geo/; accession number GSE36895), which is supported by the Affymetrix Human genome U133 plus 2.0 array (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA) (16). This dataset contains 29 tumor samples and 23 adjacent normal samples.
In order to investigate human miRNA-target regulatory associations, miRNA target information were collected from 7 databases, which includes miRanda 2010 (17), miRcode 11 (18), miRTarBase 6.0 (19), starBase V2.0 (20), TargetScan 7.1 (21), miRecords 2013 (22) and lnCeDB 2014 (23). Following redundancy analysis and standardization using R version 3.3.3, the databases were processed and 1,698,634 miRNA-mRNA interactions and 613,523 miRNA-lncRNA interactions were obtained. These interactions contained 18,013 mRNAs, 12,154 lncRNAs and 2,586 miRNAs. In addition, 93 KIRC-associated miRNAs were collected from the Human microRNA Disease Database V2.0 (24).
Construction of the ceRNA network
In order to identify ceRNA interactions, the miRNA target interactions were considered as well as the expression associations of the miRNA-mRNA, miRNA-lncRNA and lncRNA-mRNA. In this case two principles, explained below, were followed to identify ceRNA interactions.
Firstly, a hypergeometric test was used to compute the significance of shared miRNAs for each possible lncRNA-mRNA pair. The P-value was calculated according to:
Where N represents the total number of human miRNAs; K represents the total number of miRNAs targeting mRNAs; M represents the total number of miRNAs targeting lncRNAs; and r represents the number of common miRNAs between lncRNA-mRNA pairs. All P-values were subjected to false discovery rate (FDR) correction and pairs with FDR<0.01 were considered as candidate ceRNA interaction pairs.
The Pearson correlation coefficient (R) was computed for each candidate ceRNA pair mentioned above, to identify the active ceRNA pairs in KIRC and normal tissues. Candidate ceRNA pairs with R>0 and P-adjusted<0.01 were identified as ceRNA interactions.
Following assembling all identified ceRNA pairs, the ceRNA network for KIRC and normal tissue was separately generated.
Identification of dysregulated ceRNA interactions
To identify dysregulated ceRNA interactions, the altered R was estimated for the expression of the ceRNA pair in the KIRC samples compared with normal samples. The altered R was estimated as follows:
CorrKIRC (A, B) is the R of a ceRNA pair in the KIRC samples and Corrnormal (A, B) is the R of certain ceRNA pairs in the normal samples.
Based on the assumption that the ΔR of a certain ceRNA pair is significant, the cancer/normal labels 1,000 times were randomly permuted. The significant P-value of each ΔR was given as follows:
When Si =1, it represents that the value of random ΔR for a certain ceRNA pair was greater than the real one, otherwise Si=0.
Identification of KIRC-associated dysregulated ceRNA modules
The edges of the dysregulated ceRNA network were weighted by ΔR. Based on value of ΔR, dysregulated ceRNAs interactions were divided into two patterns, gain ceRNAs interactions (ΔR>0) and loss ceRNAs interactions (ΔR<0). Then the Markov Cluster Algorithm clustering computational method was applied to identify the dysregulated ceRNA modules in GraphWeb (25) (biit.cs.ut.ee/graphweb/; Inflate parameter=1.8, count of nodes >10).
The classification performance of dysregulated modules through unsupervised hierarchical cluster analysis were visualized and 1- R was used as a distance measure between two samples.
Differential expression analysis
Differential expression analysis was performed to identify the genes associated with KIRC. This was achieved by comparing the RNA expression between KIRC samples and normal samples. Differential expression was detected by fold change (FC) and using the limma method (FDR<0.01, logFC>0.5).
Functional enrichment analysis
The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis was performed using an online analysis tool (26) (Gene set enrichment analysis; software.broadinstitute.org/gsea/index.jsp; FDR<0.05).
Survival analysis
The univariate Cox regression was performed in order to evaluate the association between survival and the expression level of each lncRNA. Then a risk score was obtained by combining the Cox regression coefficients and lncRNA expression for each of the patients. The median risk score was used as the cut-off point and the patients were divided into high-risk and low-risk groups. The differences in the survival times were analyzed through the use of a log rank test.
Statistical analysis
Student's t-test was used to compare the difference of degree distribution between mRNA and lncRNA in KDCCNet (27). And the differences of the expression level, the transcript length and the number of exons of dysregulated and non-dysregulated lncRNAs were also compared based on the t-test. P<0.05 was considered to indicate a statically significant difference. The statistical analyses were performed using R version 3.3.3.
Results
Construction and global properties of KDCCNet
Regulatory interactions of ceRNAs are present in normal and cancer cells. In the present study, a two-stage analysis method was introduced for identifying ceRNA regulatory interactions in normal and KIRC tissues. As described in the Materials and methods, 585 and 3,646 ceRNA regulatory interactions were identified in normal tissue and KIRC tissue, respectively. Out of the ceRNA regulatory interactions in normal tissue, there were 67 lncRNAs and 415 mRNAs, while there were 127 lncRNAs and 1,706 mRNAs ceRNA regulatory interactions in KIRC tissue.
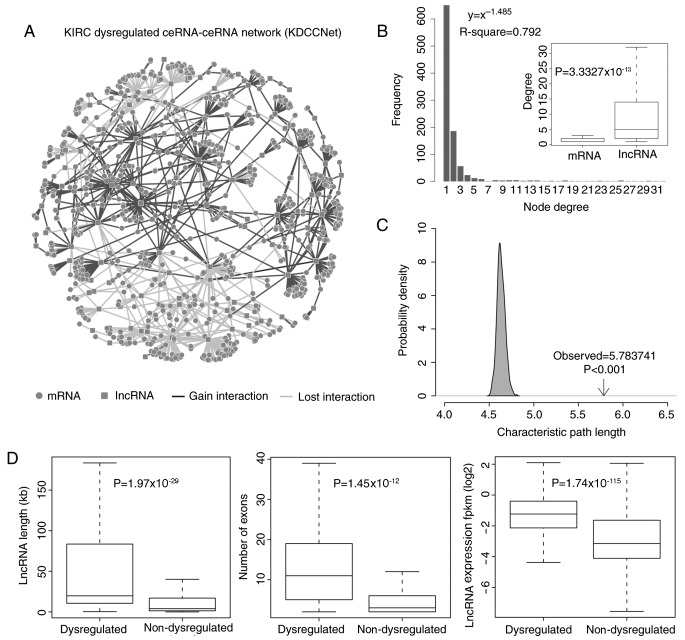
KIRC transformation from normal cells occurs depending upon the dynamics of multiple molecular and physiological alterations, including regulatory interactions of dysregulated ceRNAs (28). To evaluate the roles of dysregulated ceRNA in KIRC, a KDCCNet was constructed using a computational method based on the expression data of 517 KIRC patients and 71 adjacent normal pairs from TCGA. As described above, the KDCCNet was obtained (Fig. 1A). The KDCCNet contained 862 mRNAs, 123 lncRNAs and 1,195 edges. A total of two dysregulated patterns of ceRNAs interaction (gain and loss) exist in KDCCNet. In KDCCNet, 695 gains and 500 losses were detected (Table I). Therefore, certain normal ceRNA regulations may disappear or be reduced, while novel ceRNA regulations may be generated. These results suggest that dysregulated ceRNAs may serve an important role in KIRC progression.
Figure 1.
Layout of KDCCNet and its structural features. (A) The KDCCNet generated in the present study. A circular node marks an mRNA, a square node marks an lncRNA, a dark gray edge represents a gain dysregulation between ceRNAs and a light gray edge represents a lost dysregulation between ceRNAs. (B) Degree distribution of the KDCCNet. Degree distribution of lncRNAs and mRNAs (t test, P-value=3.33×10−13). (C) The characteristic path length of dysregulated network is increased when compared with the random networks. (D) The boxplot depicts the expression level, the transcript length and the number of exons of dysregulated and non-dysregulated lncRNAs. Significance tests based on the Students t-test. lncRNA, long noncoding RNA; ceRNA, competitive endogenous RNA; KDCCNet, clear cell kidney carcinoma dysregulated ceRNA-ceRNA network; fpkm, fragments per kilobase of transcript per million mapped reads.
Table I.
Statistics of the nodes and edges in the competitive endogenous RNAs dysregulated network.
| Type | mRNA | lncRNA | Edges |
|---|---|---|---|
| Dysregulated network | 862 | 123 | 1,195 |
| Gain | 520 | 75 | 695 |
| Loss | 367 | 61 | 500 |
Lnc, long non-coding.
By topological feature analysis, the topological features of the KDCCNet were revealed. Examination of the degree of distribution of KDCCNet revealed the power law distribution (R=0.792). In the KDCCNet, the majority of ceRNAs had few interacting dysregulated ceRNA partners, while a small subset of ceRNAs regarded as hubs possessed a relatively large number of interacting dysregulated ceRNA partners (Fig. 1B). Furthermore, it was demonstrated that the characteristic path length of KDCCNet was significantly increased when compared with random linked networks (P<0.001; Fig. 1C).
LncRNAs as topological key components involved in KIRC progression
In the KDCCNet, there are two types of dysregulated ceRNAs: LncRNAs and mRNAs. To analyze the roles of dysregulated ceRNA in more detail, the contribution degree of dysregulated ceRNAs in the two groups was investigated. It was demonstrated that the degree distribution of lncRNAs was significantly increased compared with the mRNA in KDCCNet (t-test, P=3.33×10−13; Fig. 1B). This result indicated that lncRNAs covered the majority of hubs and lncRNAs were topological key nodes in the KDCCNet associated with KIRC progression. In addition, a recent study demonstrated that lncRNAs were key regulators of the ceRNA network (29).
As dysregulated lncRNAs serve more important roles in KIRC progression, it was hypothesized that there are significant differences between dysregulated lncRNAs and non-dysregulated lncRNAs. To further analyze dysregulated lncRNAs, the characteristics of dysregulated lncRNAs and non-dysregulated lncRNAs were compared. As dysregulated lncRNAs were expected to contribute to the KIRC aberrant transcriptome, the expression levels of dysregulated lncRNAs and non-dysregulated lncRNAs were investigated first. The results demonstrated that the expression level of dysregulated lncRNAs was significantly increased compared with non-dysregulated lncRNAs (P=1.74×10−115, t-test; Fig. 1D). Secondly, the distribution of lengths for dysregulated lncRNAs and non-dysregulated lncRNAs on chromosomes was analyzed. It was revealed that the length of dysregulated lncRNAs on chromosomes was significantly increased compared with non-dysregulated lncRNAs (P=1.97×10−29, t-test; Fig. 1D). Finally, the counts of exons of dysregulated lncRNAs and non-dysregulated lncRNAs were compared, which also demonstrated that the counts of exons of dysregulated lncRNAs was significantly increased compared with non-dysregulated lncRNAs (P=1.45×10−12, t-test; Fig. 1D).
These results demonstrated that lncRNAs are topological key nodes of the KDCCNet associated with KIRC progression. Dysregulated lncRNAs exhibited increased expression in KIRC tissues, longer chromosome lengths and larger numbers of exons. This makes dysregulated lncRNAs more suitable as molecular sponges of miRNAs in KIRC.
Module analysis of the KDCCNet
It is generally thought that cancer occurs through the interactive activity of a number of genes. Functionally coherent modules can be regarded as biomarkers for distinguishing cancer from normal tissue (30). Therefore, the Markov Cluster Algorithm clustering computational method was applied to identify dysregulated ceRNA modules in the KDCCNet. As a result, 18 dysregulated ceRNA modules were obtained (Inflate=1.8, count of ceRNAs >10). The modules were enriched with known KIRC miRNAs in the Human microRNA Disease Database. An unsupervised hierarchical cluster analysis was conducted to visualize the classification performance of the modules. Modules that could distinguish cancer from normal tissue in TCGA and validation dataset were selected (area under the curve>0.65). Finally, five dysregulated ceRNA modules were obtained.
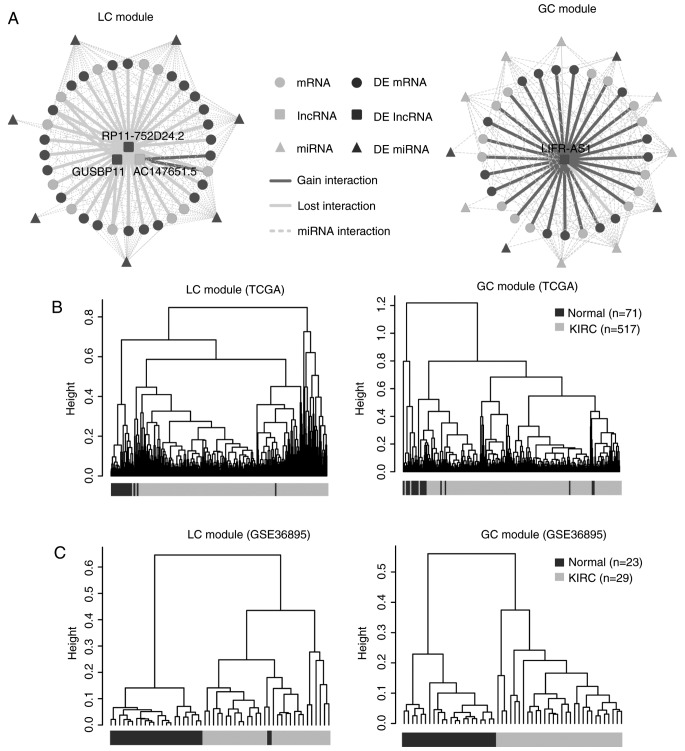
The distribution of the two patterns of ceRNA interactions was analyzed and two typical modules (95% loss interactions and 97% gain interactions) were observed (Fig. 2A). They were defined as loss ceRNA interactions module (LC module) and gain ceRNA interactions module (GC module), respectively (Table II).
Figure 2.
KIRC-associated dysregulated ceRNA modules. (A) The layout of the LC and GC modules. (B) The cluster trees of the LC and GC modules in The Cancer Genome Atlas dataset. The dark gray color represents the normal samples and the light gray color represents the KIRC samples. (C) The cluster trees of LC and GC modules in the validation dataset. The dark gray color represents the normal samples and the light gray color represents the KIRC samples. KIRC, clear cell kidney carcinoma; ceRNA, competitive endogenous RNA; GC, gain ceRNA; LC, loss ceRNA; DE, differential expression; GUSBP11, glucuronidase b pseudogene 11; LIFR-AS1, LIFR antisense RNA 1.
Table II.
A detailed description of the LC and GC modules.
| Module | Nodes | Edges | Lost | Gain | ΔR | mRNA | lncRNA | miRNA |
|---|---|---|---|---|---|---|---|---|
| LC module | 37 | 39 | 0.95 | 0.05 | 0.7334 | 34 | 3 | 7 |
| GC module | 30 | 29 | 0.03 | 0.97 | 0.7401 | 29 | 1 | 12 |
Lnc, long non-coding; GC, competitive endogenous RNA gains; LC, competitive endogenous RNA losses; ΔR, change in the Pearson correlation coefficient; miRNA, microRNA.
Typical dysregulated ceRNA modules contribute to KIRC
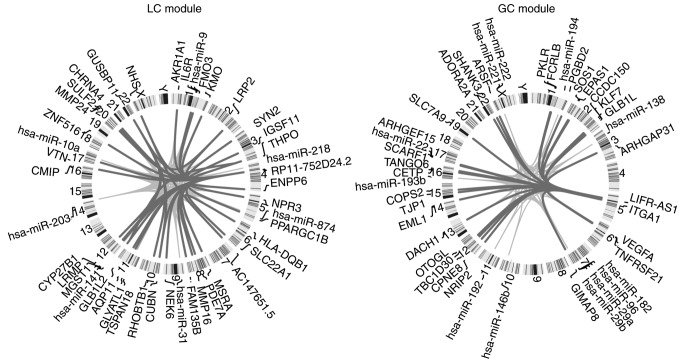
Firstly, unsupervised hierarchical cluster analysis demonstrated that the two typical modules as aforementioned could distinguish the normal samples from the cancer samples (Fig. 2B and C). The differentially expressed ceRNAs covered 67 and 50% in the LC and GC modules, respectively (Fig. 2A). Therefore, the two typical modules contributed to KIRC progression. In addition, the dysregulated ceRNAs of the two typical modules and their miRNA regulators were distributed on different chromosomes (Fig. 3).
Figure 3.
Chromosomal location of the ceRNA and miRNA regulators of typical modules. Chromosomal ideograms of ceRNAs and miRNA regulators of the two typical modules demonstrated that they were distributed on different chromosomes. A dark gray linking line represents a positive correlation between two RNAs, while a light gray linking line represents a negative correlation between two RNAs. miRNA, microRNA; ceRNA, competitive endogenous RNA; GC, gain ceRNA; LC, loss ceRNA.
Secondly, the GO and KEGG functional enrichment analysis of the LC and GC modules was investigated. Through functional enrichment analysis, it was demonstrated that the LC module was significantly enriched in vitamin D metabolic processes (FDR q-value=2.70×10−4; data not shown). Vitamin D metabolic processes have been demonstrated to inhibit the cell cycle and induce apoptosis in various types of cancer, including KIRC (31). This suggested that the LC module may contribute to KIRC progression by influencing vitamin D metabolic processes via loss dysregulated ceRNA interactions. Additionally, functional enrichment analysis revealed that the GC module was significantly enriched in the renal cell carcinoma pathway (FDR q-value=6.23×10−3; data not shown), which is associated with KIRC.
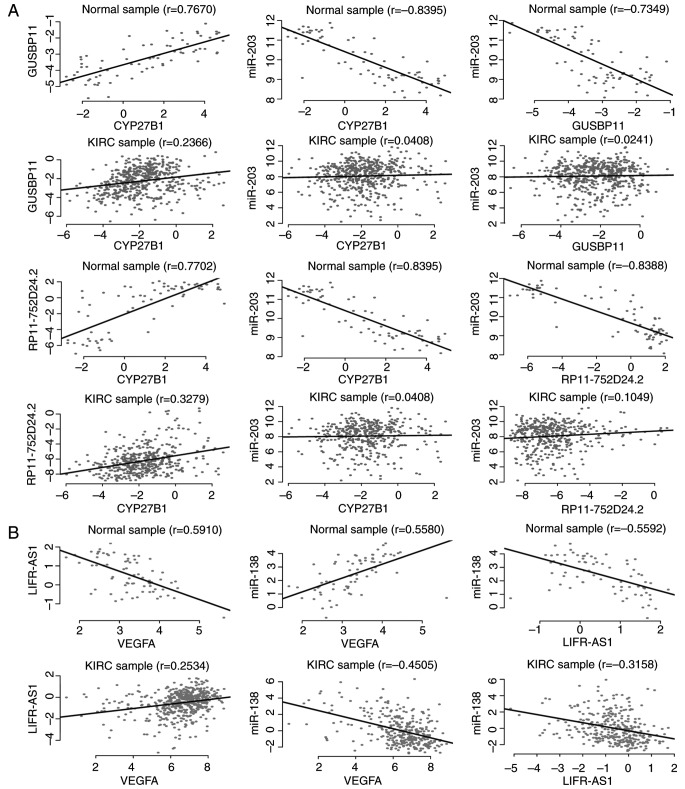
ceRNA contributes to cancer progression by influencing the expression of mRNAs in cancer associated GO biological processes or the KEGG pathway (32). It was hypothesized that if the two ceRNAs within the pair were differentially expressed simultaneously and at least one ceRNA was in KIRC-associated pathways, then they may exhibit more important effects on the dysregulated ceRNA module than the other ceRNAs which have no significant difference in expression or are independent of KIRC. It was demonstrated that two loss ceRNA interactions [glucuronidase β pseudogene 11 (GUSBP11) vs. cytochrome P450 family 27 subfamily B member 1 (CYP27B1), and RP11-752D24.2 vs. CYP27B1; Fig. 4A) composed of three differentially expressed ceRNAs within the LC module, influenced the vitamin D metabolic processes via CYP27B1 and one gain ceRNA interaction [LIFR antisense RNA 1 (LIFR-AS1) vs. vascular endothelial growth factor A (VEGFA); Fig. 4B] composed of two differentially expressed ceRNAs within the GC module, influenced the renal cell carcinoma pathway via VEGFA. Blomberg et al (33) demonstrated that differential expressed CYP27B1 was associated with KIRC, however the mechanism is unknown. The results of the present study further complemented the suggestion that CYP27B1 may be upregulated by ceRNAs (GUSBP11 and RP11-752D24.2) in KIRC. The above indicated that the three-dysregulated ceRNA interaction pairs may serve a key role via affecting the KIRC-associated pathways.
Figure 4.
Dysregulations of KIRC-associated ceRNA. (A) Expression variation across individuals (x- and y-axis) of two KIRC-associated ceRNA pairs (GUSBP11 vs. CYP27B1 and RP11-752D24.2 vs. CYP27B1) in the LC module reveals a loss ceRNA interaction in KIRC. (B) Expression variation across individuals (x- and y-axis) of a KIRC-associated ceRNA pairs (LIFR-AS1 vs. VEGFA) in the GC module reveals a gain ceRNA interaction in KIRC. KIRC, clear cell kidney carcinoma; VEGFA, vascular endothelial growth factor A; ceRNA, competitive endogenous RNA; GC, gain ceRNA; LC, loss ceRNA; GUSBP11, glucuronidase β pseudogene 11; LIFR-AS1, LIFR antisense RNA 1.
As lncRNAs are the key components for the typical modules, attention was paid to lncRNAs of the three ceRNA pairs mentioned above. Based on survival analysis, two lncRNAs were demonstrated to be associated with the survival of patients with KIRC (GUSBP11, P=1.78×10−13; LIFR-AS1, P=1.32×10−3). In fact, GUSBP11 has been discovered to be associated with the survival of patients with head and neck squamous cell carcinoma (34). Also, LIFR-AS1 was predicted to mediate the inhibitory action of leukemia inhibitory factor, a cytokine that is involved in embryonic uterine development and is associated with the tumor size of the uterine fibroid (35). The results of the present study established the link between KIRC, and GUSBP11 and LIFR-AS1 via dysregulated ceRNA analysis.
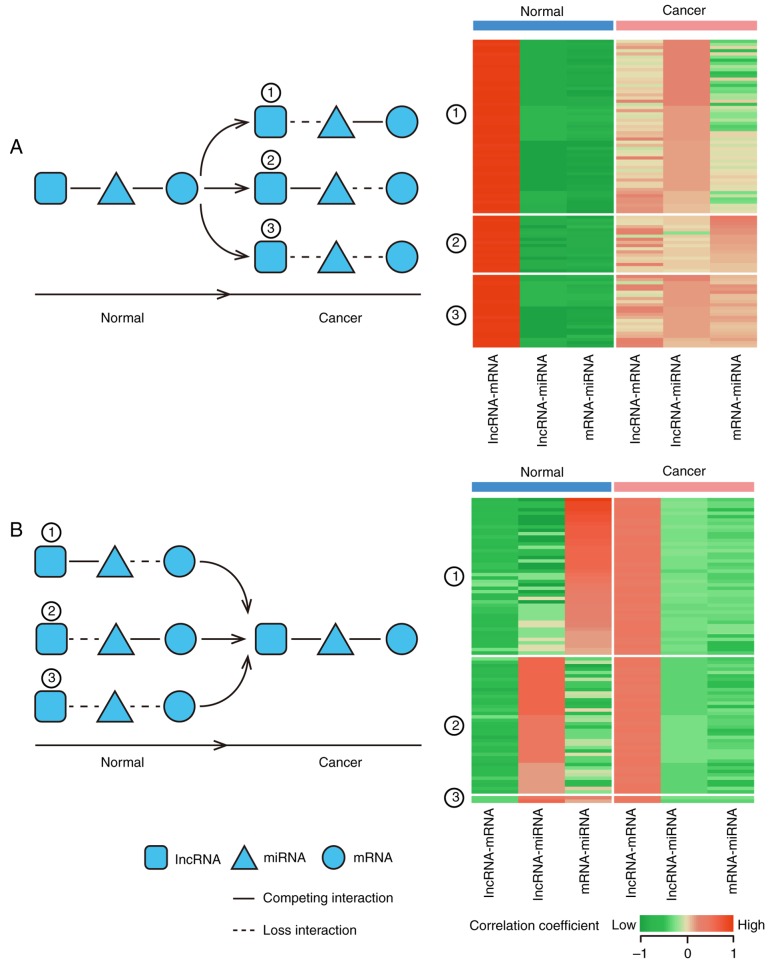
In addition, miRNAs mediate competitive interactions, therefore expression pattern alterations of miRNAs and ceRNAs may cause dysregulation of ceRNA interactions. The correlations between miRNAs and ceRNAs in GC and LC module are shown in Fig. 5 and miRNA regulators of the dysregulation modules were evaluated based on the Human microRNA Disease Database (HMDD). It was found that 4 miRNAs regulators of the LC module (miRNA-141, miRNA-203, miRNA-9 and miRNA-218) and 2 miRNA (miRNA-138 and miRNA-146b) regulators of the GC module were involved in KIRC from HMDD. For example, miR-203 inhibits renal cancer cell proliferation, migration and invasion (36). miR-203 was also a regulator of the LC module, and therefore it was hypothesized that this dysregulated ceRNA interaction may be involved in the mechanism of KIRC.
Figure 5.
Layout of miRNA mediated dysregulated ceRNAs patterns. A circular node marks an mRNA, a square node marks an lncRNA and a triangle node marks a miRNA. Corresponding expression profiles in KIRC and normal samples are presented in the right panel. Highly correlated RNAs pairs are indicated in red and low correlation pairs are presented in green. (A) The layout of miRNA describes the loss ceRNA interaction patterns and the heatmap is based on the correlation coefficient of lncRNA, mRNA and miRNA. (B) The layout of miRNA describes the gain ceRNAs interaction patterns and the heatmap based on the correlation coefficient of lncRNA, mRNA and miRNA. KIRC, clear cell kidney carcinoma; miRNA, microRNA; lncRNA, long non-coding RNA; ceNRA, competitive endogenous RNA; KIRC, clear cell kidney carcinoma.
Discussion
In the present study, a multidimensional integration strategy was introduced to construct the KDCCNet. Topological analysis identified that the dysregulated lncRNAs were the key components of the KDCCNet. A total of two dysregulated patterns of ceRNAs interaction exist in the KDCCNet: i) Gain ceRNAs interactions: A lncRNA-mRNA pair demonstrated no competitive interactions in normal samples but did in the KIRC samples; ii) Loss ceRNAs interactions: A lncRNA-mRNA pair demonstrated competing activities in normal samples, though in not cancer samples. Gain of KIRC ceRNA interactions and also loss of normal ceRNA interactions occurred during the initiation and progression of KIRC. The two patterns contributed to KIRC progression. Next, two KIRC-associated dysregulated ceRNA modules were identified, the LC and GC modules, which can distinguish cancer from normal samples in TCGA and the validation dataset. Furthermore, two dysregulated lncRNAs (GUSBP11 and LIFR-AS1) in the two modules demonstrated associations with the survival of KIRC patients. In addition, certain KIRC associated genes/miRNAs were observed in the modules. For example, overproduction of VEGFA in the GC module occurs early during the development of VHL−/− renal cell carcinomas [the patients who have biallelic inactivating mutations of the von Hippel-Lindau (VHL) gene] and therefore reduces the selection pressure to activate ‘collateral’ angiogenic pathways (37,38). miRNA-203 in the LC module could inhibit renal cancer cell proliferation, migration and invasion (36). The present study of the KDCCNet revealed a novel biological mechanism for KIRC and provided a novel link between the two dysregulated lncRNAs (GUSBP11 and LIFR-AS1) and KIRC.
Although genome-wide analysis was performed to investigate the potential mechanism of dysregulated ceRNA interactions in KIRC progression and identified KIRC-associated dysregulated ceRNA modules, there were certain limitations to the present study. The understanding of the functional roles of lncRNAs is limited and further studies should examine the remaining regulatory elements, including transcription factors, DNA methylation and copy number variation, which may reveal a more detailed molecular mechanism of KIRC progression.
In conclusion, a KIRC-associated dysregulated ceRNA network was developed and a method for examining the competitive interactions of RNAs that are potentially dysregulated in KIRC progression. These results increase the understanding of lncRNA characteristics and provide a foundation for leveraging publicly available genomic data to study the functions and mechanisms of dysregulated ceRNAs in KIRC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- KIRC
clear cell kidney carcinoma
- lncRNAs
long non-coding RNAs
- ceRNA
competing endogenous RNAs
- KDCCNet
KIRC dysregulated ceRNA-ceRNA network
Funding
The present study was supported in part by the Natural Science Foundation of Heilongjiang Province (grant no. B201302), National Natural Science Foundation of China (grant no. 31301094), Education Department Foundation of Heilongjiang Province (grant no. 12531227) and Health Department Foundation of Heilongjiang Province (grant no. 2012–798).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
KL and LW conceived and designed the experiments. HW, DX, HH, YC, CL, CZ, SG, LZ, XX, JX and JL collected and analyzed the data. KL and LW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
References
- 1.Ponting CP, Belgard Grant T. Transcribed dark matter: Meaning or myth? Hum Mol Genet. 2010;19:R162–R168. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker BJ, Moltke I, Roth A, Washietl S, Wen J, Kellis M, Breaker R, Pedersen JS. New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. Genome Res. 2011;21:1929–1943. doi: 10.1101/gr.112516.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 4.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Liu J, Wang W. Construction and investigation of breast-cancer-specific ceRNA network based on the mRNA and miRNA expression data. IET Syst Biol. 2014;8:96–103. doi: 10.1049/iet-syb.2013.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Li Y, Lu J, Pan T, Ding N, Wang Z, Shao T, Zhang J, Wang L, Li X. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 2015;43:8169–8182. doi: 10.1093/nar/gkv853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poliseno L, Pandolfi PP. PTEN ceRNA networks in human cancer. Methods. 2015;77–78:41–50. doi: 10.1016/j.ymeth.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Xul B, Chen S, Lu K, Liu C, Wang Y, Zhao Y, Zhang X, Liu D, Chen M. The complex roles of microRNAs in the metastasis of renal cell carcinoma. J Nanosci Nanotechnol. 2013;13:3195–3203. doi: 10.1166/jnn.2013.6712. [DOI] [PubMed] [Google Scholar]
- 12.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Wang Y, Wang L, Xu W. Identifying prognostic biomarkers based on aberrant DNA methylation in kidney renal clear cell carcinoma. Oncotarget. 2017;8:5268–5280. doi: 10.18632/oncotarget.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan Y, Guo W, Zhang Y, Wang Q, Xu XJ, Zhu L. A five-gene signature predicts prognosis in patients with kidney renal clear cell carcinoma. Comput Math Methods Med. 2015;2015:842784. doi: 10.1155/2015/842784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linehan WM. Genetic basis of kidney cancer: Role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22:2089–2100. doi: 10.1101/gr.131110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, Leng N, Pavía-Jiménez A, Wang S, Yamasaki T, Zhrebker L, Sivanand S, Spence P, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng0912-1072b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeggari A, Marks DS, Larsson E. miRcode: A map of putative microrna target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. MiRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, Ghosal S, Sen R, Chakrabarti J. lnCeDB: Database of human long noncoding RNA acting as competing endogenous RNA. PLoS One. 2014;9:e98965. doi: 10.1371/journal.pone.0098965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, Cui Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–D1074. doi: 10.1093/nar/gkt1023. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reimand J, Tooming L, Peterson H, Adler P, Vilo J. GraphWeb: Mining heterogeneous biological networks for gene modules with functional significance. Nucleic Acids Res. 2008;36:W452–W459. doi: 10.1093/nar/gkn230. (Web Server Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Statistician William Sealy Gosset (Student): The probable error of a mean. Biometrika. 1908;6:1–25. doi: 10.2307/2331554. [DOI] [Google Scholar]
- 28.Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang LK, Chen XF, He DD, Li Y, Fu J. Dissection of functional lncRNAs in Alzheimer's disease by construction and analysis of lncRNA-mRNA networks based on competitive endogenous RNAs. Biochem Biophys Res Commun. 2017;485:569–576. doi: 10.1016/j.bbrc.2016.11.143. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Xuan J, Clarke R, Ressom HW. Module-based breast cancer classification. Int J Data Min Bioinform. 2013;7:284–302. doi: 10.1504/IJDMB.2013.053309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Park SH. Tumor-suppressive activity of 1,25-dihydroxyvitamin D3 against kidney cancer cells via up-regulation of FOXO3. Biosci Biotechnol Biochem. 2016;80:1947–1953. doi: 10.1080/09168451.2016.1184561. [DOI] [PubMed] [Google Scholar]
- 32.Zhou M, Diao Z, Yue X, Chen Y, Zhao H, Cheng L, Sun J. Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget. 2016;7:56383–56394. doi: 10.18632/oncotarget.10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen Blomberg M, Andersen CB, Nielsen JE, Bagi P, Jørgensen A, Juul A, Leffers H. Expression of the vitamin D receptor, 25-hydroxylases, 1alpha-hydroxylase and 24-hydroxylase in the human kidney and renal clear cell cancer. J Steroid Biochem Mol Biol. 2010;121:376–382. doi: 10.1016/j.jsbmb.2010.03.069. [DOI] [PubMed] [Google Scholar]
- 34.Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T, Chen WT, Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. doi: 10.1016/j.oraloncology.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 35.Aissani B, Zhang K, Wiener H. Genetic determinants of uterine fibroid size in the multiethnic NIEHS uterine fibroid study. Int J Mol Epidemiol Genet. 2015;6:9–19. [PMC free article] [PubMed] [Google Scholar]
- 36.Xu M, Gu M, Zhang K, Zhou J, Wang Z, Da J. miR-203 inhibition of renal cancer cell proliferation, migration and invasion by targeting of FGF2. Diagn Pathol. 2015;10:24. doi: 10.1186/s13000-015-0255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaelin WG., Jr The von Hippel-Lindau tumor suppressor protein and clear cell renal carcinoma. Clin Cancer Res. 2007;13:S680–S684. doi: 10.1158/1078-0432.CCR-06-1865. [DOI] [PubMed] [Google Scholar]
- 38.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.