 Alkyne–azide cycloaddition derivatives of DHZ (1) were synthesized and screened for cytotoxic potential in which the derivatives, 3, 6, 7, 8, 9 and 15 displayed most potent with IC50 value ranging from 1.8–3.0 μM.
Alkyne–azide cycloaddition derivatives of DHZ (1) were synthesized and screened for cytotoxic potential in which the derivatives, 3, 6, 7, 8, 9 and 15 displayed most potent with IC50 value ranging from 1.8–3.0 μM.
Abstract
Herein, we report the isolation and synthetic modification of dehydrozingerone (DHZ, 1), a secondary metabolite present in the rhizome of Zingiber officinale. We synthesized O-propargylated dehydrozingerone, which was subsequently coupled by alkyne–azide cycloaddition (3–20) using click chemistry. The compounds (1–20) were evaluated for their in vitro cytotoxic activity in a panel of three cancer cell lines. Among all the DHZ derivatives, 3, 6, 7, 8, 9 and 15 displayed potent cytotoxic potential with an IC50 value ranging from 1.8–3.0 μM in MCF-7, PC-3 and HCT-116 cell lines. Furthermore, compound 7 has proven to be the most potent cytotoxic compound in all the three distinct cancer cell lines and also demonstrated significant anti-invasive potential in prostate cancer. The mechanistic study of compound 7 showed that it not only suppressed the AKT/mTOR signalling which regulates nuclear transcription factor-NF-kB but also augmented the expression of anti-invasive markers E-cadherin and TIMP. Compound 7 significantly decreased the expression of pro-invasive markers vimentin, MMP-2 and MMP-9, respectively. This study underscores an efficient synthetic approach employed to evaluate the structure–activity relationship of dehydrozingerone (1) in search of potential new anticancer agents.
1. Introduction
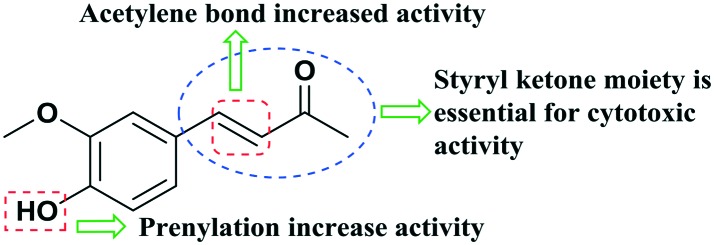
Dehydrozingerone (4-hydroxy-3-methoxybenzalacetone, DHZ, Fig. 1) is isolated from rhizomes of ginger (Zingiber officinale)1 and can be synthesized by simple aldol condensation of vanillin and acetone.2 DHZ, which is a recognized biosynthetic intermediate and degradant of curcumin,3,4 shows a structural resemblance to curcumin, as both bear styryl ketone moieties with similar substitutions on the phenyl ring.5 DHZ (1) has shown significant therapeutic potential with profound biological properties, such as anti-inflammatory, antioxidant6,7 antifungal8 and anti-mycobacterial activity.9 Although few preliminary reports unveiled the antiproliferative effect of 1 against cancer cells,10,11 so far, no molecular mechanisms of its action on cell growth/invasion have been described. Motohashi et al.12–14 have investigated the antimutagenic activities of DHZ and its synthetic analogs and reported that 4-O-benzalacetone substituted analogues of DHZ revealed the strongest antimutagenic activity. These results disclosed that ring substitution with a group such as 4-hydroxyl, methoxyl or methyl increased the antimutagenic activity. In light of the above studies, we selected the modification in the aromatic core of DHZ for the development of the SAR. We have synthesized 4-O-propargylated dehydrozingerone by alkyne–azide cycloaddition (3–20) demonstrating moderately high yields (85–93%) by means of click chemistry. This study aimed to investigate the in vitro cytotoxic potential of dehydrozingerone derivatives15 in prostate cancer cells.
Fig. 1. Pharmacophore of (DHZ) 1 for antimutagenic/cytotoxic activity.
Prostate cancer is considered as one of the leading causes of all cancer deaths in most developed countries.16 The phosphatidylinositol 3-kinase (PI3K)/AKT pathway is constitutively hyperactivated in prostate cancer prognosis and is an effective target for therapeutic development.17 By modulating diverse signaling pathways, AKT is implicated in the metastasis progression in an advanced condition of the disease.18 The direct target of hyperactive AKT, nuclear transcription factor-KB (NF-kB), has been correlated with increased metastatic potential in prostate cancer and is associated with metastasis and invasion in many tumors.19 The recent literature demonstrates that NF-kB is constitutively activated in most types of prostate cancers. NF-kB promotes the migration and invasion of prostate cancer cells by a number of NF-kB-regulated gene products.20 The current study demonstrates the anti-invasive and anti-motility activity of dehydrozingerone derivative 7 in the human prostate cancer line (PC-3).
2. Results and discussion
2.1. Chemistry
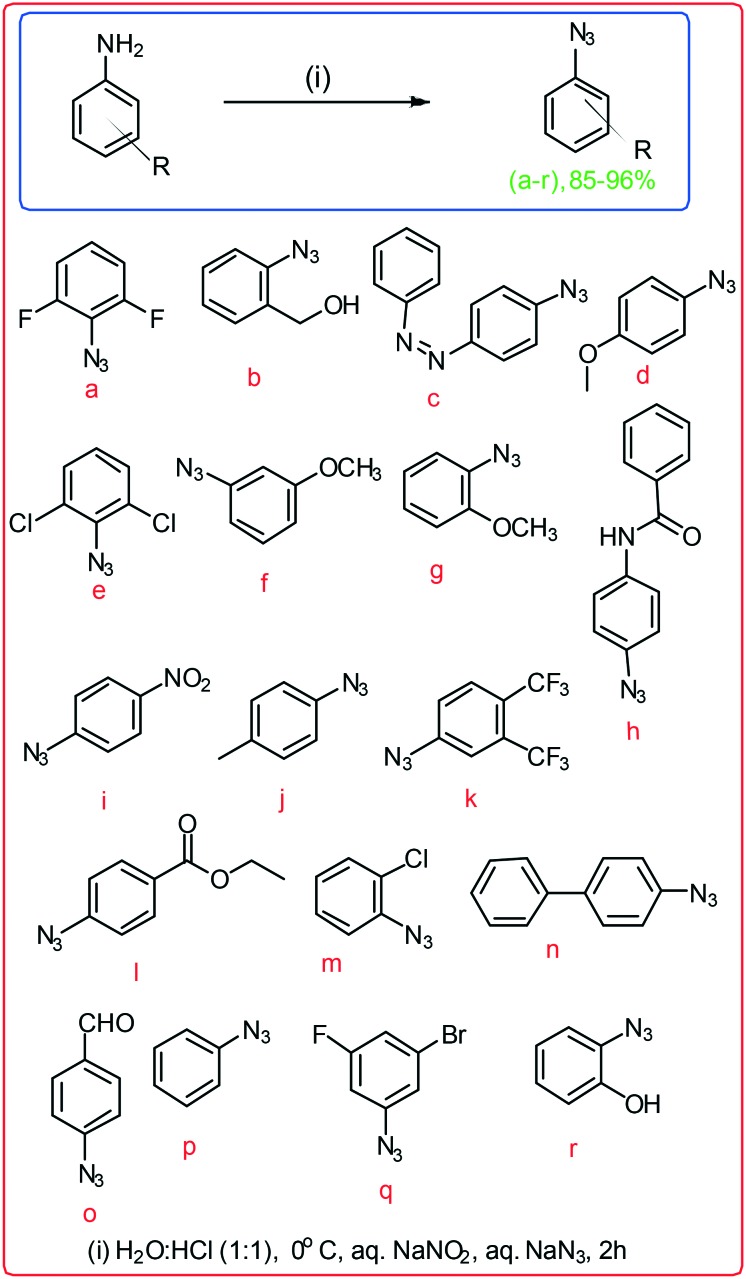
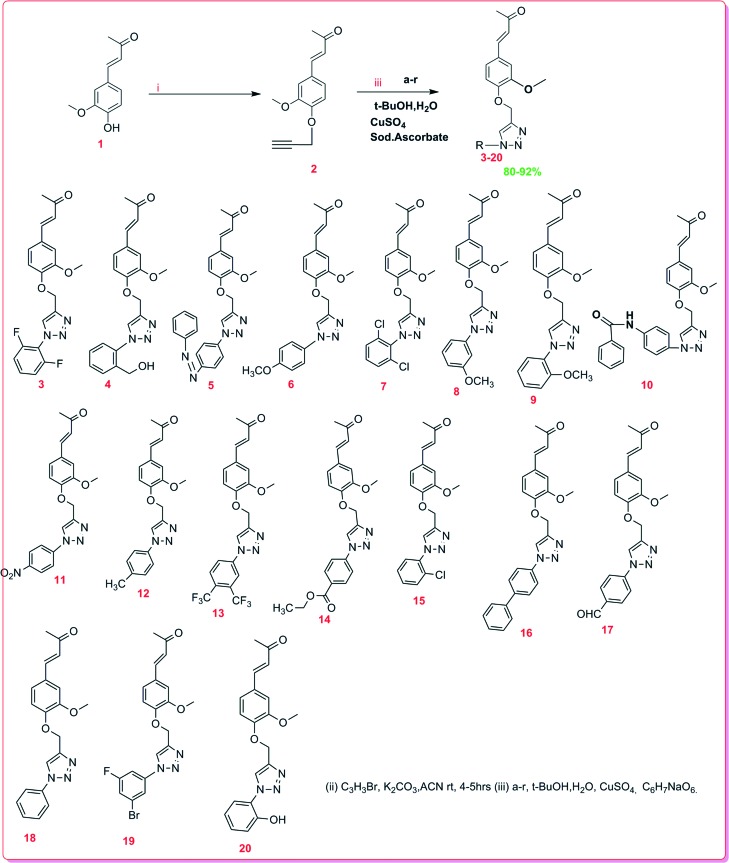
Dehydrozingerone (DHZ, 1) is known as feruloylmethane which displays versatile therapeutic activities against various types of diseases and is used as anti-infective, anticancer, antiviral, and anti-hypertensive agents.4–6,9 Due to the profound biological potential of 1, we explored its chemistry and biological activities by modifications of its structure and synthesised the C4-aryl substituted 1,2,3-triazoles of 1 by alkyne–azide cycloaddition. The aromatic ring substituted 1-azido derivatives (a–r) were synthesised viaScheme 1.15
Scheme 1. Synthesis of substituted 1-azido derivatives.
Synthesis of alkyne–azide cycloaddition derivatives of 1
The compound 1 C4-hydroxyl was O-propargylated, affording intermediate 2 which was further coupled with different substituents of azide by employing facile click chemistry15 to obtain distinct derivatives of 1,2,3-triazol-4-yl-dehydrozingerone (3–20). The appearance of 1H NMR signals at δH 4.81 (2H) and 2.54 (1H) confirmed the condensation of propargyl bromide. The signals at δC 56.57 are attributed to the carbon of the active methylene inserted after O-propargylation of 1. 1H NMR of all the triazole derivatives (3–20) has shown a characteristic singlet for the triazole proton in the range δH 7.8–8.44. In addition, 13C NMR spectra exhibited characteristic signals for the triazole ring carbon at δC 119.5–122.7 and 149.3–144.6, respectively. The synthetic yields, molecular weight, HPLC data and NMR spectra of all the compounds have been described in the experimental section.
For SAR studies, compounds (2–20) were screened for their in vitro cytotoxicities against three human cancer cell lines. As is evident from Table 1, 4-O-substituted alkyne–azide cycloaddition derivatives 3, 6, 7, 8, 9 and 15 showed the highest cytotoxic potential probably due to the change in pharmacophore, which facilitated the binding of the ligand in the receptor pocket.
Table 1. The IC50 values (μM) of dehydrozingerone derivatives in a panel of cancer cell lines. Data were compared with untreated control, and IC50 values were expressed as the mean ± SD of three independent experiments.
| S. no | PC-3 (IC50) ± SD | HCT-116 (IC50) ± SD | MCF-7 (IC50) ± SD |
| 1 | 43.21 ± 0.5 | 34.78 ± 0.5 | 54.65 ± 0.1 |
| 2 | 40.23 ± 0.5 | 49.74 ± 0.4 | 65.38 ± 0.4 |
| 3 | 24.22 ± 0.3 | 37.67 ± 0.1 | 2.028 ± 0.5 |
| 4 | 50.34 ± 0.2 | >100 | 65.78 ± 0.3 |
| 5 | 42.11 ± 0.2 | >100 | 75.85 ± 0.1 |
| 6 | 2.013 ± 0.3 | 21.34 ± 0.1 | 33.67 ± 0.2 |
| 7 | 3.073 ± 0.3 | 2.002 ± 0.1 | 26.58 ± 0.1 |
| 8 | 31.56 ± 0.4 | 29.55 ± 0.2 | 1.8 ± 0.2 |
| 9 | 2.045 ± 0.2 | 25.65 ± 0.3 | 29.14 ± 0.5 |
| 10 | 76.81 ± 0.2 | >100 | 86.69 ± 0.5 |
| 11 | 46.74 ± 0.2 | 96.34 ± 0.5 | >100 ± 0.3 |
| 12 | 58.12 ± 0.2 | 76.57 ± 0.5 | >100 ± 0.1 |
| 13 | 67.23 ± 0.1 | 57.78 ± 0.5 | 87.37 ± 0.2 |
| 14 | 59.22 ± 0.5 | 78.9 ± 0.1 | >100 ± 0.2 |
| 15 | 47.98 ± 0.5 | 39.00 ± 0.1 | 2.19 ± 0.2 |
| 16 | 56.81 ± 0.3 | 65.25 ± 0.4 | 65.38 ± 0.5 |
| 17 | 80.67 ± 0.4 | 76.22 ± 0.5 | 86.37 ± 0.5 |
| 18 | 78.56 ± 0.3 | 67.33 ± 0.3 | >100 |
| 19 | 56,89 ± 0.5 | 45.36 ± 0.5 | 65.48 ± 0.3 |
| 20 | 59.23 ± 0.2 | 76.36 ± 0.3 | >100 |
Biological assay
2.2. Anti-proliferative activity of compound 7
The cell viability assay (MTT) was performed on a panel of cancer cell lines. The cells were exposed to the corresponding derivatives for 48 h with a concentration range of 100 nM to 100 μM. A consistent cytotoxic activity was found in compound 7 against prostate cancer (PC-3) and HCT-116 (human colon cancer) cell lines with an IC50 concentration of 3.073 μM and 2.002 μM. As shown in Table 1, compounds 3, 8 and 15 showed an IC50 value of 2.028, 1.8 and 2.19 μM respectively in MCF-7 (breast cancer cell) while the IC50 values of 6 and 9 were found to be 2.013 and 2.045 μM respectively in PC-3 cell lines. In addition, to verify the cytotoxic effects of 7 on normal prostate cells (BPH-1), MTT results showed an IC50 value higher than 100 μM indicating the safety profile of this compound 7 in normal cells (data not shown).
2.3. Anti-motility and anti-invasive activities of 7
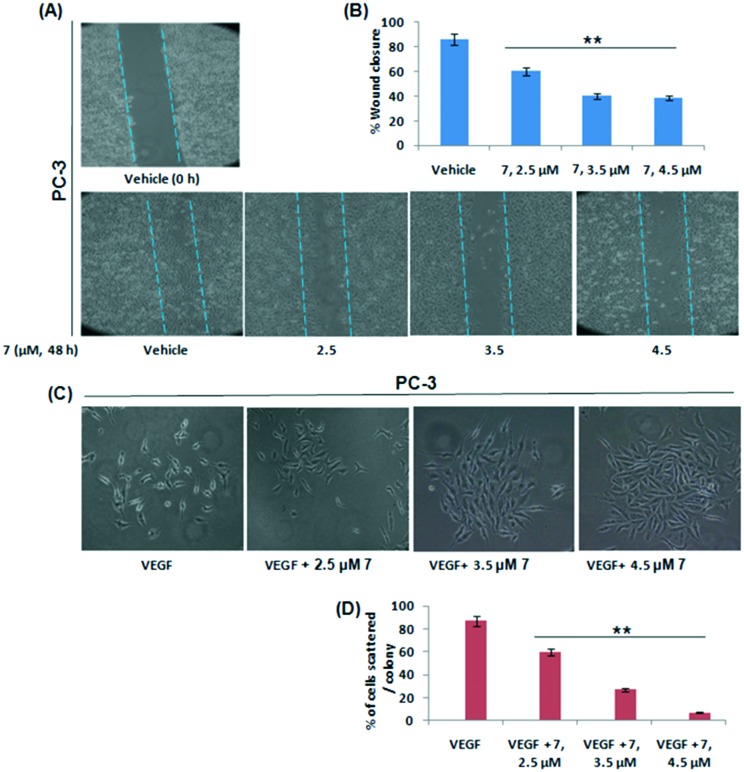
Prostate cancer invasion and metastasis remains the most critical determinant of resectability and hence survival.21 Here, we aimed to study the effect of 7 on the invasive potential of PC-3 cells in vitro. To examine any effect of 7 on the motility of PC-3 cells, a wound healing assay was performed. It was observed that 7 significantly inhibited the cell motility in a concentration dependent manner and there was a characteristic wound closure due to the migration of PC-3 cells in vehicle treated wells (Fig. 2A and B). Cell scattering is an inherent incident associated with metastatic cancer cells.22 Here, we evaluated the effects of 7 on PC-3 cell scattering and the results demonstrated that 7 efficiently blocked the cell scattering in the presence of growth factor VEGF (Vascular endothelial growth factor) (Fig. 2C and D).
Fig. 2. Effects of compound 7 on motility and cell scattering aptitude of PC3 cells. (A and B) The wound healing assay was performed by treating PC-3 cells with increasing concentrations of 7 which were consecutively assessed for the degree of wound healing. Photographs of scratched areas were taken at 20× magnification (C and D). PC-3 cells were treated with indicated concentrations of 7 along with or without VEGF to check its effects on cell scattering. Following 24 h of incubation with 7, individual colonies were observed under an inverted microscope and were photographed. The data shown are representative of three independent experiments. SD ± **P < 0.01.
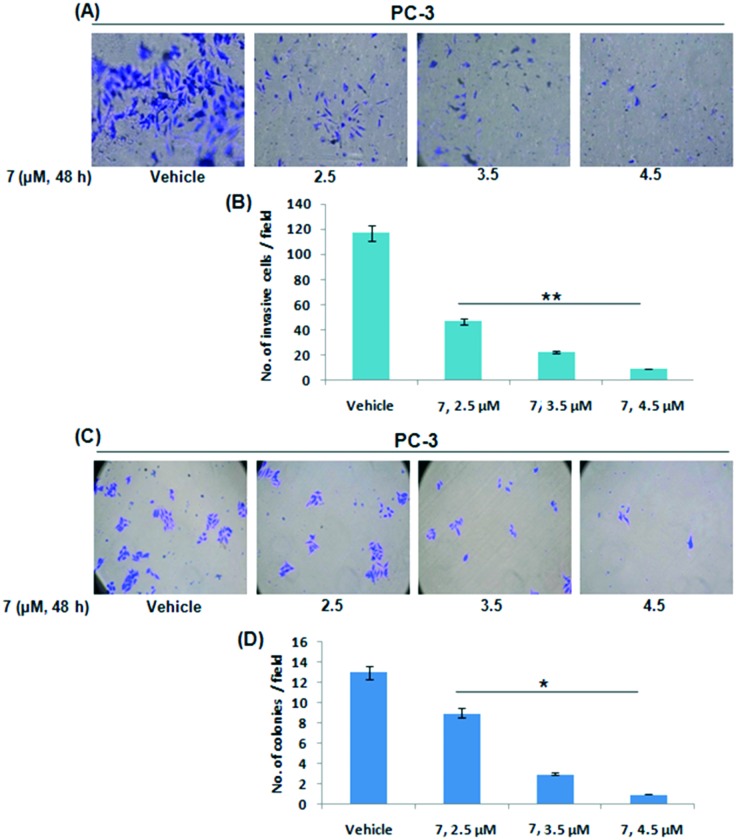
Since the PC-3 cell line is considered as an aggressive cell line due to PTEN mutation and having the ability to invade through the extracellular matrix to colonize at the secondary tumor sites,23 we sought to inspect the effect of 7 on cell invasion through the Boyden chamber invasion assay. The results showed that 7 subdued the cell invasion potential of PC-3 cells in a dose dependent manner (Fig. 3A and B). Colony formation was totally restrained by 7 in PC-3 cells as well (Fig. 3C and D). Further, we investigated the apoptosis inducing property of compound 7 in PC-3 cells by studying the mitochondrial membrane potential loss and casapase-3/7 activation through JC-1 staining and the Caspase-Glo assay, respectively. The results showed that compound 7 induced apoptosis in a concentration-dependent manner, as suggested by increased JC-1 monomer formation and decreased JC-1 aggregate formation (mitochondrial membrane potential loss). The JC-1 results were further validated by an increase in caspase 3/7 activity (Fig. S1A and B†). These collective results clearly support the anti-metastatic, anti-invasive and anti-proliferative potential of 7 in PC-3 cells.
Fig. 3. Effects of compound 7 on the invasive and colony formation ability of PC-3 cell lines. (A) Boyden chamber assay was performed to check the effect of 7 on the invasive ability of PC-3 cells. Following incubation with compound 7 for the indicated time point, invaded cells were counted and images were captured at 20× magnification. (B) Bar graphs are representative of the number of invasive cells per field. (C and D) To check the effects on colony formation, PC-3 cells were exposed to different concentrations of 7 and incubated for five days. After the incubation, the cells were stained with crystal violet; stained colonies were counted randomly, quantified and images were taken under an inverted microscope at 20× magnification. Data from three independent experiments were subjected to statistical analysis. SD ± **P < 0.01, *p < 0.05.
2.4. Compound 7 suppresses Akt-mTOR signalling
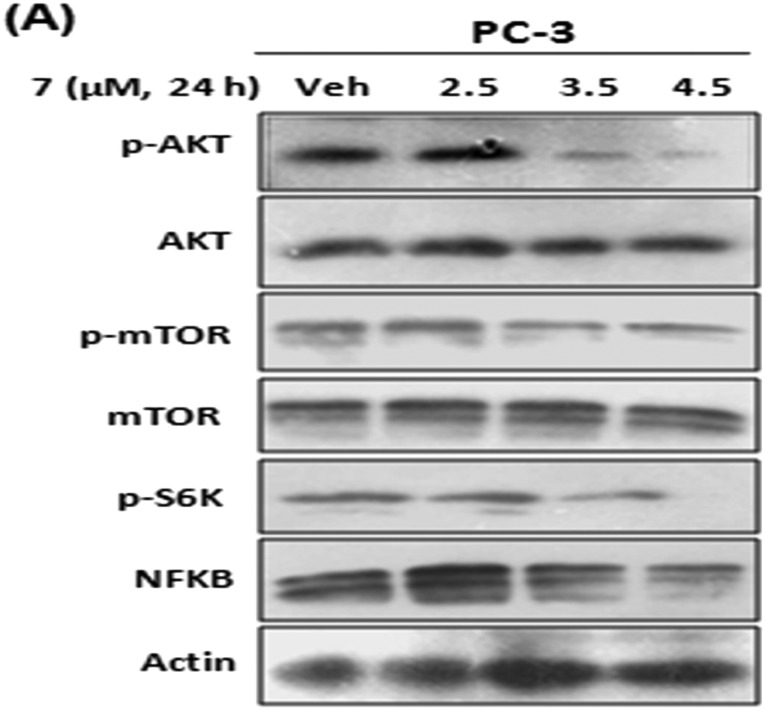
In prostate cancer, AKT is considered as a major player for tumor growth and metastasis. Also in the majority of prostate cancers, NF-kB through phosphatidylinositol 3-kinase (PI3K) dependent activation of IKK is constitutively activated, leading to the stimulation of direct downstream MMPs via transcriptional regulation.24 Therefore, by western blotting, we analyzed the effect of 7 on the phosphorylation of AKT and the corresponding NF-kB levels. The results showed that the expression of NF-kB was significantly attenuated in a dose dependent manner and the phosphorylation of AKT was strongly impeded by 7 treatments but showed negligible effects on total AKT (Fig. 4A). This was in contrast to the insignificant effects of the parent compound 1 on the phosphorylation of AKT (Fig. S1C†).
Fig. 4. Compound 7 inhibits PI3K/Akt/NF-KB signaling. (A and B) PC-3 cells were treated with indicated concentrations of 7. Whole cell lysates were checked for the expressions of indicated proteins by western blotting. β-Actin was used as a loading control and the data shown are representative of three independent experiments.
Furthermore, it is well known that NF-kB is regulated by the PI3K/AKT pathway via mTOR activation.19 So here, we studied the effects of 7 on mTOR and S6K activation through western blotting. The results revealed that the phosphorylation of mTOR was attenuated by 7 along with the downregulation of S6K, and no characteristic change in the expression of total mTOR was observed.
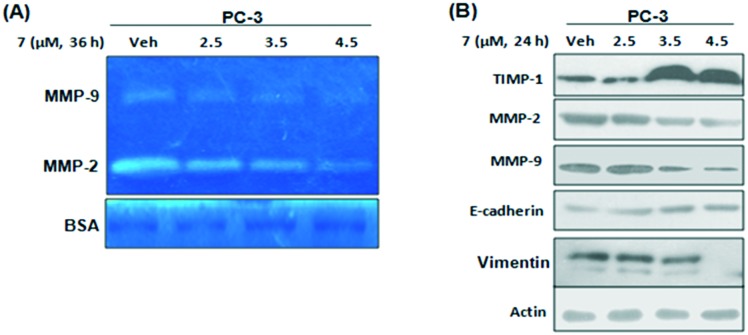
2.5. Compound 7 abrogates MMP 2 and MMP 9 gelatinase activity as well as their expression
Matrix metalloproteinases (MMPs) play a major role in metastasis and invasion.25 To check the consequences of 7 on MMP activation, zymography was performed with decreasing concentrations of 7. The results showed that matrix gelatin degradation was inhibited significantly in aggressive PC-3 cells with increased concentrations of 7 (Fig. 5A). Among all MMPs, MMP-2 and MMP-9 play a critical role in cancer progression; so in order to check the effect of 7 on the expression of MMPs, western blotting experiments were performed and the results showed that MMP-2 and MMP-9 expressions were significantly attenuated by 7. TIMP-1 is a well known marker involved in the negative regulation of pro-invasive and pro-metastatic MMPs. So, by employing western blotting we demonstrated that 7 strongly amplified the expression of TIMP-1 in a dose dependent manner (Fig. 5B). In higher incidences of metastasis and advanced tumors, the expression and localization of E-cadherin to the cell membrane are lost; both loss of E-cadherin and overexpression of MMPs contribute to sturdy tumors.26 Our results demonstrated that treatment of PC-3 cells with 7 elevated the expression of E-cadherin, an epithelial marker, along with a steady decline in the expression of vimentin, a mesenchymal marker (Fig. 5B).
Fig. 5. Compound 7 abrogates MMP-2 and MMP-9 gelatinase activity and expression. (A) Gelatin zymography was carried out to check the effects of 7 on MMP-2 and MMP-9 activity. BSA was used as a loading control. (B) PC-3 cells were seeded in six-well plates and treated with indicated concentrations of 7. Whole cell lysates were checked for the expressions of indicated proteins by western blotting. β-Actin was used as loading control and the data shown are representative of three independent experiments.
3. Conclusions
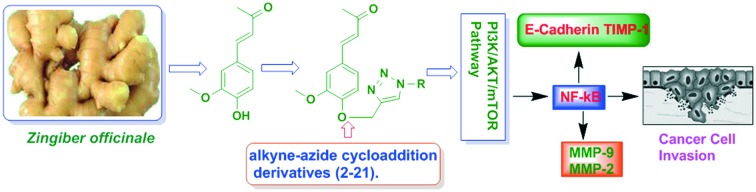
Distinct dehydrozingerone derivatives (2–20) were synthesized via the alkyne–azide cycloaddition reaction with good yield. All these synthetic derivatives were evaluated for their in vitro cytotoxicities in three distinct human cancer cell lines. The compounds containing 4-O-substituted alkyne–azide on the aromatic ring attached to 2,6-difluorophenyl (3), 4-methoxyphenyl (6), 3-methoxyphenyl (8), 2-methoxyphenyl (9), and 2-chlorophenyl (15) derivatives were shown to be 16–18-fold more potent than 1 against human cancer cell lines, viz. PC-3 and MCF-7, while 2,6-dichlorophenyl (7) showed better potency than 1 in all the cancer cell lines tested. Compounds of this class appear amenable to further modifications and will prove useful for prevention and treatment of cancer proliferation. Collectively, it can be concluded from the above studies that compound 7 blocks invasion and metastasis of PC-3 cells by abrogating NF-KB dependent expression and activation of MMP-2 and MMP-9 via the suppression of the PI3K/AKT pathway. So, it is implied that derivative 7 of dehydrozingerone could be explored as an anti-metastatic/anti-invasive agent against malignant prostate cancer.
4. Experimental section
4.1. Chemistry
High resolution mass spectra were obtained using an Agilent 6540 (Q-TOF) high resolution mass spectrometer in electrospray (ESI MS) mode. 1H NMR spectra were recorded (Bruker Avance) at 400 and 500 MHz while 13C NMR spectra were recorded at 125 MHz in CDCl3 and CD3OD. The chemical shift values were reported in δ (ppm) and coupling constants in Hertz. Tetramethylsilane (TMS) was used as an internal standard. All reactions were monitored by thin layer chromatography (TLC) on silica gel 60F254 (0.25 mm thick, Merck) with spots visualized by UV (254 and 366 nm) and anisaldehyde reagent was used as the development agent. Dehydrozingerone (DHZ 1) was isolated from the rhizome of Zingiber officinale while other reagents were purchased commercially and used without further purification, unless otherwise stated.
4.2. Synthesis of compounds (2–20)
Compound 1 (1 g, 5.2 mmol) was dissolved in dry acetone (50 mL), and then anhydrous Na2CO3 (2.1 g, 20 mmol) was added and stirred for 15 minutes at 0 °C under a nitrogen atmosphere. Propargyl bromide (1.4 equiv.) was gradually added (Scheme 1) and stirred for 4–5 h. Subsequently, the reaction mixture was diluted with brine water and extracted thrice with ethyl acetate. Finally, the combined extract passed through Na2SO4 and evaporated under vacuum afforded the crude product, which was further purified using a silica column to obtain intermediate compound 2 in excellent yield (92%). Furthermore, compound 2 was dissolved in a mixture of t-butyl alcohol and water (3 mL, 1 : 1) followed by the successive addition of substituted 1-azido derivatives (1.2 equiv.), sodium ascorbate (0.5 equiv.) and CuSO4·5H2O (0.1 equiv.). The reaction mixture was stirred at room temperature for 1–2 h (Scheme 2). Subsequently, the mixture was diluted with water (10 mL) and extracted with ethyl acetate (3 × 10 mL) dried over Na2SO4, and concentrated under vacuum, affording the crude product, which was subjected to purification by flash column chromatography leading to the isolation of alkyne–azide cycloaddition derivatives (2–20).
Scheme 2. Synthesis of alkyne–azide cycloaddition derivatives (3–20).
4.3. Cell culture and antibodies
PC3, MCF-7 and HCT-116 cell lines were purchased from the European Collection of Cell Culture (ECACC). Cells were cultured in RPMI 1640/DMEM containing 10% FBS, 70 mg L–1 penicillin plus 0.1 g L–1 streptomycin and were incubated at 37 °C with 95% air and 5.0% CO2. Antibodies were obtained from the following commercial sources: anti-AKT, anti-NF-kB, anti-MMP-2, anti-MMP-9, anti-E-cadherin, anti-mTOR, anti-S6 K and anti-TIMP-1 from Santa Cruz Biotechnology (Santa Cruz, CA), anti-p-AKT and anti p-mTOR from Cell Signaling (Cell Signaling Technologies, MA) and anti-beta actin from Sigma Chemical (St Louis, MO).
4.4. Cell viability assay
MCF-7, PC-3 and HCT-116 cells (2.5 × 103 cells per well) were plated in a 96-well tissue culture plate. The cells were treated with different concentrations of indicated compounds by the standard MTT dye uptake method according to a procedure described previously.27
4.5. Clonogenic assay
The clonogenic assay was performed according to a previously described method.21 Briefly, PC-3 cells (1 × 103 cells per well) were plated in 6-well tissue culture grade plates. The culture medium was changed after 24 h and then a new medium was added. Cells were then exposed to various concentrations of conjugate 7 along with the vehicle DMSO for 5 days.
4.6. Scratch motility (wound healing) assay
The assay was performed as described previously.28 Briefly, in a 6-well plate PC-3 cells (5 × 105 cells per well) were seeded and allowed to form a confluent monolayer for 24 h, and then serum starved for another 24 h. After that, the monolayer was scratched with a sterile pipette tip (200 μL), washed with a serum-free medium and photographed (time 0 h). Cells were successively treated with medium containing low serum (1.0%) in the presence of different concentrations of 7 along with the vehicle DMSO for 24 h. Scratched areas were progressively photographed with a Nikon D3100 inverted microscope camera (20× magnification). The percentage of wound closure was estimated using the following equation: % wound closure = [1 – (wound area at t1/wound area at t0) × 100%], where t1 is the time after wounding and t0 is the time immediately after wounding.
4.7. Matrigel invasion assay
The effect of 7 on cell invasion was determined using a BD Biocoat Tumor Invasion Assay System (BD Bioscience, Bedford, MA) according to the instruction of the manufacturer. Briefly, PC-3 cells (1.2 × 106 cells per well) were treated with different concentrations of 7 or the vehicle DMSO. The Matrigel invasion assay was performed following a previously described method.29
4.8. Cell scattering assay
The cell scattering assay was performed as described previously.28 Briefly, PC-3 cells were seeded at a density of 1000 cells per well in 6-well plates. After 3–4 days of incubation, small, cohesive, and discrete colonies were formed. Cells were then stimulated with VEGF (20 mg ml–1) in the presence of various concentrations of 7 in growth medium. Following overnight incubation, colonies were observed under a bright field microscope. Photographs were taken for individual colonies.
4.9. Immunoblotting
PC-3 cells were exposed to the indicated concentrations of 7 along with DMSO. Cells were then lysed with lysis buffer and an equal amount of protein from each sample was subjected to western blot analysis as described previously.30
4.10. Gelatin zymography
The gelatinase activity of MMP-2 was assessed by a method standardized previously.29 Briefly, PC-3 cells in a subconfluent culture (70–80% cell density of confluent culture) were refreshed with a new medium and kept for incubation with increasing concentrations of 7 for 48 h. The conditional media obtained from both treated and untreated samples were employed for protein estimation and equal amounts of total proteins (20 mg) were mixed with sample buffer (2.0% SDS, 25% glycerol, 0.1% bromophenol blue and 60 mM Tris-HCl, pH 6.8). Gelatin zymography of the samples was carried out using 7.5% SDS-polyacrylamide gels containing 0.1% gelatin at 100 V for 3 h at 4 °C. The gels were rinsed accordingly with washing buffer (2.0% Triton X-100 in dd water) at room temperature to remove SDS, followed by incubation overnight at 37 °C in TCNB buffer (50 mM Tris-HCl, pH 8.0, 10 mM CaCl2, 0.02% NaN3). The gels were stained with Coomassie blue R-250 (Sigma) (0.125% Coomassie blue R-250, 50% methanol, 10% acetic acid) for 1–1.5 h and destained with a destaining solution (20% methanol, 10% acetic acid, 70% dd water). The gelatinase activity was detected by observing unstained bands on a blue background on Coomassie stained gels.
4.11. JC-1 staining
For JC-1 staining, treated cells (1 × 106 cells per well) were washed with chilled PBS and then 1 mM JC-1 (Thermo Fisher) solution was added to the cells so that the final concentration of JC-1 was 1 μM. The cells were accordingly incubated at 37 °C for 30 min and analysed under fluorescence microscope for JC-1 aggregates and monomers according to the standard protocol as per manufacturer's instructions.
4.12. Caspase3/7-Glo assay
Briefly, cells were seeded in a 96-well plate at a density of 5 × 103 cells per well for 24 h. The caspase 3/7 reagent and substrate mix was prepared as per the manufacturer's protocol (Promega). After proper treatment of the cells, approximately 100 μl of the caspase 3/7 reagent and substrate mix was added and incubated for 25 min. After the incubation, the luminescence was read using a Tecan plate reader.
4.13. Statistical analysis
Data were expressed as the mean ± standard error of three independent experiments. Statistical analyses were performed using Graph Pad Prism version 5. Comparisons between two groups were conducted using unpaired Student's t-test. ‘p’ values <0.05 were considered as statistically significant.
4.14. Characterization of synthesized compounds
4.14.1. (E)-4-(4-Hydroxy-3-methoxyphenyl)but-3-en-2-one (1)
TLC, Hex : EtOAc (7 : 3), Rf 0.4, yellowish crystalline solid. 1H NMR (400 MHz, CDCl3) δ 7.45 (d, J = 16.2 Hz, 1H), 7.12–7.03 (m, 2H), 6.93 (d, J = 8.1 Hz, 1H), 6.59 (d, J = 16.2 Hz, 1H), 5.93 (s, 1H), 3.94 (s, 3H), 2.37 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 198.57, 148.45, 147.04, 143.91, 126.88, 124.92, 123.52, 114.95, 109.53, 55.98, 27.26.
4.14.2. (E)-4-(3-Methoxy-4-(prop-2-yn-1-yloxy)phenyl)but-3-en-2-one (2)
TLC, Hex : EtOAc (7 : 3), Rf 0.6, colourless amorphous solid, yield: 1.1 g (92.19%).1H NMR (400 MHz, CDCl3) δ 7.49 (t, J = 15.8 Hz, 1H), 7.13 (dd, J = 8.3, 1.9 Hz, 1H), 7.09 (d, J = 1.9 Hz, 1H), 7.04 (d, J = 8.3 Hz, 1H), 6.68–6.52 (m, 1H), 4.81 (d, J = 2.4 Hz, 2H), 3.92 (s, 3H), 2.54 (t, J = 2.4 Hz, 1H), 2.38 (s, 3H); 13C NMR (126 MHz, CDCl3) δ 198.06, 149.63, 149.36, 143.21, 128.52, 125.89, 122.38, 113.41, 110.25, 77.96, 76.35, 56.57, 55.94, 27.80. HRMS (ESI+) calculated for C14H15O3 [M + H]+: 231.1016, found: 231.1013. HPLC purity: 99.0%.
4.14.3. (E)-4-(4-((1-(2,6-Difluorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxyphenyl)but-3-en-2-one (3)
TLC, CHCl3 : MeOH (95 : 5), Rf (0.4), white crystalline solid, yield: 81.79 mg (81.57%). 1H NMR (400 MHz, CDCl3) δ 7.96 (s, 1H), 7.57–7.45 (m, 2H), 7.21–7.10 (m, 5H), 6.64 (d, J = 16.2 Hz, 1H), 5.46 (s, 2H), 3.94 (s, 3H), 2.40 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 198.36, 149.86, 143.73, 143.33, 131.46, 128.37, 125.71, 122.81, 113.95, 113.40, 112.67, 112.64, 112.51, 112.48, 110.28, 91.41, 77.28, 77.02, 76.77, 66.07, 62.95, 59.10, 55.96, 53.43, 50.88, 44.31, 27.36. HRMS (ESI+) calculated for C20H32F2N2O3 [M + H]+: 386.1311, found: 368.2380. Purity: 94.78%.
4.14.4. (E)-4-(4-((1-(2-(Hydroxymethyl)-phenyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxy-phenyl)but-3-en-2-one (4)
TLC, CHCl3 : MeOH (95 : 5), Rf (0.3) yellowish solid, yield: 85.29 mg (86.42%). 1H NMR (400 MHz, CDCl3) δ 8.05 (s, 1H), 7.63 (d, J = 7.9 Hz, 1H), 7.48 (ddd, J = 16.2, 13.3, 5.2 Hz, 3H), 7.38 (d, J = 7.7 Hz, 1H), 7.12 (s, 2H), 7.09 (s, 1H), 6.61 (d, J = 16.2 Hz, 1H), 5.41 (s, 2H), 4.47 (s, 2H), 3.90 (s, 3H), 2.36 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 198.73, 149.77, 143.25, 135.86, 135.60, 131.61, 130.18, 129.20, 128.40, 125.75, 124.45, 122.78, 113.77, 110.27, 62.91, 61.85, 55.97, 29.70, 27.38. HRMS (ESI+) calculated for C21H22N3O4 [M + H]+: 380.1605, found: 380.1598. HPLC purity: 99.0%.
4.14.5. (3E)-4-(3-Methoxy-4-((1-(4-(phenyldiazenyl)phenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)but-3-en-2-one (5)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.4, orange colour solid, yield: 105.2 mg (89.19%). 1H NMR (400 MHz, CDCl3) δ 8.18 (s, 1H), 8.09 (d, J = 8.7 Hz, 2H), 7.95 (dd, J = 8.0, 1.5 Hz, 2H), 7.91 (d, J = 8.8 Hz, 2H), 7.57–7.51 (m, 3H), 7.48–7.43 (m, 1H), 7.13 (s, 2H), 7.10 (s, 1H), 6.64–6.59 (m, 1H), 5.43 (s, 2H), 3.93 (s, 3H), 2.37 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 198.45, 152.34, 149.93, 144.68, 143.24, 138.37, 131.75, 129.23, 128.35, 125.74, 124.39, 123.11, 122.81, 121.16, 120.98, 113.56, 110.12, 62.92, 56.00, 27.31. HRMS (ESI+) calculated for C26H24N5O3 [M + H]+: 454.1874, found: 454.1879. HPLC purity: 94.61%.
4.14.6. (E)-4-(3-Methoxy-4-((1-(4-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)but-3-en-2-one (6)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.5, creamy white solid, yield: 87.37 mg (88.52%). 1H NMR (400 MHz, CDCl3) δ 7.99 (d, J = 11.4 Hz, 1H), 7.61 (d, J = 8.5 Hz, 2H), 7.45 (d, J = 16.2 Hz, 1H), 7.17–7.09 (m, 2H), 7.08 (d, J = 7.6 Hz, 1H), 7.03–6.99 (m, 2H), 6.60 (d, J = 16.2 Hz, 1H), 5.40 (s, 2H), 3.90 (d, J = 4.9 Hz, 3H), 3.86 (d, J = 0.9 Hz, 3H), 2.36 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 198.30, 159.98, 149.98, 149.75, 144.19, 143.32, 130.39, 128.11, 125.64, 122.82, 122.27, 121.47, 114.83, 113.63, 110.22, 62.96, 55.97, 55.65, 27.37. HRMS (ESI+) calculated for C21H22N3O4 [M + H]+: 380.1605, found: 380.1599. HPLC purity: 99.0%.
4.14.7. (E)-4-(4-((1-(2,6-Dichlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxyphenyl)but-3-en-2-one (7)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.4, yellow crystalline solid, yield: 97.22 mg (89.52%). 1H NMR (400 MHz, CDCl3) δ 7.80 (s, 1H), 7.51 (d, J = 1.8 Hz, 1H), 7.49 (s, 1H), 7.46 (d, J = 7.2 Hz, 1H), 7.43 (t, J = 2.5 Hz, 1H), 7.10 (s, 2H), 7.09 (s, 1H), 6.61 (d, J = 16.2 Hz, 1H), 5.47 (s, 2H), 3.91 (s, 3H), 2.37 (s, 3H).; 13C NMR (100 MHz, CDCl3) δ 198.28, 150.06, 149.82, 143.94, 143.29, 133.88, 131.84, 128.89, 128.49, 125.75, 125.26, 122.75, 114.59, 110.46, 63.33, 56.00, 27.37. HRMS (ESI+) calculated for C20H18Cl2N3O3 [M + H]+: 418.0714, found: 418.0727. HPLC purity: 99.0%.
4.14.8. (E)-4-(3-Methoxy-4-((1-(3-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)but-3-en-2-one (8)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.6, creamy white solid, yield: 90.57 mg (91.77%). 1H NMR (400 MHz, CDCl3) δ 8.08 (s, 1H), 7.50–7.37 (m, 2H), 7.33 (t, J = 2.2 Hz, 1H), 7.26–7.23 (m, 1H), 7.13 (t, J = 4.8 Hz, 2H), 7.09 (s, 1H), 6.98 (dd, J = 8.4, 1.7 Hz, 1H), 6.61 (d, J = 16.2 Hz, 1H), 5.41 (s, 2H), 3.92 (s, 3H), 3.88 (s, 3H), 2.37 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 198.68, 160.53, 149.63, 144.18, 142.94, 137.78, 130.46, 128.17, 125.28, 122.37, 121.42, 115.01, 113.40, 112.78, 110.25, 106.39, 62.77, 55.97, 55.66, 27.53. HRMS (ESI+) calculated for C21H22N3O4 [M + H]+: 380.1605, found: 380.1602.
4.14.9. (E)-4-(3-Methoxy-4-((1-(2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)but-3-en-2-one (9)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.6, creamy white solid, yield: 82.90 mg (84.0%). 1H NMR (400 MHz, CDCl3) δ 8.23 (s, 1H), 7.78 (d, J = 7.9 Hz, 1H), 7.44 (dd, J = 15.7, 12.1 Hz, 2H), 7.17 (t, J = 7.5 Hz, 1H), 7.10 (dd, J = 14.5, 7.7 Hz, 4H), 6.61 (d, J = 16.2 Hz, 1H), 5.43 (s, 2H), 3.90 (d, J = 14.2 Hz, 6H), 2.37 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 198.47, 151.14, 150.11, 149.87, 143.35, 143.02, 130.16, 128.11, 125.60, 125.48, 125.41, 122.83, 121.36, 113.99, 112.22, 110.37, 62.64, 55.66, 27.03. HRMS (ESI+) calculated for C21H22N3O4 [M + H]+ 380.1605, found: 380.1599. HPLC purity: 99.0%.
4.14.10. (E)-N-(4-(4-((2-Methoxy-4-(3-oxobut-1-en-1-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)phenyl) benzamide (10)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.4, white solid, yield: 110.71 mg (90.84%). 1H NMR (400 MHz, MeOD) δ 8.31 (s, 1H), 7.99–7.92 (m, 4H), 7.78–7.72 (m, 2H), 7.60–7.48 (m, 5H), 7.21–7.14 (m, 3H), 6.65 (d, J = 16.2 Hz, 1H), 5.39 (s, 2H), 3.94 (s, 3H), 2.40 (s, 3H). 13C NMR (125 MHz, MeOD) δ 199.99, 167.59, 149.92, 149.66, 144.45, 144.05, 139.50, 134.43,, 132.59, 131.83, 128.50, 128.12, 127.46, 125.17, 123.05, 121.98, 121.61, 121.10, 113.49, 110.40, 62.41, 55.74, 26.82. HRMS (ESI+) calculated for C27H25N4O4 [M + H]+: 469.1870, found: 469.1874. HPLC purity: 85.24%.
4.14.11. (E)-4-(3-Methoxy-4-((1-(4-nitrophenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)but-3-en-2-one (11)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.4, yellow crystalline solid, yield: 90.81 mg (88.28%). 1H NMR (400 MHz, CDCl3) δ 8.44–8.38 (m, 2H), 8.20 (s, 1H), 8.00–7.94 (m, 2H), 7.44 (d, J = 16.2 Hz, 1H), 7.13–7.06 (m, 3H), 6.60 (d, J = 16.2 Hz, 1H), 5.41 (s, 2H), 3.91 (s, 3H), 2.36 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 198.18, 149.83, 143.06, 125.87, 125.59, 122.70, 121.03, 120.60, 113.75, 110.41, 62.84, 56.00, 27.41. HRMS (ESI+) calculated for C20H19N4O5 [M + H]+: 395.1350, found: 395.1353. Purity: 90.06%.
4.14.12. (E)-4-(3-Methoxy-4-((1-(p-tolyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)but-3-en-2-one (12)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.5, white solid, yield: 73.33 mg (77.57%). 1H NMR (400 MHz, CDCl3) δ 8.05 (s, 1H), 7.59 (d, J = 8.4 Hz, 2H), 7.45 (d, J = 16.2 Hz, 1H), 7.31 (d, J = 8.3 Hz, 2H), 7.13 (dd, J = 6.8, 4.9 Hz, 2H), 7.10–7.07 (m, 1H), 6.60 (d, J = 16.2 Hz, 1H), 5.40 (s, 2H), 3.90 (d, J = 5.1 Hz, 3H), 2.42 (s, 3H), 2.36 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 202.14, 153.71, 153.57, 148.09, 147.14, 142.93, 138.49, 134.06, 132.03, 129.47, 126.64, 125.09, 124.36, 117.47, 114.04, 66.79, 59.85, 31.14, 24.92. HRMS (ESI+) calculated for C21H22N3O3 [M + H]+: 364.1656, found: 364.1653. HPLC purity: 96.70%.
4.14.13. (E)-4-(4-((1-(3,4-bis(Trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxy phenyl)but-3-en-2-one (13)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.4, white solid, yield: 108.55 mg (85.94%). 1H NMR (400 MHz, CDCl3) δ 8.24 (d, J = 7.6 Hz, 3H), 7.96 (s, 1H), 7.45 (d, J = 16.2 Hz, 1H), 7.15–7.06 (m, 3H), 6.62 (d, J = 16.2 Hz, 1H), 5.43 (s, 2H), 3.93 (s, 3H), 2.37 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 149.61 (s), 143.05 (s), 128.65 (s), 125.89 (s), 122.68 (s), 122.35 (s), 120.97 (s), 120.45 (s), 113.82 (s), 110.46 (s), 77.33 (s), 77.02 (s), 76.70 (s), 62.88 (s), 55.99 (s), 27.39 (s). HRMS (ESI+) calculated for C22H18F6N3O3 [M + H]+: 486.1247, found: 486.1236. HPLC purity: 99.0%.
4.14.14. Ethyl(E)-4-(4-((2-methoxy-4-(3-oxobut-1-en-1-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)benzoate (14)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.45, white solid, yield: 89.22 mg (81.38%). 1H NMR (400 MHz, CDCl3) δ 8.24–8.18 (m, 2H), 8.16 (s, 1H), 7.83 (d, J = 8.8 Hz, 2H), 7.45 (d, J = 16.2 Hz, 1H), 7.12 (s, 2H), 7.09 (s, 1H), 6.61 (d, J = 16.2 Hz, 1H), 5.42 (s, 2H), 4.42 (q, J = 7.1 Hz, 2H), 3.91 (d, J = 5.9 Hz, 3H), 2.37 (s, 3H), 2.17 (s, 2H), 1.59 (s, 2H), 1.42 (t, J = 7.1 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 198.10, 165.24, 149.94, 145.06, 144.97, 143.25, 143.16, 139.90, 139.79, 131.33, 130.86, 128.45, 125.78, 122.74, 121.06, 119.96, 113.57, 110.41, 62.94, 61.22, 55.65, 27.03, 14.10. HRMS (ESI+) calculated for C23H24N3O5 [M + H]+: 422.1710, found: 422.1716. HPLC purity: 99.0%.
4.14.15. Synthesis of (E)-4-(4-((1-(2-chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxyphenyl)but-3-en-2-one (15)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.3, white solid, yield: 89.92 mg (90.16%). 1H NMR (400 MHz, CDCl3) δ 8.09 (s, 1H), 7.74–7.39 (m, 5H), 7.11 (d, J = 15.7 Hz, 3H), 6.61 (d, J = 16.2 Hz, 1H), 5.44 (s, 2H), 3.91 (s, 3H), 2.37 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 198.15, 150.00, 143.60, 143.18, 134.84, 130.80, 130.71, 128.57, 128.30, 127.94, 127.78, 125.71, 125.13, 122.74, 114.06, 110.43, 63.07, 55.82, 27.19. HRMS (ESI+) calculated for C20H19ClN3O3 [M + H]+: 384.1109, found: 384.1114. HPLC purity: 99.0%.
4.14.16. (E)-4-(4-((1-([1,1′-Biphenyl]-4-yl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxyphenyl)-but-3-en-2-one (16)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.6, white solid, yield: 91.58 mg (82.75%). 1H NMR (400 MHz, CDCl3) δ 8.16 (s, 1H), 7.80 (dd, J = 26.0, 8.6 Hz, 6H), 7.65 (d, J = 7.2 Hz, 3H), 7.51 (t, J = 7.5 Hz, 3H), 7.18–7.11 (m, 3H), 6.64 (d, J = 16.2 Hz, 1H), 5.46 (s, 2H), 3.95 (d, J = 4.8 Hz, 4H), 2.40 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 198.23, 149.84, 144.56, 143.25, 142.01, 139.58, 136.03, 129.01, 128.40, 128.03, 127.10, 125.73, 122.80, 121.17, 120.90, 113.79, 110.36, 63.03, 56.08, 56.01, 55.16, 40.62, 29.78, 27.36. HRMS (ESI+) calculated for C15H6F2N2O2 [M + H]+: 285.0470, found: 285.0482. HPLC purity: 86.04%.
4.14.17. (E)-4-(4-((2-Methoxy-4-(3-oxobut-1-en-1-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)benzaldehyde (17)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.35, creamy white solid, yield: 89.71 mg (91.38%). 1H NMR (400 MHz, CDCl3) δ 10.09 (s, 1H), 8.22 (d, J = 5.0 Hz, 1H), 8.08 (d, J = 8.6 Hz, 2H), 7.97 (d, J = 8.5 Hz, 2H), 7.47 (d, J = 16.2 Hz, 1H), 7.16–7.10 (m, 3H), 6.63 (d, J = 16.2 Hz, 1H), 5.44 (s, 2H), 3.94 (s, 3H), 2.39 (s, 3H); 13C NMR (125 MHz, CDCl3) δ 198.32, 190.58, 149.73, 149.67, 145.11, 143.21, 140.82, 136.14, 131.37, 128.39, 125.74, 122.75, 121.10, 120.56, 113.59, 110.25, 62.78, 55.95, 29.70, 27.39. HRMS (ESI+) calculated for C21H20N3O4 [M + H]+ 378.1448, found: 378.1444. HPLC purity: 94.64%.
4.14.18. (E)-4-(3-Methoxy-4-((1-phenyl-1H-1,2,3-triazol-4-yl)methoxy)phen-yl)but-3-en-2-one (18)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.5, white crystalline solid, yield: 75.92 mg (83.53%). 1H NMR (400 MHz, CDCl3) δ 8.07 (d, J = 10.6 Hz, 1H), 7.71 (d, J = 8.1 Hz, 2H), 7.52 (dd, J = 8.3, 7.0 Hz, 2H), 7.47–7.39 (m, 2H), 7.11 (dd, J = 12.8, 11.2 Hz, 3H), 6.60 (d, J = 16.2 Hz, 1H), 5.41 (s, 2H), 3.91 (s, 3H), 2.36 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 198.13, 149.88, 144.50, 143.26, 136.98, 129.81, 128.96, 128.30, 125.71, 122.80, 121.25, 120.65, 113.75, 110.32, 63.14, 56.08, 27.48. HRMS (ESI+) calculated for C20H20N3O3 [M + H]+: 350.1494, found: 350.1500. Purity: 95.70%.
4.14.19. (E)-4-(4-((1-(3-Bromo-5-fluorophenyl)-1H-1,2,3-triazol-4-yl)meth-oxy)-3-methoxy-phenyl)-but-3-en-2-one (19)
TLC, CHCl3 : MeOH (95 : 5), Rf 0.5, creamy white solid, yield: 99.27 mg (85.66%). 1H NMR (400 MHz, CDCl3) δ 7.96 (s, 1H), 7.57–7.45 (m, 2H), 7.21–7.10 (m, 5H), 6.64 (d, J = 16.2 Hz, 1H), 5.46 (s, 2H), 3.94 (s, 3H), 2.40 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 198.36, 149.86, 143.73, 143.33, 131.46, 128.37, 125.71, 122.81, 113.95, 113.40, 112.67, 112.64, 112.51, 112.48, 110.28, 91.41, 77.28, 77.02, 76.77, 66.07, 62.95, 59.10, 55.96, 53.43, 50.88, 44.31, 27.36. HRMS (ESI+) calculated for C20H18BrFN3O3 [M + H]+: 446.0510, found: 446.0506. Purity: 97.53%.
4.14.20. (E)-4-(4-((1-(2-Hydroxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)-3-methoxyphenyl)but-3-en-2-one (20)
TLC, CHCl3 : MeOH (9 : 1), Rf 0.5, yellowish white solid, yield: 82.17 mg (86.44%). 1H NMR (400 MHz, CDCl3) δ 8.48 (d, J = 5.1 Hz, 1H), 7.69 (dd, J = 8.0, 1.6 Hz, 1H), 7.55 (t, J = 8.1 Hz, 1H), 7.37–7.27 (m, 1H), 7.19 (dt, J = 13.2, 4.9 Hz, 3H), 7.02 (ddd, J = 15.4, 8.3, 4.2 Hz, 2H), 6.65 (d, J = 16.2 Hz, 1H), 5.38 (s, 2H), 3.91 (d, J = 5.2 Hz, 3H), 2.40 (d, J = 5.2 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 198.17, 149.89, 149.66, 149.32, 144.06, 143.10, 129.94, 125.84, 122.67, 120.95, 120.30, 119.57, 119.52, 113.92, 110.49, 62.89, 55.98, 27.35. HRMS (ESI+) calculated for C20H19N3O4 [M + H]+: 366.1448, found: 366.1450. HPLC purity: 99.0%.
Conflicts of interest
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We are thankful to the Ministry of AYUSH for the financial assistance in the GAP-2143 project. We are also thankful to Deepika Singh for NMR spectral data analysis. The manuscript bears the Institutional publication No. IIIM/2164/2017.
Footnotes
†Electronic supplementary information (ESI) available: 1D and 2D NMR and HRESIMS spectra of all new compounds. NMR spectral data of compounds (1–20) along with HRMS spectral data. See DOI: 10.1039/c7md00267j
References
- de Bernardi M., Vidari G., Vita-Finzi P. Phytochemistry. 1976;15(11):1785–1786. [Google Scholar]
- Elias G., Rao M. N. A. Eur. J. Med. Chem. 1988;23(4):379–380. [Google Scholar]
- Wang Y.-J., Pan M.-H., Cheng A.-L., Lin L.-I., Ho Y.-S., Hsieh C.-Y., Lin J.-K. J. Pharm. Biomed. Anal. 1997;15(12):1867–1876. doi: 10.1016/s0731-7085(96)02024-9. [DOI] [PubMed] [Google Scholar]
- Hampannavar G. A., Karpoormath R., Palkar M. B., Shaikh M. S. Bioorg. Med. Chem. 2016;24(4):501–520. doi: 10.1016/j.bmc.2015.12.049. [DOI] [PubMed] [Google Scholar]
- Rajakumar D. V., Rao M. N. A. Mol. Cell. Biochem. 1994;140(1):73–79. doi: 10.1007/BF00928368. [DOI] [PubMed] [Google Scholar]
- Parihar V. K., Dhawan J., Kumar S., Manjula S. N., Subramanian G., Unnikrishnan M. K., Rao C. M. Chem.-Biol. Interact. 2007;170(1):49–58. doi: 10.1016/j.cbi.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Liu Y., Dolence J., Ren J., Rao M. N. A., Sreejayan N. J. Cardiovasc. Pharmacol. 2008;52(5):422–429. doi: 10.1097/FJC.0b013e31818aed93. [DOI] [PubMed] [Google Scholar]
- Kubra I. R., Murthy P. S., Rao L. J. Food Sci. 2013;78(1):M64–M69. doi: 10.1111/j.1750-3841.2012.03009.x. [DOI] [PubMed] [Google Scholar]
- Hampannavar G. A., Karpoormath R., Palkar M. B., Shaikh M. S., Chandrasekaran B. ACS Med. Chem. Lett. 2016;7(7):686–691. doi: 10.1021/acsmedchemlett.6b00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuzaki J., Taniguchi M., Bastow K. F., Nakagawa-Goto K., Morris-Natschke S. L., Itokawa H., Baba K., Lee K.-H. Bioorg. Med. Chem. 2007;15(18):6193–6199. doi: 10.1016/j.bmc.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuzaki J., Bastow K. F., Nakagawa-Goto K., Nakamura S., Itokawa H., Lee K.-H. J. Nat. Prod. 2006;69(10):1445–1449. doi: 10.1021/np060252z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N., Yamagami C., Tokuda H., Konoshima T., Okuda Y., Okuda M., Mukainaka T., Nishino H., Saito Y. Cancer Lett. 1998;134(1):37–42. doi: 10.1016/s0304-3835(98)00239-0. [DOI] [PubMed] [Google Scholar]
- Tatsuzaki J., Nakagawa-Goto K., Tokuda H., Lee K.-H. J. Asian Nat. Prod. Res. 2010;12(3):227–232. doi: 10.1080/10286021003591617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N., Yamagami C., Tokuda H., Okuda Y., Ichiishi E., Mukainaka T., Nishino H., Saito Y. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2000;464(2):247–254. doi: 10.1016/s1383-5718(99)00198-9. [DOI] [PubMed] [Google Scholar]
- Kumar S., Pathania A. S., Satti N. K., Dutt P., Sharma N., Mallik F. A., Ali A. Eur. J. Med. Chem. 2015;92:236–245. doi: 10.1016/j.ejmech.2014.12.047. [DOI] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Ca-Cancer J. Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Courtney K. D., Corcoran R. B., Engelman J. A. J. Clin. Oncol. 2010;28(6):1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J. R., Konicek B. W., McNulty A. M., Wang Z., Houck K., Allen S., Paul J. D., Hbaiu A., Goode R. G., Sandusky G. E. J. Biol. Chem. 2000;275(32):24500–24505. doi: 10.1074/jbc.M003145200. [DOI] [PubMed] [Google Scholar]
- Dolcet X., Llobet D., Pallares J., Matias-Guiu X. Virchows Arch. 2005;446(5):475–482. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- Shukla S., MacLennan G. T., Marengo S. R., Resnick M. I., Gupta S. Prostate. 2005;64(3):224–239. doi: 10.1002/pros.20217. [DOI] [PubMed] [Google Scholar]
- Ur Rasool R., Rah B., Amin H., Nayak D., Chakraborty S., Rawoof A., Mintoo M. J., Yousuf K., Mukherjee D., Kumar L. D. Sci. Rep. 2015;6:1–16. doi: 10.1038/srep18800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J. P. Nat. Rev. Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Hudson B. D., Kulp K. S., Loots G. G. Briefings Funct. Genomics. 2013;12:397–410. doi: 10.1093/bfgp/elt021. [DOI] [PubMed] [Google Scholar]
- Lee Y.-C., Lin H.-H., Hsu C.-H., Wang C.-J., Chiang T.-A., Chen J.-H. Eur. J. Pharmacol. 2010;632(1):23–32. doi: 10.1016/j.ejphar.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Curran S., Murray G. I. Eur. J. Cancer. 2000;36(13):1621–1630. doi: 10.1016/s0959-8049(00)00156-8. [DOI] [PubMed] [Google Scholar]
- Umbas R., Isaacs W. B., Bringuier P. P., Schaafsma H. E., Karthaus H. F. M., Oosterhof G. O. N., Debruyne F. M. J., Schalken J. A. Cancer Res. 1994;54(14):3929–3933. [PubMed] [Google Scholar]
- Rah B., Lone S. H., Rasool R. U., Farooq S., Nayak D., Chikan N. A., Chakraborty S., Behl A., Mondhe D. M., Goswami A. J. Med. Chem. 2015;58(8):3432–3444. doi: 10.1021/jm501942m. [DOI] [PubMed] [Google Scholar]
- Amin H., Nayak D., Chakraborty S., Kumar A., Yousuf K., Sharma P. R., Ahmed Z., Sharma N., Magotra A., Mukherjee D. Mol. Carcinog. 2016;55(5):864–881. doi: 10.1002/mc.22328. [DOI] [PubMed] [Google Scholar]
- Rah B., Amin H., Yousuf K., Khan S., Jamwal G., Mukherjee D., Goswami A. PLoS One. 2012;7(9):e44039. doi: 10.1371/journal.pone.0044039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rah B., Rasool R. U., Nayak D., Yousuf S. K., Mukherjee D., Kumar L. D., Goswami A. Autophagy. 2015;11(2):314–331. doi: 10.1080/15548627.2015.1017182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.