Abstract
The aim of the present study was to investigate the key genes, miRNAs and pathways in hypopharyngeal squamous cell carcinoma (HPSCC) and to elucidate the mechanisms underlying HPSCC development. The gene and microRNA (miRNA) expression profiles of HPSCC tissues and adjacent normal tissues from three subjects were obtained. Differentially expressed genes (DEGs) and differentially expressed miRNAs were identified in HPSCC. Functional annotation and protein-protein interaction (PPI) network were conducted to elucidate the biological functions of DEGs. A total of 160 DEGs (16 upregulated and 144 downregulated genes) and 79 differentially expressed miRNAs (48 upregulated and 31 downregulated miRNAs) were identified in HPSCC. The deregulated genes were significantly involved in spliceosome, cell cycle and RNA degradation. In the PPI network, S-phase kinase associated protein 1 (SKP1), non-POU domain containing octamer binding (NONO) and zinc activated ion channel (ZACN) were identified as hub proteins. On the whole, the present study may help to gain a comprehensive understanding of tumorigenesis in HPSCC and provide valuable information for early diagnosis and drug design of HPSCC in future research.
Keywords: hypopharyngeal squamous cell carcinoma, RNA-sequencing, differentially expressed genes, protein-protein interaction network, expression profiling
Introduction
Hypopharyngeal squamous cell carcinoma (HPSCC) is a common type of head and neck cancer that is associated with a high invasiveness and a poor prognosis. Recently, surgical resection along with chemoradiation therapy have been indicated as a good modality for HPSCC treatment; however, the majority of patients with HPSCC have an unfavorable prognosis due to diagnosis at an advanced stage with the presence of distant metastasis and therefore, the opportunity for radical treatment is lost. It has been reported that the overall survival rate of patients with HPSCC is only 15–45% (1,2). The etiological factors of HPSCC are unclear. A series of recent studies have described that smoking (active and passive), alcohol consumption and exposure to the human papilloma virus may be risk factors for the development of HPSCC (3–5).
The pathophysiological mechanisms of HPSCC are not yet well understood. Previous studies have demonstrated that ectopically expressed genes, microRNAs (miRNAs or miRs) and long non-coding RNAs were involved in the initiation, development and prognosis of HPSCC. Tumor necrosis factor receptor 1 (TNFR1) is a membranous receptor that anchors on the surface of the cell membrane, which belongs to the TNF receptor superfamily (6,7). The upregulation of TNFR1 has been shown to be associated with clinical staging, T stage, cervical lymph node metastasis and histological grade in HPSCC (8). The immunoglobulin heavy chain binding protein (BiP)/glucose-regulated protein 78 (GRP78) plays an important role in the endoplasmic reticulum stress and is known to be highly expressed in various human neoplasms. A decreased expression of GRP78/BiP has been reported to predict a poor overall survival and progression-free survival of patients with advanced HPSCC (9). The downregulation of miR-140-5p has also been shown to be associated with tumor stage and lymph node metastasis, and the restoration of miR-140-5p inhibits cell migration and invasion in HPSCC by targeting ADAM metallopeptidase (ADAM10), which is involved in the Notch1 signaling pathway (10). The expression of AB209630, a long non-coding RNA, has been shown to be markedly lower in HPSCC cell lines and tumor tissues compared with the normal cells and tissues, and functions as a tumor suppressor in HPSCC. A high expression of AB209630 significantly has been shown to inhibit the growth, metastasis and invasion of HSPCC in vitro, and a decreased expression of AB209630 predicts a poorer prognosis (11). In addition, abnormal methylation is associated with tumorigenesis in HSPCC. The hypermethylation of G protein-coupled receptor kinase 6 (GRK6) has been observed in HPSCC tissues compared to the matched adjacent normal tissues. GRK6 is also associated with tumor invasion and TNM stage in HPSCC (12).
HPSCC is a common malignancy with a poor prognosis. To the best of our knowledge, the expression profiling of mRNAs and miRNAs via RNA-sequencing has not yet been documented in HPSCC. In the present study, we used high-throughput RNA-sequencing to examine gene and miRNA expression and to investigate the functional significance of aberrantly expressed genes and miRNAs in HSPCC, with the aim of providing the groundwork for the elucidation of HPSCC tumorigenesis and potential biomarkers for the early diagnosis of HPSCC.
Materials and methods
Patients and samples
A total of 3 patients diagnosed with HPSCC were enrolled in this study from the First People's Hospital of Jining from August, 2015 to January, 2016. The mean age of the 3 patients with HPSCC was 60.7 years (range, 56–64 years). All 3 patients with HPSCC patients with squamous cell carcinoma were male. All patients had lymph node metastasis tumor loci. Primary HPSCC tissues and adjacent normal tissues of patients were obtained through surgical resection. The TNM stage and tumor grade of the patients are presented in Table I. The study was approved by the Ethics Committee of the First People's Hospital of Jining, and informed written consent was obtained from all patients. Primary tumors and adjacent normal tissues of patients with HPSCC were obtained based on the Declaration of Helsinki.
Table I.
Basic characteristics of patients with HPSCC.
| Patient ID | Age (years) | Sex | Cigarettes per day | Years of smoking | Alcohol consumption each day (g) | Years of alcohol consumption | TNM stage | Histological feature | Tumor grade |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 62 | Male | 20 | 40 | 100 | 30 | T2N1M0 | SCC | Medium |
| 2 | 56 | Male | 20 | 30 | 250 | 30 | T2N1M0 | SCC | Medium |
| 3 | 64 | Male | 20 | 30 | No details | No details | T3N1M0 | SCC | Medium |
HPSCC, hypopharyngeal squamous cell carcinoma.
Isolation of RNA
A total of 6 specimens from the 3 patients were used for RNA isolation using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions. RNA quality and quantity were assessed using the NanoDrop 2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific) and Agilent 2100 bioanalyzer (Agilent Technologies GmbH, Waldron, Germany). The average RNA quantity for the specimens was 73.42 µg (0.84–174.98 µg) with RNA integrity number (RIN) values between 1.0 and 7.6.
Library preparation and RNA sequencing
The Illumina TruSeq RNA Sample Prep kit (Illumina, Inc., San Diego, CA, USA) was used for the library preparation according to the manual. PolyT oligo-conjugated magnetic beads were used to enrich the RNA with polyA+ tail, followed by mRNA fragmentation, first strand cDNA synthesis, second strand cDNA synthesis and end repair. Subsequenlty, the product was ligated to Illumina TruSeq adaptors. Following PCR amplification, enriched cDNA libraries were sequenced using the Illumina HiSeq 2500 sequencing platform (Illumina).
Data preprocessing
The raw data from high-throughput RNA-sequencing was translated into raw FASTQ sequence data by base calling. Raw RNA-sequencing data were filtered and trimmed using FASTx-Tool SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle). Clean and trimmed FASTQ reads were aligned to the human hg19 genome by using TopHat (version 1.3.1; Center for Computational Biology, Johns Hopkins University, Baltimore, MD, USA) (13). The aligned read files were then processed using Cufflinks version 1.2.1 software package (Trapnell Lab, Seattle, WA, USA) which assessed the abundance of genes (14). Fragments per kilobase of transcript per million mapped reads (FPKM) was used to determine the transcription abundance of each gene.
Differentially expressed genes (DEGs) and miRNAs
The DEGs between HPSCC tumor and adjacent normal tissues were carried out using paired t-tests and limma package in R (15). The false discovery rate (FDR) was performed; multiple testing corrections of raw P-values were performed using the Benjamini and Hochberg method (16,17). The genes with an FDR <0.05 and abs (log2 fold change) >1 were screened as DEGs. A two-way hierarchical clustering analysis was conducted to assess the similarity of gene expression patterns between samples, and the results were displayed in a heatmap using the ‘pheatmap’ package (18). In addition, DEGseq (http://bioconductor.org/packages/DEGseq/) was applied to quantify the expression of miRNAs. miRNAs with P-value <0.001 and abs (log2FC) >2 were screened as differentially expressed miRNAs.
Analysis of the miRNA target genes
Identifying the target genes of miRNAs is a key step towards studying the function of miRNAs in specific tissues and cells. In the present study, 6 miRNA-target prediction tools (DIANAmT, miRanda, miRDB, miRWalk, PICTAR5 and TargetScan) were applied to predict the target genes of differentially expressed miRNAs. The miRNA-targets that were predicted by >4 algorithms or verified by experiment in the miRWalk database were identified. All miRNA-target pairs were obtained, which were not only predicted by algorithms (or verified by experiment), but also negatively associated in the expression level. The miRNA-target regulatory network was then constructed, which was visualized using Cytoscape (19).
Functional annotation of DEGs
The Gene Ontology (GO) function was used to elucidate the biological function of DEGs in biological process, molecular function and cellular component using the GeneCodis 3 online software (http://genecodis.cnb.csic.es/analysis) (20). P<0.01 was set as the cut-off for selecting significantly enriched functional GO terms. KOBAS version 2.0 software (http://kobas.cbi.pku.edu.cn) was used to enrich the signaling pathways of DEGs (21). P<0.05 was set as the threshold for determining significantly enriched pathways.
Protein-protein interaction network (PPI)
In order to elucidate the protein interactions between upregulated and downregulated DEGs in HPSCC, the BioGRID database was used to screen pairs of interacting proteins (22). The PPI network was visualized using Cytoscape (http://cytoscape.org/) (23). In the network, nodes represent proteins and edges represent interaction between two proteins.
Validation of the expression of selected DEGs in The Cancer Genome Atlas (TCGA)
TCGA dataset was used to validate the expression of selected DEGs in HPSCC patients with a large sample size by using the online software SurvExpress (http://bioinformatica.mty. itesm.mx:8080/Biomatec/SurvivaX.jsp).
Results
RNA sequencing of HPSCC specimens
Total RNA was extracted from HPSCC tissues and adjacent normal controls from 3 patients. A total of 5 out of 6 samples, including 1C, 1N, 3C, 3N and 2C passed the assessment for RNA quality and quantity. Sample 2N did not pass the assessment, as the RIN value was not applicable (Table II). A total of 5 samples were used for library construction and RNA-sequencing. For each sample, RNA-sequencing reads were generated and yielded 34–44 million sequencing reads per sample (Table III). All sequencing reads were aligned to the human hg19 genome by using TopHat and FPKM and the relative expression of genes was determined using the Cufflinks tool.
Table II.
RNA quantity and quality measurements of 6 samples.
| Sample ID | RNA volume (µl) | RNA concentration (ng/µl) | RNA amount (µg) | RIN |
|---|---|---|---|---|
| 1C | 38 | 2,723 | 103.47 | 4 |
| 2C | 65 | 2,692 | 174.98 | 7.6 |
| 3C | 27 | 3,288 | 88.78 | 7.5 |
| 1N | 14 | 334 | 4.68 | 5.7 |
| 2N | 12 | 70 | 0.84 | N/A |
| 3N | 38 | 1,784 | 67.79 | 4.7 |
C, primary tissues of hypopharyngeal squamous cell carcinoma; N, adjacent normal tissues; RIN, RNA integrity number; N/A, not available.
Table III.
RNA-sequencing of the specimens.
| Sample_ID | Total reads | Total bases | Q20% | Q30% | Average coverage |
|---|---|---|---|---|---|
| 1C | 4.4×107 | 5.6×109 | 96.66 | 93.47 | 99.1863479139598 |
| 2C | 3.5×107 | 4.3×109 | 96.61 | 93.33 | 99.4073847620807 |
| 3C | 3.4×107 | 4.2×109 | 96.68 | 93.41 | 99.2552832336352 |
| 1N | 4.3×107 | 5.4×109 | 96.27 | 92.79 | 99.348671268904 |
| 3N | 3.5×107 | 4.4×109 | 96.58 | 93.32 | 99.2810150213976 |
C, primary tissues of hypopharyngeal squamous cell carcinoma; N, adjacent normal tissues; Q20, the percentage of bases with quality value >20; Q30, the percentage of bases with quality value >30.
DEGs and differentially expressed miRNAs between primary HPSCC and adjacent normal controls
A total of 160 DEGs, including 16 upregulated and 144 downregulated genes, were identified in the HPSCC tumors compared to the adjacent normal controls using the screening criteria FDR<0.05 and abs (log2FC) >1 (data not shown). In addition, a total of 79 differentially expressed miRNAs (48 upregulated and 31 downregulated miRNAs) were identified in the HPSCC tumors compared to adjacent normal controls using the screening criteria P<0.001 and abs (log2FC) >2 (data not shown).
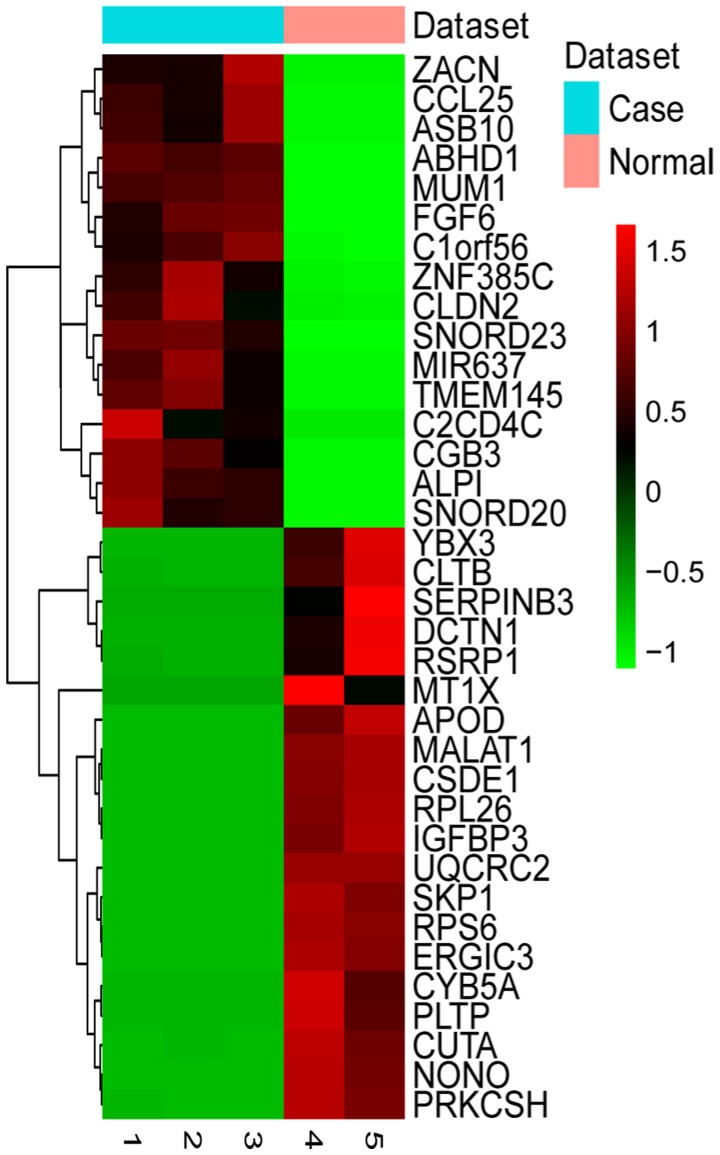
As shown in Table IV, zinc activated ion channel (ZACN), chorionic gonadotropin β subunit 3 (CGB3) and zinc finger protein 385C (ZNF385C) were the top 3 upregulated genes. Ribosomal protein L26 (RPL26), ribosomal protein S6 (RPS6) and S-phase kinase associated protein 1 (SKP1) were the top 3 downregulated genes in HPSCC. The expression pattern of the top 20 upregulated and downregulated DEGs was examined by two-way hierarchical clustering analysis (Fig. 1). Compared to the normal control, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was a significantly downregulated long non-coding RNA in HPSCC based on our RNA-sequencing data (FDR=0.014705). The top 20 differentially expressed miRNAs are shown in Table V. As shown in Table V, hsa-let-7i-5p, hsa-miR-136-5p and hsa-miR-143-5p were the top 3 upregulated miRNAs in HPSCC, and hsa-miR-15a-5p, hsa-miR-2278 and hsa-miR-125a-5p were the top 3 downregulated miRNAs in HPSCC.
Table IV.
DEGs in HPSCC.
| Gene ID | Gene symbol | Log2FC | P-value | FDR |
|---|---|---|---|---|
| Upregulated genes | ||||
| 353174 | ZACN | 6.288149 | 8.86E-05 | 0.038292 |
| 1082 | CGB3 | 5.765826 | 4.12E-05 | 0.034011 |
| 201181 | ZNF385C | 5.310858 | 0.000127 | 0.039757 |
| 126567 | C2CD4C | 5.256596 | 0.000222 | 0.049962 |
| 84696 | ABHD1 | 5.10952 | 4.07E-05 | 0.034011 |
| 6370 | CCL25 | 5.035685 | 5.76E-05 | 0.035804 |
| 693222 | MIR637 | 4.830281 | 6.51E-05 | 0.035863 |
| 692091 | SNORD23 | 4.628719 | 3.74E-05 | 0.033502 |
| 9075 | CLDN2 | 4.580931 | 0.000195 | 0.047531 |
| 6082 | SNORD20 | 4.49704 | 8.54E-05 | 0.038072 |
| 284339 | TMEM145 | 4.395233 | 8.36E-05 | 0.038072 |
| 2251 | FGF6 | 3.962287 | 6.55E-05 | 0.035863 |
| 136371 | ASB10 | 3.961177 | 0.000138 | 0.040535 |
| 248 | ALPI | 3.959662 | 7.79E-05 | 0.038041 |
| 54964 | C1orf56 | 3.849454 | 0.000166 | 0.044327 |
| 84939 | MUM1 | 3.145926 | 9.19E-05 | 0.038292 |
| Downregulated genes | ||||
| 6154 | RPL26 | −10.6837 | 1.06E-06 | 0.014705 |
| 6194 | RPS6 | −9.89489 | 1.14E-06 | 0.014705 |
| 6500 | SKP1 | −7.78013 | 3.26E-06 | 0.030707 |
| 8531 | YBX3 | −7.37473 | 2.26E-05 | 0.033502 |
| 347 | APOD | −7.36529 | 6.56E-06 | 0.033502 |
| 51614 | ERGIC3 | −7.28305 | 0.000122 | 0.03936 |
| 4501 | MT1X | −6.78769 | 0.000215 | 0.049189 |
| 3486 | IGFBP3 | −6.56019 | 6.70E-06 | 0.033502 |
| 7812 | CSDE1 | −6.49789 | 5.73E-06 | 0.033502 |
| 4841 | NONO | −6.45686 | 8.40E-06 | 0.033502 |
| 6317 | SERPINB3 | −6.45555 | 0.000213 | 0.049086 |
| 5360 | PLTP | −6.29414 | 2.00E-05 | 0.033502 |
| 1528 | CYB5A | −6.28125 | 2.21E-05 | 0.033502 |
| 51596 | CUTA | −6.24975 | 0.000199 | 0.048247 |
| 57035 | RSRP1 | −6.20271 | 0.000205 | 0.048326 |
| 7385 | UQCRC2 | −6.01731 | 8.71E-06 | 0.033502 |
| 1639 | DCTN1 | −5.79807 | 0.000143 | 0.040725 |
| 1212 | CLTB | −5.70891 | 0.000204 | 0.048326 |
| 5589 | PRKCSH | −5.58244 | 8.30E-05 | 0.038072 |
| 6194 | RPS6 | −9.89489 | 1.14E-06 | 0.014705 |
| 340277 | FAM221A | −3.26588 | 1.30635 | −17.5303 |
DEGs, differentially expressed genes; HPSCC, hypopharyngeal squamous cell carcinoma; FDR, false discovery rate.
Figure 1.
Hierarchical clustering analysis based on the expression profile of the top 20 DEGs in HPSCC compared to adjacent normal tissues. Turquoise and pink indicate the HPSCC and adjacent normal tissues, respectively. Red represents the relative expression level of genes that was higher than the mean expression level, and green represents the relative expression of genes that was lower than the mean expression level. DEGs, differentially expressed genes; HPSCC, hypopharyngeal squamous cell carcinoma.
Table V.
Top 20 differentially expressed miRNAs in HPSCC.
| miRNA symbol | Log2FC | P-value |
|---|---|---|
| Upregulated miRNAs | ||
| hsa-let-7i-5p | 2.122547495 | 0 |
| hsa-miR-136-5p | 2.444466751 | 0 |
| hsa-miR-143-5p | 2.976688335 | 0 |
| hsa-miR-370-3p | 3.344940583 | 0 |
| hsa-miR-140-3p | 3.395556571 | 0 |
| hsa-let-7g-5p | 9.933859336 | 8.71E-85 |
| hsa-miR-106b-3p | 9.933859336 | 8.71E-85 |
| hsa-miR-127-3p | 9.933859336 | 8.71E-85 |
| hsa-miR-148b-3p | 9.933859336 | 8.71E-85 |
| hsa-miR-187-3p | 9.933859336 | 8.71E-85 |
| Downregulated miRNAs | ||
| hsa-miR-15a-5p | −12.79407766 | 0 |
| hsa-miR-2278 | −12.05711064 | 1.56E-243 |
| hsa-miR-125a-5p | −10.472141 | 3.27E-107 |
| hsa-miR-1269a | −10.472141 | 3.27E-107 |
| hsa-miR-135b-5p | −10.472141 | 3.27E-107 |
| hsa-miR-18a-3p | −10.472141 | 3.27E-107 |
| hsa-miR-19b-3p | −10.472141 | 3.27E-107 |
| hsa-miR-200b-3p | −10.472141 | 3.27E-107 |
| hsa-miR-203b-3p | −10.472141 | 3.27E-107 |
| hsa-miR-20a-5p | −10.472141 | 3.27E-107 |
HPSCC, hypopharyngeal squamous cell carcinoma.
miRNA target gene interactions
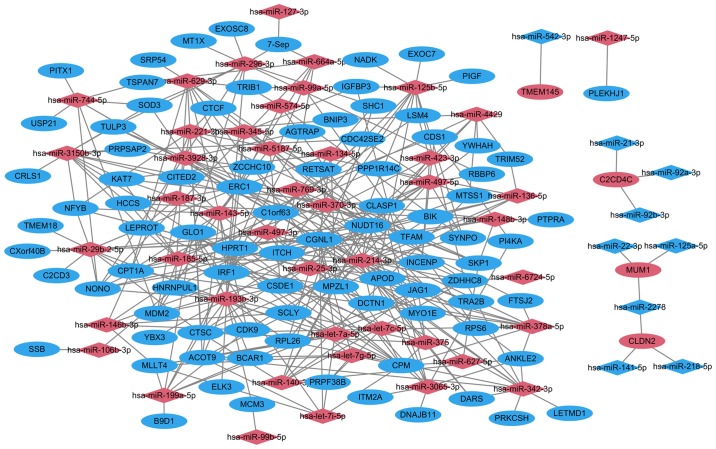
Following target correlation analysis, a total of 339 miRNA-target pairs were obtained in HPSCC. A total of 228 upregulated miRNA-target pairs and 9 downregulated miRNA-target pairs were predicted using 6 prediction tools. A total of 101 upregulated miRNAs-target pairs and 1 downregulated miRNAs-target pair were validated in the miRWalk dataset. The top 10 miRNAs that targeted the majority of genes are listed in Table VI. The miRNA-target network between DEGs and differentially expressed miRNAs is presented in Fig. 2.
Table VI.
Top 10 miRNAs that targeted the majority of genes.
| miRNA | Up/downregulated | Count of targets | Target genes |
|---|---|---|---|
| hsa-miR-193b-3p | Upregulated | 18 | CDK9, HNRNPUL1, ELK3, ACOT9, GLO1, HPRT1, IRF1, MCM3, NONO, SCLY, RPL26, YBX3, CTSC, ERC1, MDM2, CSDE1,CGNL1, BCAR1 |
| hsa-miR-214-3p | Upregulated | 17 | ERC1, CDK9, SYNPO, NUDT16, DCTN1, CLASP1, ZDHHC8, APOD, INCENP, MYO1E, BIK, TRA2B, TFAM, ITCH, CGNL1, MPZL1, JAG1 |
| hsa-miR-497-5p | Upregulated | 14 | RBBP6, YWHAH, CDS1, CTSC, SYNPO, NUDT16, INCENP, IRF1, CSDE1, PPP1R14C, CGNL1, MPZL1, MTSS1, CDC42SE2 |
| hsa-miR-29b-2-5p | Upregulated | 13 | TMEM18, NUDT16, CPM, CPT1A, ERC1, C2CD3, NFYB, NONO, CXorf40B, LEPROT, TFAM, CGNL1, CTSC |
| hsa-miR-125b-5p | Upregulated | 12 | EXOC7, LSM4, PIGF, SYNPO, DCTN1, ZDHHC8, IGFBP3, IRF1, NADK, CSDE1, ITCH, MPZL1 |
| hsa-miR-3150b-3p | Upregulated | 12 | KAT7, NUDT16, CPM, ERC1, HCCS, NFYB, CRLS1, LEPROT, PRPSAP2, CDC42SE2, SOD3, TULP3 |
| hsa-miR-185-5p | Upregulated | 11 | KAT7, INCENP, MDM2, NUDT16, CPM, CPT1A, ERC1, HCCS, LEPROT, PPP1R14C, MPZL1 |
| hsa-miR-342-3p | Upregulated | 11 | DARS, ERC1, ACOT9, LETMD1, PRKCSH RPL26, NUDT16, ZDHHC8, CGNL1, MPZL1 |
| hsa-miR-378a-5p | Upregulated | 11 | IRF1, RPS6, CTSC, CPM, CPT1A, ERC1, ANKLE2, FTSJ2, NFYB, TULP3, ITCH |
| hsa-miR-629-3p | Upregulated | 11 | HNRNPUL1, ACOT9, TRIB1, CDK9, CTSC, NUDT16, CPM, CPT1A, ERC1, SRP54, TFAM |
Figure 2.
miRNA target network between DEGs and differentially expressed miRNAs in HPSCC. Diamond and ellipse shapes represent the miRNAs and target genes, respectively. Red and blue represent upregulation and downregulation, respectively. DEGs, differentially expressed genes; HPSCC, hypopharyngeal squamous cell carcinoma; miRNA, microRNA.
Functional annotation of DEGs
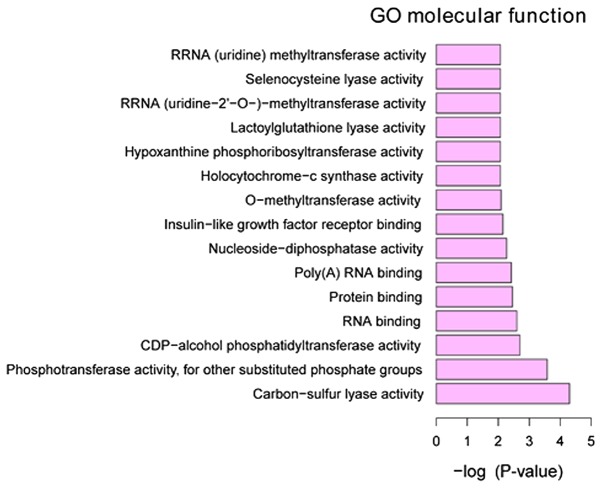
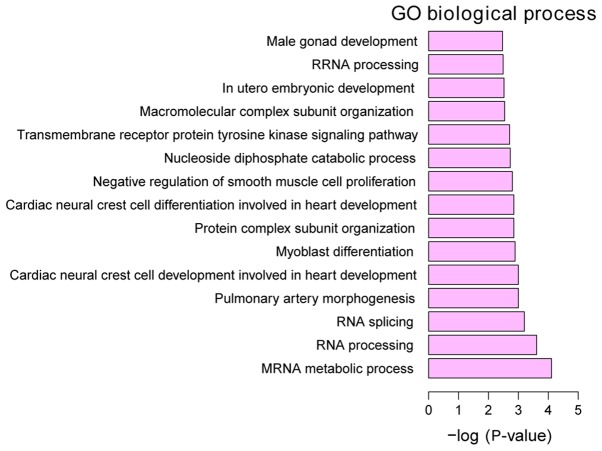
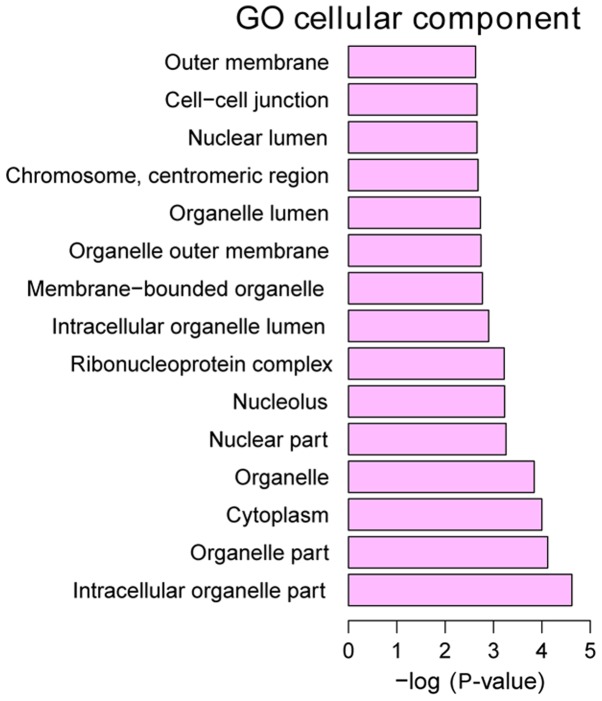
GO annotation of the 160 identified DEGs in HPSCC was performed to elucidate the biological roles of DEGs using the GeneCodis 3 online software. P<0.05 was used to determine significantly enriched GO terms. The most enriched ‘molecular functions’ of the DEGs were carbon-sulfur lyase activity (GO, 0016846), phosphotransferase activity, for other substituted phosphate groups (GO, 0016780) and CDP-alcohol phosphatidyltransferase activity (GO, 0017169). The most enriched ‘cellular components’ were intracellular organelle part (GO, 0044446), organelle part (GO, 0044422) and cytoplasm (GO, 0005737). The most enriched ‘biological processes’ were mRNA metabolic process (GO, 0016071), RNA processing (GO, 0006396) and RNA splicing (GO, 0008380) (Figs. 3–5).
Figure 3.
Top 15 significantly enriched molecular functions of DEGs in HPSCC. DEGs, differentially expressed genes; HPSCC, hypopharyngeal squamous cell carcinoma.
Figure 5.
Top 15 significantly enriched biological processes of DEGs in HPSCC. DEGs, differentially expressed genes; HPSCC, hypopharyngeal squamous cell carcinoma.
KEGG pathway enrichment
We performed KEGG pathway enrichment analysis for DEGs using KOBAS version 2.0. P<0.05 was used as the criteria for pathway enrichment. The most enriched pathways were spliceosome (hsa03040), cell cycle (hsa04110) and RNA degradation (hsa03018) (Fig. 6).
Figure 6.
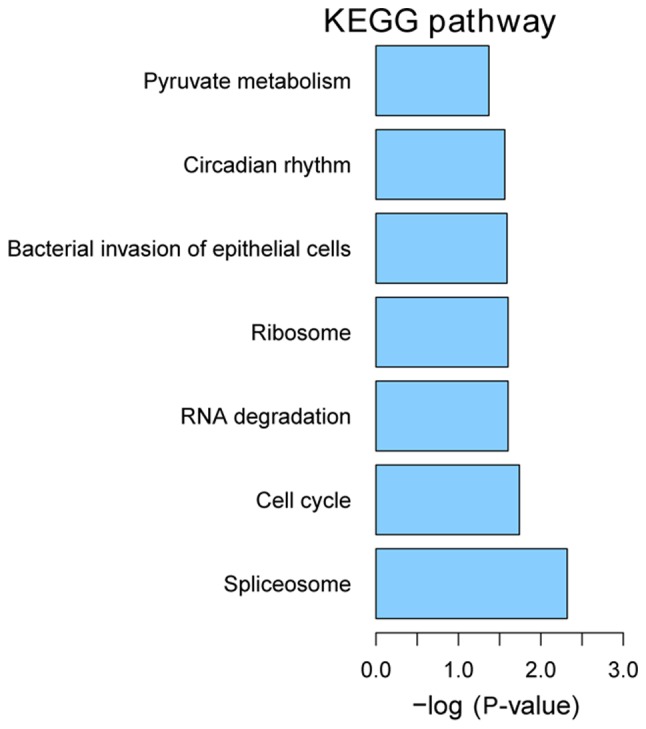
Significantly enriched KEGG pathways of DEGs in HPSCC. DEGs, differentially expressed genes; HPSCC, hypopharyngeal squamous cell carcinoma; KEGG, Kyoto Encyclopedia of Genes and Genomes.
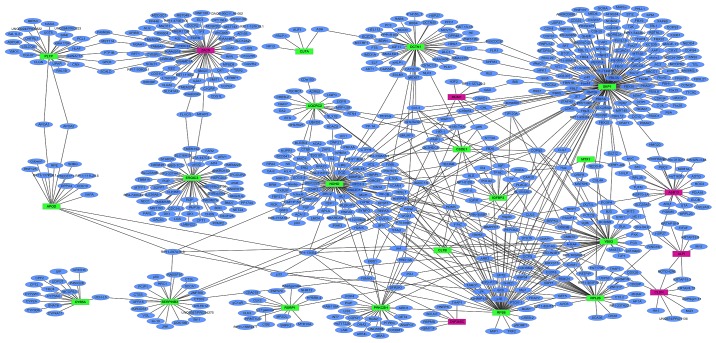
PPI network of DEGs in primary HPSCC compared to adjacent normal controls
PPI networks of the top 20 upregulated and downregulated DEGs in HPSCC compared to adjacent normal controls were determined using Cytoscape, which consisted of 673 nodes and 812 edges (data not shown). The proteins that had high connectivity with other proteins were hub proteins. In the PPI network, SKP1, NONO and ZACN were the hub proteins, which interacted with 166, 101 and 70 proteins, respectively (Fig. 7).
Figure 7.
Protein-protein interaction networks of the top 20 upregulated and downregulated DEGs in HPSCC. Red and green rectangle nodes represent upregulated and downregulated DEGs, respectively. Blue circular nodes represent proteins that interact with the proteins encoded by DEG. The solid line indicates the interactions between proteins.
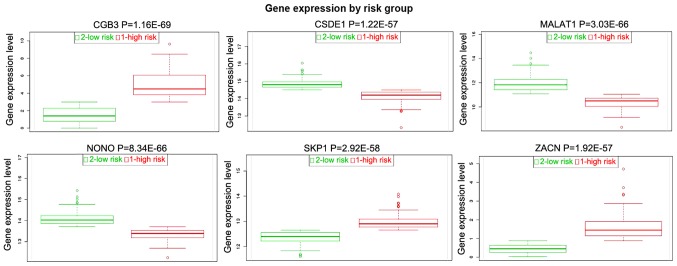
Expression validation of selected DEGs in TCGA
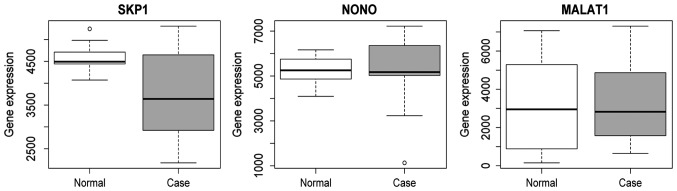
A total of 5 DEGs [SKP1, ZACN, NONO, cold shock domain containing E1 (CSDE1), chorionic gonadotropin β subunit 3 (CGB3) and MALAT1] were selected to perform the tumor risk evaluation in HPSCC by using the head and neck squamous cell carcinoma datasets (283 samples) in TCGA. CGB3, SKP1 and ZACN were upregulated while MALAT1, NONO and CSDE1 were downregulated in the HPSCC high-risk group (Fig. 8). Additionally, we also performed validation of SKP1, NONO and MALAT1 using the HPSCC dataset in TCGA. The result showed that the expression of SKP1, NONO and MALAT1 was decreased in HPSCC tissues compared to the normal control, which was consistent with the RNA-sequencing analysis (Fig. 9).
Figure 8.
Tumor risk evaluations of SKP1, ZACN, NONO, CSDE1, CGB3 and MALAT1 in the TCGA dataset. CGB3, chorionic gonadotropin beta subunit 3; CSDE1, cold shock domain containing E1; MALAT1, metastasis associated in lung adenocarcinoma transcript 1; NONO, non-POU domain containing octamer binding; SKP1, S-phase kinase associated protein 1; TCGA, The Cancer Genome Atlas; ZACN, zinc activated ion channel.
Figure 9.
Box plots of SKP1, NONO and MALAT1 in the TCGA dataset. MALAT1, metastasis associated in lung adenocarcinoma transcript 1; NONO, non-POU domain containing octamer binding; SKP1, S-phase kinase associated protein 1; TCGA, The Cancer Genome Atlas.
Discussion
At present, there is a lack of early detection biomarkers in clinical practice for HPSCC. To the best our knowledge, this is the first study to report the expression pattern of mRNAs and miRNAs in HPSCC via high-throughput RNA sequencing.
SKP1 was significantly downregulated in HPSCC compared to adjacent normal controls (Table IV). SKP1 had the highest connectivity with 193 proteins (Fig. 7). According to KEGG signaling pathway enrichment, SKP1 was significantly enriched in cell cycle and circadian rhythm. In homo sapiens, SKP1 encodes S-phase kinase-associated protein 1, which is a scaffold protein of the ubiquitin E3 ligase Skp1/Cullin1/Rbx1/F-box protein complex (SCF complex). The SCF complex catalyzes the ubiquitination of proteins, which are involved in cell-cycle progression, signal transduction and transcription (24). SKP1 was overexpressed in 56.3% of the non-small cell lung cancer (NSCLC) specimens and elevated SKP1 was associated with a poor prognosis (25). The other component of the SCF complex, cullin, is highly expressed in NSCLC tissues and significantly associated with histological differentiation and clinical stage (26). Our finding indicated that SKP1 may play vital roles in the development of HSPCC, which indicates that it may serve as a diagnostic and prognostic biomarker for HSPCC.
Apart from SKP1, ZACN (also termed L2, ZAC) was also a hub protein according to the HSPCC-specific PPI network. ZACN is located at chromosome 17, and it encodes zinc-activated ion channel, which belongs to the cysteine-loop superfamily of ligand-gated ion channels. ZACN mRNA is expressed in various organs and tissues, including the brain, pancreas, liver, lung, heart, kidney and skeletal muscle (27). The overexpression of ZACN was implicated in transient neonatal diabetes mellitus, which is a rare inherited diabetic syndrome apparent in neonate and again during early adulthood. In a transgenic mouse model, the expression of human ZACN and hydatidiform mole associated and imprinted (HYMAI) impaired the development of the endocrine pancreas and the function of β cells (28). In the present study, ZACN was the most significantly upregulated gene in the HPSCC dataset compared to the adjacent normal control. The overexpression of ZACN in HPSCC was first reported in this study. The biological roles of ZACN overexpression in HPSCC warrant further investigation in in vitro and in vivo studies.
NONO encodes an RNA-binding protein and functions to regulate transcription and RNA splicing in the nucleus. NONO has been implicated in the progression of various tumor types, including breast and colorectal cancer and melanoma. NONO was also reported to regulate lipid metabolism by binding to SREBP-1A in breast cancer (29).
Gastric adenocarcinoma associated, positive CD44 regulator, long intergenic non-coding RNA (GAPLINC) promotes cell invasion by targeting snail family transcriptional repressor 2 (SNAI2) via binding with NONO in colorectal cancer (30). As a hub protein of the HSPCC-specific PPI network, NONO was significantly downregulated in HPSCC in this study. In addition, NONO was targeted by hsa-miR-29b-2-5p (Table VI). It has been reported that miR-29b-2-5p was potentially associated with survival in high-grade serous ovarian carcinoma (31). Furthermore, miR-29b-2-5p has been shown to be downregulated in gallbladder cancer (32). It was suggested that NONO may be involved in HPSCC under the regulation of hsa-miR-29b-2-5p.
CGB3 is a member of the glycoprotein hormone β chain family and encodes the β 3 subunit of chorionic gonadotropin (33). Previous studies have indicated that the overexpression of CGB3 plays a role in carcinogenesis (34,35). The synthesis of human chorionic gonadotropin β subunit has been detected in 30–50% of various malignant tumors (36,37). In addition, upregulated CGB3 has been found to be closely associated with the metastatic cancer phenotype, resistance to therapy, as well as with a poor prognosis (37,38). To date, to the best of our knowledge, no study has reported the association between CGB3 and HSPCC. In the present study, an increased CGB3 expression was detected in patients with HSPCC. Hence, it can be concluded that CGB3 may be involved in the pathogenesis of HSPCC; however, its precise function warrants further investigation.
MALAT1 was the first long non-coding RNA prognostic biomarker identified for early-stage NSCLC (39). The deregulation of MALAT1 has been found in various types of cancer, including lung, breast, pancreas, colon, prostate and liver cancer (40). Previous studies have demonstrated that MALAT1 plays a role in cell proliferation, invasion and tumorigenesis (41–43). In addition, the prognostic value of MALAT1 for cancer metastasis of cancer has been reported (44). The expression of MALAT1 was significantly downregulated in HSPCC in this study, which suggested that MALAT1 may be a key regulator of tumorigenesis in HSPCC. In addition, long-term studies are required in order to identify whether MALAT1 is a predictor for metastasis of HSPCC.
CSDE1 (also known as UNR) encodes the RNA-binding protein, which is found to be upregulated in melanoma tumors, and promotes invasion and metastasis (45). The upregulation of CSDE1 has also been identified in breast cancer (46), which suggests that CSDE1 may be involved in tumorigenesis. In the present study, the expression of CSDE1 was downregulated in HSPCC, which may be due to the difference in the type of cancer tissues used. Additionally, this study demonstrated that CSDE1 is under the regulation of hsa-miR-497-5p and hsa-miR-125b-5p (Table VI). The expression of hsa-miR-497-5p has been reported to be significantly decreased in triple-negative breast cancer (47) and to be negatively associated with survival in oropharyngeal squamous cell carcinoma (48). hsa-miR-125b-5p is involved in regulating the proliferation, migration and invasion of tumor cells. The overexpression of hsa-miR-125b-5p has been shown to inhibit cell proliferation, migration and invasion in esophageal squamous cell carcinoma (49). The underlying molecular mechanism of CSDE1 under the regulation of hsa-miR-497-5p and hsa-miR-125b-5p in HPSCC need to be elaborated in future studies.
In differentially expressed miRNAs, we observed that hsa-let-7i-5p, hsa-miR-136-5p and hsa-miR-143-5p were the top 3 upregulated miRNAs in HPSCC. hsa-miR-15a-5p, hsa-miR-2278 and hsa-miR-125a-5p were the top 3 downregulated miRNAs in HPSCC. hsa-let-7i-5p has been shown to be upregulated in triple-negative breast cancer cells, colon cancer cells and non-muscle invasive bladder cancer cells (50–52). The upregulation of hsa-let-7i-5p is associated with an advanced stage, a high grade and therefore, with the progression of clear cell renal cell carcinoma (53). hsa-miR-136-5p has been shown to be significantly upregulated in osteosarcoma and colorectal adenocarcinoma (54,55). hsa-miR-143-5p acts as a strong tumor suppressive factor. It has been confirmed that hsa-miR-143-5p inhibits the proliferation, invasion and migration of cervical cancer cells (56). It has also been shown that hsa-miR-15a-5p acted as a tumor suppressor by silencing the expression of growth promoting oncogenes (57–59). In chronic myeloid leukemia, hsa-miR-15a-5p has been shown to suppress cell growth and metastasis (60). In addition, hsa-miR-15a-5p has been shown to be associated with patient survival in lung adenocarcinoma (61). hsa-miR-2278 has been shown to be associated with survival in both rectal and colon cancer (62). hsa-miR-125a-5p is known to act as a tumor suppressor and is downregulated in various malignant tumors, such as laryngeal carcinoma and verrucous carcinoma of the head and neck (63,64). In multiple myeloma cells, the inhibition of hsa-miR-125a-5p has been shown to reduce cell growth, increased cell apoptosis and attenuated cell migration (65). On the whole, these differentially expressed miRNAs may play an important role in the development of HPSCC. In the present study, spliceosome, cell cycle and RNA degradation were significantly enriched signal pathways in HPSCC. Deregulated genes regulated the progression and development of cancer via the cell cycle in various cancer types, including lung adenocarcinoma, colorectal and head and neck cancer (44–46), which indicated that the cell cycle pathway was implicated in the tumorigenesis of HPSCC.
In conclusion, in the present study, we identified 160 DEGs and 79 differentially expressed miRNAs in HPSCC tissues compared to adjacent normal tissues. The top 20 upregulated and downregulated genes in HPSCC were used to construct PPI networks, where SKP1, NONO and ZACN were the hub proteins. DEGs were significantly enriched in the spliceosome, cell cycle and RNA degradation. The present study was the first to investigate the gene and mRNA expression profiles in HPSCC. Our findings may provide the groundwork for the identification of early diagnosis biomarkers for HPSCC and the mechanisms that underlie its pathogenesis, as well as make a contribution for future drug design. However, there are limitations to this study. Firstly, the sample size in the RNA-sequencing was small. Therefore, large numbers of HPSCC tumor samples are needed for further research. Secondly, although deregulated genes, miRNAs and pathways in HPSCC were identified, the biological functions of these genes, miRNAs and pathways were not investigated in this study. In vitro and in vivo experiments are essential for the elucidation of the biological roles of DEGs and differentially expressed miRNAs in HPSCC in future investigations.
Figure 4.
Top 15 significantly enriched cellular components of DEGs in HPSCC. DEGs, differentially expressed genes; HPSCC, hypopharyngeal squamous cell carcinoma.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ADAM10
ADAM metallopeptidase
- CGB3
chorionic gonadotropin beta subunit 3
- CSDE1
cold shock domain containing E1
- DEGs
differentially expressed genes
- FDR
false discovery rate
- FPKM
fragments per kilobase of exon per million fragments mapped
- GRK6
G protein-coupled receptor kinase 6
- GAPLINC
gastric adenocarcinoma associated, positive CD44 regulator, long intergenic non-coding RNA
- GO
Gene Ontology
- GRP78
glucose-regulated protein 78
- HYMAI
hydatidiform mole associated and imprinted
- HPSCC
hypopharyngeal squamous cell carcinoma
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- NONO
non-POU domain containing octamer binding
- NSCLC
non-small cell lung cancers
- PPI
protein-protein interaction
- RPL26
ribosomal protein L26
- RPS6
ribosomal protein S6
- RIN
RNA integrity number
- SNAI2
snail family transcriptional repressor 2
- SKP1
S-phase kinase associated protein 1
- TCGA
The Cancer Genome Atlas
- TNFR1
tumor necrosis factor receptor 1
- ZACN
zinc activated ion channel
- ZNF385C
zinc finger protein 385C
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FW, YF and YL collected the data. FL, WL and XX analyzed and interpreted the data. HL and PG were major contributors in writing the manuscript. HL organized all the data and finally revised the manuscript. PG designed the project. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First People's Hospital of Jining, and informed written consent was obtained from all patients. Primary tumors and adjacent normal tissues of HPSCC patients were obtained based on the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/jco.2011.29.15_suppl.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Monsjou HS, Balm AJ, van den Brekel MM, Wreesmann VB. Oropharyngeal squamous cell carcinoma: A unique disease on the rise? Oral Oncol. 2010;46:780–785. doi: 10.1016/j.oraloncology.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Hashibe M, Brennan P, Chuang SC, Boccia S, Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova E, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18:541–550. doi: 10.1158/1055-9965.EPI-08-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26:612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 5.Gillison ML, Alemany L, Snijders PJ, Chaturvedi A, Steinberg BM, Schwartz S, Castellsagué X. Human papillomavirus and diseases of the upper airway: Head and neck cancer and respiratory papillomatosis. Vaccine. 2012;30(Suppl 5):F34–F54. doi: 10.1016/j.vaccine.2012.05.070. [DOI] [PubMed] [Google Scholar]
- 6.Tartaglia LA, Goeddel DV. Two TNF receptors. Immunol Today. 1992;13:151–153. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- 7.Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 8.Ma X, Li X, Lu X, Jia L, Li H, Song Q. Interaction between TNFR1 and TNFR2 dominates the clinicopathologic features of human hypopharyneal carcinoma. Tumour Biol. 2015;36:9421–9429. doi: 10.1007/s13277-015-3684-8. [DOI] [PubMed] [Google Scholar]
- 9.Kaira K, Toyoda M, Shimizu A, Imai H, Sakakura K, Nikkuni O, Suzuki M, Iijima M, Asao T, Chikamatsu K. Prognostic significance of GRP78/BiP expression in patients with stage III/IV hypopharyngeal squamous cell carcinoma. Neoplasma. 2016;63:477–483. doi: 10.4149/319_151002N513. [DOI] [PubMed] [Google Scholar]
- 10.Jing P, Sa N, Liu X, Liu X, Xu W. MicroR-140-5p suppresses tumor cell migration and invasion by targeting ADAM10-mediated Notch1 signaling pathway in hypopharyngeal squamous cell carcinoma. Exp Mol Pathol. 2016;100:132–138. doi: 10.1016/j.yexmp.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Li M, Yu W, Li W, Wang J, Xiang X, Li G, Pan X, Lei D. AB209630, a long non-coding RNA decreased expression in hypopharyngeal squamous cell carcinoma, influences proliferation, invasion, metastasis, and survival. Oncotarget. 2016;7:14628–14638. doi: 10.18632/oncotarget.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu X, Chen J, Zhang Z, You Y, Wang Z. Aberrant GRK6 promoter methylation is associated with poor prognosis in hypopharyngeal squamous cell carcinoma. Oncol Rep. 2016;35:1027–1033. doi: 10.3892/or.2015.4469. [DOI] [PubMed] [Google Scholar]
- 13.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh S, Chan CK. Analysis of RNA-Seq data using tophat and cufflinks. Methods Mol Biol. 2016;1374:339–361. doi: 10.1007/978-1-4939-3167-5_18. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reiner-Benaim A. FDR control by the BH procedure for two-sided correlated tests with implications to gene expression data analysis. Biom J. 2007;49:107–126. doi: 10.1002/bimj.200510313. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate-a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;57:289–300. [Google Scholar]
- 18.Gołębiowski M, Sosnowska A, Puzyn T, Boguś MI, Wieloch W, Włóka E, Stepnowski P. Application of two-way hierarchical cluster analysis for the identification of similarities between the individual lipid fractions of Lucilia sericata. Chem Biodivers. 2014;11:733–748. doi: 10.1002/cbdv.201300294. [DOI] [PubMed] [Google Scholar]
- 19.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochs C, Perl Y, Halper M, Geller J, Lomax J. Quality assurance of the gene ontology using abstraction networks. J Bioinform Comput Biol. 2016;14:1642001. doi: 10.1142/S0219720016420014. [DOI] [PubMed] [Google Scholar]
- 21.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O'Donnell L, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keaton MA. Morgan DO: The cell cycle: Principles of control (Primers in Biology) Cell Division. 2007;2:27. doi: 10.1186/1747-1028-2-27. [DOI] [Google Scholar]
- 25.Liu YQ, Wang XL, Cheng X, Lu YZ, Wang GZ, Li XC, Zhang J, Wen ZS, Huang ZL, Gao QL, et al. Skp1 in lung cancer: Clinical significance and therapeutic efficacy of its small molecule inhibitors. Oncotarget. 2015;6:34953–34967. doi: 10.18632/oncotarget.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu M, Yang X, Zhao J, Zhang J, Zhang S, Huang H, Liu Y, Liu J. High expression of Cullin1 indicates poor prognosis for NSCLC patients. Pathol Res Pract. 2014;210:397–401. doi: 10.1016/j.prp.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Houtani T, Munemoto Y, Kase M, Sakuma S, Tsutsumi T, Sugimoto T. Cloning and expression of ligand-gated ion-channel receptor L2 in central nervous system. Biochem Biophys Res Commun. 2005;335:277–285. doi: 10.1016/j.bbrc.2005.07.079. [DOI] [PubMed] [Google Scholar]
- 28.Ma D, Shield JP, Dean W, Leclerc I, Knauf C, Burcelin R Ré, Rutter GA, Kelsey G. Impaired glucose homeostasis in transgenic mice expressing the human transient neonatal diabetes mellitus locus, TNDM. J Clin Invest. 2004;114:339–348. doi: 10.1172/JCI200419876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Z, Zhao X, Zhao L, Yang H, Liu L, Li J, Wu J, Yang F, Huang G, Liu J. p54nrb/NONO regulates lipid metabolism and breast cancer growth through SREBP-1A. Oncogene. 2016;35:1399–1410. doi: 10.1038/onc.2015.197. [DOI] [PubMed] [Google Scholar]
- 30.Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183–42194. doi: 10.18632/oncotarget.9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vilming Elgaaen B, Olstad OK, Haug KB, Brusletto B, Sandvik L, Staff AC, Gautvik KM, Davidson B. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC cancer. 2014;14:80. doi: 10.1186/1471-2407-14-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandra V, Kim JJ, Mittal B, Rai R. MicroRNA aberrations: An emerging field for gallbladder cancer management. World J Gastroenterol. 2016;22:1787–1799. doi: 10.3748/wjg.v22.i5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubiczak M, Walkowiak GP, Nowak-Markwitz E, Jankowska A. Human chorionic gonadotropin beta subunit genes CGB1 and CGB2 are transcriptionally active in ovarian cancer. Int J Mol Sci. 2013;14:12650–12660. doi: 10.3390/ijms140612650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hotakainen K, Lintula S, Jarvinen R, Paju A, Stenman J, Rintala E, Stenman UH. Overexpression of human chorionic gonadotropin beta genes 3, 5 and 8 in tumor tissue and urinary cells of bladder cancer patients. Tumour Biol. 2007;28:52–56. doi: 10.1159/000097703. [DOI] [PubMed] [Google Scholar]
- 35.Hotakainen K, Lintula S, Ljungberg B, Finne P, Paju A, Stenman UH, Stenman J. Expression of human chorionic gonadotropin beta-subunit type I genes predicts adverse outcome in renal cell carcinoma. J Mol Diagn. 2006;8:598–603. doi: 10.2353/jmoldx.2006.060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iles RK. Ectopic hCGbeta expression by epithelial cancer: Malignant behaviour, metastasis and inhibition of tumor cell apoptosis. Mol Cell Endocrinol. 2007;260-262:264–270. doi: 10.1016/j.mce.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 37.Iles RK, Delves PJ, Butler SA. Does hCG or hCGβ play a role in cancer cell biology? Mol Cell Endocrinol. 2010;329:62–70. doi: 10.1016/j.mce.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Hotakainen K, Ljungberg B, Haglund C, Nordling S, Paju A, Stenman UH. Expression of the free beta-subunit of human chorionic gonadotropin in renal cell carcinoma: Prognostic study on tissue and serum. Int J Cancer. 2003;104:631–635. doi: 10.1002/ijc.11000. [DOI] [PubMed] [Google Scholar]
- 39.Wu Y, Lu W, Xu J, Shi Y, Zhang H, Xia D. Prognostic value of long non-coding RNA MALAT1 in cancer patients. Tumour Biol. 2016;37:897–903. doi: 10.1007/s13277-015-3870-8. [DOI] [PubMed] [Google Scholar]
- 40.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 41.Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 44.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wurth L, Papasaikas P, Olmeda D, Bley N, Calvo GT, Guerrero S, Cerezo-Wallis D, Martinez-Useros J, García-Fernández M, Hüttelmaier S, et al. UNR/CSDE1 drives a post-transcriptional program to promote melanoma invasion and metastasis. Cancer Cell. 2016;30:694–707. doi: 10.1016/j.ccell.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Fang H, Yue X, Li X, Taylor JS. Identification and characterization of high affinity antisense PNAs for the human unr (upstream of N-ras) mRNA which is uniquely overexpressed in MCF-7 breast cancer cells. Nucleic Acids Res. 2005;33:6700–6711. doi: 10.1093/nar/gki968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH, Chang YC, Lin WC, Shen CY, Huang CS, Hsieh FJ, et al. Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing. Mol Cancer. 2015;14:36. doi: 10.1186/s12943-015-0301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong N, Khwaja SS, Baker CM, Gay HA, Thorstad WL, Daly MD, Lewis JS, Jr, Wang X. Prognostic microRNA signatures derived from The Cancer Genome Atlas for head and neck squamous cell carcinomas. Cancer Med. 2016;5:1619–1628. doi: 10.1002/cam4.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mei LL, Wang WJ, Qiu YT, Xie XF, Bai J, Shi ZZ. miR-125b-5p functions as a tumor suppressor gene partially by regulating HMGA2 in esophageal squamous cell carcinoma. PLoS One. 2017;12:e0185636. doi: 10.1371/journal.pone.0185636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qattan A, Intabli H, Alkhayal W, Eltabache C, Tweigieri T, Amer SB. Robust expression of tumor suppressor miRNA's let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer. 2017;17:799. doi: 10.1186/s12885-017-3776-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choo KB, Soon YL, Nguyen PN, Hiew MS, Huang CJ. MicroRNA-5p and −3p co-expression and cross-targeting in colon cancer cells. J Biomed Sci. 2014;21:95. doi: 10.1186/s12929-014-0095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer. 2015;14:194. doi: 10.1186/s12943-015-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gowrishankar B, Ibragimova I, Zhou Y, Slifker MJ, Devarajan K, Al-Saleem T, Uzzo RG, Cairns P. MicroRNA expression signatures of stage, grade, and progression in clear cell RCC. Cancer Biol Ther. 2014;15:329–341. doi: 10.4161/cbt.27314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lauvrak SU, Munthe E, Kresse SH, Stratford EW, Namløs HM, Meza-Zepeda LA, Myklebost O. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br J Cancer. 2013;109:2228–2236. doi: 10.1038/bjc.2013.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng G, Du L, Yang X, Zhang X, Wang L, Yang Y, Li J, Wang C. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111:1985–1992. doi: 10.1038/bjc.2014.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin X, Chen X, Hu Y, Ying F, Zou R, Lin F, Shi Z, Zhu X, Yan X, Li S, Zhu H. LncRNA-TCONS_00026907 is involved in the progression and prognosis of cervical cancer through inhibiting miR-143-5p. Cancer Med. 2017;6:1409–1423. doi: 10.1002/cam4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pekarsky Y, Croce CM. Role of miR-15/16 in CLL. Cell Death Differ. 2015;22:6–11. doi: 10.1038/cdd.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang E, Liu R, Chu Y. miRNA-15a/16: As tumor suppressors and more. Future Oncol. 2015;11:2351–2363. doi: 10.2217/fon.15.101. [DOI] [PubMed] [Google Scholar]
- 60.Chen D, Wu D, Shao K, Ye B, Huang J, Gao Y. MiR-15a-5p negatively regulates cell survival and metastasis by targeting CXCL10 in chronic myeloid leukemia. Am J Transl Res. 2017;9:4308–4316. [PMC free article] [PubMed] [Google Scholar]
- 61.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 62.Slattery ML, Pellatt AJ, Lee FY, Herrick JS, Samowitz WS, Stevens JR, Wolff RK, Mullany LE. Infrequently expressed miRNAs influence survival after diagnosis with colorectal cancer. Oncotarget. 2017;8:83845–83859. doi: 10.18632/oncotarget.19863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao XD, Li P, Wang JS. MicroRNA differential expression spectrum and microRNA-125a-5p inhibition of laryngeal cancer cell proliferation. Exp Ther Med. 2017;14:1699–1705. doi: 10.3892/etm.2017.4685. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Odar K, Boštjančič E, Gale N, Glavač D, Zidar N. Differential expression of microRNAs miR-21, miR-31, miR-203, miR-125a-5p and miR-125b and proteins PTEN and p63 in verrucous carcinoma of the head and neck. Histopathology. 2012;61:257–265. doi: 10.1111/j.1365-2559.2012.04242.x. [DOI] [PubMed] [Google Scholar]
- 65.Leotta M, Biamonte L, Raimondi L, Ronchetti D, Di Martino MT, Botta C, Leone E, Pitari MR, Neri A, Giordano A, et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J Cell Physiol. 2014;229:2106–2116. doi: 10.1002/jcp.24669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.