SUMMARY
The antitumor effector T helper 1 (Th1) and Th17 cells represent two T cell paradigms: short-lived cytolytic Th1 cells and “stem cell-like” memory Th17 cells. We report that Th9 cells represent a third paradigm—they are less-exhausted, fully cytolytic, and hyperproliferative. Only tumor-specific Th9 cells completely eradicated advanced tumors, maintained a mature effector cell signature with cytolytic activity as strong as Th1 cells, and persisted as long as Th17 cells in vivo. Th9 cells displayed a unique Pu.1-Traf6-NF-κB activation-driven hyperproliferative feature, suggesting a persistence mechanism rather than an antiapoptotic strategy. Th9 antitumor efficacy depended on interleukin-9 and upregulated expression of Eomes and Traf6. Thus, tumor-specific Th9 cells are a more effective CD4+ T cell subset for adoptive cancer therapy.
Graphical Abstract
In Brief
Lu et al. report that adaptively transferred tumor-specific CD4+ Th9 cells eradicate large established murine tumors and protect surviving mice against tumor rechallenge. Th9 cells maintain a mature effector cell signature with cytolytic activity as strong as Th1 cells and persist as long as Th17 cells in vivo.
INTRODUCTION
Adoptive cell therapy (ACT) using tumor-specific T cells has focused primarily on CD8+ cytotoxic T lymphocytes (CTLs) (Restifo et al., 2012). However, treatment of large established tumors with CD8+ CTLs expanded ex vivo has only yielded limited promising results, and systemic administration of interleukin-2 (IL-2), which is required for survival of effector CD8+ T cells (Klebanoff et al., 2011), may inhibit infiltration of transferred T cells into tumor tissues (Kjaergaard et al., 2001) and stimulate suppressive effects of regulatory T (Treg) cells (Zhang et al., 2005).
Although CD8+ T cells are potent mediators of antitumor immunity, the role of CD4+ T cells in tumor immunity remains underappreciated. Nevertheless, recent observations revealed that CD4+ T cell recognition of neoantigens is frequent, underscoring the potential clinical relevance of targeting MHC class II-restricted neoantigens by transfer of CD4+ T cells (Linnemann et al., 2015). Also encouraging is the emergence of cytotoxic T helper 1 CD4+ T (Th1) cells as a physiologically relevant and therapeutically useful T cell lineage for ACT to treat tumors in the clinic (Hunder et al., 2008). However, improvements to this approach are needed because ex-vivo-generated tumor-specific Th1 cells display an exhausted phenotype, and transferred cells have a disappointing lack of persistence (Hunder et al., 2008). Current advances in ACT also suggest that T cells with an early memory and/or a stem cell-like phenotype (Th17 paradigm) and reduced cytolytic function in vitro outperform their short-lived, terminal/end-effector-like counterparts (Th1 paradigm) in vivo (Muranski et al., 2011). Thus, identification of CD4+ T cell subsets that possess a mature effector and less-exhausted phenotype, and persist significantly longer remains a critical challenge to advancing cancer immunotherapy. To our knowledge, such a T cell subset has not yet been discovered.
Recently, using mouse models of cancer, we (Lu et al., 2012) and others (Purwar et al., 2012; Vegran et al., 2014) have characterized IL-9-producing CD4+ Th (Th9) cells as an antitumor T cell subset. Furthermore, subsequent elegant studies also demonstrated the potential for triggering endogenous antitumor Th9 responses in vivo (Kim et al., 2015; Liu et al., 2015; Zhao et al., 2016b), by both an antigen-nonspecific manner via glucocorticoid-induced tumor necrosis factor (TNF) receptor-related protein costimulation and by an antigen-specific manner via vaccination. However, the T cell features of Th9 cells beyond IL-9 production and whether these cells can be used to cure late-stage advanced tumors (a scenario more like that seen clinically) have not been explored. Therefore, we carried out this study to uncover the T cell features of Th9 cells related to cancer adoptive immunotherapy.
RESULTS
Transfer of Th9 Cells Eradicates Advanced Late-Stage Tumor and Leads to Long-Term Survival
Tumor-specific Th9 cells were generated by priming OT-II or tyrosinase-related protein 1 (TRP-1) naive CD4+CD62L+ T cells with peptide-loaded antigen-presenting cells (APCs) (irradiated, T cell-depleted splenocytes) for 5 days in Th9-polarized medium. As Figures S1A–S1C show, differentiated Th9 cells typically were more than 55% IL-9-expressing CD4+ T cells, with limited production of interferon γ (IFN-γ), IL-4, or IL-17 (Lu et al., 2012). In addition, we generated (cultured 5 days) Th1 cells as a control because cytotoxic Th1 cells are therapeutically useful CD4+ T cells for ACT in the clinic (Hunder et al., 2008). We also generated (cultured 5 days) Th17 cells as an additional control because these cells represent the T cell lineage that may possess the highest antitumor efficacy among CD4+ T cell subsets tested so far (Muranski et al., 2011).
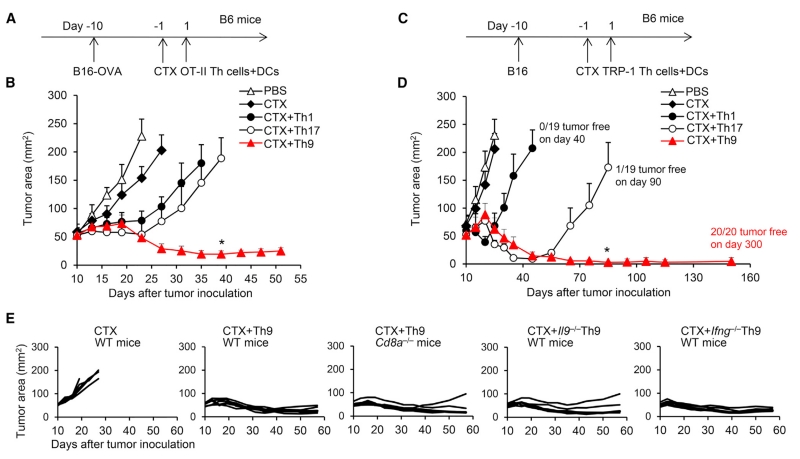
To test our central hypothesis that Th9 cells can be utilized as a potential CD4+ T cell subset for ACT of cancer, we performed studies by transferring ovalbumin (OVA)-specific CD45.1+ OT-II Th1, Th17, or Th9 cells into CD45.2+ wild-type (WT) C57BL/6 (B6) mice bearing large (~8 × 7 mm), established B16-OVA melanoma (Figure 1A). One day before T cell transfer, B6 mice were given one dose of cyclophosphamide (CTX) (200 mg/kg) to induce temporary lymphopenia, which is frequently induced as part of clinical ACT protocols to promote homeostatic proliferation of transferred T cells (North, 1982). Mice also received adjuvant OVA peptide-pulsed dendritic cell (DC) vaccination on the day of transfer, which is frequently used to boost the antitumor responses during ACT (Chodon et al., 2014; Lu et al., 2014). Surprisingly, only Th9 cells mediated significant tumor regression that resulted in long-term survival, whereas Th1, Th17, and Th2 cell treatment induced only temporary tumor regression, which was followed by aggressive recurrence (Figures 1B and S1D).
Figure 1. Transfer of Tumor-Specific Th9 Cells Eradicates the Large Established Tumor.
(A) OVA-specific Th1, Th9, or Th17 cells (CD45.1+, 2.5 × 106) were transferred intravenously (i.v.) into CD45.2+ B6 mice bearing 10-day large established B16-OVA tumors (1 × 106 B16-OVA cells challenged subcutaneously [s.c.] 10 days before T cell transfer). Adjuvant cyclophosphamide (CTX) (intraperitoneally [i.p.]) was administered as indicated 1 day before T cell transfer. DC vaccination (2.5 × 105, i.v.) was given to mice that received CTX.
(B) Tumor responses to OT-II T cell transfer are shown (n = 5/group).
(C) TRP-1-specific Th1, Th9, or Th17 cells (CD45.2+, 2.5 × 106) were transferred i.v. into CD45.1+ B6 mice bearing 10-day large established B16 (1 × 106 B16 cells challenged s.c. 10 days before T cell transfer). Adjuvant CTX (i.p.) was administered as indicated 1 day before T cell transfer. DC vaccination (2.5 × 105, i.v.) was given to mice that received CTX.
(D) Representative tumor responses to TRP-1 T cell transfer are shown (n = 5/group). The description of tumor-free survival is summarized from several independent studies.
(E) Tumor responses to OT-II T cell transfer are shown (n = 5/group). WT or Cd8a−/− mice received CTX and DC vaccination and transfer of WT, Il9−/−, or Ifng−/− Th9 cells. Control mice received no Th9 transfer. Representative results of one from at least two repeated experiments are shown (total mice/group ≥10). Data are mean ± SD; *p < 0.05, compared with Th17 cells.
See also Figure S1.
Because OT-II cells target artificial antigen, we next used the TRP-1 model of adoptive immunotherapy, which reproduces the clinical challenge of targeting gp75 tumor/self-antigen in the poorly immunogenic B16 melanoma (Muranski et al., 2008). CD45.2+ TRP-1-Th1, Th17, or Th9 cells were transferred into CD45.1+ B6 mice bearing large established B16 melanomas (~8 ± 7 mm) in conjunction with CTX administration and DC vaccination (Figure 1C). Similar to previous reports (Muranski et al., 2011), Th17 cells more potently induced tumor rejection compared with Th1 cells (Figure 1D). Surprisingly, Th9 cell transfer eradicated these advanced late-stage tumors, with all treated mice remaining tumor free at 300 days, whereas Th1 and Th17 cell-treated mice suffered relapse by 3 and 8 weeks, respectively (Figure 1D). DC vaccination seemed to be required for optimal antitumor responses of Th9 cells (Figure S1E). In addition, Th9 cells also exerted stronger antitumor activity compared with pathogenic Th17 cells or other types of “Th17” cells generated by different polarizing conditions (Figure S1F), and protected the mice against three sequential rechallenges with B16 tumor cells starting at 150 days after Th9 transfer (Figures S1G and S1H).
We have reported that, in tumor prevention models with low tumor burden, Th9 cells promote CD8+ CTL-mediated antitumor immune response (Lu et al., 2012). However, in a more clinical scenario (e.g., late-stage advanced tumor burden and lymphodepleting conditions during ACT), the relative contributions of transferred Th9 cells versus induced host CD8+ CTLs in eradicating large tumors is unclear. Although we also observed that Th9 cells induced a significant increase in tumor-infiltrating tumor (OVA)-specific CD8+ T cells (Figure S1I), deficiency in host CD8+ T cells only slightly affected the antitumor efficacy of Th9 cell transfer compared with that in WT mice under lymphopenic conditions (Figure 1E), indicating that tumor-specific Th9 cells may be the major effector responsible for eradicating tumor cells in vivo. In addition, we found that Il9 deficiency in Th9 cells also only marginally affected their efficacy (Figure 1E). Finally, our results suggested that Th9 cells were not a significant IFN-γ producer, nor did they require IFN-γ to exert their antitumor function because Ifng−/− Th9 cells exerted intact antitumor immunity (Figures 1E and S1J). Taken together, these results suggested that tumor-specific Th9 cells might be an ideal T cell subset for ACT, whereas IL-9 production and the induced CD8+ CTL responses are required only for their optimal antitumor function.
Th9 Cells Are Distinct Mature Effector T Cells
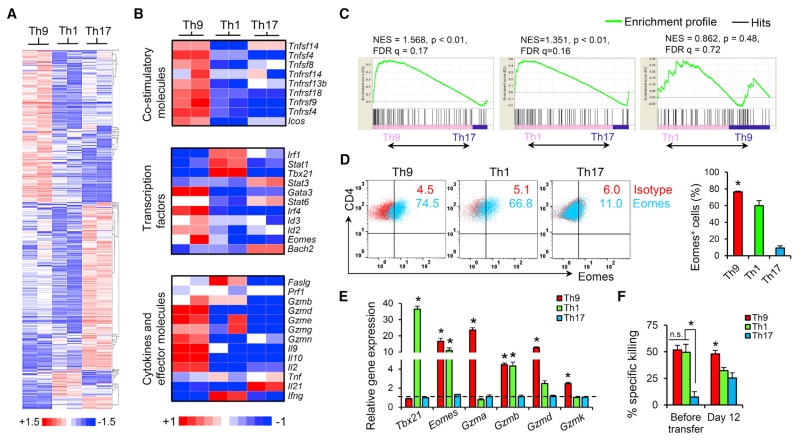
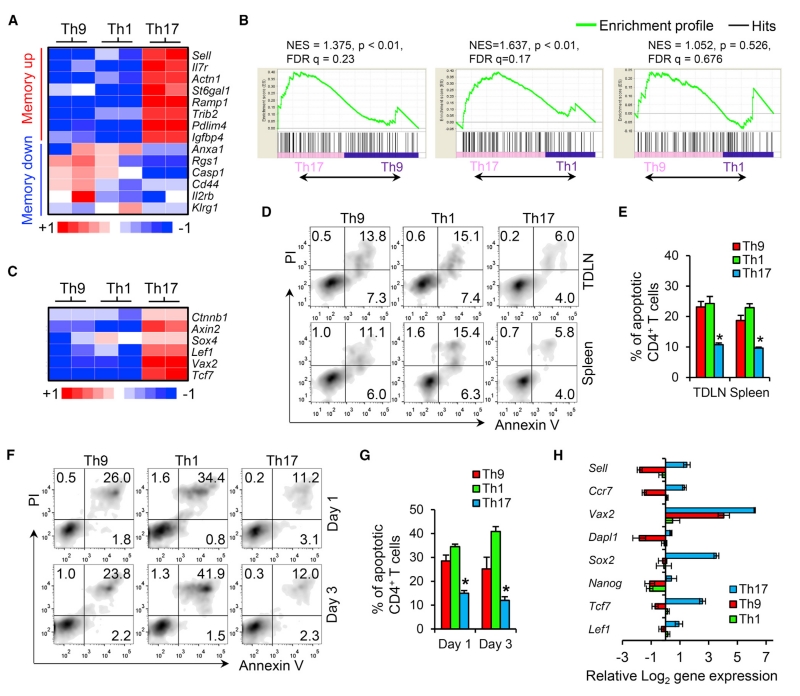
Current advances suggest that T cells exhibiting the long-lived early memory and/or stem cell-like paradigm (Th17 paradigm) should be selected for ACT (Berger et al., 2008; Gattinoni et al., 2009). Th17 cells are endowed with an enhanced capacity to survive/self-renew, generate Th1-like effector progeny, and enter the memory pool with an antitumor efficacy superior to that of short-lived terminally differentiated Th1 cells (Th1 paradigm) for cancer therapy (Muranski et al., 2011). However, the T cell features of Th9 cells beyond IL-9 production have yet to be studied, and the extraordinary ability of Th9 cells to completely cure large advanced tumors prompted us to explore the T cell features of these cells. We sorted CD45.1+ OVA-specific Th9-, Th17-, and Th1-derived cells from the spleens of tumor-bearing CD45.2+ WT B6 mice 12 days after transfer. The global transcriptional profile of the T cells was analyzed in duplicate by gene array. Analysis revealed that the Th9 cell gene profile was distinct from that of Th1 and Th17 cells (Figure 2A). Strikingly, Th9 cells expressed higher levels of several costimulatory molecules (Figure 2B). These three subsets of Th cells also differed in gene expression of effector molecules, transcriptional factors, and cytokines (Figures 2B, S2A, and S2B). Th1 cells highly expressed Th1-related transcriptional factor genes (Irf1, Stat1, and Tbx21) and several effector molecule genes (Faslg and Gzmb), but did not express Il2, suggesting a more terminally differentiated state (Hinrichs et al., 2006). Particularly interesting is that Th9 cells had greater gene expression of Id2 and Eomes, which encode transcriptional factors that suggest effector cell development (Pearce et al., 2003; Yang et al., 2011), and increased gene expression of a granzyme panel (Gzmb, Gzmd, Gzme, Gzmg, and Gzmn). On the other hand, transferred Th9 cells were also enriched in the gene expression of Id3 and Il2 (Hinrichs et al., 2006; Yang et al., 2011), which suggests that transferred Th9 cells are neither terminally differentiated nor short-lived. Finally, we found that Bach2, which promotes the differentiation of long-lived memory cells and Treg cells, but restrains effector cell development (Roychoudhuri et al., 2013), was highly expressed only in Th17 cells (Figure 2B), confirming the memory feature and reduced cytolytic function of Th17 cells. The increased Bach2 expression in Th17 cells may also account for their partial conversion into Foxp3+ Treg-like cells in vivo (Figure S2C), because Bach2 is known to promote formation and stabilization of Treg cells (Roychoudhuri et al., 2013).
Figure 2. Th9 Cells Are Distinct Cytolytic Effector T Cells.
Mice were treated as shown in Figure 1A. CD45.1+ OT-II Th cells were sorted from the spleens of tumor-bearing mice 12 days after the transfer. RNA (biological samples from two mice) was extracted for gene microarray.
(A) Global transcriptional profiles revealed by microarray of purified Th-derived cells from spleens of tumor-bearing mice 12 days after transfer. The heatmap shows the log2-fold change relative to the global average of the top upregulated and downregulated genes, with a cutoff of change in expression >1.5-fold and a p value < 0.05.
(B) Heatmaps illustrating the relative expression of gene sets as indicated (data are log scaled).
(C) GSEA of the mature effector gene signature. NES, normalized enrichment score; FDR, false discovery rate.
(D) In vitro (5-day) cultured OT-II-Th cells were stained for Eomes expression by FACS. Representative (left) and summarized (right) data for Eomes+ cells are shown.
(E) RT-PCR results for expression of the indicated genes in Th cells before transfer (5-day culture, n = 3 mice).
(F) Specific killing assay of OT-II-Th cells before transfer (5-day culture) or CD45.1+ OT-II Th cells sorted from spleens of tumor-bearing mice (n = 3) was performed against B16-OVA cells. Representative results from one of two repeated experiments are shown. Data are mean ± SD; *p < 0.05, compared with Th1 or Th17 cells.
See also Figure S2.
To more accurately assess effector T cell development of the Th9 cells in an unbiased manner, we performed gene set enrichment analysis (GSEA) to generate an enrichment plot for a mature T cell effector gene signature set (Wirth et al., 2010). GSEA of the gene array data obtained from Th cells 12 days after transfer revealed that the mature effector gene signature was significantly enriched in both Th9 and Th1 cells, but not in Th17 cells, and enrichment did not differ between Th1 and Th9 cells (Figure 2C). These results again suggested that Th9 may be equal to Th1 cells in terms of mature effector T cell development. Intriguingly, before transfer, significantly upregulated Eomes expression could be detected in Th9 cells by RT-PCR and intracellular staining, which was even greater than that in classic cytolytic Th1 cells (Figure 2D). As Eomes is the effector master regulator that controls granzyme expression (Pearce et al., 2003), Th9 cells also expressed markedly increased Gzma, Gzmb, Gzmd, and Gzmk among the tested T cell subsets and expressed a similar level of Gzmb as compared with Th1 cells (Figure 2E). The Th9 Eomes and Granzyme expression patterns prompted us to directly test the cytolytic function of these cells. As shown in Figures 2F and S2D, Th9 cells generated in vitro and sorted from tumor-bearing mice 12 days after transfer had the highest tumor-specific killing activity compared with Th1, Th17, and other Th cells. We also observed that Th9-mediated-specific killing was primarily granzyme dependent, and particularly required granzyme B activity (Figures S2D–S2F). Our data thus far indicate that Th9 cells display a core molecular signature that suggests they are programmed as mature effector T cells.
Th9 Cells Do Not Display Exhausted or Terminally Differentiated T cell Phenotype
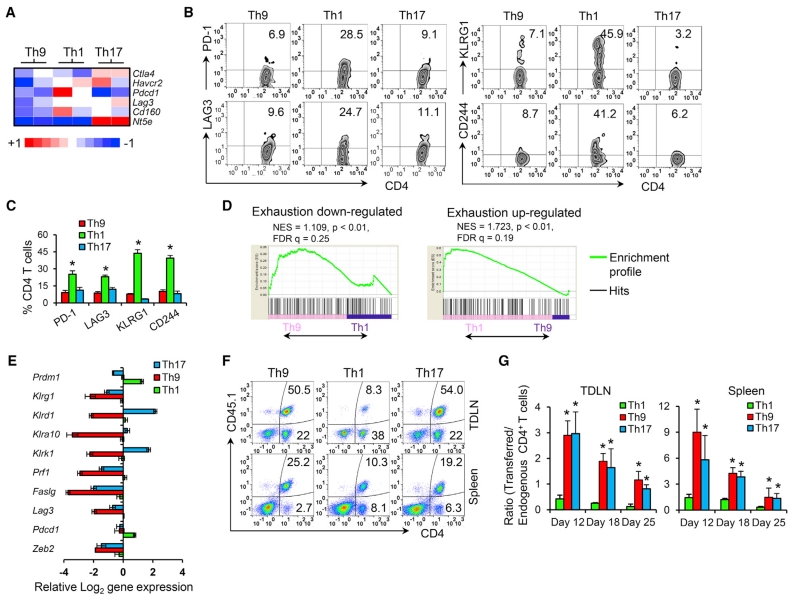
A key feature of the classic cytolytic Th1 cells is that these cells display exhausted profiles, which greatly limits their antitumor function. Although our results suggested Th9 cells to be distinct effector cells, we wondered whether Th9 cells also display an exhaustion feature. Gene array data analysis showed that Th9 cells at 12 days after transfer expressed the lowest levels of inhibitory receptors (Ctla4, Havcr2, Pdcd1, Lag3, Cd160, and Nt5e; Figure 3A). As confirmation, fluorescence-activated cell sorting (FACS) suggested that only Th1 cells upregulated these molecules, including PD-1, LAG3, KLRG1, and CD244 (Figures 3B and 3C). We further assessed the Th9 exhaustion profile by GSEA (Quigley et al., 2010), and found that Th9 cells were significantly enriched in the exhaustion-downregulated gene signature, whereas Th1 cells were significantly enriched in an exhaustion-upregulated gene signature (Figure 3D), suggesting that Th1 cells, but not Th9 cells, carry the molecular signature of the T cell exhaustion phenotype.
Figure 3. Th9 Cells Are a Less-Exhausted Effector with Long-Term Persistence Capacity.
Mice were treated as shown in Figure 1A.
(A) Heatmap illustrating the relative expression of genes (data are log scaled).
(B–C) Expression of indicated exhaustion markers by transferred cells 12 days after transfer was determined by FACS (gated on CD45.1+CD4+ cells). Representative data (B) and summarized results (C) are shown. *p < 0.05, compared with Th9 or Th17 cells.
(D) GSEA was performed to compare exhaustion-associated gene enrichment in Th1 or Th9 cells.
(E) RT-PCR for expression of the indicated genes. Shown are the relative log2 expression of selected differentially expressed genes encoding phenotypic markers of terminal differentiation and end-effector function in Th-derived cells recovered from spleens 12 days after transfer (n = 3 mice/group).
(F) FACS analysis of the presence of transferred Th cells in tumor-draining lymph nodes (TDLNs) and spleens of mice 10 days after transfer. Representative data are shown.
(G) The relative ratio of transferred cells (CD45.1+CD4+ cells) to endogenous CD4+ cells (CD45.2+CD4+ cells) summarized from (F) (n = 3 mice/group). Representative results of one from two repeated experiments are shown. Data are mean ± SD; *p < 0.05, compared with Th1 cells.
See also Figure S3.
High T-bet expression is closely associated with terminal differentiation and drives short-lived T cell development (Joshi et al., 2007), which seriously hampers the antitumor potential of Th1 cells. Because Th9 cells had low Tbx21 but high Il2 expression (Figure 2B), we hypothesized that Th9 cells are not terminally differentiated or late-stage short-lived Th1-like cells. Indeed, only polarized Th1 cells had increased expression of Prdm1 and Klrg1, the hallmarks of terminal differentiation (Rutishauser et al., 2009) and T cell senescence (Reiley et al., 2010), respectively (Figure 3E). Moreover, Th1, but not Th9 or Th17 cells, highly expressed inhibitory molecules and other end-effector function markers (Klrd1, Klra10, KlrK1, Prf1, Faslg, Lag3, Pdcd1, and Zeb2, Figure 3E) that have been reported to be associated with terminal differentiation and have less-effective in vivo antitumor activity upon transfer (Gattinoni et al., 2005).
T cell subsets possessing great persistence in vivo are essential for successful ACT, which is the key reason why long-lived Th17 cells outperform terminally differentiated short-lived Th1 cells for ACT (Muranski et al., 2011). We, therefore, determined the persistence capacity of these less-exhausted effector Th9 cells. Noticeably, Th1 cells had the lowest number of surviving transferred cells in spleen and tumor-draining lymph nodes (TDLNs) over time (Figures 3F, 3G, S3A, and S3B), confirming their short-lived terminally differentiated signature. In striking contrast, Th9 cells had extraordinary persistence equal to, if not better than, the “stem cell-like” early memory Th17 cells (Figures 3F, 3G, S3A, and S3B). The long-term persistence of TRP-1 Th9 and Th17 cells also resulted in far greater autoimmune phenomena than Th1 cells, including the development of vitiligo and uveitis (Figures S3C and S3D). Thus, Th9 cells may represent an unidentified effector T cell phenotype that is distinct from the classic cytolytic Th1 cells: a less-exhausted fully cytolytic effector function and exceptional persistence after transfer.
Th9 Cells Do Not Have Memory or Stem Cell-like Features
Early memory T cells are classically associated with prolonged peripheral persistence after ACT, so we first hypothesized that Th9 cells may also fit into this early memory classification. However, analysis of the gene profile that governs early memory development suggested that only Th17 cells carry a core molecular signature of a less-differentiated memory subset (Muranski et al., 2011) (Figure 4A), which was confirmed by FACS analysis of some commonly used phenotypic markers of memory T cells (Figure S4A). GSEA further demonstrated that, compared with Th9 or Th1 cells, Th17 cells were significantly enriched in early memory features that are characteristic of memory precursor cells that survive and give rise to long-lived memory cells (Wirth et al., 2010) (Figure 4B). In contrast, Th9 cells resembled the Th1 effector-type T cells, which were skewed away from early memory lineage development (Figures 4A and 4B).
Figure 4. Th9 Cells Do Not Acquire a Gene Signature Associated with Early Memory or Stem Cell-like Feature.
Mice were treated as shown in Figure 1A.
(A) Heatmap illustrating the relative expression of genes that have been reported in the literature to be associated with T cell memory subsets (data are log scaled).
(B) GSEA of the early memory gene signature.
(C) Heatmap illustrating the relative expression of genes that have been reported in the literature to be associated with self-renewal and hematopoietic stem cell maintenance (data are log scaled).
(D and E) FACS analysis of apoptotic transferred Th cells (gated on CD45.1+CD4+ cells) in TDLNs and spleens of tumor-bearing mice (n = 3/group) 12 days after transfer. Representative data (D) and summarized results for annexin V+ cells (E) are shown.
(F and G) FACS analysis of apoptotic Th cells (polarized in vitro for 5 days, n = 3 mice/group) restimulated with antigen-pulsed APCs in vitro. Representative data (F) and summarized results for annexin V+ cells (G) are shown.
(H) RT-PCR for expression of the indicated genes. Shown is the relative log2 expression in Th cells before transfer (polarized in vitro for 5 days, n = 3 mice/group) of selected differentially expressed genes encoding phenotypic markers of early memory/stem cell-like T cells. Representative results from one of two repeated experiments are shown. Data are mean ± SD; *p < 0.05, compared with Th1 or Th17 cells.
See also Figure S4.
T cell acquisition of “stemness” can also allow transferred T cells to persist long term (Gattinoni et al., 2009), so we next hypothesized that Th9 cells may be “stem cell-like” T cells, and analyzed the hallmark gene targets of the Wnt-β-catenin signaling axis (e.g., Ctnnb1, Axin2, Sox4, Lef1, Vax2, and Tcf7), a pathway required for the maintenance of stemness in T cells (Muranski et al., 2011). As shown in Figure 4C, only Th17 cells carried these stemness hallmark genes, confirming the previous observation of the stem cell-like nature of Th17 cells (Muranski et al., 2011). We also assessed the central stemness functional properties of these T cell subsets by analyzing their resistance to apoptosis. The results showed that the apoptotic rate of Th9 cells was similar to that of Th1 cells in both spleen and TDLN (Figures 4D, 4E, S4B, and S4C). In addition, when restimulating the in-vitro-differentiated Th cells with antigen-pulsed APCs, we observed that Th9 cells still had no greater antiapoptotic capacity than Th1 cells (Figures 4F and 4G). On the other hand, Th17 cells demonstrated the lowest apoptotic rate both in vivo and in vitro, which is consistent with their early memory/stem cell-like properties (Figures 4D–4G).
Indeed, these gene expression patterns existed before Th9 cell transfer, as shown in Figures 4H and S4D. After polarization in vitro for 5 days, we detected by RT-PCR that Th9 cells existed as a transcriptionally distinct population: they had lower expression levels (much lower than even short-lived Th1 cells) of genes for memory markers (Sell and Ccr7), early T cell development (Vax2 and Dapl1) (Muranski et al., 2011), and stemness (Sox2, Nanog, Tcf7, and Lef1) (Gattinoni et al., 2009). Considering that Th9 cells also expressed the lowest level of terminally differentiated end-effector function markers (even much lower than long-lived Th17 cells, see Figure 3E), it appears that the current understanding of a stem cell/early memory Th17 paradigm versus a terminal/end-effector Th1 paradigm is insufficient to explain the exceptional persistent capacity and antitumor effectiveness of Th9 cells.
Th9 Cells Display a Hyperproliferative Feature Mediated by the Hyperactivation of Late-Phase NF-κB Signaling
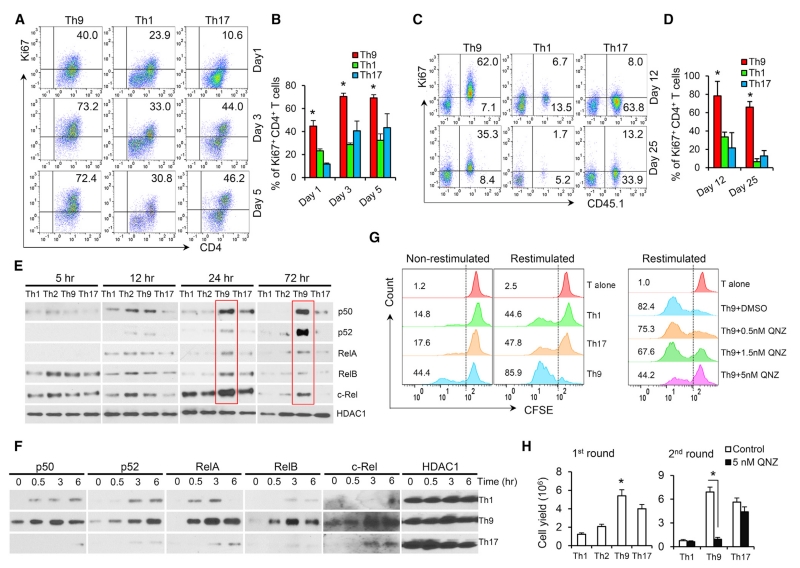
Because Th9 cells do not seem to have an enhanced antiapoptotic advantage, and the current literature does not clearly explain their prolonged persistence after transfer, we sought to determine Th9 proliferative capacity. We first reactivated Th1, Th9, and Th17 cells with antigen-pulsed APCs in vitro, and assessed Ki67 expression as a readout for proliferating cells. Surprisingly, the percentage of Ki67+ cells was significantly greater in Th9 cells compared with Th1 and Th17 cells (Figures 5A and 5B). To verify our finding, we also assessed Th9 cell proliferation over time in tumor-bearing mice. We found a large population (>80% on day 12 and ~70% on day 25) of proliferating OT-II-Th9 cells in the TDLNs, whereas Th1 and Th17 cells showed limited proliferation over time (Figures 5C and 5D). Moreover, ~150 days after transfer of TRP-1-Th9 cells, ~10% of splenic CD4+ T cells in the mice were transferred Th9-derived cells that exhibited cytolytic activity, a less-exhausted profile, and greater proliferation (Figures S1G and S5A–S5C). These results suggested that Th9 cells possess a unique hyperproliferative advantage over other antitumor Th cells, which may be responsible for the observed antitumor features of Th9 cells.
Figure 5. The Hyperactivation of Late-Phase NF-κB Signaling Drives the Hyperproliferative Feature in Th9 Cells.
(A and B) FACS determination of Ki67+ proliferative Th cells (polarized in vitro for 5 days, n = 3 mice) restimulated with antigen-pulsed APCs in vitro. Representative flow data (A) and summarized results (B) are shown.
(C and D) Mice were treated as shown in Figure 1A. FACS analysis of Ki67+ proliferating transferred Th cells in TDLNs of tumor-bearing mice (n = 3) 12 and 25 days after transfer. Representative data (C) and summarized results (D) are shown. *p < 0.05, compared with Th9 or Th17 cells.
(E) Naive CD4+ T cells were differentiated for 5–72 hr with plate-bound anti-CD3 monoclonal antibodies (mAbs) and soluble anti-CD28 mAbs, and NF-κB nuclear translocation was analyzed by western blot (nuclear fraction). Red solid lines indicate upregulated nuclear translocation of NF-κB in Th9 cells.
(F) OT-II-Th cells (polarized in vitro for 5 days) were restimulated with plate-bound αCD3 mAbs and soluble αCD28 mAbs, and NF-κB nuclear translocation was analyzed by western blot (nuclear fraction).
(G) OT-II-Th cells (polarized in vitro for 5 days) were labeled with CFSE and cocultured with unpulsed APCs (non-restimulated), OT-II peptide-pulsed APCs (restimulated), or OT-II peptide-pulsed APCs (restimulated) in the presence of QNZ for 48 hr. T alone is Th cells fixed with paraformaldehyde immediately after CFSE labeling. The percentage of CFSElow proliferative cells was determined by FACS.
(H) Th cell yields after the first activation round (day 5, n = 3 mice) and after restimulation for an additional 2 days (second round, equal number of OT-II-Th cells was collected for the restimulation). QNZ is a specific NF-κB inhibitor. Representative results from one of two repeated experiments are shown. Data are mean ± SD; *p < 0.05, compared with Th1 or Th2 cells.
See also Figure S5.
Next, we comprehensively analyzed the T cell receptor (TCR) signaling in Th9 and other Th cell subsets. Upon activation, we noted that all these Th cells displayed similar TCR-proximal signaling events, including phosphorylation of the protein tyrosine kinases Lck, LCγ1, Src, and Zap70, and their downstream signaling events, such as the MAP kinase Erk, the kinase Akt, and nuclear translocation of NFAT (Figure S5D).
Because nuclear factor κB (NF-κB) signaling is pivotal in controlling T cell proliferation (Paul and Schaefer, 2013), we systematically analyzed NF-κB signaling activation in Th cells. First, we detected no decrease in cytosolic proteins that are involved in negative regulation of TCR-to-NF-κB signaling (such as A20 and CYLD) in Th9 cells (Figure S5D). However, a striking difference is the hyperactivation of NF-κB, detected by nuclear translocation of p50, RelA, RelB, p52, and c-Rel in Th9, but not in Th1, Th2, or Th17 cells. This occurred at late time points after cell stimulation (24–72 hr), whereas during the early phase (<12 hr), NF-κB activation was similar across all T cell subsets assessed (Figure 5E). These results highlight that Th9 cells possess a unique hyperproliferative feature, possibly mediated by hyperactivation of late-phase NF-κB. We also found a similar re-hyperactivation of NF-κB signaling only in Th9 cells when they were restimulated with anti-CD3 plus anti-CD28 monoclonal antibodies on day 5 after the first-round activation (Figure 5F).
Moreover, we further confirmed the increased proliferative capacity of Th9 cells by CFSE-dilution assay and by calculating the cell yields after the first-round activation and the subsequent reactivation (Figures 5G and 5H). Importantly, inhibition of NF-κB signaling by a specific inhibitor (QNZ) did not induce Th9 cell apoptosis (Figure S5E), but significantly arrested Th9 hyperproliferative activity (Figures 5G and 5H) with minimal effect on proliferation of Th17 (Figure S5F). Taken together, these data indicate that the hyperactivation of late-phase NF-κB drives the hyperproliferative feature in Th9 cells, a unique feature that has not been described in any other known Th cells.
Increased Traf6 Production Drives Hyperactivation of NF-κB Signaling to Promote Th9 Hyperproliferation
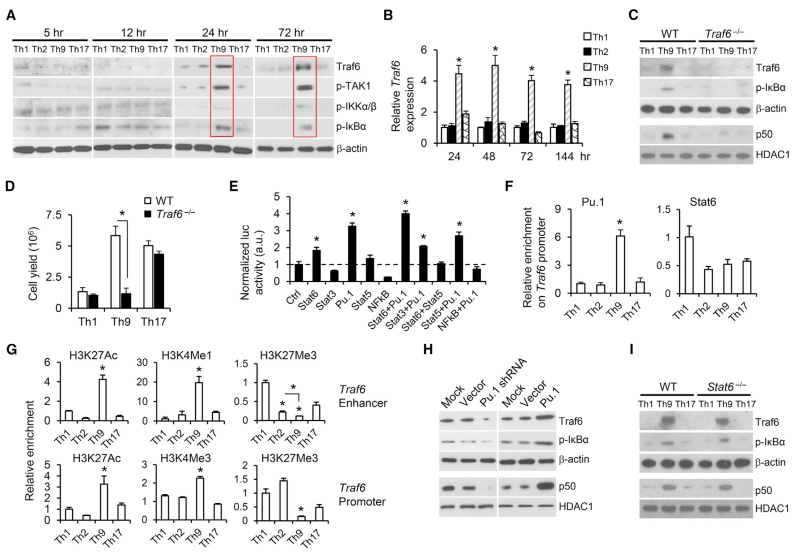
To understand the mechanism that drives the late-phase NF-κB hyperactivation in Th9 cells, we systematically analyzed the NF-κB signaling activation in these cells. Although no significant changes occurred in regard to the TCR-proximal signaling events and cytosolic proteins that are involved in negative regulation of TCR-to-NF-κB signaling, NF-κB upstream signaling protein (Traf6, pTAK1, pIKKα/β, and pIκBα) levels substantially increased in Th9 cells (Figures 6A and S6A). This intriguing difference highlights that Traf6 might be responsible for the hyperactivation of late-phase NF-κB because both Traf6 protein and mRNA were upregulated significantly (Figures 6A and 6B), and the recruitment of Traf proteins is a key step in the activation of NF-κB in T cells (Hildebrand et al., 2011). To test the importance of Traf6 in Th9 cells, we generated Traf6−/− Th9 cells from Traf6flox/floxCD4cre mice. Traf6 deficiency completely abolished hyperactivation of NF-κB signaling and the hyperproliferative potential of Th9 cells compared with WTTh9 cells (Figures 6C and 6D), further demonstrating that Traf6 is critical in regulating the Th9 hyperproliferative feature.
Figure 6. Traf6 Is Required for the Late-Phase NF-κB Hyperactivation in Th9 Cells.
(A) Naive CD4+ T cells were differentiated with plate-bound αCD3 mAbs and soluble αCD28 mAbs, and the indicated proteins were analyzed by western blot (cytoplasmic fraction). Red solid lines indicate upregulated NF-κB upstream signaling in Th9 cells.
(B) RT-PCR determination of relative Traf6 mRNA expression in OT-II-Th cells polarized in vitro.
(C) WT and Traf6−/− OT-II-Th1, Th9, and Th17 cells were differentiated for 72 hr, and the indicated proteins were analyzed by western blot (cytoplasmic fraction: Traf6, p-IκBα, and β-actin; nuclear fraction: p50 and HADC1).
(D) Cell yields of WT and Traf6−/− OT-II-Th1, Th9, and Th17 cells after the first activation round (day 5, n = 3 mice).
(E) Luciferase reporter assay for the activation of Traf6 promoter. *p < 0.05, compared with control.
(F) Chromatin immunoprecipitation (ChIP) assay of Pu.1 and Stat6 binding to the Traf6 promoter regions in OT-II-Th cells after the first activation round (24 hr, n = 3 mice).
(G) ChIP assay for H3K27Ac, H3k4Me1, H3K4Me3, and H3K27Me3 modification of Traf6 loci (enhancer or promoter) in OT-II-Th cells after the first activation round (24 hr, n = 3 mice).
(H) OT-II-Th9 cells (24 hr after the first-round activation) were treated with mock, control-vector, Pu.1-shRNA, or Pu.1-expression vector transfection. The indicated molecules were analyzed by western blot 48 hr after treatment (cytoplasmic fraction: Traf6, p-IκBα, and β-actin; nuclear fraction: p50 and HADC1).
(I) WT and Stat6−/− OT-II-Th1, Th9, and Th17 cells were differentiated for 72 hr. The indicated molecules were analyzed by western blot 48 hr after treatments (cytoplasmic fraction: Traf6, p-IκBα, and β-actin; nuclear fraction: p50 and HADC1). Representative results from one of two repeated experiments are shown.
Data are mean ± SD; *p < 0.05, compared with Th1, Th2, or Th17 cells.
These results also prompted us to determine what regulates Traf6 upregulation in Th9 cells. By analyzing the promoter region of Traf6, we predicted several binding sites for transcriptional factors, such as Stat3, Stat5, Stat6, Pu.1 (Spi1), and NF-κB, that might have been activated in Th9 cells (Table S1). To determine whether these molecules could activate the Traf6 promoter, we performed luciferase reporter assays. Stat3, Stat5, and NF-κB did not activate the Traf6 promoter, whereas Pu.1, and to a lesser extent, Stat6, did (Figure 6E). Considering that Pu.1 and Stat6 are two crucial molecules that are involved in Th9 cell development (Chang et al., 2010; Goswami et al., 2012), we performed a chromatin immunoprecipitation (ChIP) assay, and, as shown in Figure 6F, only Pu.1 bound the Traf6 promoter region in Th9 cells. To gain further insight, we determined epigenetic changes at the Traf6 locus and observed striking differences in the acetylation and methylation status (Figure 6G), suggesting active Traf6 enhancer and promoter regions in Th9 cells. Specifically, the “permissive” histone marks (H3k4Me1 on enhancer, H3k4Me3 on promoter, and H3K27Ac on both enhancer and promoter) (Shlyueva et al., 2014) were highly increased on the Th9 Traf6 locus. Conversely, Th9 cells had the least H3K27 trimethylation (H3K27Me3) on both the enhancer and promoter of Traf6, which is a “non-permissive” histone mark associated with repressed genes (Shlyueva et al., 2014) (Figure 6G). These chromatin modifications might affect Traf6 locus accessibility to transcription factors, such as Pu.1, in Th9 cells.
To obtain direct evidence that Pu.1 contributes to Traf6 production in Th9 cells, we compared Traf6 expression levels in WT Th9, Ctrl-shRNA-transduced Th9, Pu.1-shRNA-transduced Th9, GFP-retrovector-transduced Th9, Pu.1-retrovector-tranduced Th9, Il9r−/− Th9, and Stat6−/− Th9 cells. Results clearly showed that Traf6 levels and NF-κB signaling were significantly upregulated in Th9 cells overexpressing Pu.1 and downregulated in Pu.1 knockdown Th9 cells (Figure 6H), whereas in Stat6−/− Th9 and Il9r−/− Th9 cells, Traf6 level was similar to that in WT Th9 cells (Figures 6I, S6B, and S6C). These data pinpoint the importance of Pu.1 in transcription of Traf6 in Th9 cells, whereas IL-9 signaling seems not to be required for Th9 cell Traf6 expression, NF-κB signaling, or hyperproliferation.
Eomes and Traf6 Dictate the Antitumor Efficacy of Th9 Cells
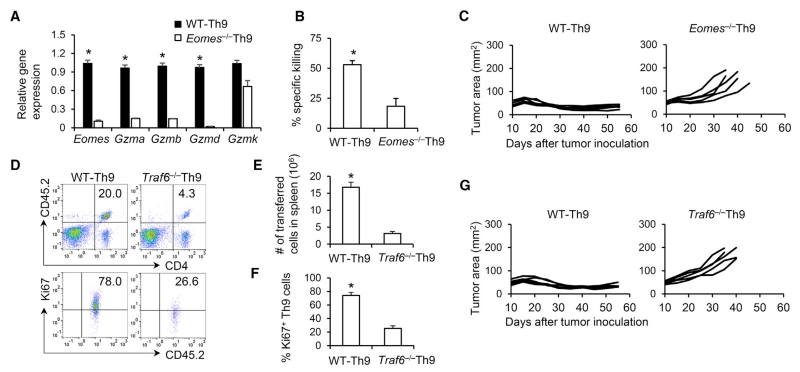
To ascertain the contributions of the effector and hyperproliferative properties of Th9 cells in the Th9-mediated eradication of large established tumors, we examined key factors involved in determining the feature and function of Th9 cells. First, we further dissected the role of Eomes in Th9 cells for their cytotoxic effector development. As Eomes is the effector master transcriptional factor (Pearce et al., 2003), Eomes deficiency abolished granzyme expression by Th9 cells (Figure 7A) and subsequently extinguished their cytolytic activity in a direct in vitro killing assay (Figure 7B), but did not significantly change the Traf6 gene expression and proliferative capacity of Th9 cells (Figures S7A-S7C). Accordingly, Eomes−/− Th9 cell transfer failed to mediate sustained antitumor responses compared with WT Th9 cells (Figure 7C). In addition, we investigated whether Traf6 was essential for the observed superior antitumor performance of Th9 cells. We observed significantly lower frequencies (Figures 7D and 7E) and decreased proliferation (Figures 7D, 7F, and S7D–S7F) of Traf6−/− Th9 cells after transfer, and this insufficient persistence of Traf6−/− Th9 cells also nullified their antitumor ability without altering cytolytic function of Th9 cells (Figures 7G, S7G, and S7H). This effect appeared to apply only to Th9 cells because antitumor function was similar between WT Th17 and Traf6−/− Th17 cells in vivo (Figure S7I). Collectively, our data provide functional confirmation of the observed molecular program of effector development and hyperproliferation displayed by tumor-specific Th9 cells.
Figure 7. Traf6 and Eomes Dictate the Antitumor Function of Th9 Cells.
(A) WT and Eomes−/− OT-II-Th9 cells were differentiated for 5 days and the relative gene expression was determined by RT-PCR (n = 3 mice).
(B) Specific killing assay of WT and Eomes−/− OT-II-Th9 cells before transfer (5-day culture, n = 3 mice) was performed against B16-OVA cells.
(C) WT and Eomes−/− OT-II-Th9 cells (CD45.2+, 2.5 × 106) were transferred i.v. into CD45.1+ B6 mice bearing B16-OVA tumors (treated similarly to Figure 1A). Tumor responses are shown (n = 5 mice/group).
(D–G) WT and Traf6−/− OT-II-Th9 cells were differentiated for 5 days (CD45.2+, 2.5 × 106) and transferred i.v. into CD45.1+ B6 mice bearing B16-OVA tumors (treated similarly to Figure 1A). (D) Representative FACS analysis for the presence of transferred Th9 cells and percentage of Ki67+ Th9 cells in spleens of mice 12 days after transfer. (E) Total number of splenic CD45.2+CD4+ Th9 cells was calculated from (D). (F) Percentage of Ki67+ cells summarized from (D). (G) Tumor responses are shown (n = 5 mice/group). Representative results from one of two repeated experiments are shown. Data are mean ± SD; *p < 0.05.
See also Figure S7.
DISCUSSION
To date, although extensive studies have focused on the transcriptional network controlling Th9 differentiation and IL-9 production (Kaplan et al., 2015), the T cell “identity” of Th9 cells beyond IL-9 secretion remains unclear, especially in the context of ACT. Our previous study (Lu et al., 2012) revealed a distinct role for Th9 cells in provoking CD8+ CTL-mediated antitumor immunity in an IL-9-dependent manner, which has been confirmed by multiple studies (Kim et al., 2015; Vegran et al., 2014; Zhao et al., 2016b). However, all these studies used early-stage tumor models, which may not represent clinically relevant scenario for ACT. It remains unclear whether Th9 cells can serve as direct effector T cells that, in combination with the lymphodepleting chemotherapy and active vaccination regimens frequently included in clinical ACT protocols, will completely eliminate the large established tumors often seen in clinic. Although we still observed that Th9 cells were more “helpful” in eliciting host CD8+ CTL responses than Th1 or Th17 cells, endogenous CD8+ CTL responses and IL-9 production by Th9 cells are required only for optimal Th9-mediated complete elimination of large tumors. Conversely, modulating Th9 cell cytotoxicity by knockout of Eomes or persistence capacity by knockout of Traf6 profoundly impaired the antitumor function of Th9 cells. Thus, the present study has revealed cellular and molecular mechanisms by which tumor-specific Th9 cells promote tumor regression in a highly realistic and clinically relevant ACT scenario.
Two paradigms have emerged in determining the functionality of T cells for ACT, based on the fact that antitumor efficacy inversely correlates with advanced maturational state through limitation of the capacity to self-renew and survive in vivo. The Th1 paradigm focuses on the terminal effectors prone to apoptosis, whereas the Th17 paradigm focuses on less-differentiated subsets capable of superior persistence and functionality in vivo. Taking into consideration the global gene expression profile, we used GSEA to determine the phenotypic features of Th9 cells by comparing their characteristics with those of these two existing T cell paradigms. Regarding the effector maturational status, GSEA suggested that a core molecular signature might be shared between Th9 and Th1 cells. In agreement with this result, Th9 cells generated in vitro and sorted from tumor-bearing mice always had the highest tumor-specific killing activity. Intriguingly, although Th9 cells are mature effector T cells, their phenotype is distinct from that of the classic Th1 effectors. First, GSEA suggested that, unlike Th1 cells, Th9 cells are not enriched in the molecular exhaustion signature. Second, unlike Th1 cells, Th9 cells do not display the features of terminally differentiated late effectors. Third, and most strikingly, Th9 cells had extraordinary persistence, whereas Th1 cells are short-lived T cells. These observed differences might be explained by Eomes upregulation, rather than T-bet, which drives the effector development of Th9 cells. On the contrary, high expression of T-bet, a master transcriptional factor for Th1 cells, has been closely associated with terminal differentiation and drives the short-lived T cell development (Joshi et al., 2007), which seriously hampers the antitumor potential of Th1 cells. In addition, we observed that a significantly increased granzyme production was responsible for Th9-mediated killing. However, Th9 cells did not possess significantly upregulated perforin expression compared with other Th cell subsets; it may be that Th9 cells have already produced enough perforin, or that Th9 cell-activated CD8+ T cells may produce perforin as an additional source (Osinska et al., 2014). Thus, these results suggest that Th9 and Th1 cells are two transcriptionally and phenotypically distinct effector populations, and that Th9 cells cannot be classified into the Th1 paradigm.
Prolonged persistence has been previously attributed only to less-differentiated early memory T cells, and T cell subsets possessing great persistence is crucial for successful ACT. This is the key reason why long-lived Th17 cells outperform the terminally differentiated short-lived Th1 cells for ACT (Muranski et al., 2011). Although Th9 cells persist equally well to early memory Th17 cells, GSEA showed that Th17 cells, but not Th9 or Th1 cells, retained the molecular signature of early memory T cells. Furthermore, Th9 cells did not acquired stemness, nor did they display enhanced resistance to apoptosis in vivo. This counterintuitive finding can be explained by the observed hyperproliferation of Th9 cells in vivo. Molecular mechanistic studies uncovered that Pu.1, an important transcription factor for Th9 cell development, bound and transcribed Traf6 in Th9 cells. This accumulated Traf6 may serve as a critical adaptor molecule that links to the MALT1-CARMA1-Bcl-10 complex downstream of TCR, and may function directly or indirectly by forming a complex with Ubc13/ Uev1a as a ubiquitin ligase in order to attach ubiquitin chains to target proteins, including itself and IKKg, which enable the formation of complexes by recruiting TAB2/3-TAK1 and then continuously activate the NF-κB signaling pathway (Walsh et al., 2015). This Pu.1-Traf6-NF-κB pathway pinpoints an alternative mechanism that drives the extraordinary persistence of Th9 cells, which differs from the antiapoptotic strategy seen in Th17 cells (Muranski et al., 2011).
Although it is not clear whether the hyperproliferation-mediated persistence of T cells possesses any advantage over the antiapoptotic Th17 behavior, a potential explanation of the Th9 superior functionality compared with Th17 cells is that the less-differentiated Th17 cells may not have developed into fully mature effector T cells, whereas newly polarized and transferred Th9 cells all have highly cytolytic activity. The plasticity of Th17 cells, on the one hand, allows a portion of transferred Th17 cells to convert into Th1-like effector cells. On the other hand, Th17-to-Treg conversion has been suggested (Obermajer et al., 2014), and a portion of Th17 cells also converted into Foxp3+ Treg-like cells upon transfer in our experimental conditions, possibly due to the upregulated expression of Bach2, which promotes efficient formation and stabilization of Treg cells but restrains effector cell development (Roychoudhuri et al., 2013). These results suggest that the potential for “stem cell-like” CD4+ T cells to convert into Treg cells in vivo might negatively affect their antitumor efficacy.
Our work thus far has revealed that tumor-specific Th9 cells have a less-exhausted and long-lived effector profile, which represents a unique Th9 paradigm for ACT. This Th9 paradigm may challenge our current understanding of T cell selection criteria for ACT: (1) pre-acquisition of a maturational effector state in vitro may not limit antitumor functionality in vivo; (2) capacity for long-term persistence may not be associated with stem cell-like or early memory properties; and (3) IFN-γ and TNF-a production may not be required from the transferred T cells. In the Th9 paradigm, T cells possess a unique phenotype that is a combination of Th1 cytolytic and Th17 stem cell-like persistence characteristics, and thus they have significant implications for the design of future ACT therapies.
STAR★METHODS
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC, e450, PE anti-mouse CD45.1 | BioLegend | 110714, 110722, 110708 |
| APC, e450, PE anti-mouse CD45.2 | BioLegend | 109814, 109820, 109808 |
| BV570, APC, e450 anti-mouse CD4 | BioLegend | 100541, 100412, 100428 |
| E450 anti-mouse CD44 | BioLegend | 103002 |
| PE anti-mouse CD62L | BioLegend | 104408 |
| PE anti-mouse IL-2Rβ | BioLegend | 123210 |
| PE anti-mouse CCR7 | BioLegend | 120106 |
| PE anti-mouse IL-7Rα | BioLegend | 135010 |
| PE anti-mouse Ki67 | BioLegend | 652404 |
| APC anti-mouse KLRG1 | BioLegend | 138412 |
| PE anti-mouse PD-1 | BioLegend | 135206 |
| PE anti-mouse LAG-3 | BioLegend | 125208 |
| FITC anti-mouse CD244 | BioLegend | 133504 |
| PE anti-mouse IL-9 | BioLegend | 514104 |
| APC anti-mouse IFN-γ | BioLegend | 505810 |
| PE anti-mouse IL-17A | BioLegend | 506904 |
| PE anti-mouse IL-2 | BioLegend | 503808 |
| APC anti-mouse Granzyme B | BioLegend | 372204 |
| PE anti-mouse Eomes | eBioscience | 12-4875-82 |
| Fc BLOCK | BioLegend | 101320 |
| Fixable Viability Dye eFlour 450 | eBioscience | 65-0863-14 |
| APC, FITC Annexin V | BioLegend | 640941, 640945 |
| PE anti-mouse CD8a | BioLegend | 100708 |
| PE anti-mouse Foxp3 | BioLegend | 320008 |
| anti-mouse IL-4 | BioXCell | 11B11 |
| anti-mouse IL-2 | BioXCell | JES6-1A12 |
| anti-mouse IFNg | BioXCell | XMG1.2 |
| anti-mouse FasL | BioLegend | 106805 |
| Kb tetramer carrying the OVA257-264 | Beckman Coulter | ts-m542-1 |
| anti-mouse p-CD3-ζ | Santa Cruz Biotechnology | sc-9975 |
| anti-mouse TRAF6 | Santa Cruz Biotechnology | sc-7221 |
| anti-mouse IκB-β | Santa Cruz Biotechnology | sc-945 |
| anti-mouse p-IKKα/β (S180/181) | Santa Cruz Biotechnology | sc-23470-R |
| anti-mouse Stat6 | Santa Cruz Biotechnology | sc-981 |
| anti-mouse IKKγ | Santa Cruz Biotechnology | sc-8256 |
| anti-mouse Src | Cell Signaling | No. 2108 |
| anti-mouse RelA | Cell Signaling | No. 4764 |
| anti-mouse RelB | Cell Signaling | No.4954 |
| anti-mouse p52 | Cell Signaling | No. 4882 |
| anti-mouse NFAT1 | Cell Signaling | No. 5862 |
| anti-mouse NFAT2 | Cell Signaling | No. 8032 |
| anti-mouse HDAC1 | Cell Signaling | No. 2062 |
| anti-mouse CYLD | Cell Signaling | No. 8462 |
| anti-mouse p-TAK1 | Cell Signaling | No. 9339 |
| anti-mouse Zap70 | Cell Signaling | No. 3165 |
| anti-mouse p-Zap70 (Y319/Y352) | Cell Signaling | No. 2717 |
| anti-mouse Lck | Cell Signaling | No. 2752 |
| anti-mouse p-Lck (Y505) | Cell Signaling | No. 2751 |
| anti-mouse CD3-ζ | Cell Signaling | No. 4443 |
| anti-mouse PLCγ | Cell Signaling | No. 2822 |
| anti-mouse and β-actin | Cell Signaling | No. 4970 |
| anti-mouse p-PLCγ (Y783) | Cell Signaling | No. 14008 |
| anti-mouse p-Src (Y416) | Cell Signaling | No. 6943 |
| anti-mouse p-LAT (Y191) | Cell Signaling | No. 3584 |
| anti-mouse p-IκB-α (S32) | Cell Signaling | No. 2859 |
| anti-mouse p50 | eBioscience | 14-6732-81 |
| anti-mouse c-Rel | eBioscience | 14-6111-82 |
| Critical Commercial Assays | ||
| Foxp3 staining kit | BioLegend | 136803 |
| Annexin V-FITC Apoptosis Detection Kit | eBioscience | BMS500FI-100 |
| NE-PER Nuclear and Cytoplasmic Extraction kit | ThermoScientific | 78833 |
| ELISA kits mouse GM-CSF | eBioscience | 50-173-42 |
| ELISA kits mouse granzym B | eBioscience | 50-174-75 |
| ELISA kits mouse IL-9 | eBioscience | 50-112-5217 |
| ELISA kits mouse IL-10 | eBioscience | 50-112-8654 |
| ELISA kits mouse IL-6 | eBioscience | 50-112-8808 |
| ELISA kits mouse IL-21 | eBioscience | 50-174-80 |
| ELISA kits mouse TNF-α | eBioscience | 50-112-8899 |
| ELISA kits mouse IFN-γ | eBioscience | 50-112-9023 |
| ELISA kits mouse granzyme A | MyBioSource | MBS704766 |
| ChIP assay kit | Millipore | 17-295 |
| Secrete-Pair Dual Luminescence Assay Kit | GeneCopoeia | LF032 |
| Recombinant DNA | ||
| MSCV-PIG-Pu.1 | Addgene | 66982 |
| MSCV-PIG | Addgene | 18751 |
| pLKO.1-GFP-Pu.1 shRNA | Sigma | N/A |
| pLKO.1-GFP | Addgene | 30323 |
| Negative control clone | Genecopoeia | NEG-PG04 |
| pcDNA3.1 | Qing Yi | N/A |
| pcDNA 3.1_Stat6 | Qing Yi | N/A |
| pcDNA3.1_Stat5 | Qing Yi | N/A |
| pcDNA3.1_Stat3 | Qing Yi | N/A |
| pcDNA3.1_Pu.1 | Qing Yi | N/A |
| pcDNA3.1_p50 | Qing Yi | N/A |
| pcDNA3.1_p52 | Qing Yi | N/A |
| pcDNA3.1_RelA | Qing Yi | N/A |
| pcDNA3.1_RelB | Qing Yi | N/A |
| pcDNA3.1_c-Rel | Qing Yi | N/A |
| Recombinant Proteins | ||
| Mouse GM-CSF | R&D Systems | Q14AD9 |
| Mouse TNF-α | R&D Systems | P06804 |
| Mouse IL-1β | R&D Systems | NP_032387 |
| Mouse IL-4 | R&D Systems | P07750 |
| Mouse IL-6 | R&D Systems | P08505 |
| Mouse IL-2 | R&D Systems | P04351 |
| Mouse IL-12 | R&D Systems | P43432 |
| Mouse IL-23 | R&D Systems | P43432 |
| Mouse IL-21 | R&D Systems | Q9ES17.1 |
| Human TGF-β1 | R&D Systems | P01137 |
| Oligonucleotides | ||
| mTbx21 F: 5′-CAACAACCCCTTTGCCAAAG-3′ | Sigma | N/A |
| mTbx21 R: 5′-TCCCCCAAGCAGTTGACAGT-3′ | Sigma | N/A |
| mEomes F: 5′-TTCCGGGACAACTACGATTCA-3′ | Sigma | N/A |
| mEomes R: 5′-ACGCCGTACCGACCTCC-3′ | Sigma | N/A |
| mGzmA F: 5′-CCTGAAGGAGGCTGTGAAAG-3′ | Sigma | N/A |
| mGzmA R: 5′-GTTACAGTGGGCAGCAGTCA-3′ | Sigma | N/A |
| mGrzB F: 5′-AGGGGGTACAAGGTCACAGA-3′ | Sigma | N/A |
| mGrzB R: 5′-CAAGAGTGTTGTCCTTGCTCTCT-3′ | Sigma | N/A |
| mGzmD F: 5′-TAACGAATGCCATGTAGGGG-3′ | Sigma | N/A |
| mGzmD R: 5′-TGACCCTACTTCTGCCTCTCA-3′ | Sigma | N/A |
| mGzmK F: 5′-CCGTGGTTTTAGGAGCACAT-3′ | Sigma | N/A |
| mGzmKR: 5′-TTTTTGGATCCCAGGTGAAG-3′ | Sigma | N/A |
| mPrdm1 F: 5′-GACAGAGGCCGAGTTTGAAG-3′ | Sigma | N/A |
| mPrdm1 R: 5′-GGCATTCTTGGGAACTGTGT-3′ | Sigma | N/A |
| mKlrg1 F: 5-CCTCTGGACGAGGAATGGTA-3′ | Sigma | N/A |
| mKlrg1 R: 5-ACCTCCAGCCATCAATGTTC-3′ | Sigma | N/A |
| mKlrd1 F: 5′-CTATGGGAGGATGGCACAGT-3′ | Sigma | N/A |
| mKlrd1 R: 5′-CCGTGGACCTTCCTTGTCTA-3′ | Sigma | N/A |
| mKlra10 F: 5′-CCATAACTGCAGCAACATGC-3′ | Sigma | N/A |
| mKlra10 R: 5′-ATTTAACACCTCCGCCTGTG-3′ | Sigma | N/A |
| mKlrk1 F: 5′-CACCTTGATTTCCTCCCAGA-3′ | Sigma | N/A |
| mKlrk1 R: 5′-GGAAGTGAGGCAAGAACTG-3′ | Sigma | N/A |
| mPrf1 F: 5′-AATATCAATAACGACTGGCGTGT-3′ | Sigma | N/A |
| mPrf1 R: 5′-CATGTTTGCCTCTGGCCTA-3′ | Sigma | N/A |
| mFasl F: 5′-CATCACAACCACTCCCACTG-3′ | Sigma | N/A |
| mFasl R: 5′-TACTGGGGTTGGCTATTTGC-3′ | Sigma | N/A |
| mLag3 F: 5′-GCCATCTCGTTCTCGTTCTC-3′ | Sigma | N/A |
| mLag3 R: 5′-GTCTCCAGTTCTCGCTCCAG-3′ | Sigma | N/A |
| mPdcd1 F: 5′-GGAGCAGAGCTCGTGGTAAC-3′ | Sigma | N/A |
| mPdcd1 R: 5′-GCTCCTCCTTCAGAGTGTCG-3′ | Sigma | N/A |
| mZeb2 F: 5′-CCACCAGCCCTTTAGGTGTA-3′ mZeb2 R: 5′-CCCTTGTTCTTCTGGCTGAG-3′ |
Sigma | N/A |
| mSell F: 5′-ACCCACTCTCTTGGAGCTGA-3′ | Sigma | N/A |
| mSell R: 5′-GTTGGGCAAGTTAAGGAGCA-3′ | Sigma | N/A |
| mCcr7 F: 5′-AGTCTTCCAGCTGCCCTACA-3′ | Sigma | N/A |
| mCcr7 R: 5′-CAGCCCAAGTCCTTGAAGAG-3′ | Sigma | N/A |
| mVax2 F: 5′-TTGGTTGACCCCAGAAACTC-3′ | Sigma | N/A |
| mVax2 R: 5′-CAAGTGTCACACAGGGATGG-3′ | Sigma | N/A |
| mDapl1 F: 5′-CGAAAAAGACAGGCTTGGAG-3′ | Sigma | N/A |
| mDapl1 R: 5′-TGGCTGTGTTTTCTGTCCTG-3′ | Sigma | N/A |
| mSox2 F: 5′-CACAACTCGGAGATCAGCAA-3′ | Sigma | N/A |
| mSox2 R: 5′-CTCCGGGAAGCGTGTACTTA-3′ | Sigma | N/A |
| mNanog F: 5′-AAGCAGAAGATGCGGACTGT-3′ | Sigma | N/A |
| mNanog R: 5′-GTGCTGAGCCCTTCTGAATC-3′ | Sigma | N/A |
| mTcf7 F: 5′-GCCAGAAGCAAGGAGTTCAC-3′ | Sigma | N/A |
| mTcf7 R: 5′-TACACCAGATCCCAGCATCA-3′ | Sigma | N/A |
| mLef1 F: 5′-TCACTGTCAGGCGACACTTC-3′ | Sigma | N/A |
| mLef1 R: 5′-TGAGGCTTCACGTGCATTAG-3′ | Sigma | N/A |
| mTraf6 F: 5′-GATCGGGTTGTGTGTGTCTG-3′ | Sigma | N/A |
| mTraf6 R: 5′-AGACACCCCAGCAGCTAAGA-3′ | Sigma | N/A |
| mIL17a F: 5′-TCCAGAAGGCCCTCAGACTA-3′ | Sigma | N/A |
| mIL17a R: 5′-AGCATCTTCTCGACCCTGAA-3′ | Sigma | N/A |
| mGAPDH F: 5′-AGCTTGTCATCAACGGGAAG-3′ | Sigma | N/A |
| mGAPDH R: 5′-TTTGATGTTAGTGGGGTCTCG-3′ | Sigma | N/A |
| Pu.1 binding site at the Traf6 promoter region 1: 5′-CTCTCCCGTGACAATGTTGGA-3′ |
Sigma | N/A |
| Pu.1 binding site at the Traf6 promoter region 2: 5′-CTCCACGCTGAAGCCTTACC-3′ |
Sigma | N/A |
| Pu.1 binding site at the Traf6 promoter region 3: 5′-TGTTGGAGAATGGGATCATGC-3′ |
Sigma | N/A |
| Pu.1 binding site at the Traf6 promoter region 4: 5′-CTCGCTAGGAGCAGCAAGG-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 promoter 1: 5′-GGAGGGGACAGCTATACGCA-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 promoter 2: 5′-TGTGTGCTCATCACGCAGTT-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 promoter 3: 5′-AGCTCTCCCGTGACAATGTT-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 promoter 4: 5′-TTCCTCGGACCAGTGCAAAA-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 promoter 5: 5′-TCTACTTACCTTACCTAACAGCCT-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 promoter 6: 5′-GCACAATGCAATAGATGCCCA-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 enhancer 1: 5′- AAGGGACTCACCAAGAACCT-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 enhancer 2: 5′- GCTCCAAATACAAGAGCAGCC-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 enhancer 3: 5′- TACTGACTGCTGTGTTAGCTGGAA-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 enhancer 4: 5′- GCAGAGATGCACTGTTCCCT-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 enhancer 5: 5′- TGGACAGGGGCACTAAGACT-3′ |
Sigma | N/A |
| chromatin modification status of mouse Traf6 enhancer 6: 5′- GAGCTCTGGGCTGTCTCTTC-3′ |
Sigma | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 | Jackson Laboratories | 000664 |
| Cd8a−/− (B6.129S2-Cd8atm1Mak/J) | Jackson Laboratories | 002665 |
| Stat6−/− (B6.129S2(C)-Stat6tm1Gru/J) | Jackson Laboratories | 005977 |
| Cd4-Cre (Tg(Cd4-cre)1Cwi/BfluJ) | Jackson Laboratories | 017336 |
| OT-II (B6.Cg-Tg(TcraTcrb)425Cbn/J) | Jackson Laboratories | 004194 |
| TRP-1 (B6.Cg-Rag1tm1Mom Tyrp1B-wTg (Tcra,Tcrb)9Rest/J) mice |
Jackson Laboratories | 008684 |
| CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ), | Jackson Laboratories | 002014 |
| Eomesfl/fl (B6.129S1(Cg)-Eomestm1.1Bflu/J) | Jackson Laboratories | 017293 |
| Ifng−/− (B6.129S7-Ifngtm1Ts/J), | Jackson Laboratories | 002287 |
| CD45.1 OT-II mice | Qing Yi | N/A |
| Eomesfl/flCd4-Cre OT-II mice | Qing Yi | N/A |
| Traf6fl/flCd4-Cre OT-II mice | Qing Yi | N/A |
| Il9r−/− CD45.1 OT-II mice | Qing Yi | N/A |
| Il9−/− CD45.1 OT-II mice | Qing Yi | N/A |
| Ifng−/− CD45.1 OT-II mice | Qing Yi | N/A |
| Other | ||
| The NFκB-specific inhibitor QNZ | Santa Cruz Biotechnology | sc-200675 |
| Granzyme B specific inhibitor Z-AAD-CMK | Enzo Life Sciences | BML-P165-0001 |
| 3,4 Dichloroisocoumarin (DCI), inhibits granzymes A, B, and H |
Santa Cruz Biotechnology. | sc-3502 |
| MHC class II–restricted TRP1 (SGHNCGTCRPGWRGAACNQKILTVR) |
GenScript | N/A |
| MHC class II–restricted OT-II (ISQAVHAAHAEINEAGR) | GenScript | N/A |
| Deposited Data and software | ||
| Th1, Th9, Th17 cell gene array | NCBI Gene Expression Omnibus | GSE97087 |
| Mature T cell effector gene signature set | NCBI Gene Expression Omnibus | GSE21360 |
| T cell exhaustion signature set | NCBI Gene Expression Omnibus | GSE24081 |
| Early memory T cell gene signature set | NCBI Gene Expression Omnibus | GSE21360 |
| GSEA v2.2.2 | Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Qing Yi (yiq@ccf.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6, Cd8a−/− (B6.129S2-Cd8atm1Mak/J), Ifng−/− (B6.129S7-Ifngtm1Ts/J), Eomesfl/fl (B6.129S1(Cg)-Eomestm1.1Bflu/J), Stat6−/− (B6.129S2(C)-Stat6tm1Gru/J), Cd4-Cre (B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ), OT-II (C57BL/6-Tg(TcraTcrb)425Cbn/J), CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ), and TRP-1 (B6.Cg-Rag1tm1Mom Tyrp1B-wTg(Tcra,Tcrb)9Rest/J) mice were purchased from Jackson Laboratory. Traf6fl/fl and Il9r−/−mice on the B6 background were generated as described previously(King et al., 2006; Steenwinckel et al., 2007). Il9−/− mice on the B6 background were provided by Dr. Dong Chen from Tsinghua University. CD45.1-OT-II, Ifng−/−-CD45.1-OT-II, Il9−/−-CD45.1-OT-II, Eomesfl/fl-Cd4-Cre-OT-II, and Traf6fl/fl-Cd4-Cre-OT-II mice were generated by crossing and backcrossing the existing mice above. Male and female 6- to 8 -week-old mice were used for each animal experiment. The studies were approved by the Institutional Animal Care and Use Committee of the Cleveland Clinic Foundation and Wake Forest School of Medicine.
Cell Lines
WT B16 and B16 melanoma cell lines (ATCC) were transfected with OVA (B16-OVA) and cultured in Iscove’s Modified Dulbecco’s Medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Thermo Scientific), 100 U/ml penicillin-streptomycin, and 2 mM L-glutamine (both from Invitrogen).
METHOD DETAILS
In Vitro Th Cell Differentiation
Naïve CD4+CD62L+ T cells were purified from spleens of OT-II or TRP-1 mice and differentiated into Th1, Th9, or Th17 cells according to established methods(Ghoreschi et al., 2010; Lu et al., 2012; Muranski et al., 2011). OVA- or TRP-1-specific naïve CD4+ T cells were cultured for 3 days with irradiated splenic APCs from C57BL/6 mice in the presence of OVA323-339 peptide or TRP-1106-133 (5 μg/ml) with:
-
(a)
Th9-polarized medium supplemented with IL-4 (10 ng/ml), TGF-β1 (1 ng/ml), and anti-IFN-γ monoclonal antibodies (mAbs; 10 μg/ml);
-
(b)
Th1-polarized medium supplemented with IL-2 (30 ng/ml), IL-12 (4 ng/ml), and anti-IL-4 mAbs (10 μg/ml);
-
(c)
Th17-polarized medium supplemented with IL-6 (30 ng/ml), TGF-β1 (2.5 ng/ml), and anti-IFN-γ mAbs (10 μg/ml);
-
(d)
Th2-polarized medium supplemented with IL-4 (10 ng/ml) and anti-IFN-γ mAbs (10 μg/ml);
-
(e)
pTh17-polarized medium supplemented with IL-6 (30 ng/ml), IL-1β (20 ng/ml), IL-23 (50 ng/ml), and anti-IFN-γ mAbs (10 μg/ml);
-
(f)
Th17 (αIL-2+IL-23)-polarized medium supplemented with IL-6 (30 ng/ml), IL-1β (20 ng/ml), TGF-β1 (2.5 ng/ml), IL-21 (100 ng/ml), anti-IL-4 mAbs (10 μg/ml), anti-IL-2 mAbs (10 μg/ml) and anti-IFN-γ mAbs (10 μg/ml);
-
(g)
pTh17 (lowTGFβ)-polarized medium supplemented with IL-6 (30 ng/ml), IL-1β (20 ng/ml), IL-23 (50 ng/ml), TGF-β1 (0.25 ng/ml) and anti-IFN-γ mAbs (10 μg/ml).
After the initial 3-day culture, cells were provided with IL-2 (5 ng/ml), except Th17 (αIL-2+IL-23) cells which received IL-2 (5 ng/ml) plus IL-23 (50 ng/ml). After culture for a total of 5 days, differentiated Th cells were depleted of dead cells and used in animal studies. In some experiments, cells were restimulated for 5 hr with OVA-peptide in presence of a protein transport inhibitor (GolgiPlug, BD Biosciences) before ICS using a Cytofix/Cytoperm kit (BD Biosciences). In some experiments, naïve CD4+CD62L+ T cells may be activated as indicated in the polarized condition with plate-bound anti-CD3 mAbs (2 μg/ml, clone 17A2, eBioscience) and soluble anti-CD28 mAbs (1 μg/ml, clone 37.51, eBioscience).
Viral Production and Transduction
Viruses were packaged in 293T cells transfected with Lipofectamine 2000 (Life Science). Viral supernatant was harvested from day 1 to day 3, filtered with a 0.45-mm filter, concentrated with PEG-itVirus Precipitation Solution, and stored at −80°C until use. For the transfection, naïve CD4+CD62L+ T cells were activated in the polarized condition for 24 hr and then were mixed with the virus and 10 μg/ml protamine sulfate (Sigma), followed by centrifugation for 120 min at 1,800 rpm at 32°C. GFP+ T cells were sorted for some experiments.
Real-Time PCR
Total RNA was extracted from T cells using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Genes were expressed with specific primers and analyzed by using SYBR green real-time PCR (Applied Biosystems). Expression was normalized to the expression of the housekeeping gene Gapdh.
Tumor Models and Adoptive Transfer
Mice received subcutaneous (s.c.) abdominal injection with 1 × 106 B16 or B16-OVA tumor cells. At 10 days after tumor injection, mice (5/group) were treated with adoptive transfer of 2.5 × 106 Th1, Th9, or Th17 cells, followed by intravenous (i.v.) injection of 2.5 × 105 peptide-pulsed bone marrow-derived dendritic cells generated as previously described (Hong et al., 2012). Cyclophosphamide (CTX, Sigma) was administrated intraperitoneally (i.p.) as a single dose at 200 mg/kg 1 day before T-cell transfer. Mice were sacrificed at indicated days, and tumor-draining lymph nodes and splenocytes were analyzed. The number of transferred cells in spleens was calculated by multiplying the total number of viable splenocytes by the percentages of transferred populations. In some experiments, transferred T cells were sorted from splenocytes for indicated analyses.
Flow Cytometry and Western Blot Analysis
FITC-, PE, APC or PerCP-conjugated mAbs (1:100 dilution) were used for staining after Fc blocking, and analyzed using a FACS For-tessa flow cytometer or MACSQuant. Ki67 staining was performed using a Foxp3 staining kit with anti-Ki67 mAbs.
For Western blot, mAbs from Santa Cruz Biotechnology were used at a 1:500 dilution. mAbs from Cell Signaling and used at a 1:1000 dilution. For some experiments, we prepared cytoplasmic and nuclear extracts from cells using the NE-PER Nuclear and Cytoplasmic Extraction kit.
CFSE Labeling and Cytotoxicity Assay
In some experiments, Th cells were incubated for 5 minutes at 37°C with 1 mM CFSE in PBS, and then washed extensively. We measured proliferation of T cells by the relative CFSE dilution method after stimulation or transfer into tumor-bearing mice. In the cytotoxicity assay, B16-OVA target cells or B16 non-target cells for OT-II T cells were labeled with 5 mM CFSE. B16-OVA target cells or B16 non-target control cells were incubated alone in triplicate with the OT-II T cells at a 1:10 effector-to-target ratio. For TRP-1 T cells, B16 target cells or MC38 non-target control cells were used. After 18 hr, CFSE+ tumor cells from each target and control well were stained using FVD and analyzed by FACS. FVD+ tumor cells were considered as dead cells. The percent specific lysis was calculated as (FVD+ target - FVD+ control) × 100%.
Chromatin Immunoprecipitation
ChIP assay was performed with a ChIP assay kit (Millipore) according to the manufacturer’s instructions. Chromatin was extracted from OT-II-Th1, Th2, Th9, and Th17 cells differentiated for 3 days and fixed with formaldehyde. For the chromatin immunoprecipitation, anti-Pu.1 (sc-390659) and anti-Stat6 (sc-981X) were purchased from Santa Cruz Biotechnology and used at a 1:20 dilution and isotype-matched control antibodies were from Cell Signaling and used at a 1:20 dilution. As the predicted Stat6 binding site is adjacent to the Pu.1 binding site, the precipitated DNA was analyzed by RT-PCR with the following two primer sets surrounding the Pu.1 binding site at the Traf6 promoter region:
5′-CTCTCCCGTGACAATGTTGGA-3′ and 5′-CTCCACGCTGAAGCCTTACC-3′
5′-TGTTGGAGAATGGGATCATGC-3′ and 5′-CTCGCTAGGAGCAGCAAGG-3′
To evaluate chromatin modification status, tri-acetyl-histone H3 (K27), mono-methyl-histone H3 (K4), tri-methyl-histone H3 (K4), tri-methyl-histone H3 (K27) mAbs (all from Cell Signaling, 1:20 dilution) were used for the chromatin immunoprecipitation. The precipitated DNA was analyzed by RT-PCR with the following primer sets in the region of mouse Traf6 promoter:
5′-GGAGGGGACAGCTATACGCA-3′ and 5′-TGTGTGCTCATCACGCAGTT-3′
5′-AGCTCTCCCGTGACAATGTT-′3 and 5′-TTCCTCGGACCAGTGCAAAA-′3
5′-TCTACTTACCTTACCTAACAGCCT-′3 and 5′-GCACAATGCAATAGATGCCCA-3′;
the following primer sets in the region of mouse Traf6 enhancer(Zhao et al., 2016a):
5′-AAGGGACTCACCAAGAACCT-3′ and 5′- GCTCCAAATACAAGAGCAGCC-3′
5′- TACTGACTGCTGTGTTAGCTGGAA-′3 and 5′- GCAGAGATGCACTGTTCCCT-′3
5′- TGGACAGGGGCACTAAGACT-′3 and 5′- GAGCTCTGGGCTGTCTCTTC-3′
Values were subtracted from the amount of IgG control and were normalized to the corresponding input control.
Luciferase Reporter Assays
Using the University of California Santa Cruz Genome Browser, we identified and analyzed the genetic sequence 1 Kb upstream of the mouse Traf6 promoter. Potential transcription factor binding sites were predicted using the following online bioinformatics tools: TRANSFAC, Patch, and GPMiner. High confidence binding sites (87.5% likelihood cutoff) were accepted for additional analysis. Using these 3 tools, we manually identified 19 transcription factors as shown in Table S1.
HEK 293T cells were transiently transfected with a 1256-bp mouse luciferase reporter vector pEZX-PG04 (mTraf6-PG04) inserted into the Traf6 promoter (Genecopoeia) or control vector (NEG-PG04) along with expression vectors for Stat6, Stat5, Stat3, Pu.1, and NF-κB molecules (p50, p52, RelA, RelB and c-Rel, Addgene) by Lipofectamine 2000 (Invitrogen). Promoter activity was measured with the Secrete-Pair Dual Luminescence Assay Kit (GeneCopoeia) according to the manufacturer’s instructions. Values are expressed as the mean ± S.D. of relative luciferase units normalized to the internal control.
Microarray Analysis
Total RNA was extracted with the RNeasy Mini kit (Qiagen) from CD45.1+CD4+ Th cells sorted from spleens of tumor-bearing mice 12 days after transfer. RNA samples were sent to the Cleveland Clinic Genomics Core for quality evaluation using an Agilent Bio-analyzer. Samples with intact 18S and 28S ribosomal RNA bands with RIN >8.5 were processed for microarray analysis performed with a Mouse Ref-8 v2.0 Expression BeadChip Kit in the Cleveland Clinic Genomics Core. GSEA was run for each cell subset in pre-ranked list mode with 1000 permutations (nominal p value cutoff <0.01). The early memory signature gene set was selected from an existing publication(Wirth et al., 2010) of genes differentially expressed by >2 fold in primary versus quaternary cells (GSE21360). The mature effector gene set was selected from the same study (Wirth et al., 2010) of genes differentially expressed by >2 fold in quaternary versus primary cells (GSE21360). The T cell exhaustion-associated signature gene sets (down and up) from the Broad Institute Molecular Signature Database were used: (GSE24081_CONTROLLER_VS_PROGRESSOR_HIV_SPECIFIC_CD8_TCELL_DN) and (GSE24081_CONTROLLER_VS_PROGRESSOR_HIV_SPECIFIC_CD8_TCELL_UP).
Statistical Analyses
For statistical analysis, Student’s t-test was used. A p value less than 0.05 was considered statistically significant. Results are presented as mean ± S.D. unless otherwise indicated.
DATA AND SOFTWARE AVAILABILITY
The data reported in this paper is deposited in the Gene Expression Omnibus (GEO) database under accession number GSE97087. Data used for GSEA (v2.2.2; http://software.broadinstitute.org/gsea/index.jsp) are also available in GEO database under accession numbers GSE21360 and GSE24081.
Supplementary Material
Significance.
Adoptive cell transfer (ACT) using tumor-specific T cells is a promising method for cancer treatment. Despite the successes in obtaining objective clinical responses in patients, durable complete responses are rare. Recent observations revealed that CD4+ T cell recognition of tumor antigens is frequent, and transfer of cytolytic CD4+ T helper cell subsets may be a solution. Here we demonstrated that transfer of tumor-specific CD4+ Th9 cells eradicated large established murine tumors, leading to long-term survival of tumor-bearing mice and protection against rechallenges of tumor. Unlike Th1 and Th17 cells, Th9 cells displayed a less-exhausted, fully cytolytic, and hyperproliferative phenotype. Therefore, this study identifies Th9 cells as a potential powerful effector T cells for ACT of human cancers.
Highlights.
TRP-1-Th9 but not Th1 or Th17 cells exerted a complete antitumor response in vivo
Th9 cells are less-exhausted cytolytic effectors with upregulated Eomes expression
A hyperproliferative feature enables Th9 cells to persist long in vivo
Pu.1-Traf6-NF-κB pathway is critical for Th9 cell antitumor function in vivo
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Institute (R01 CA138398, R01 CA163881, R01 CA200539, R01 CA211073, K99CA190910, and 4R00CA190910-03) and the Leukemia and Lymphoma Society (6469-15). This study was also supported by the National Cancer Institute’s Cancer Center Support Grant award number P30CA012197 issued to the Wake Forest Baptist Comprehensive Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.ccell.2018.05.004.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
Conceptualization, Y.L. and Q.Y.; Investigation, Y.L. and Q.W.; Methodology, G.X., E.B., X.M., and J.Q.; Visualization, Y.L. and Q.W.; Resources, A.W. and C.D.; Writing – Original Draft, Y.L. and Q.Y.; Supervision, Y.L. and Q.Y.; Funding Acquisition, Y.L. and Q.Y.
REFERENCES
- Berger C Jensen MC Lansdorp PM Gough M Elliott C Riddell SR Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates J. Clin. Invest 2008. 118 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC Sehra S Goswami R Yao W Yu Q Stritesky GL Jabeen R McKinley C Ahyi AN Han L et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation Nat. Immunol 2010. 11 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodon T Comin-Anduix B Chmielowski B Koya RC Wu Z Auerbach M Ng C Avramis E Seja E Villanueva A et al. Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma Clin. Cancer Res 2014. 20 2457–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L Klebanoff CA Palmer DC Wrzesinski C Kerstann K Yu Z Finkelstein SE Theoret MR Rosenberg SA Restifo NP Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells J. Clin. Invest 2005. 115 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L Zhong XS Palmer DC Ji Y Hinrichs CS Yu Z Wrzesinski C Boni A Cassard L Garvin LM et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells Nat. Med 2009. 15 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K Laurence A Yang XP Tato CM McGeachy MJ Konkel JE Ramos HL Wei L Davidson TS Bouladoux N et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling Nature 2010. 467 967–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R Jabeen R Yagi R Pham D Zhu J Goenka S Kaplan MH STAT6-dependent regulation of Th9 development J. Immunol 2012. 188 968–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JM Yi Z Buchta CM Poovassery J Stunz LL Bishop GA Roles of tumor necrosis factor receptor associated factor 3 (TRAF3) and TRAF5 in immune cell functions Immunol. Rev 2011. 244 55–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs CS Gattinoni L Restifo NP Programming CD8+ T cells for effective immunotherapy Curr. Opin. Immunol 2006. 18 363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S Li H Qian J Yang J Lu Y Yi Q Optimizing dendritic cell vaccine for immunotherapy in multiple myeloma: tumour lysates are more potent tumour antigens than idiotype protein to promote anti-tumour immunity Clin. Exp. Immunol 2012. 170 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunder NN Wallen H Cao J Hendricks DW Reilly JZ Rodmyre R Jungbluth A Gnjatic S Thompson JA Yee C Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1 N. Engl. J. Med 2008. 358 2698–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NS Cui W Chandele A Lee HK Urso DR Hagman J Gapin L Kaech SM Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor Immunity 2007. 27 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MH Hufford MM Olson MR The development and in vivo function of T helper 9 cells Nat. Rev. Immunol 2015. 15 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IK Kim BS Koh CH Seok JW Park JS Shin KS Bae EA Lee GE Jeon H Cho J et al. Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Nat. Med 2015. 21 1010–1017 [DOI] [PubMed] [Google Scholar]
- King CG Kobayashi T Cejas PJ Kim T Yoon K Kim GK Chiffoleau E Hickman SP Walsh PT Turka LA Choi Y TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis Nat. Med 2006. 12 1088–1092 [DOI] [PubMed] [Google Scholar]
- Kjaergaard J Peng L Cohen PA Drazba JA Weinberg AD Shu S Augmentation versus inhibition: effects of conjunctional OX-40 receptor monoclonal antibody and IL-2 treatment on adoptive immunotherapy of advanced tumor J. Immunol 2001. 167 6669–6677 [DOI] [PubMed] [Google Scholar]
- Klebanoff CA Gattinoni L Palmer DC Muranski P Ji Y Hinrichs CS Borman ZA Kerkar SP Scott CD Finkelstein SE et al. Determinants of successful CD8+ T-cell adoptive immunotherapy for large established tumors in mice Clin. Cancer Res 2011. 17 5343–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann C van Buuren MM Bies L Verdegaal EM Schotte R Calis JJ Behjati S Velds A Hilkmann H Atmioui DE et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma Nat. Med 2015. 21 81–85 [DOI] [PubMed] [Google Scholar]
- Liu JQ, Li XY, Yu HQ, Yang G, Liu ZQ, Geng XR, Wang S, Mo LH, Zeng L, Zhao M. et al. Tumor-specific Th2 responses inhibit growth of CT26 colon-cancer cells in mice via converting intratumor regulatory T cells to Th9 cells. Sci. Rep. 2015;5:10665. doi: 10.1038/srep10665. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lu Y Hong B Li H Zheng Y Zhang M Wang S Qian J Yi Q Tumor-specific IL-9-producing CD8+ Tc9 cells are superior effector than type-I cytotoxic Tc1 cells for adoptive immunotherapy of cancers Proc. Natl. Acad. Sci. USA 2014. 111 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y Hong S Li H Park J Hong B Wang L Zheng Y Liu Z Xu J He J et al. Th9 cells promote antitumor immune responses in vivo J. Clin. Invest 2012. 122 4160–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranski P Boni A Antony PA Cassard L Irvine KR Kaiser A Paulos CM Palmer DC Touloukian CE Ptak K et al. Tumor-specific Th17-polarized cells eradicate large established melanoma Blood 2008. 112 362–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranski P Borman ZA Kerkar SP Klebanoff CA Ji Y Sanchez-Perez L Sukumar M Reger RN Yu Z Kern SJ et al. Th17 cells are long lived and retain a stem cell-like molecular signature Immunity 2011. 35 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North RJ Cyclophosphamide-facilitated adoptive immunotherapy of an established tumor depends on elimination of tumor-induced suppressor T cells J. Exp. Med 1982. 155 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermajer N Popp FC Soeder Y Haarer J Geissler EK Schlitt HJ Dahlke MH Conversion of Th17 into IL-17A(neg) regulatory T cells: a novel mechanism in prolonged allograft survival promoted by mesenchymal stem cell-supported minimized immunosuppressive therapy J. Immunol 2014. 193 4988–4999 [DOI] [PubMed] [Google Scholar]
- Osinska I Popko K Demkow U Perforin: an important player in immune response Cent. Eur. J. Immunol 2014. 39 109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S Schaefer BC A new look at T cell receptor signaling to nuclear factor-kappa B Trends Immunol 2013. 34 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL Mullen AC Martins GA Krawczyk CM Hutchins AS Zediak VP Banica M DiCioccio CB Gross DA Mao CA et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin Science 2003. 302 1041–1043 [DOI] [PubMed] [Google Scholar]
- Purwar R Schlapbach C Xiao S Kang HS Elyaman W Jiang X Jetten AM Khoury SJ Fuhlbrigge RC Kuchroo VK et al. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells Nat. Med 2012. 18 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley M Pereyra F Nilsson B Porichis F Fonseca C Eichbaum Q Julg B Jesneck JL Brosnahan K Imam S et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF Nat. Med 2010. 16 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley WW Shafiani S Wittmer ST Tucker-Heard G Moon JJ Jenkins MK Urdahl KB Winslow GM Woodland DL Distinct functions of antigen-specific CD4 T cells during murine Mycobacterium tuberculosis infection Proc. Natl. Acad. Sci. USA 2010. 107 19408–19413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restifo NP Dudley ME Rosenberg SA Adoptive immunotherapy for cancer: harnessing the T cell response Nat. Rev. Immunol 2012. 12 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhuri R Hirahara K Mousavi K Clever D Klebanoff CA Bonelli M Sciume G Zare H Vahedi G Dema B et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis Nature 2013. 498 506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser RL Martins GA Kalachikov S Chandele A Parish IA Meffre E Jacob J Calame K Kaech SM Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties Immunity 2009. 31 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D Stampfel G Stark A Transcriptional enhancers: from properties to genome-wide predictions Nat. Rev. Genet 2014. 15 272–286 [DOI] [PubMed] [Google Scholar]
- Steenwinckel V Louahed J Orabona C Huaux F Warnier G McKenzie A Lison D Levitt R Renauld JC IL-13 mediates in vivo IL-9 activities on lung epithelial cells but not on hematopoietic cells J. Immunol 2007. 178 3244–3251 [DOI] [PubMed] [Google Scholar]
- Vegran F Berger H Boidot R Mignot G Bruchard M Dosset M Chalmin F Rebe C Derangere V Ryffel B et al. The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells Nat. Immunol 2014. 15 758–766 [DOI] [PubMed] [Google Scholar]
- Walsh MC Lee J Choi Y Tumor necrosis factor receptor-associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system Immunol. Rev 2015. 266 72–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth TC Xue HH Rai D Sabel JT Bair T Harty JT Badovinac VP Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation Immunity 2010. 33 128–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY Best JA Knell J Yang E Sheridan AD Jesionek AK Li HS Rivera RR Lind KC D’Cruz LM et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets Nat. Immunol 2011. 12 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H Chua KS Guimond M Kapoor V Brown MV Fleisher TA Long LM Bernstein D Hill BJ Douek DC et al. Lymphopenia and interleukin-2 therapy alter homeostasis of CD4+CD25+ regulatory T cells Nat. Med 2005. 11 1238–1243 [DOI] [PubMed] [Google Scholar]
- Zhao C, Li X, Hu H. PETModule: a motif module based approach for enhancer target gene prediction. Sci. Rep. 2016a;6:30043. doi: 10.1038/srep30043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chu X, Chen J, Wang Y, Gao S, Jiang Y, Zhu X, Tan G, Zhao W, Yi H. et al. Dectin-1-activated dendritic cells trigger potent anti-tumour immunity through the induction of Th9 cells. Nat. Commun. 2016b;7:12368. doi: 10.1038/ncomms12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.