Abstract
MicroRNA (miR)-1 is associated with various human malignancies through repressing tumor growth, migration and angiogenesis. Recently, high-throughput transcriptional profiling confirmed that miR-1 is markedly downregulated in metastatic colorectal cancer; however, its biological functions and the specific underlying mechanisms in colorectal cancer (CRC) require further investigation. In this study, the expression of miR-1 in 111 CRC and paired normal tissue samples was measured using quantitative polymerase chain reaction analysis, and the association between miR-1 expression and clinical characteristics was evaluated. miR-1 was found to be significantly downregulated in CRC tissues compared with paired normal tissues, and in CRC cell lines compared with non-cancer cells (P<0.001), and was negatively associated with tumor size (P=0.001), differentiation (P=0.011), lymph node metastasis (P=0.001) and TNM stage (P=0.001). Further experiments revealed that miR-1 inhibited the migration and invasion of HCT116 and ClonA1 cells, and inhibited cell proliferation by affecting the cell cycle. Vascular endothelial growth factor (VEGF) was found to be a potential target of miR-1 by biological prediction, and further investigation confirmed that miR-1 significantly inhibited the expression and paracrine function of VEGF. In CRC tissues, the expression of VEGF was negatively correlated with miR-1. The low expression of miR-1 in CRC may be one of the reasons for the abnormally high expression of VEGF; the upregulation of miR-1 expression may inhibit cancer progression by downregulating VEGF. These findings indicate that treatment with miR-1 may be a novel method of tumor suppression, and provide a theoretical and experimental basis for the further targeted treatment of CRC through the regulation of miR-1 and VEGF expression.
Keywords: colorectal cancer, miR-1, VEGF, oncosuppressive
Introduction
The incidence of colorectal cancer (CRC) is increasing, and CRC currently represents a major cause of cancer-related morbidity and mortality worldwide, with high incidence rates in Westernized societies and increasing rates in developing countries (1,2). As the majority of the patients present with advanced disease, such as the presence of liver metastases, at the time of diagnosis, the scope of therapeutic intervention is significantly limited (3). Various factors have been confirmed to participate in this progression, such as the silencing of tumor-suppressor genes, the hyperactivation or overexpression of proto-oncogenes, and the dysregulation of genes that are associated with cell growth, apoptosis or transformation (4–6).
MicroRNAs (miRNAs/miRs) are a recently characterized class of small non-coding RNA molecules of 20–22 nucleotides. Mature miRNAs can specifically bind to the 3′-untranslated region (3′-UTR) of target cell mRNAs, resulting in mRNA degradation or the inhibition of translation. Post-transcriptional regulation of gene expression by miRNAs is an important characteristic of the cell differentiation process, and it has been predicted that there are numerous as yet undiscovered miRNAs in the genome of humans and other higher vertebrates (7,8). In recent decades, studies have validated that miRNAs are key regulators of diverse cellular processes, including apoptosis, proliferation, differentiation, metabolism and immunity (9–11).
miR-1 is a muscle-enriched miRNA that inhibits the proliferation of progenitor cells and promotes myogenesis (12,13). The downregulated expression of miR-1 has also been identified in lung, liver, breast, prostate and kidney cancer. The restoration of miR-1 expression in cancer cell lines was found to markedly reversed their tumorigenic properties, such as growth, clone formation, migration, invasion and tumor formation ability in nude mice (14,15). The decreased expression of miR-1 has been suggested to be associated with liver metastasis, whereas its function and the underlying mechanism in colon cancer require further investigation (16,17). Vascular endothelial growth factor (VEGF)-A and VEGF-C/D are major factors affecting angiogenesis and/or lymphangiogenesis (18). Angiogenesis plays a crucial role in prenatal development, wound healing, chronic inflammation, angiogenesis and lymphangiogenesis, and promotes the metastasis and progression of various carcinomas (19–21). The ectopic expression of VEGF has been demonstrated to be closely associated with cell proliferation, invasion and the metastatic potential of colon cancer cells, which contributes to cancer progression, whereas anti-VEGF-based antiangiogenic drugs, including bevacizumab, aflibercept, ramucirumab and tyrosine kinase inhibitors, are routinely used for the treatment of various types of tumor (22).
In the present study, we examined the expression of miR-1 in CRC tissues and cell lines using immunohistochemistry and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analyses. The effect of miR-1 on cell growth, apoptosis, migration and invasion were assessed by several assays. Furthermore, VEGF was predicted to be a target protein of miR-1 by bioinformatic analysis, and a negative association of miR-1 and VEGF expression was observed, which suggests that miR-1 downregulates the expression of VEGF. It is well established that high expression of VEGF promotes biological behaviors such as cell proliferation, invasion and metastasis in colon cancer, leading to cancer progression. Collectively, this study demonstrated that decreased expression of miR-1 may cause increased expression of VEGF, which sets a theoretical and practical basis for miR-1 as a biological marker for CRC diagnosis and targeted therapy.
Materials and methods
Patients and cell lines
A total of 111 samples of cancer and paracancerous tissues were collected, cut into small sections, immediately frozen in liquid nitrogen and stored at −80°C. The patients included 45 men and 66 women (median age, 61.3 years; range, 27–85 years). All tissues were obtained from patients who underwent surgery for CRC between 2007 and 2012 at the First Hospital of China Medical University (Shenyang, China). All diagnoses were histologically confirmed. The study was approved by the Ethics Committee of China Medical University (Shenyang, China), and written informed consent was obtained prior to sample collection. In addition, the human peritoneal mesothelial cell line HMrSV5, and colorectal cancer cell lines, including HCT-116, CL-187, ClonA1, HT-29 and SW-620, were cultured in HyClone RPMI-1640 medium (GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) and maintained at 37°C in 5% CO2. We failed to identify a normal colon epithelial cell line with cell identification. Therefore, the human peritoneal mesothelial cell line (HMrSV5) was used as a control. The HMrSV5 cell line was derived from normal peritoneal cells and it did not display malignant biological behavior. The HCT-116, SW-480, SW-620 and HT-29 cell lines were obtained from the Cell Repository of the Typical Culture Preservation Committee of the Chinese Academy of Sciences/The Cell Resource Center of the Shanghai Academy of Life Sciences, Chinese Academy of Sciences. The ClonA1, CL-187 and HMrSV5 cell lines were gifts from the Department of Cell Biology, China Medical University (Shenyang, China).
RNA extraction and RT-qPCR
Total RNA was isolated from tumor tissues or cells with Invitrogen™ TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the miRNAs were then isolated using the Ambion® mirVana miRNA Isolation kit (Thermo Fisher Scientific, Inc.). In order to detect the expression of miR-1, stem-loop RT-qPCR was performed. The primers used in this study were as follows: RT primer for miR-1, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATACATAC-3′; RT primer for U6 snRNA: 5′-AACGCTTCACGAATTTGCGT-3′; miR-1 forward primer, 5′-ACACTCCAGCTGGGTGGAATGTAAAGAAGT-3′; miR-1 reverse primer, 5′-TGGTGTCGTGGAGTCG-3′; U6 forward primer, 5′-CTCGCTTCGGCAGCACA-3′, U6 reverse primer, 5′-AACGCTTCACGAATTTGCGT-3′. The expression of VEGF was normalized to GAPDH, and the primers used in RT-qPCR were as follows: VEGF forward primer, 5′-CAAGGCCAGCACATAGGAGAG3-3′, VEGF reverse primer, 5′-CCTCGGCTTGTCACATCTTGC-3′; GAPDH forward primer, 5′-GACTGTGGATGGCCCCTCCGG-3′, GAPDH reverse primer, 5′-AGGTGGAGGAGTGGGTGTCGC-3′. The RT-qPCR procedure in this study was performed as previously described (23).
Cell transfection
miR-1-overexpressing and control cell lines were constructed by infecting the cells with lentiviral miR-1 and control vector (Shanghai Genechem Co., Ltd., Shanghai, China), according to the manufacturer's instructions. The expression of miR-1 in the stable cell lines was determined with RT-qPCR analysis.
ELISA
miR-1-overexpressing or control cells (1×105) were seeded in 6-well plates. After 36 h, the supernatants were collected and the levels of VEGF were assessed with a human VEGF ELISA kit (R&D Systems, Inc., Minneapolis, MN, USA) according to the manufacturer's instructions.
Western blotting
Harvested cells were lysed in RIPA lysis buffer containing protease inhibitor. Total cellular protein (30 mg/lane) was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk and incubated with rabbit anti-VEGF antibody at a dilution of 1:1,000 (cat. no. ab155944; Abcam, Cambridge, MA, USA), or a rabbit anti-GAPDH monoclonal antibody at a dilution of 1:3,000 (cat. no. ab181602; Abcam), or a rabbit anti-VEGF receptor 2 (VEGFR-2) antibody at a dilution of 1:500 (cat. no. 26415-1-AP; ProteinTech Group, Inc., Chicago, IL, USA). A secondary antibody was then incubated with the membrane for 2 h. Bound proteins were visualized using electrochemiluminescence (Pierce; Thermo Fisher Scientific, Inc.) and detected with a bio-imaging system (DNR Bio-Imaging Systems, Israel).
Immunohistochemistry
Paraffin-embedded sections (4-µm) were deparaffinized in three xylene washes, and rehydrated through a graded alcohol series. Following antigen retrieval with 10 mM sodium citrate buffer, the sections were blocked with goat serum for 1 h at room temperature. Then sections were incubated with a VEGF antibody (Abcam) overnight at 4°C or a rabbit anti-VEGFR-2 antibody (1:500) (Proteintech), and the expression of VEGF and VEGFR-2 were examined with the UltraSensitive™ SP kit (Maixin-Bio, Fuzhou, Fujian, China) according to the manufacturer's instructions.
Migration and invasion assays
Cell migration was assessed using 24-well 8-µm-pore Transwell chambers (Corning Costar, Corning, NY, USA) according to the manufacturer's instructions. For the invasion assay, the insert membranes were coated with diluted BD Matrigel™ (1:20) (BD Biosciences, Franklin Lakes, NJ, USA). A total of 2×105 cells in 100-µl serum-free medium were added to the upper chamber, and 600 µl 10% FBS medium was added to the corresponding lower chamber. After 24 h of incubation, the cells were fixed with methanol and stained with 0.1% crystal violet. Non-invading cells in the upper surface of the chamber were removed with a cotton swab, and invading cells on the lower membrane surface were photographed with an inverted fluorescence microscope (Olympus DP80; Olympus Corp., Tokyo, Japan) and counted. Six random fields at a magnification of ×100 for each insert were counted. Measurements were conducted in triplicate in three separate experiments. For the migration assay, the procedures were similar, except that 2×105 cells were added to the inserts without Matrigel pre-coating.
Wound scratch assay
miR-1-overexpressing or control cells (1×104) were seeded in 24-well plates. After cells had grown to a confluence of 80–90%, a scratch was produced in the cell monolayer using a 200-µl pipette. The medium was discarded and cells were rinsed with phosphate-buffered saline (PBS) three times to remove the cell debris, followed by the addition of fresh culture medium. Wound areas were marked and photographed at different time points (0, 24 and 48 h) using a phase-contrast microscope and an Olympus DP74 color camera (Olympus Corp).
CCK-8 assay and cell growth curve
miR-1-overexpressing or control cells (5×103) were seeded in 96-well plates, and the effect of miR-1 on cell proliferation was measured by the colorimetric water-soluble tetrazolium salt (WST) assay from a CCK-8 kit (Beyotime Institute of Biotechnology, Haimen, China). The absorbance at 450 nm was measured with a microplate reader. The extent of proliferation was evaluated every 24 h for 5 days.
Cell cycle distribution analysis
miR-1-overexpressing or control cells (1×105) were seeded in 6-well plates. After 36 h, the cells were harvested by trypsinization, collected and washed with PBS, and fixed with ethanol for 1 h at 4°C. After washing with PBS three times, the cells were resuspended in 0.2 ml RNase A buffer (1 mg/ml) at 37°C for 30 min and stained with 0.3 ml propidium iodide (PI) buffer (50 µl/ml). The stained cells were then analyzed for DNA content by FACS flow cytometry.
Assessment of apoptosis by Annexin V-FITC
miR-1- overexpressing or control cells (1×105) were seeded in 6-well plates. After 36 h, the cells were digested with trypsin and collected by centrifugation. The cells were washed with cold PBS three times, resuspended in 100 µl binding buffer, and stained with 5 µl FITC-conjugated Annexin V (10 mg/ml) and 10 µl PI (50 mg/ml) in the dark for 20 min at room temperature. Then, 300 µl binding buffer was added and apoptotic cells were analyzed with FACS flow cytometry.
Statistical analysis
The data from three independent experiments are expressed as the mean ± standard deviation and were processed using SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA). The expression levels of miR-1 in CRC and paired adjacent normal tissue samples were compared by Wilcoxon's paired test. The Student's t-test was used to evaluate the association between the RT-qPCR results for miR-1 expression and clinicopathological factors as documented in Table I. Chi-square tests were used to determine the association between the expression of miR-1 and VEGF in primary CRC as shown in Table II. A P-value of <0.05 was considered to indicate a statistically significant difference.
Table I.
Relationship between the clinicopathological parameters and miR-1 expression in primary CRC cases (N=111).
| Expression of miR-1a | ||||
|---|---|---|---|---|
| Characteristics | No. of cases | Mean | 95% CI | P-value |
| Age (years) | ||||
| <60 | 48 | −2.441 | −3.42–1.65 | 0.076 |
| ≥60 | 63 | −0.903 | ||
| Sex | ||||
| Male | 45 | −1.303 | −1.09–2.39 | 0.458 |
| Female | 66 | −1.956 | ||
| Tumor size (cm) | ||||
| <5 | 78 | −2.594 | −5.21–1.70 | 0.001 |
| ≥5 | 33 | 0.858 | ||
| Differentiation | ||||
| Well + moderate | 33 | 0.094 | 0.55–4.18 | 0.011 |
| Poor | 78 | −2.271 | ||
| Lymph node status | ||||
| Negative | 63 | −3.082 | −5.01–1.91 | 0.001 |
| Positive | 48 | 0.419 | ||
| Metastasis | ||||
| Negative | 95 | −1.543 | −2.26–2.61 | 0.888 |
| Positive | 16 | −1.717 | ||
| TNM stage | ||||
| I + II | 55 | −3.122 | −4.69–1.47 | 0.001 |
| III + IV | 56 | −0.042 | ||
Mean of the relative expression of miR-1 is expressed as log2 (fold change) value and 95% confidence interval (CI). CRC, colorectal cancer; microRNA-1, miR-1.
Table II.
Association between the expression of miR-1 and VEGF in the primary CRC cases (N=111).
| Expression of miR-1 | |||
|---|---|---|---|
| VEGF | High (n=28) | Low (n=83) | P-value |
| Positive | 5 | 72 | <0.001 |
| Negative | 23 | 9 | |
CRC, colorectal cancer; microRNA-1, miR-1; VEGF, vascular endothelial growth factor.
Results
miR-1 expression in colon cancer is low
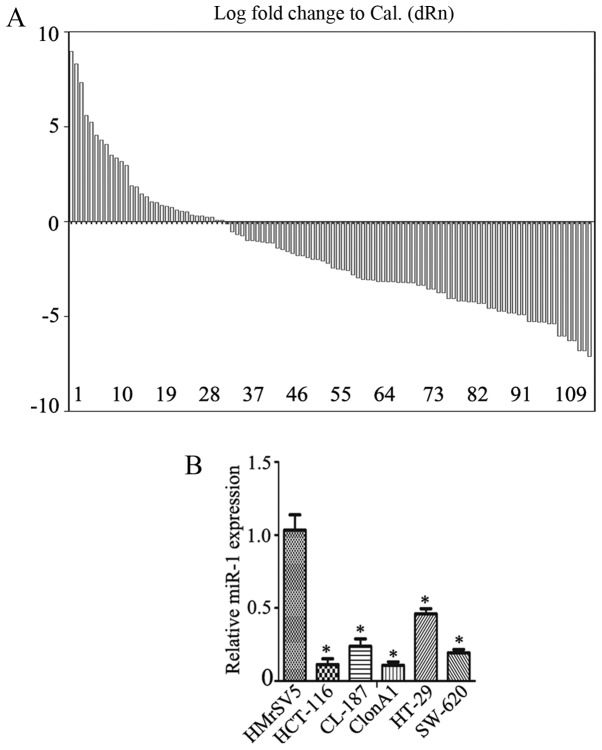
First, the expression of miR-1 in colorectal cancer tissues and their pair-matched adjacent non-tumor tissues was examined using RT-qPCR. The data revealed a decrease in miR-1 expression in 70.3% of cases, with a median change of ~1.57-fold (Fig. 1A; P<0.01), a fold change relative to the adjacent normal tissue. miR-1 expression was also measured in several colorectal cancer cell lines, including HCT-116, CL-187, ClonA1, HT-29 and SW-620, using RT-qPCR. The data revealed that the expression of miR-1 was significantly downregulated in HCT-116 (0.07±0.02-fold), CL-187 (0.41±0.03-fold), ClonA1 (0.04±0.01-fold), HT-29 (0.60±0.01-fold) and SW-620 (0.38±0.04-fold) cells compared with the human peritoneal mesothelial cell line HMrSV5 (Fig. 1B), one-way ANOVA was used to analyze data in Fig. 1B.
Figure 1.
Analysis of miR-1 expression in CRC. (A) The relative expression levels of miR-1 in 111 pairs of human CRC specimens were detected by RT-qPCR. (B) The expression of miR-1 in 5 CRC cell lines (HCT-116, CL-187, ClonA1, HT-29 and SW-620) and a normal peritoneal mesothelial cell line (HMrSV5) was examined by RT-qPCR (*P<0.05 compared with the HMrSV5 cells). miR-1, microRNA-1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CRC, colorectal cancer.
The miR-1 expression and the clinicopathological characteristics of the CRC specimens are summarized in Table I. The expression of miR-1 was significantly correlated with tumor size, degree of differentiation, lymph node metastasis and TNM stage (P=0.001, P=0.011, P=0.001 and P=0.001, respectively). There were no significant correlations with the other clinicopathological characteristics considered, including age and gender. These results suggest that miR-1 may act as a tumor suppressor in CRC.
Ectopic expression of miR-1 inhibits the growth of CRC cells
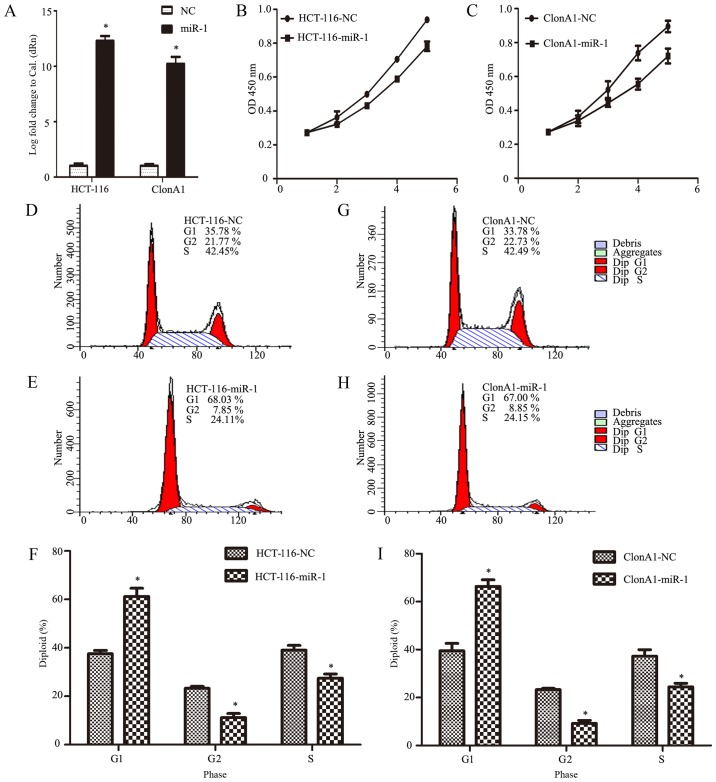
To investigate the biological function of miR-1 in the development and progression of CRC, HCT-116 and ClonA1 cells were infected with an miR-1 lentivirus to generate miR-1-overexpressing cells. Fig. 2A shows that the expression of miR-1 was effectively altered, achieving an increase of >10,000-fold (P<0.0001, n=3, Fig. 2A).
Figure 2.
Ectopic expression of miR-1 inhibits the growth of CRC HCT-116 and ClonA1 cells. (A) The relative expression levels of miR-1 in stable cell lines were determined by RT-qPCR. miR-1 expression levels were calculated by the log10 (2−ΔΔCt) method and normalized to U6 small nuclear RNA. Data represent the means from three independent measurements. (B and C) Effect of miR-1 on cell proliferation, as assessed by a Cell Counting Kit-8 assay. (D-I) Flow cytometry was applied to assess the difference in cell cycle distribution between miR-1-overexpressing and NC cells (*P<0.05 compared with NC). miR-1, microRNA-1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CRC, colorectal cancer; NC negative control.
To determine the role of miR-1 in CRC, we investigated the effect of miR-1 on the proliferation of cells with CCK-8 and cell count assays at different time points in vitro. The results demonstrated that the growth of both HCT-116 and ClonA1 miR-1-overexpressing cells was markedly impaired compared with the negative control cells (Fig. 2B and C). To further confirm the reason for this change, we assessed the cell cycle phase distribution with flow cytometric analysis. It was observed that, when ectopically elevating miR-1 expression in HCT-116 and ClonA1 cells, the transition from the G1 to the S phase was significantly inhibited. As shown in Fig. 2D-I, 27.51 and 24.41% of miR-1-overexpressing HCT-116 and ClonA1 cells, respectively, were in the S phase, whereas 39.12 and 37.23% of the HCT-116 and ClonA1 control cells, respectively, were in the S phase. Moreover, the cell population in the G1 phase of the cell cycle was markedly larger in the miR-1-overexpressing cells (61.28 and 66.37%, respectively) compared with that in the negative control cells (37.58 and 39.48%, respectively). Therefore, these results indicate that miR-1 arrested the cells at the G1 phase of the cell cycle.
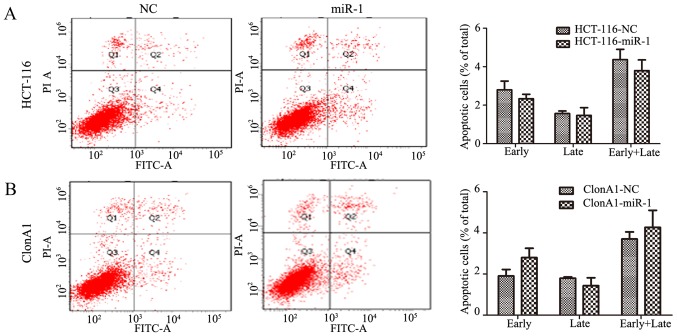
Considering that apoptosis is also a key factor affecting tumor growth, Annexin V and PI staining was performed to assess whether miR-1 increased the rate of apoptosis. Interestingly, no obvious change was observed in miR-1-overexpressing HCT-116 and ClonA1 cells compared with the negative control cells (P>0.05, Fig. 3). This finding indicated that the effect of miR-1 on tumor proliferation is mainly produced through inhibiting cell cycle transition.
Figure 3.
Annexin V-FITC apoptosis assay to assess the differences between miR-1-overexpressing and NC cells in (A) HCT-116 and (B) ClonA1 cell lines. miR-1, microRNA-1; NC, negative control.
Overexpression of miR-1 suppresses tumor cell migration and motility in vitro
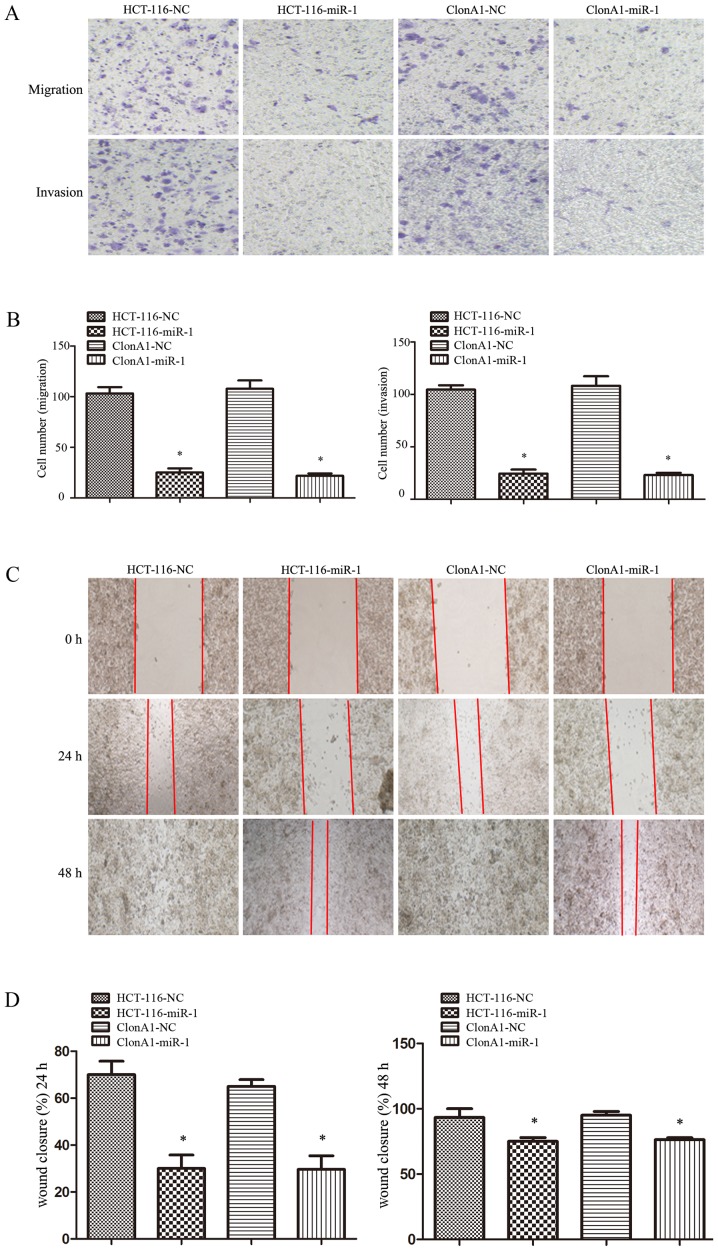
The ability to invade and migrate through the basement membrane is the most important property of metastatic cancer cells. To assess the effect of miR-1 on tumor cell invasion and motility, Transwell and wound-healing assays were performed. For the Transwell assay, miR-1-overexpressing or control cells were seeded on the upper chamber of 24-well Transwell plate. The migrated cells on the lower chamber of the insert were fixed, stained, photographed and counted after incubation for 24 h. As shown in Fig. 4A and B, the migration rate of miR-1-overexpressing HCT-116 and ClonA1 cells was inhibited by 76 and 68%, respectively, compared with the control cells. A corresponding effect on invasion ability was also observed in a parallel invasion assay.
Figure 4.
miR-1 inhibits the migration and invasion of CRC cells. (A and B) Transwell migration (lower panel) and invasion (lower panel) assays were performed to investigate the effects of miR-1. (C and D) A wound-healing assay was performed to assess the effect of miR-1 on cell motility (*P<0.05 compared with NC). miR-1, microRNA-1; CRC, colorectal cancer; NC, negative control.
For the wound-healing assay, the motility of cells after scratching was monitored under a microscope at different time points. The results demonstrated that the edges of the wound had completely fused within 72 h in the HCT-116 and ClonA1 control cells; however, miR-1-overexpressing cells exhibited a slower wound closure rate (Fig. 4C and D). All these results demonstrated that increasing the miR-1 expression in CRC cells markedly inhibited their migration and motility abilities.
miR-1 targets and suppresses VEGF expression in CRC cells
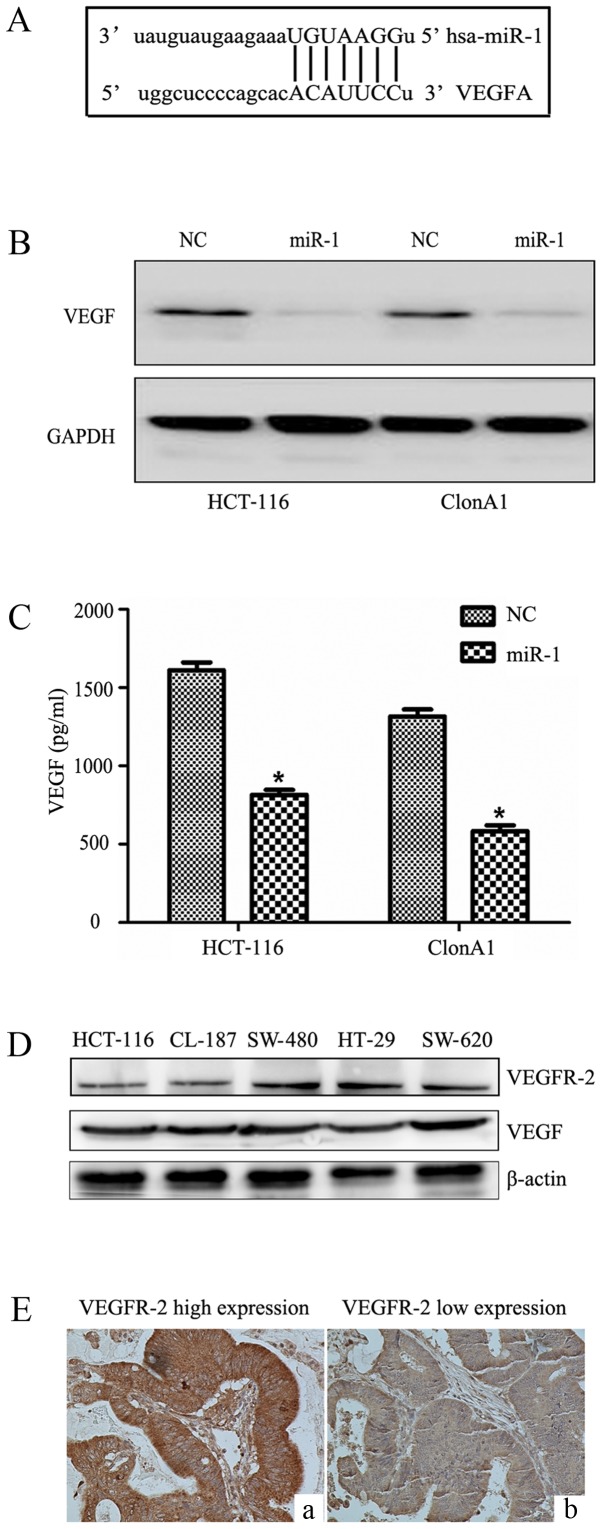
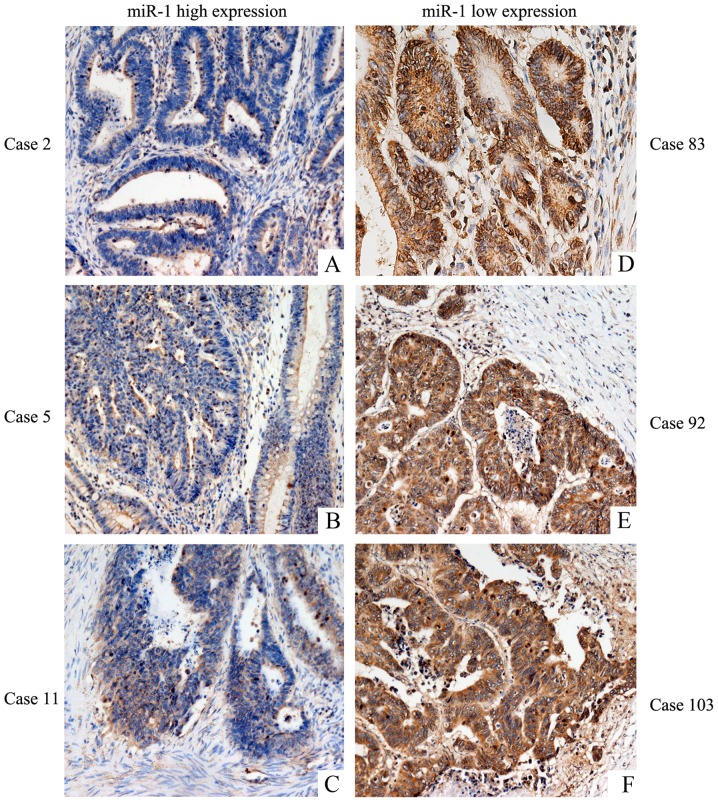
We performed bioinformatic analysis with the TargetScan, PicTar and Miranda algorithms, and identified a potential target site in the 3′UTR of VEGF that may interact with miR-1 (Fig. 5A). To further determine how miR-1 regulates VEGF, western blotting and ELISA were performed. We observed that miR-1 overexpression significantly reduced VEGF protein expression in the HCT-116 and ClonA1 cells (Fig. 5B), whereas the levels of VEGF in the culture supernatants of the two cell lines were also downregulated (Fig. 5C). The expression of VEGFR-2 and VEGF were examined in five CRC cell lines (Fig. 5D) by western blotting. Furthermore, the expression of VEGFR-2 in tumor tissues (Fig. 5E) was assessed by immunohistochemistry. The results revealed that CRC cell lines and tumor tissues express VEGFR-2. The expression of VEGF was low in tissues highly expressing miR-1 (Fig. 6A-C), and VEGF was highly expressed in tissues exhibiting low miR-1 expression (Fig. 6D-F). These results strongly indicated that miR-1 downregulates the expression of VEGF at the post-transcriptional level. The results regarding the negative correlation between the expression of miR-1 and VEGF are presented in Table II (P<0.001).
Figure 5.
miR-1 downregulates the expression of VEGF at the post-transcriptional level. (A) The interaction between miR-1 and the 3′-untranslated region of VEGF was predicted by TargetScan software. (B) Western blot analysis of VEGF expression following transfection with the LV-miR-NC and LV-miR-1 vectors. (C) The VEGF levels (pg/ml per 106 tumor cells) in the culture supernatants were determined by ELISA. (D) The expression of VEGF receptor 2 (VEGFR-2) and VEGF in five CRC cell lines were determined by western blotting. (E) The expression of VEGFR-2 in the tumor tissues was evaluated by immunohistochemistry. miR-1, microRNA-1; VEGF, vascular endothelial growth factor; CRC, colorectal cancer; NC, negative control. *P<0.05 compared with NC.
Figure 6.
Association between VEGF and miR-1 expression levels in CRC tissues. (A-C) The low expression of VEGF was observed in tissues highly expressing miR-1 (cases 2, 5 and 11). (D-F) The high expression of VEGF was observed in tissues exhibiting low miR-1 expression (cases 83, 92 and 103). miR-1, microRNA-1; VEGF, vascular endothelial growth factor; CRC, colorectal cancer.
Discussion
Colorectal cancer (CRC) is the third most common type of cancer worldwide. It is characterized by various hallmarks, including excessive proliferation, high invasion ability, cellular heterogeneity, hypoxia, angiogenesis, necrosis and infiltration by inflammatory cells (2). Despite major advances in diagnostics and treatment, the prognosis for CRC patients has not significantly improved over the past decades, and it remains a major public health concern (1–5). Therefore, the elucidation of the molecular mechanisms underlying tumor metastasis and progression is crucial for improving the treatment and prognosis of CRC patients.
miRNAs are important non-coding RNAs that participate in the regulation of numerous cellular pathways by controlling the protein expression of target genes. miR-1, which is abundant in cardiac tissues and muscles, regulates numerous functions in the heart, such as cardiac morphogenesis, electrical conduction and cell cycle control (24). Moreover, miR-1 has also been found to be a tumor-suppressor miRNA in lung, thyroid and gastric cancer, as well as head and neck squamous cell carcinoma, by inhibiting cancer cell proliferation and metastasis (25–28). In esophageal squamous cell carcinoma and renal cell carcinoma, miR-1 was found to repress cell proliferation by targeting CDK4 (29). In CRC, miR-1 and miR-374 have been identified as potential biomarkers (16). Recently, miRNA profiling revealed that miR-1 is downregulated in liver metastases from CRC, indicating that miR-1 is a potential regulator of angiogenesis (30,31), although the function of miR-1 and its underlying mechanism require further investigation. In this study, we examined the expression of miR-1 in 111 pairs of human CRC and adjacent tissues and 5 CRC cell lines, and found that the expression of miR-1 was significantly downregulated. Our data also revealed that a decreased miR-1 level is negatively associated with increased malignant behavior of CRC. Further experiments demonstrated that the restoration of miR-1 expression in CRC cell lines reduced cell proliferation, and induced G0/G1 phase arrest, cell invasion and motility, which provides a theoretical and practical basis for miR-1 to be used as a biological marker in diagnosis, and a target for CRC therapy.
Our further experiments revealed that VEGF is a target gene of miR-1 in CRC cell lines, indicating that miR-1 can inhibit the protein expression of VEGF-A and its paracrine effects. There was a negative correlation between miR-1 and VEGF expression in 111 colon cancer tissues, indicating that the biological effect of miR-1 may be mediated by inhibiting the VEGF pathway. VEGF-A is a pivotal angiogenic factor (32,33), and angiogenesis is involved in a wide variety of physiological and pathological processes, including organogenesis, development, inflammation, wound healing and carcinogenesis. Various transcriptional factors, including HIF1α (34), CREB (35) and NF-κB (36), may promote the expression of VEGF in tumors. Several studies have reported that knockdown of VEGF-A in cancer cells suppresses tumor proliferation and invasion. Previous studies have also demonstrated that the high protein expression of VEGF in CRC is associated with a relatively poor clinical outcome and liver metastasis (37,38). In addition, several drugs targeting the VEGF pathway, including vandetanib, avastin, ranibizumab and zaltrap, are widely used to treat CRC (39–41). Furthermore, a recent study demonstrated that miR-1 inhibited the epithelial-mesenchymal transition and metastasis via regulating the MAPK and PI3K/AKT pathways (42). As the MAPK and PI3K/AKT pathways are upregulated and activated by VEGF, our results suggest that miR-1 may affect the MAPK and PI3K/AKT pathways via VEGF downregulation. The mechanism underlying the effects of miR-1 may be complex, and may involve cross-interactions of additional factors and signaling pathways, which requires further investigation.
In conclusion, the present study confirmed that miR-1 acts as a tumor suppressor; miR-1 was found to be significantly downregulated in CRC cell lines and tissue specimens. Furthermore, the restoration of miR-1 expression inhibited tumor proliferation, cell cycle transition, migration and motility by affecting VEGF expression. These findings indicate a novel mechanism of tumor suppression by miR-1, and provide a theoretical and experimental basis for the development of targeted treatments for CRC through miR-1 and VEGF.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Shenyang Science and Technology Project of China (grant no. F14-158-9-35) and the Special-Term Professor from the Educational Department of Liaoning Province, China (Liao Cai Zhi Jiao no. 2012-512).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
DZ, YS and JZ conceived and supervised the study; DZ, YS, DZ, GJ and XZ designed experiments; DZ and MD performed experiments; MD and JZ analyzed data; D Z and DZ wrote the manuscript; MD and JZ made manuscript revisions. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the China Medical University, and written informed consent was obtained prior to sample collection.
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing interests.
References
- 1.Clarke SJ, Karapetis CS, Gibbs P, Pavlakis N, Desai J, Michael M, Tebbutt NC, Price TJ, Tabernero J. Overview of biomarkers in metastatic colorectal cancer: Tumour, blood and patient-related factors. Crit Rev Oncol Hematol. 2013;85:121–135. doi: 10.1016/j.critrevonc.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Sigurdsson JA, Getz L, Sjönell G, Vainiomäki P, Brodersen J. Marginal public health gain of screening for colorectal cancer: Modelling study, based on WHO and national databases in the Nordic countries. J Eval Clin Pract. 2013;19:400–407. doi: 10.1111/j.1365-2753.2012.01845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar R, Jain K, Beeke C, Price TJ, Townsend AR, Padbury R, Roder D, Young GP, Richards A, Karapetis CS. A population-based study of metastatic colorectal cancer in individuals aged ≥80 years: Findings from the South Australian clinical registry for metastatic colorectal cancer. Cancer. 2013;119:722–728. doi: 10.1002/cncr.27802. [DOI] [PubMed] [Google Scholar]
- 4.Melucci E, Cosimelli M, Carpanese L, Pizzi G, Izzo F, Fiore F, Golfieri R, Giampalma E, Sperduti I, Ercolani C, et al. Decrease of survivin, p53 and Bcl-2 expression in chemorefractory colorectal liver metastases may be predictive of radiosensivity after radioembolization with yttrium-90 resin microspheres. J Exp Clin Cancer Res. 2013;32:13. doi: 10.1186/1756-9966-32-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawson JB, Bapat B. Epigenetic biomarkers in colorectal cancer diagnostics. Expert Rev Mol Diagn. 2012;12:499–509. doi: 10.1586/erm.12.39. [DOI] [PubMed] [Google Scholar]
- 6.Miller S, Steele S. Novel molecular screening approaches in colorectal cancer. J Surg Oncol. 2012;105:459–467. doi: 10.1002/jso.21704. [DOI] [PubMed] [Google Scholar]
- 7.Akbari Moqadam F, Pieters R, den Boer ML. The hunting of targets: Challenge in miRNA research. Leukemia. 2013;27:16–23. doi: 10.1038/leu.2012.179. [DOI] [PubMed] [Google Scholar]
- 8.Okayama H, Schetter AJ, Harris CC. MicroRNAs and inflammation in the pathogenesis and progression of colon cancer. Dig Dis. 2012;30(Suppl 2):S9–S15. doi: 10.1159/000341882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanematsu S, Tanimoto K, Suzuki Y, Sugano S. Screening for possible miRNA-mRNA associations in a colon cancer cell line. Gene. 2014;533:520–531. doi: 10.1016/j.gene.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Katoh M. Therapeutics targeting angiogenesis: Genetics and epigenetics, extracellular miRNAs and signaling networks (Review) Int J Mol Med. 2013;32:763–767. doi: 10.3892/ijmm.2013.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Roitbak T, Bragina O, Padilla JL, Pickett GG. The role of microRNAs in neural stem cell-supported endothelial morphogenesis. Vasc Cell. 2011;3:25. doi: 10.1186/2045-824X-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusakabe R, Tani S, Nishitsuji K, Shindo M, Okamura K, Miyamoto Y, Nakai K, Suzuki Y, Kusakabe TG, Inoue K. Characterization of the compact bicistronic microRNA precursor, miR-1/miR-133, expressed specifically in Ciona muscle tissues. Gene Expr Patterns. 2013;13:43–50. doi: 10.1016/j.gep.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Sarver AL, Alamgir S, Subramanian S. Downregulation of microRNAs miR-1, −206 and −29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab Invest. 2012;92:571–583. doi: 10.1038/labinvest.2012.10. [DOI] [PubMed] [Google Scholar]
- 15.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–333405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Wu X, Li S, Xu X, Wu S, Chen R, Jiang Q, Li Y, Xu Y. The potential value of miR-1 and miR-374b as biomarkers for colorectal cancer. Int J Clin Exp Pathol. 2015;8:2840–2851. [PMC free article] [PubMed] [Google Scholar]
- 17.Sayagués JM, Corchete LA, Gutiérrez ML, Sarasquete ME, Del Mar Abad M, Bengoechea O, Fermiñán E, Anduaga MF, Del Carmen S, Iglesias M, et al. Genomic characterization of liver metastases from colorectal cancer patients. Oncotarget. 2016;7:72908–72922. doi: 10.18632/oncotarget.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Chen X, Fang J, Yang C. Overexpression of both VEGF-A and VEGF-C in gastric cancer correlates with prognosis, and silencing of both is effective to inhibit cancer growth. Int J Clin Exp Pathol. 2013;6:586–597. [PMC free article] [PubMed] [Google Scholar]
- 19.Martins SF, Garcia EA, Luz MA, Pardal F, Rodrigues M, Filho AL. Clinicopathological correlation and prognostic significance of VEGF-A, VEGF-C, VEGFR-2 and VEGFR-3 expression in colorectal cancer. Cancer Genomics Proteomics. 2013;10:55–67. [PubMed] [Google Scholar]
- 20.Ekinci D, Kargi A, Yalcin AD, Savas B. The role of VEGF and other parameters in tracking the clinical course in metronomic chemotherapy. J BUON. 2013;18:245–252. [PubMed] [Google Scholar]
- 21.Zhao SF, Yang XD, Lu MX, Sun GW, Wang YX, Zhang YK, Pu YM, Tang EY. Prognostic significance of VEGF immunohistochemical expression in oral cancer: A meta-analysis of the literature. Tumour Biol. 2013;34:3165–3171. doi: 10.1007/s13277-013-0886-9. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Zhang Y, Iwamoto H, Hosaka K, Seki T, Andersson P, Lim S, Fischer C, Nakamura M, Abe M, et al. Discontinuation of anti-VEGF cancer therapy promotes metastasis through a liver revascularization mechanism. Nat Commun. 2016;7:12680. doi: 10.1038/ncomms12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D, Zhou J, Dong M. Dysregulation of microRNA-34a expression in colorectal cancer inhibits the phosphorylation of FAK via VEGF. Dig Dis Sci. 2014;59:958–67. doi: 10.1007/s10620-013-2983-4. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, Zeng J, Li H, Chen K, Yu G, Hu J, Tang K, Zhou H, Huang Q, Li A, et al. MiR-1 downregulation correlates with poor survival in clear cell renal cell carcinoma where it interferes with cell cycle regulation and metastasis. Oncotarget. 2015;6:13201–13215. doi: 10.18632/oncotarget.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leone V, D'Angelo D, Rubio I, de Freitas PM, Federico A, Colamaio M, Pallante P, Medeiros-Neto G, Fusco A. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1alpha. J Clin Endocrinol Metab. 2011;96:E1388–E1398. doi: 10.1210/jc.2011-0345. [DOI] [PubMed] [Google Scholar]
- 27.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nohata N, Sone Y, Hanazawa T, Fuse M, Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, et al. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget. 2011;2:29–42. doi: 10.18632/oncotarget.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang S, Zhao C, Yang X, Li X, Pan Q, Huang H, Wen X, Shan H, Li Q, Du Y, Zhao Y. miR-1 suppresses the growth of esophageal squamous cell carcinoma in vivo and in vitro through the downregulation of MET, cyclin D1 and CDK4 expression. Int J Mol Med. 2016;38:113–122. doi: 10.3892/ijmm.2016.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slattery ML, Herrick JS, Pellatt DF, Mullany LE, Stevens JR, Wolff E, Hoffman MD, Wolff RK, Samowitz W. Site-specific associations between miRNA expression and survival in colorectal cancer cases. Oncotarget. 2016;7:60193–60205. doi: 10.18632/oncotarget.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grahnén A, Sjöholm I. Spectropolarimetric determination of unconjugated bilirubin in human serum. Z Klin Chem Klin Biochem. 1974;12:220. [PubMed] [Google Scholar]
- 32.Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat Rev Cancer. 2013;13:871–882. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chekhonin VP, Shein SA, Korchagina AA, Gurina OI. VEGF in tumor progression and targeted therapy. Curr Cancer Drug Targets. 2013;13:423. doi: 10.2174/15680096113139990074. [DOI] [PubMed] [Google Scholar]
- 34.Weijts BG, Bakker WJ, Cornelissen PW, Liang KH, Schaftenaar FH, Westendorp B, de Wolf CA, Paciejewska M, Scheele CL, Kent L, et al. E2F7 and E2F8 promote angiogenesis through transcriptional activation of VEGFA in cooperation with HIF1. EMBO J. 2012;31:3871–3884. doi: 10.1038/emboj.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang Z, Zhu H, Luan H, Han F, Jiang W. Curculigoside A induces angiogenesis through VCAM-1/Egr-3/CREB/VEGF signaling pathway. Neuroscience. 2014;267:232. doi: 10.1016/j.neuroscience.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 36.Anna L, Eugene K, Mayumi J, Matter ML. Stretch-induced Hypertrophy activates NFkB-mediated VEGF secretion in adult cardiomyocytes. Plos One. 2011;6:e29055. doi: 10.1371/journal.pone.0029055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goos JA, de Cuba EM, Coupé VM, Diosdado B, Delis-Van Diemen PM, Karga C, Beliën JA, Menke-Van der Houven van Oordt CW, Geldof AA, Meijer GA, et al. Glucose Transporter 1 (SLC2A1) and vascular endothelial growth factor A (VEGFA) predict survival after resection of colorectal cancer liver metastasis. Ann Surg. 2016;263:138–145. doi: 10.1097/SLA.0000000000001109. [DOI] [PubMed] [Google Scholar]
- 38.Cao D, Hou M, Guan YS, Jiang M, Yang Y, Gou HF. Expression of HIF-1alpha and VEGF in colorectal cancer: Association with clinical outcomes and prognostic implications. BMC Cancer. 2009;9:432. doi: 10.1186/1471-2407-9-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai Z, Yu X, Yang B, Zhang Y, Zhang L, Li X, Sun H. Colorectal cancer heterogeneity and targeted therapy: Clinical implications, challenges and solutions for treatment resistance. Semin Cell Dev Biol. 2017;64:107–115. doi: 10.1016/j.semcdb.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 40.Loupakis F, Cremolini C, Yang D, Salvatore L, Zhang W, Wakatsuki T, Bohanes P, Schirripa M, Benhaim L, Lonardi S, et al. Prospective validation of candidate SNPs of VEGF/VEGFR pathway in metastatic colorectal cancer patients treated with first-line FOLFIRI plus bevacizumab. PLoS One. 2013;8:e66774. doi: 10.1371/journal.pone.0066774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fakih M. The evolving role of VEGF-targeted therapies in the treatment of metastatic colorectal cancer. Expert Rev of Anticancer Ther. 2013;13:427–438. doi: 10.1586/era.13.20. [DOI] [PubMed] [Google Scholar]
- 42.Yim E, Vivas A, Maderal A, Kirsner RS. Neuropathy and ankle mobility abnormalities in patients with chronic venous disease. JAMA Dermatol. 2014;150:385–389. doi: 10.1001/jamadermatol.2013.6723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.