Abstract
Asthma, the most common chronic respiratory tract disease in children, is characterized by allergy, recurring airway obstruction and bronchospasm. The aim of the present study was to screen critical differentially expressed genes (DEGs) involved in asthma in children. Gene expression in different tissues was compared between asthmatic children and healthy control subjects in order to identify DEGs associated with asthma. Protein-protein interaction (PPI) networks were constructed for the DEGs and weighted gene co-expression network analysis methods were used to further determine the functional modules associated with DEGs in different tissue samples. In addition, the gene co-expression network was constructed. Gene Ontology function analysis and pathway analysis were conducted to identify critical DEGs. The results identified numerous DEGs from the different tissue samples, including 1,662 DEGs from nasal-epithelium tissue samples, 572 DEGs from peripheral blood (PB) samples and 146 DEGs from PB mononuclear cells samples. In the PPI network, F-box only protein 6 (FBXO6), histone deacetylase 1 (HDAC1) and amyloid β precursor protein (APP) were hub genes and served an important role in the process of asthma. In addition, proliferating cell nuclear antigen (PCNA), integrin α-4 (ITGA4), catenin α-1 (CTNNA1), nuclear factor-κB1 (NF-κB1) and mechanistic target of rapamycin (MTOR) may be critical DEGs involved in the progression of asthma in children. These results suggested that FBXO6, HDAC1 and APP may interact with PCNA, ITGA4, CTNNA1, NF-κB1 and mTOR in the progression of asthma in children.
Keywords: differentially expressed genes, microarray, asthma, respiratory epithelia
Introduction
Asthma is a common chronic respiratory tract disease in children, and it affects more than 6.1 million American children in recent years (1). Minority and low-income children were reported that suffered from a higher ratio of asthma morbidity and mortality (2). Asthma is characterized by allergy, recurring airway obstruction, and bronchospasm (3). It is mainly caused by a combination of genetic and environmental factors, such as air pollution, allergens, viral infection, aspirin and beta blockers (4). Although great scientific advances have improved our understanding of asthma and promoted abilities to control asthma effectively, the mechanism of asthma is still unclear and needs to be explored. Exploring the progressive of asthma and assessing asthma severity, especially asthma in children, have been defined as urgent and persistent task to initiating appropriate therapy.
Recent studies on asthma in children have focused on genetic factors, especially critical genes and mRNAs in the pathogenesis of childhood asthma (5). β2-AR agonist is now the most important bronchodilator for the treatment of asthma in clinical, and the polymorphism of the ADRB2 response to inhaled beta-agonists in children with asthma (6). Besides, imbalance of CD4+ T cell subgroup were also major factors resulting in asthma, and study showed that differentiation of Th2 cell, regulatory T cells (Treg) were involved in the occurrence of asthma (7). To date, the pathogenetic basis for the relationship between potential gene expression changes and asthma has not been clearly elucidated.
In 2017, Yang et al (8) identified the DNA methylation and gene expression changes in nasal-epithelium (NE) tissue associated with childhood asthma. They demonstrated that the methylation marks in the nasal epithelia of children with allergic asthma were associated with gene expression changes. Based on the microarray data deposited by Yang et al (8), as well as other three microarray data of asthma downloaded from GEO database, we identified many differentially expressed genes (DEGs) from different tissue samples associated with asthma in children. Furthermore, we constructed protein-protein interaction (PPI) network for the DEGs and investigated the functional modules of DEGs in NE, peripheral blood mononuclear cells (PBMC) and peripheral blood (PB) samples. Gene Ontology (GO) functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed. In addition, gene co-expression network was constructed to identify the critical DEGs in asthmatic children. Our study might provide novel diagnostic biomarkers and therapeutic target molecules in progression of asthma in children.
Materials and methods
Data source
The four microarray datasets associated with asthma in children [access nos. GSE65204 (8), GSE40732 (9), GSE40888 (10), and GSE35571 (11)] were downloaded from National Center of Biotechnology Information (NCBI) GEO database. GSE65204 dataset was sequenced on the platform of GPL14550 Agilent-028004 SurePrint G3 Human GE 8× 60K Microarray (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL14550), including NE tissue samples from 36 asthmatic and 33 non-asthmatic children (Table I). GSE40732 dataset included PBMCs from 97 atopic asthmatic and 97 nonatopic nonasthmatic children, which was sequenced on platform of GPL16025 NimbleGen Homo sapiens Expression Array [(100718_HG18_opt_expr) www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL16025]. GSE40888 dataset was sequenced on the platform of GPL6244 (HuGene-1_0-st) Affymetrix Human Gene 1.0 ST Array, including PBMCs samples from 41 asthmatic and 24 non-asthmatic children. GSE35571 dataset contained PB samples from 60 asthmatic and 64 non-asthmatic children based on the platform of GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array.
Table I.
Microarray databases associated with asthmatic children, including GSE65204, GSE40732, GSE40888 and GSE35571.
DEGs screening
The DEGs between disease groups and control groups were screened by the limma package (12) in R software. The adjusted P-value <0.05 and |log2 fold-change (FC)|>1 were considered as cut-off criteria.
The microarray data from GSE40732 and GSE40888 datasets were analyzed to identify DEGs in PBMCs samples between asthmatic children and healthy groups. Besides, GSE35571 dataset was used to identify DEGs in PB samples and GSE65204 dataset were used to identify DEGs in NE tissue samples.
Identification of co-expression modules
To identify the gene co-expression modules related to different tissue samples, we used weighted gene co-expression network analysis (WGCNA) (13) package to further mine the modules. The WGCNA provided topology properties of co-expression network, as well as the correlation of two node genes and relevant other genes. Besides, we changed the connection coefficient into weight coefficient and screened significant co-expression modules that related to different tissue samples using WGCNA analysis.
PPI network construction
For the identified DEGs, we screened the PPI relationship pairs of DEGs from some common databases. The databases included HPRD (www.hprd.org/), BIOGRID (thebiogrid.org/), DIP (dip.doe-mbi.ucla.edu/dip/Main.cgi), MINT (mint.bio.uniroma2.it/mint/Welcome.do), menthe (mentha.uniroma2.it/index.php), PINA (cbg.garvan.unsw.edu.au/pina/), InnateDB (www.innatedb.com/), Instruct (instruct.yulab.org/index.html). We then obtained the regression coefficient value of each relationship pair in different conditions through linear regression analysis.
To investigate the changes of adjust power under two conditions, we proposed a method to calculate the changes of adjust power associated with screened PPI relationship pairs. The equation is as follows:
C1coefi and C2coefi represent regression coefficient value of each relationship pairs in condition 1 and condition 2, respectively; C1sdi and C2csdi represent standard deviation (SD) values responding to regression coefficient in condition 1 and condition 2, respectively; M represents compensation coefficient; If a relationship pair is only presented in the condition of disease, then its regression coefficients and SD value would be zero in other conditions; dDRl ranging from positive to negative value represents that the regulative relations are increased or decreased from condition 1 to 2. The higher the absolute values of dDRl, the greater change of adjust power.
In our study, we mainly compared the gene relationships in three types of tissues: NE-Normal (from normal to NE), PB-Normal (from normal to PB) and PBMC-Normal (from normal to PBMC). After calculating dDRl values of the whole relationships, we constructed the weighted correlation network. In the network, the dDRl values represented edge weight. The higher the absolute values of dDRl, the greater significance of relationship pairs. Besides, we performed the KEGG pathway and GO analyses to identify the enrichment functions of DEGs in the network.
Identification of specific genes
In the weighted correlation network, the dDRl values were used to evaluate the adjusted power of DEGs. For the specific gene g, we calculated its adjusted power through the following formula:
g represents the specific genes; k represents the number of relationships corresponding with the specific gene; the dDRl can be displayed as dDRli (i=1, 2, . . ., k). The higher the dDRg values, the greater effect of the specific genes on the network. Finally, we exacted top 10 genes according to the dDRg value.
Results
Identification of DEGs
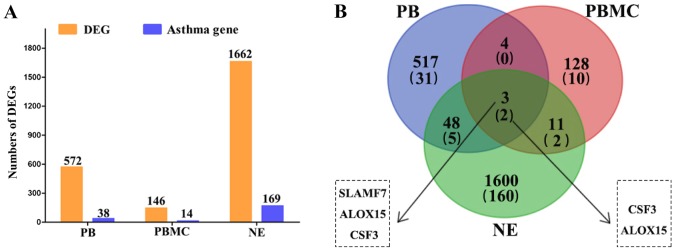
We identified many DEGs from different tissue samples, including 1,662 DEGs from NE tissue samples, 572 DEGs from PB samples, 146 DEGs from PBMC samples. Furthermore, the genes related to asthma were downloaded from DisGeNET database and the number of genes in three types of tissue was counted, respectively. A total of 169 DEGs related to asthma were screened from NE tissue, 38 DEGs were screened from PB and 14 DEGs were screened from PBMC (Fig. 1A).
Figure 1.
DEGs in three types of tissues associated with asthmatic children and the Venn diagrams. (A) The DEGs in three types of tissues associated with asthmatic children. (B) Venn diagrams presenting the number of DEGs in the different tissues of asthmatic children. DEGs, differentially expressed genes; PB, peripheral blood; PBMC, peripheral blood mononuclear cells; NE, nasal-epithelium; CSF3, colony stimulating factor 3; ALOX15, arachidonate 15-lipoxygenase; SLAMF7, SLAM family member 7.
The Venn diagram was used to visualize the DEGs screened from differential tissues (Fig. 1B). Among these genes, colony stimulating factor 3 (CSF3) and arachidonate 15-lipoxygenase (ALOX15) were significantly differentially expressed in all three types of tissue.
PPI network analysis
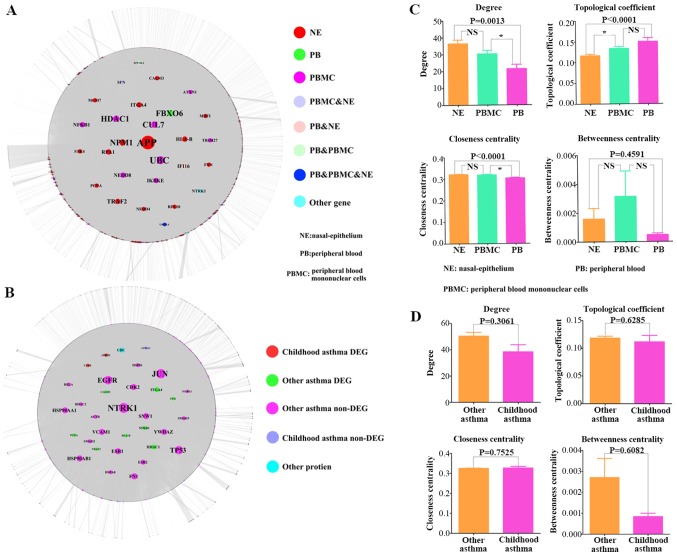
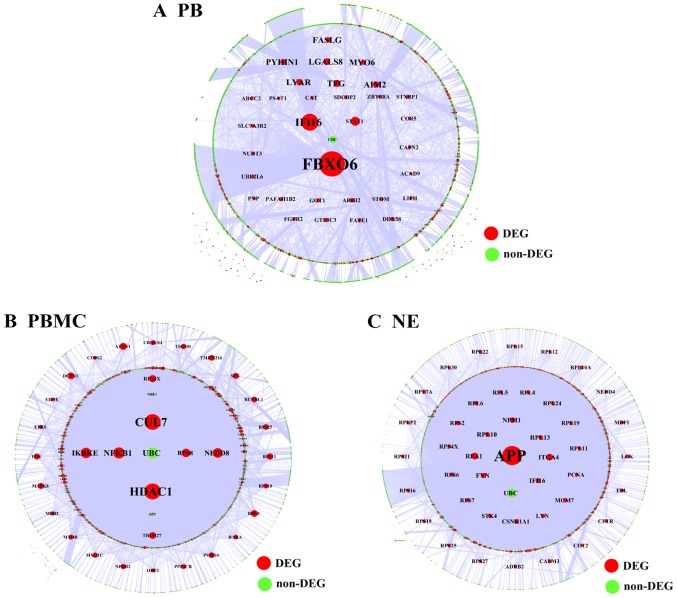
The PPI relationship pairs associated with DEGs were extracted from common databases and the PPI network was constructed. To further explore the DEGs related to PBMC tissue, we take the union set of DEGs in two databases (GSE40732 and GSE40888) to construct the PPI network. In the PPI network, a total of 13496 nodes and 71275 relationship pairs were identified (Fig. 2A). F-box only protein 6 (FBXO6) and histone deacetylase 1 (HDAC1) were hub genes and played an important role in the process of asthma. Additionally, we performed the topological centrality analysis of the network. The results showed that there were significance differences between the DEGs in NE and PB samples according to the degree, topological coefficient, and closeness centrality analyses. The values of degree in NE tissue were higher than that in PBMC tissue. The values of degree and closeness centrality in PBMC tissue were higher than that in PB tissue (Fig. 2C).
Figure 2.
PPI network construction and analysis. (A) The PPI network of DEGs. (B) The PPI network of DEGs in the different tissues of asthmatic patients. (C) The topological property comparison of the DEGs between different tissues, including degree, topological coefficient, closeness centrality and betweenness centrality. (D) The topological property comparison between the DEGs of childhood asthmatic patients and non-childhood asthmatic patients. *P<0.05, as indicated. PPI, protein-protein interaction; DEGs, differentially expressed genes; NS, not significant.
The DEGs related to asthma were downloaded from DisGeNET databases (http://www.disgenet.org/web/DisGeNET/menu/home;jsessionid=mjbtjh4yncdzuqjlnk3rnvbn) and a total of 1,403 DEGs were screened, including 104 DEGs associated with asthma in children. We integrated the PPI relationship pairs and constructed the PPI network related to asthma, including 11,960 nodes and 61,164 relationship pairs (Fig. 2B). Based on the topology properties analysis (including degree, topological coefficient, closeness centrality and betweenness centrality), we found that there was no significant difference between childhood asthma DEGs and other asthma DEGs (Fig. 2D).
Gene co-expression network analysis
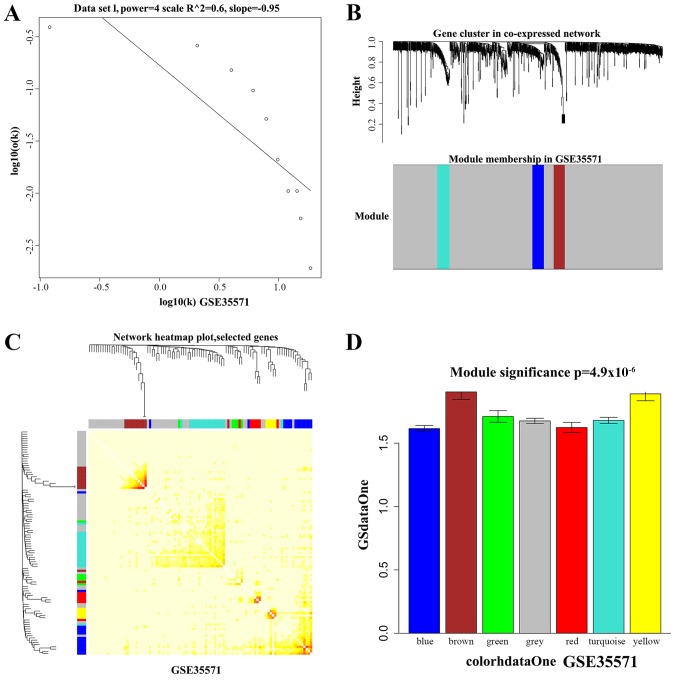
The cuttee Static Color function in the WGCNA package was used to mine the functional modules associated with different tissues. The parameters were set as follows: The X-axis represents gene degrees k, Y-axis represents the ratio of genes p (k) (Fig. 3A). Hierarchical cluster diagrams were used to visualize the functional modules associated with different tissues (Fig. 3B). A total of 100 genes were selected randomly from network modules and the correlation between these genes were further investigated (Fig. 3C).
Figure 3.
Gene co-expression modules analysis of GSE35571. (A) Soft threshold setting in the co-expression network. The x-axis represents gene degrees k, and the y-axis represents the ratio of genes p (k). (B) Hierarchical clustering heatmap of DEGs in co-expression network. (C) Hierarchical clustering heatmap showed the inner correlation between these function modules. (D) The GO enrichment analysis of DEGs in modules. The x-axis represents DEGs of modules and the y-axis represents the significance of GO analysis. DEGs, differentially expressed genes; GO, Gene Ontology.
We analyzed the P-values of each DEG in the disease groups and normal tissues. A higher degree indicated that the DEG had greater connection with other genes and might play an important role in the progression of asthma. K represents the degree of each DEG in the modules. P-values represent the significance of each DEG. The correlation between K values and -log10 (p) was calculated. According to the GO and pathway analysis, we obtained the functional modules related to different tissues. Fig. 3D showed functional modules of PB tissue, and functional modules of other tissues were also obtained with the same method (data not shown).
Two functional modules in the color of brown and yellow were screened significantly associated with PB tissue; modules of black and green were significantly related to NE tissue; functional modules in the color of green and red were significantly associated with PBMC tissue.
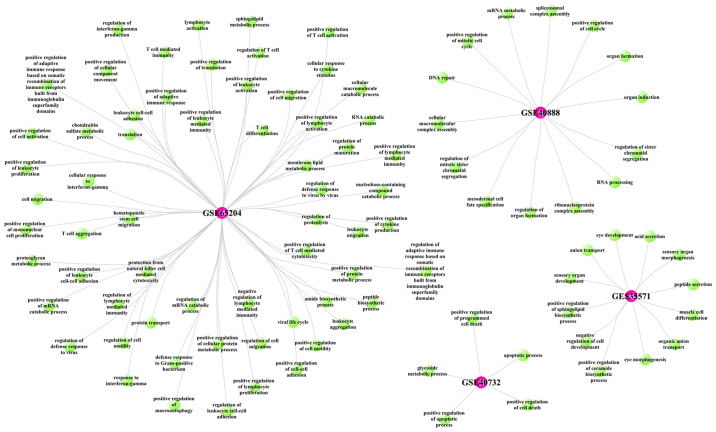
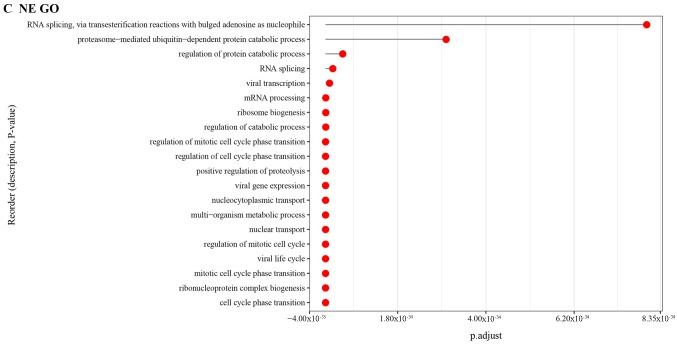
The GO biological process (BP) enrichment analysis was performed to identify the function of DEGs associated with different tissue in asthmatic children (Fig. 4). The GO functional analysis results showed that the DEGs related to PB tissue were mainly enriched in the processes of acid secretion, sensory organ development and negative regulation of cell development, etc; the DEGs related to NE tissue were mainly involved in the processes of positive regulation of cell motility, and positive regulation of cellular component movement; the DEGs in PBMC tissue were mainly enriched in the processes of positive regulation of apoptotic process, and organ induction.
Figure 4.
Gene Ontology Biological Process analysis of differentially expressed genes in different databases.
Construction of regulatory network
We constructed the regulatory network related to DEGs in different tissues (Fig. 5). As for the PBMC tissue, we took the interaction of DEGs in two databases to perform WGCNA analysis and the gene expression profile of GSE40732 dataset were used to analysis the adjust power.
Figure 5.
DEGs in the regulatory network associated with different types of tissues. (A) The DEGs in the regulatory network associated with PB. (B) The DEGs in the regulatory network associated with PBMC. (C) The DEGs in the regulatory network associated with NE tissue. DEGs, differentially expressed genes; PB, peripheral blood; PBMC, peripheral blood mononuclear cells; NE, nasal-epithelium.
The results showed that FBXO6 was a hub gene in the regulatory network associated with PB tissue, which included 3,670 nodes and 5,330 relationship pairs (Fig. 5A). HDAC1 and cullin-7 (CUL7) were hub genes in the regulatory network of PBMC tissue, which included 6,803 nodes and 18,220 relationship pairs (Fig. 5B). Additionally, the regulatory network associated with NE tissue included 6,276 nodes and 18,209 relationship pairs, and amyloid β precursor protein (APP) was a hub gene (Fig. 5C).
GO and KEGG pathway analyses of DEGs in regulatory network
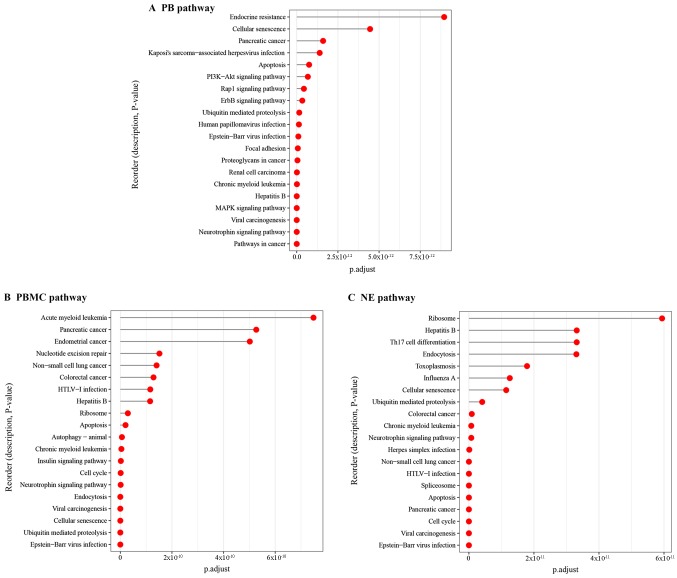
The pathway analysis results showed that the DEGs in the PB tissue were mainly enriched in MAPK signaling pathway, PI3K-Akt signaling pathway and neurotrophin signaling pathway, etc. (Fig. 6A). As for the DEGs in the PBMC tissue, the enriched pathways included neurotrophin signaling pathway, and insulin signaling pathway, etc. (Fig. 6B). The DEGs in the NE tissue were significantly enriched in ubiquitin mediated proteolysis, cell cycle and neurotrophin signaling pathway (Fig. 6C).
Figure 6.
Kyoto Encyclopedia of Genes and Genomes pathway analysis for the DEGs in the regulatory network. (A) Pathway analysis for the DEGs in the network associated with PB. (B) Pathway analysis for the DEGs in the network associated with PBMC. (C) Pathway analysis for the DEGs in the network related to NE tissue. DEGs, differentially expressed genes; PB, peripheral blood; PBMC, peripheral blood mononuclear cells; NE, nasal-epithelium.
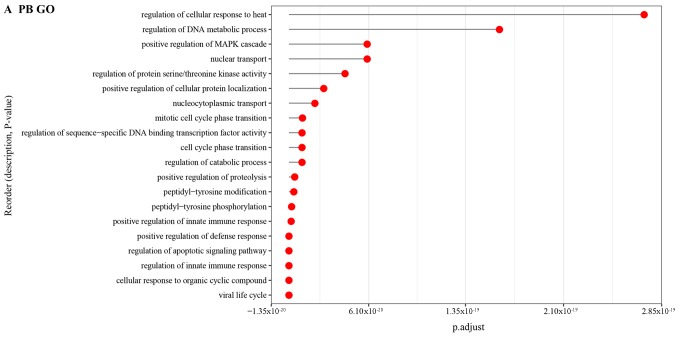
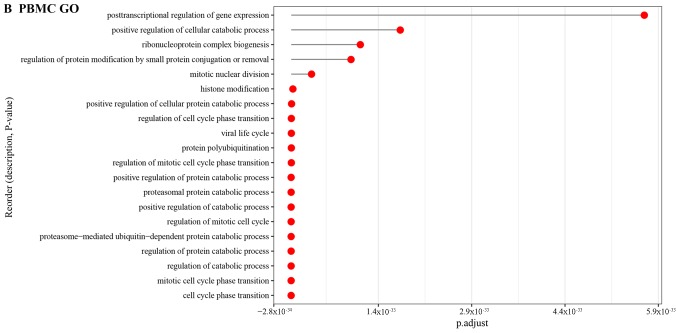
Moreover, the GO BP terms enriched by DEGs in PB tissue were mainly associated with cellular response to organic cyclic compound (Fig. 7A). As for the DEGs in PBMC together with DEGs in NE tissue samples, the GO BP terms were related to cell cycle phase transition (Fig. 7B and C).
Figure 7.
GO BP analysis for the DEGs in the regulatory network. (A) GO BP analysis for the DEGs in the regulatory network associated with PB. (B) GO BP analysis for the DEGs in the regulatory network associated with PBMC. (C) GO BP analysis for the DEGs in the regulatory network associated with NE tissue. GO, Gene Ontology; BP, biological process; DEGs, differentially expressed genes; PB, peripheral blood; PBMC, peripheral blood mononuclear cells; NE, nasal-epithelium.
Specific DEGs in the regulatory network
The DEGs in the regulatory network were sequenced by dDRg values and top 20 DEGs were selected (Table II). The results showed that proliferating cell nuclear antigen (PCNA), integrin α-4 (ITGA4), catenin α-1 (CTNNA1), nuclear factor-κB1 (NF-κB1) and mechanistic target of rapamycin (MTOR) were specific DEGs that related to asthma. Besides, these DEGs were also identified in DisGetNET database associated with asthma. Our results indicated that PCNA, ITGA4, CTNNA1, NF-κB1 and MTOR might be the potential genes related to asthma in children.
Table II.
Top 20 nodes in the regulatory network associated with the DEGs of asthmatic children.
| NE Samples | PB Samples | PBMC samples | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | DEG | dDRg | Label | DEG | dDRg | Label | DEG | dDRg | Label |
| 1 | APP | 202.5712 | No | FBXO6 | 42.82477 | No | HDAC1a | 106.7973 | Other asthma |
| 2 | SETDB1 | 78.09696 | No | ARIH2 | 36.51952 | No | RPS8 | 61.2846 | No |
| 3 | RPA1 | 68.47313 | No | IFI16 | 33.23794 | No | RPS11 | 58.52655 | No |
| 4 | STK4 | 66.04559 | No | PAFAH1B2 | 30.54515 | No | RPS4X | 39.92426 | No |
| 5 | NEDD4 | 61.91617 | No | TFG | 29.83253 | No | TRIM27 | 39.09394 | No |
| 6 | CSNK1A1 | 57.83066 | No | FGFR2 | 26.67677 | No | RPL5 | 35.9798 | No |
| 7 | EDC4 | 53.42212 | No | LGALS8 | 26.21989 | No | CUL7 | 29.2522 | No |
| 8 | CTDP1 | 49.7963 | No | NEK6 | 24.95025 | No | NEDD8 | 28.72204 | No |
| 9 | RPS2 | 43.28688 | No | LYAR | 22.42635 | No | RPS10 | 27.68076 | No |
| 10 | CALM3 | 41.95928 | No | SPINT2 | 19.52418 | No | RPS25 | 27.62278 | No |
| 11 | RPS7 | 40.06917 | No | GOT1 | 17.31375 | No | TMEM216 | 27.26486 | No |
| 12 | IFI16 | 39.24005 | No | MYO6 | 17.17874 | No | NFKB1a | 25.80718 | Other asthma |
| 13 | MID2 | 35.10338 | No | PELO | 16.25726 | No | RPL35A | 25.62394 | No |
| 14 | PCNAa | 32.54975 | Other asthma | PTK6 | 15.76188 | No | PSMD4 | 25.04536 | No |
| 15 | RPL10 | 30.89499 | No | CTNNA1a | 15.75888 | Other asthma | EIF3E | 19.85108 | No |
| 16 | RPS16 | 29.85522 | No | NTRK2 | 14.60812 | No | MTORa | 19.81246 | Other asthma |
| 17 | ITGA4a | 29.2464 | Other asthma | LIPH | 12.77759 | No | DDX24 | 19.53866 | No |
| 18 | MCM7 | 28.55749 | No | CA10 | 12.35675 | No | UBQLN4 | 19.34685 | No |
| 19 | SPTAN1 | 28.18446 | No | NUDT3 | 11.95574 | No | ATXN1 | 18.88822 | No |
| 20 | LZTS2 | 28.04549 | No | SLC9A3R2 | 11.93033 | No | IKBKE | 17.33138 | No |
The dDRg values of DEGs in the regulatory network are presented.
DEGs associated with asthma. DEGs, differentially expressed genes; PB, peripheral blood; dDRg, weighted control values; PBMC, peripheral blood mononuclear cells; NE, nasal-epithelium.
Discussion
In the current study, we identified many DEGs from three types of tissue samples, including 1,662 DEGs from NE tissue samples, 572 DEGs from PB samples, and 146 DEGs from PBMC samples. In PPI network, FBXO6, HDAC1 and APP were hub genes and might play an important role in the process of asthma. In addition, PCNA, ITGA4, CTNNA1, NF-κB1 and MTOR might be critical DEGs related to asthma in children.
FBXO6 encodes a member of the F-box protein family, which constitutes the subunit of ubiquitin protein ligase complex called SKP1-cullin-F-box (SCFs) (14). Overexpression of F-box protein FBXL19 can abrogate the inflammatory effects of IL-33 and lessen the severity of pulmonary inflammation in mouse models of pneumonia (15). However, the role of FBXO6 in asthma has not been reported. In our study, we found that FBXO6 were hub gene in the PPI network. Together with previous findings, we proposed that FBXO6 might relate to the inflammation in asthma.
HDAC1 is a member of HDAC family and highly expressed in inflammation-related diseases, such as arthritis (16). Deletion of HDAC1 increased allergic airway inflammation and promoted Th2 cytokine production in asthma mice, while asthmatic mice treated with herbal extract can resulted in significant anti-inflammatory and anti-allergic activity by increasing expression level of alveolar macrophages HDAC1 (17). Recent study showed that a polymorphism in the HDAC1 gene is associated with the response to corticosteroids in asthmatics (18). Our study revealed that HDAC1 was a hub gene in the PPI network and might play an important role in the process of asthma, which was consisting with previous studies. PCNA served as a factor to coordinate DNA replication and epigenetic inheritance, such as DNA methylation (19). A study showed that PCNA interacted with HDAC1 in human cells in vitro can co-localize in the cell nucleus, and finally led to integration of DNA replication (20). DNA methylation changes in PB were associated with childhood allergic asthma (9). These finding indicated that PCNA interacted with HDAC1 might involve in DNA methylation of asthma in children. In addition, a previous study showed that PCNA expression was associated with the epithelium thickness in corticosteroid-dependent asthma (21). The cell proliferation related molecule PCNA and cell activation related molecule NF-κB were both highly expressed in corticosteroid-dependent asthmatic subjects (21), revealing the potential role in the treatment and disease epithelium repair.
ITGA4 is also named as CD49d. A study reported that up-regulation of lysophosphatidic acid receptor 1 and down-regulation of ITGA4 can increase the number of monocytes in the PB and finally impact on immune function (22). However, the relationships between ITGA4 and asthma disease were unclear. The PPI network analysis in our study revealed that ITGA4 might be a major factor related to asthma in children. CTNNA1 is also known as αE-catenin, and it plays a major role in epithelial tissue, both at adherent junctions and in signaling pathways (23). Epithelial damage from airway inflammation during asthma may result in immune response to self-antigens, including αE-catenin and epidermal group factor receptor (EGFR); and finally contributes to the pathogenesis of asthma (24). Moreover, a replication study in a Caucasian worker population revealed that α-catenin gene variants were also associated with diisocyanate asthma (25). These results suggested that CTNNA1 might be critical gene that regulated the progressive of asthma in children.
MTOR is a serine/threonine kinase that is evolutionary conserved and can regulate lymphocyte cellular immunity by activating cytokine secreted from inflammatory cells (26). MTORC2 regulated the differentiation of naive CD4+ T cells into Th9 cells, and mTORC2 deficiency in T cells could result in less severe inflammation in the murine allergic airway inflammation model (27). Zhang et al (28) revealed that increased serum mTOR pathway activation can lead to elevated levels of Th17 cells and IL-4, following decreased Treg cells and IFN-γ. Our study demonstrated a clear relationship between MTOR in the PBMC of childhood patients with allergic asthma. These findings strongly suggested a necessary for mTOR pathway activation in asthma process. In addition, NF-κB1 is reported as a transcription factor that is activated by multiple intra-cellular and extra-cellular stimuli such as cytokines, oxidant-free radicals, and bacterial or viral products (29). Activated NF-κB can stimulate the expression of genes involved in many biological or pathological processes, including acute lung injury/acute respiratory distress syndrome and asthma. Recent study showed that the over-expression of PI3K and NF-κB in childhood asthma were negatively correlated with pulmonary functions, which indicated that PI3K and NF-κB might involve in the development of bronchial asthma in children (30). Inhibition of the NF-κB signaling pathway can improve airway inflammation in an ovalbumin-induced rat model (31). In our study, we also found that the NF-κB1 and mTOR were hub genes related to asthma in children. Considering the previous studies, we suggested that NF-κB1 interacted with mTOR might play an important role in the progression of asthma in children, such as inflammation in asthma. Additionally, a study of asthmatic mouse model indicated that mTOR is activated during asthma onset and inhibited during asthma remission, and blocking the mTOR pathway in asthmatic mice restores the Th17/Treg and Th1/Th2 cytokines balances (28). These findings strongly documented a critical role of mTOR pathway activation in asthma onset, revealing potential targets for asthma treatments.
Using the network analysis, we also identified APP hitherto not associated with asthma as important hub gene in regulatory network associated with NE tissue. APP gene encoding the amyloid beta precursor protein is known as a major player in Alzheimer's disease (AD) that have immune and inflammatory components (32). Recent studies also suggested a link between APP and asthma genes (32,33). It is revealed that APP was potentially associated with airway hyperresponsiveness through the interaction with a disintegrin and metalloproteinase (ADAM33) (33). ADAM33 is an asthma susceptibility gene with catalytic properties, functioned as a negative regulator of APP (34). In our module, APP was found potentially interacted with other asthma genes, it is speculated that these possible connections can provide new insights for exploring the functional role and relationships of these genes in asthma, as well as in AD.
In conclusion, our findings suggest that genes are differentially expressed in the three type tissue samples of asthmatic children, including NE, PB and PBMC samples. Among these DEGs, FBXO6, HDAC1 and APP interact with PCNA, ITGA4, CTNNA1, NF-κB1 and mTOR might be critical DEGs related to asthma in children.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Science and Technology Development Fund of Fengxian District, Shanghai (grant no. 20151236).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
GW and CW conceived and designed the present study. CW, HL and LC conducted the data analysis. CW and GW prepared the manuscript. All of the authors reviewed the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Center for Health Statistics: Tables of Summary Health Statistics: National Health Interview Survey. Table C-1b. 2016:3–4. [Google Scholar]
- 2.James CV, Rosenbaum S. Paying for quality care: Implications for racial and ethnic health disparities in pediatric asthma. Pediatrics. 2009;123(Suppl 3):S205–S210. doi: 10.1542/peds.2008-2233L. [DOI] [PubMed] [Google Scholar]
- 3.Lin HW, Lin SC. Environmental factors association between asthma and acute bronchiolitis in young children-a perspective cohort study. Eur J Pediatr. 2012;171:1645–1650. doi: 10.1007/s00431-012-1788-3. [DOI] [PubMed] [Google Scholar]
- 4.Martinez FD. Genes, environments, development and asthma: A reappraisal. Eur Respir J. 2007;29:179–184. doi: 10.1183/09031936.00087906. [DOI] [PubMed] [Google Scholar]
- 5.Goetghebuer T, Isles K, Moore C, Thomson A, Kwiatkowski D, Hull J. Genetic predisposition to wheeze following respiratory syncytial virus bronchiolitis. Clin Exp Allergy. 2004;34:801–803. doi: 10.1111/j.1365-2222.2004.1947.x. [DOI] [PubMed] [Google Scholar]
- 6.Finkelstein Y, Bournissen FG, Hutson JR, Shannon M. Polymorphism of the ADRB2 gene and response to inhaled beta-agonists in children with asthma: A meta-analysis. J Asthma. 2009;46:900–905. doi: 10.3109/02770900903199961. [DOI] [PubMed] [Google Scholar]
- 7.Wills-Karp M. Immunologic basis of antigen-induced airway hyperresponsiveness. Annu Rev Immunol. 1999;17:255–281. doi: 10.1146/annurev.immunol.17.1.255. [DOI] [PubMed] [Google Scholar]
- 8.Yang IV, Pedersen BS, Liu AH, O'Connor GT, Pillai D, Kattan M, Misiak RT, Gruchalla R, Szefler SJ, Hershey Khurana GK, et al. The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol. 2017;139:1478–1488. doi: 10.1016/j.jaci.2016.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang IV, Pedersen BS, Liu A, O'Connor GT, Teach SJ, Kattan M, Misiak RT, Gruchalla R, Steinbach SF, Szefler SJ, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136:69–80. doi: 10.1016/j.jaci.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raedler D, Ballenberger N, Klucker E, Böck A, Otto R, da Costa Prazeres O, Holst O, Illig T, Buch T, von Mutius E, Schaub B. Identification of novel immune phenotypes for allergic and nonallergic childhood asthma. J Allergy Clin Immunol. 2015;135:81–91. doi: 10.1016/j.jaci.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 11.Williams-DeVane CR, Reif DM, Hubal EC, Bushel PR, Hudgens EE, Gallagher JE, Edwards SW. Decision tree-based method for integrating gene expression, demographic, and clinical data to determine disease endotypes. BMC Syst Biol. 2013;7:119. doi: 10.1186/1752-0509-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Duan LH, Luo PC, Hu G, Yu X, Liu J, Lu H, Liu B. FBXO6-mediated ubiquitination and degradation of Ero1L inhibits endoplasmic reticulum stress-induced apoptosis. Cell Physiol Biochem. 2016;39:2501–2508. doi: 10.1159/000452517. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Wei J, Mialki RK, Mallampalli DF, Chen BB, Coon T, Zou C, Mallampalli RK, Zhao Y. F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat Immunol. 2012;13:651–658. doi: 10.1038/ni.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantley MD, Fairlie DP, Bartold PM, Marino V, Gupta PK, Haynes DR. Inhibiting histone deacetylase 1 suppresses both inflammation and bone loss in arthritis. Rheumatology (Oxford) 2015;54:1713–1723. doi: 10.1093/rheumatology/kev022. [DOI] [PubMed] [Google Scholar]
- 17.Grausenburger R, Bilic I, Boucheron N, Zupkovitz G, El-Housseiny L, Tschismarov R, Zhang Y, Rembold M, Gaisberger M, Hartl A, et al. Conditional deletion of histone deacetylase 1 in T cells leads to enhanced airway inflammation and increased Th2 cytokine production. J Immunol. 2010;185:3489–3497. doi: 10.4049/jimmunol.0903610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MH, Kim SH, Kim YK, Hong SJ, Min KU, Cho SH, Park HW. A polymorphism in the histone deacetylase 1 gene is associated with the response to corticosteroids in asthmatics. Korean J Intern Med. 2013;28:708–714. doi: 10.3904/kjim.2013.28.6.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 20.Milutinovic S, Zhuang Q, Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J Biol Chem. 2002;277:20974–20978. doi: 10.1074/jbc.M202504200. [DOI] [PubMed] [Google Scholar]
- 21.Vignola AM, Chiappara G, Siena L, Bruno A, Gagliardo R, Merendino AM, Polla BS, Arrigo AP, Bonsignore G, Bousquet J, Chanez P. Proliferation and activation of bronchial epithelial cells in corticosteroid-dependent asthma. J Allergy Clin Immunol. 2001;108:738–746. doi: 10.1067/mai.2001.119160. [DOI] [PubMed] [Google Scholar]
- 22.Maugeri N, Powell JE, t Hoen PA, de Geus EJ, Willemsen G, Kattenberg M, Henders AK, Wallace L, Penninx B, Hottenga JJ, et al. LPAR1 and ITGA4 regulate peripheral blood monocyte counts. Hum Mutat. 2011;32:873–876. doi: 10.1002/humu.21536. [DOI] [PubMed] [Google Scholar]
- 23.Herrenknecht K, Ozawa M, Eckerskorn C, Lottspeich F, Lenter M, Kemler R. The uvomorulin-anchorage protein alpha catenin is a vinculin homologue. Proc Natl Acad Sci USA. 1991;88:9156–9160. doi: 10.1073/pnas.88.20.9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M, Subramanian V, Christie C, Castro M, Mohanakumar T. Immune responses to self-antigens in asthma patients: Clinical and immunopathological implications. Hum Immunol. 2012;73:511–516. doi: 10.1016/j.humimm.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein DI, Kashon M, Lummus ZL, Johnson VJ, Fluharty K, Gautrin D, Malo JL, Cartier A, Boulet LP, Sastre J, et al. CTNNA3 (α-catenin) gene variants are associated with diisocyanate asthma: A replication study in a Caucasian worker population. Toxicol Sci. 2013;131:242–246. doi: 10.1093/toxsci/kfs272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Zhang L, Wang P, Su H, Wang W, Chu Z, Zhang L, Zhang X, Zhao Y. mTORC2 controls Th9 polarization and allergic airway inflammation. Allergy. 2017;72:1510–1520. doi: 10.1111/all.13152. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Jing Y, Qiao J, Luan B, Wang X, Wang L, Song Z. Activation of the mTOR signaling pathway is required for asthma onset. Sci Rep. 2017;7:4532. doi: 10.1038/s41598-017-04826-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer R, Hatada EN, Hohmann HP, Haiker M, Bartsch C, Röthlisberger U, Lahm HW, Schlaeger EJ, van Loon AP, Scheidereit C. Cloning of the DNA-binding subunit of human nuclear factor kappa B: The level of its mRNA is strongly regulated by phorbol ester or tumor necrosis factor alpha. Proc Natl Acad Sci USA. 1991;88:966–970. doi: 10.1073/pnas.88.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi HL, Liu JB, Lu AP. Expression profiles of PI3K, NF-κB, and STAT1 in peripheral blood mononuclear cells in children with bronchial asthma. Zhongguo Dang Dai Er Ke Za Zhi. 2016;18:614–617. doi: 10.7499/j.issn.1008-8830.2016.07.009. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Zheng J, Zhang N, Li C. Berberine improves airway inflammation and inhibits NF-κB signaling pathway in an ovalbumin-induced rat model of asthma. J Asthma. 2016;53:999–1005. doi: 10.1080/02770903.2016.1180530. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Brossard M, Sarnowski C, Vaysse A, Moffatt M, Margaritte-Jeannin P, Llinares-López F, Dizier MH, Lathrop M, Cookson W, et al. Network-assisted analysis of GWAS data identifies a functionally-relevant gene module for childhood-onset asthma. Sci Rep. 2017;7:938. doi: 10.1038/s41598-017-01058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vishweswaraiah S, Veerappa AM, Mahesh PA, Jayaraju BS, Krishnarao CS, Ramachandra NB. Molecular interaction network and pathway studies of ADAM33 potentially relevant to asthma. Ann Allergy Asthma Immunol. 2014;113:418–424.e1. doi: 10.1016/j.anai.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Zou J, Zhu F, Liu J, Wang W, Zhang R, Garlisi CG, Liu YH, Wang S, Shah H, Wan Y, Umland SP. Catalytic activity of human ADAM33. J Biol Chem. 2004;279:9818–9830. doi: 10.1074/jbc.M309696200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.