 Compound PBP-NC10 bearing the decylaminoethyl group was identified as a novel SERD.
Compound PBP-NC10 bearing the decylaminoethyl group was identified as a novel SERD.
Abstract
Estrogen receptors (ERs) are a family of nuclear receptors (NRs) that regulate physiological effects such as reproduction and bone homeostasis. It has been reported that approximately 70% of human breast cancers are hormone-dependent and ERα-positive. Recently, novel anti-breast cancer drugs based on different mechanisms of action have received significant attention. In this article, we have designed and synthesized a selective ER degradation inducer based on the diphenylheptane skeleton. Western blotting analysis revealed that PBP-NC10 degraded ERα through the ubiquitin–proteasome system. We also performed computational docking analysis to predict the binding mode of PBP-NC10 to ERα.
Introduction
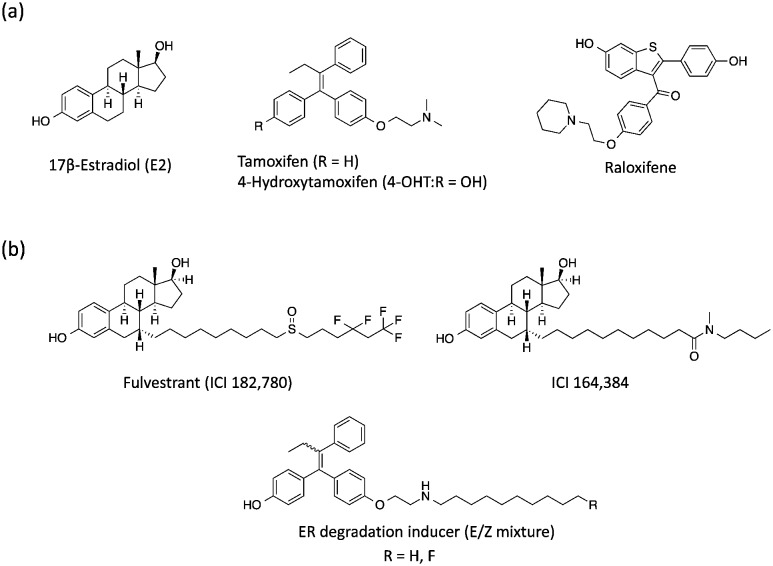
Estrogen receptor (ER), a member of the nuclear receptor (NR) family of transcription factors, relates to several biological functions such as reproduction and bone physiology.1 The binding of 17β-estradiol (E2) and its metabolites, estrone and estriol, to the ER ligand binding domain (LBD) induces the folding of Helix 12 (H12) and the subsequent interaction with co-activators along the hydrophobic groove, resulting in ER-mediated target gene transcriptions.2 The ER is divided into two subtypes, ERα and ERβ, which are encoded by distinct genes but are highly conserved in their LBD.3 It is well known that approximately 70% of human breast cancers are hormone-dependent and ERα-positive.4 Therefore, several ERα-mediated transcriptional inhibitors such as tamoxifen (ERα antagonist)5 and raloxifene6 are widely used for ERα-positive breast cancer therapy (Fig. 1a).7 Tamoxifen is metabolized to 4-hydroxytamoxifen (4-OHT) with a much higher binding affinity for ERα, and 4-OHT exerts its anti-estrogenic actions.8 However, it has been problematic that long-term administration of tamoxifen might cause drug resistance in breast cancer cells and also increases the risk factors for uterine cancer because of its agonistic effects on the uterus. Therefore, novel anti-breast cancer drugs based on different mechanisms of action have received significant attention. Recently, several methods to induce targeted protein degradation using small molecules have been reported such as PROTAC,9 SNIPER,10 and hydrophobic tag systems.11
Fig. 1. (a) Chemical structures of 17β-estradiol, anti-estrogenic compounds and (b) selective estrogen receptor downregulators.
Among these, selective ER downregulators (SERDs) have been discussed as anti-breast cancer drugs. Fulvestrant (ICI182,780) and ICI164,384 (Fig. 1b), which are E2 derivatives bearing a long alkyl chain on the 7α-position of E2, are representative SERDs and are capable of inducing ER degradation through the ubiquitin–proteasome system (UPS).12 Based on the X-ray structure analysis of ICI164,384 bound to the ER-LBD, the ER-degradation mechanism of ICI164,384 is speculated to involve binding of the long alkyl chain of ICI164,384 to the hydrophobic groove of ER, sterically preventing H12 folding in ER and the co-activator interaction with ER, resulting in destabilization of ER and subsequent degradation.13
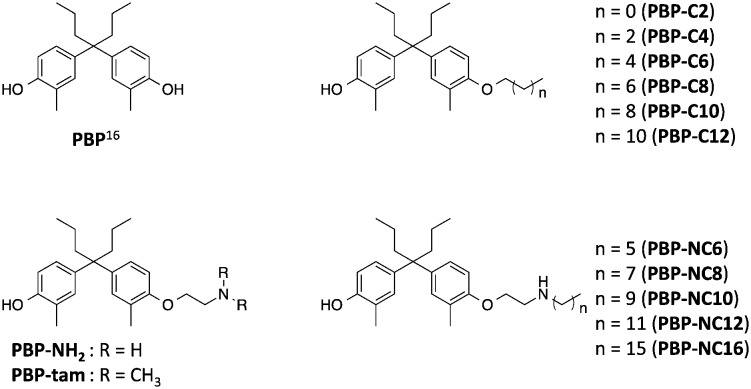
Previously, we reported that 4-OHT derivatives bearing long alkyl chains (Fig. 1b) induced ER degradation similar to fulvestrant.14 However, these synthesized compounds were obtained and assayed as a mixture of geometric isomer E and Z forms. It has been reported that the triphenylethylene skeleton of 4-OHT is easily isomerized from the active Z form to an inactive E form in solution.15 Therefore, these findings raise the possibilities that its isomerization decreases the ER-antagonistic and ER-downregulating activities. On the other hand, Hashimoto and Ishikawa et al. recently reported that the diphenylalkane skeleton acted as a steroid skeleton mimic and could be a key to the development of several NR ligands.16 Among these, the ER antagonist PBP based on the diphenylheptane skeleton has potent anti-estrogenic activity with an IC50 value of 4.5 nM from a reporter gene assay.17 Moreover, the diphenylalkane skeleton has an advantage of good chemical stability and for short-step synthesis over the triphenylethylene skeleton of 4-OHT. Herein, we described the development of PBP derivatives bearing several long alkyl chains on the hydroxy group (Fig. 2), and evaluated their biological properties such as ER degradation, binding affinity, transcriptional activity and anti-proliferation activity.
Fig. 2. Chemical structures of the diphenylheptane skeleton (PBP) and designed compounds PBP-NH2, PBP-tam, PBP-C2 to PBP-C12, and PBP-NC6 to PBP-NC16 in this study.
Results and discussion
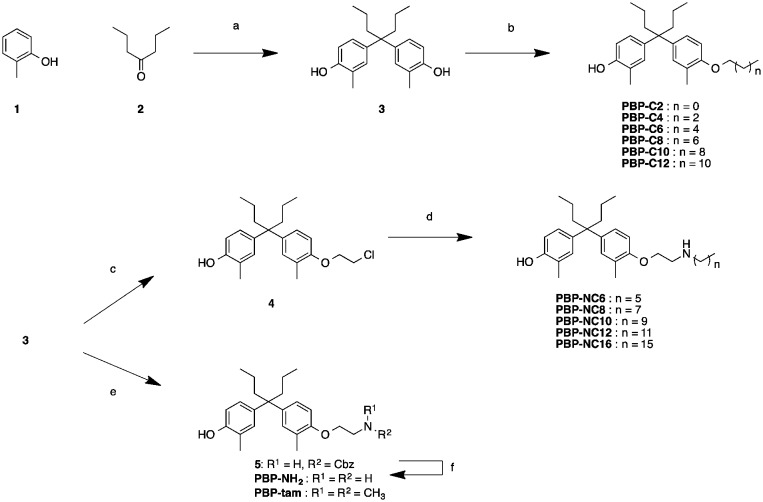
The synthetic routes to the designed compounds PBP-C2–C12, PBP-NC6–NC16, PBP-NH2, and PBP-tam are shown in Scheme 1. 4-Heptanone and o-cresol were combined under acidic conditions to give the diphenylheptane skeleton (PBP, 3). Alkylation of the 4-hydroxy group of PBP with the corresponding 1-bromoalkanes afforded alkylated products PBP-C2–C12. Compounds PBP-NC6–NC16 were synthesized by mono-alkylation of PBP with 1,2-dichloroethane and subsequent alkylation of alkylamines with compound 4. After alkylation of PBP with benzyl (2-bromoethyl)carbamate, the Cbz group was removed using Pd(OH)2 under H2 atmosphere to give PBP-NH2. PBP-tam was obtained by the treatment of PBP with 2-chloro-N,N-dimethylethylamine.
Scheme 1. Synthesis of compounds PBP-NH2, PBP-tam, PBP-C2 to PBP-C12, and PBP-NC6 to PBP-NC16. Reagents and conditions: (a) MsOH, rt, 3 days, 59%; (b) R-I (R = ethyl, n-hexyl, n-octyl, n-decyl, n-dodecyl), K2CO3, MeCN, 60–100 °C, 16–47%; (c) 1,2-dichloroethane, K2CO3, MeCN, 48 h, 19%; (d) R-NH2 (R = n-hexyl, n-octyl, n-decyl, n-dodecyl, n-hexadecyl), MeCN, microwave, 120 °C, 4 h, 20–42%; (e) benzyl (2-bromoethyl)carbamate or 2-chloro-N,N-dimethylethylamine Cs2CO3, NaI, DMF, 100 °C, 21 h, 6%; (f) Pd(OH)2, H2, MeOH, rt, 26 h, 25%.
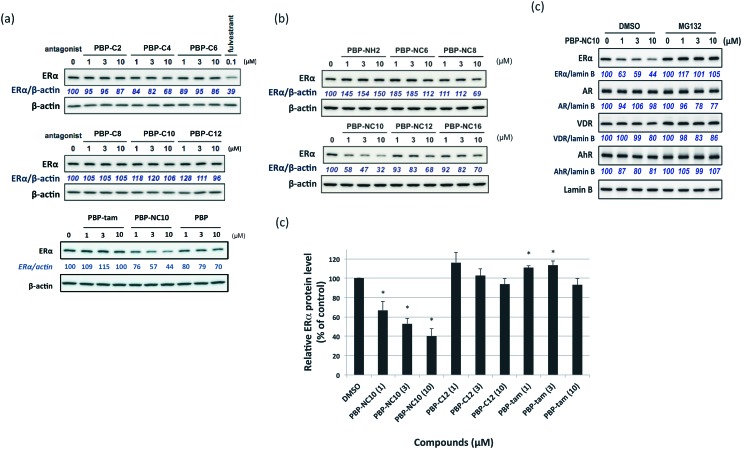
First, we investigated the effects of the synthesized compounds on ERα-downregulating activity by western blotting analysis using MCF-7 breast cancer cells. The potency of the ERα-downregulating activity is described as the ratio of ERα to standard proteins. As shown in Fig. 3, ERα downregulation was observed by treatment with PBP-NC8, PBP-NC10, and PBP-NC12 with amino alkyl chains (ERα downregulation ratio at 10 μM; PBP-NC8: 69%, PBP-NC10: 32%, and PBP-NC12: 68%). In particular, the ERα expression level was dose-dependently reduced in the presence of PBP-NC10 (ERα downregulation ratio at 1 μM: PBP-NC10: 58%). On the other hand, PBP-C2–C12, with simple alkyl chains, PBP-NC6 and PBP-NC16, with short or long alkyl chains, and PBP-tam, with a dimethylamino group, did not show ERα-downregulating activity (Fig. 3a and b). These results indicated that the nitrogen atom and the optimal length of the alkyl chain of PBP-NC derivatives play a pivotal role in the ERα-downregulating activity.
Fig. 3. (a and b) Western blotting analysis of estrogen receptor (ER) α levels in MCF-7 cells after 6 h of incubation with the synthesized compounds. (c) Quantification of the levels of ERα is provided in a histogram. Data are expressed as means ± S.E. of three independent experiments (*P < 0.05 versus control).
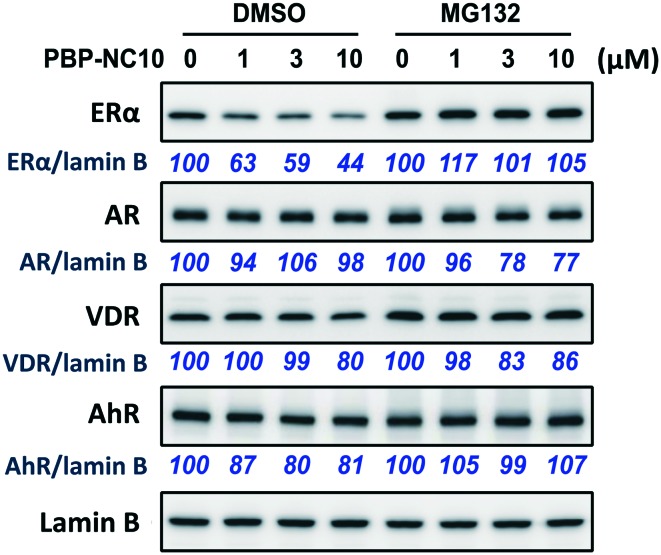
Next, we investigated the selectivity of PBP-NC10 over other NRs such as the androgen receptor (AR), vitamin D receptor (VDR), and aryl hydrocarbon receptor (AhR) because the diphenylalkane skeleton could act as several NR ligands.16 Moreover, we also performed the western blotting analysis in the presence of proteasome inhibitor MG132 to investigate whether ERα downregulation by PBP-NC10 was mediated by UPS as hypothesized. As shown in Fig. 4, ERα was downregulated by treatment with PBP-NC10 in a dose-dependent manner, whereas the other NR expression levels were unaffected. Furthermore, reductions in the expression levels of ERα induced by PBP-NC10 were suppressed by MG132, indicating that PBP-NC10 induced proteasomal degradation.
Fig. 4. The effects of PBP-NC10 on nuclear receptor levels in MCF-7 cells with or without proteasome inhibitor MG132. ER, estrogen receptor; AR, androgen receptor; VDR, vitamin D receptor; AhR, aryl hydrocarbon receptor.
Next, we investigated the effects of the synthesized compounds on the binding affinity and transcriptional activity against ERα. To evaluate the ability of the compounds to bind to ERα, a fluorescence polarization (FP)-based competitive binding assay was conducted using a PolarScreen nuclear receptor competitor assay kit (Life Technologies).14 As reported previously, 4-OHT and PBP strongly bound to ERα with IC50 values of 5.6 nM and 7.3 nM, respectively. Table 1 shows that PBP-C2 to PBP-C8 exhibited moderate binding affinity to ERα (IC50 = 40–400 nM), whereas PBP-C10 and PBP-C12 did not bind to ERα. These data indicated that the introduction of longer alkyl chains rather than an n-octyl group lowered the binding affinity. On the other hand, PBP-NC8 to PBP-NC12 showed pronounced ERα binding affinity (IC50 values of 2.4 nM for PBP-NC8, 4.9 nM for PBP-NC10, and 4.9 nM for PBP-NC12), which was similar to that of 4-OHT (IC50 value of 5.6 nM) in the FP assay. Moreover, PBP-tam exhibited lower binding affinity to ERα compared with PBP-NH2 and PBP-NC derivatives (PBP-tam: IC50 = 313 nM, PBP-NH2: IC50 = 78 nM). These results revealed that the hydrogen atom on the nitrogen atom of PBP-NC derivatives was important for binding to ERα.
Table 1. ERα binding affinity of PBP derivatives.
| Entry | Binding affinity IC50 (nM) |
| 4-OHT | 5.6 |
| PBP | 7.3 |
| PBP-C2 | 40 |
| PBP-C4 | 87 |
| PBP-C6 | 168 |
| PBP-C8 | 400 |
| PBP-C10 | NA a |
| PBP-C12 | NA a |
| PBP-tam | 313 |
| PBP-NH2 | 78 |
| PBP-NC6 | 78 |
| PBP-NC8 | 2.4 |
| PBP-NC10 | 4.9 |
| PBP-NC12 | 4.9 |
| PBP-NC16 | 20 |
aNo activity at 1 μM.
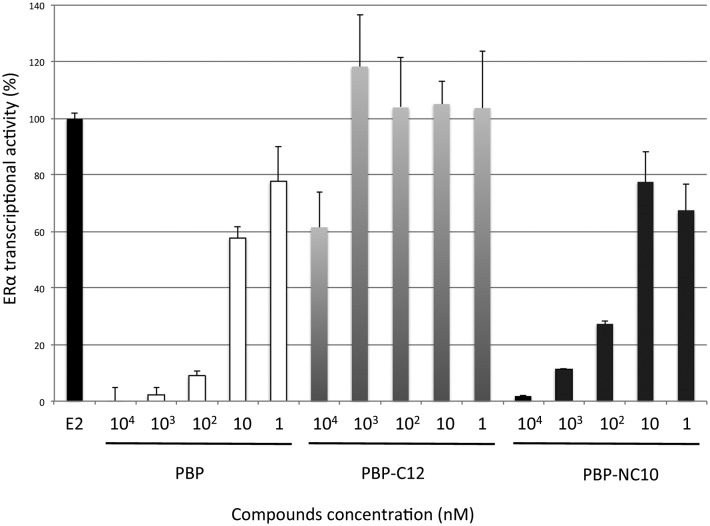
Furthermore, the ERα transcriptional activity was evaluated using a reporter gene assay with CMX-GAL4N-hERα as the recombinant receptor gene, TK-MH100x4-LUC as a reporter gene, and the CMX-β-galactosidase gene for normalization, as reported previously.18 Human Embryonic Kidney 293 (HEK293) cells were incubated with the synthesized compounds in the absence or presence of E2 (0.3 nM). After incubation, the cells were assayed for reporter gene and β-galactosidase activities. As shown in Fig. 5, it was demonstrated that PBP-NC10 exhibited potent ERα antagonistic activity, whereas PBP-C12, which has the same length of alkyl chain as PBP-NC10, showed slight ERα antagonistic activity (ERα antagonistic activity: PBP-C12, 61% inhibition at 10 μM; PBP-NC10, IC50 = 45 nM).
Fig. 5. ERα antagonistic activity of PBP, PBP-C12 and PBP-NC10 in HEK293 cells. Data are expressed as means ± S.E. of three independent experiments.
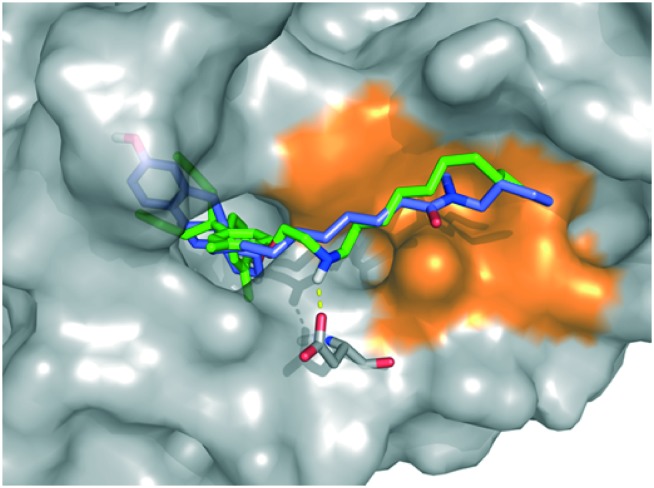
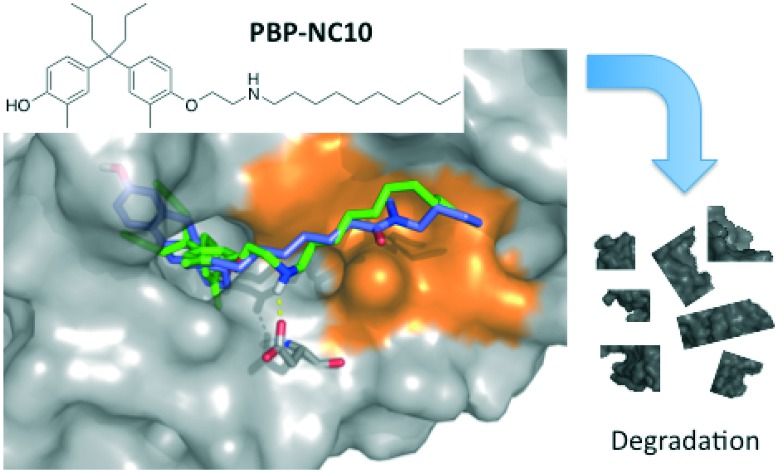
Finally, we performed a computational docking analysis to predict the binding mode of PBP-NC10 to ERα as follows; the X-ray co-crystal structure of ERα bound to 4-OHT was obtained from the Protein Data Bank (PDB: ; 3ERT) and used in docking studies.19 Docking models of PBP-NC10 bound to ERα were constructed via a conformational search analysis using Molecular Operating Environment (MOE). MMFF94S was used as a force field. In the modeled structures, the amino group of PBP-NC10 interacted with the side-chain carboxyl group of Glu351 and the alkyl chain of PBP-NC10 was included in the hydrophobic groove composed of hydrophobic amino acids such as Leu354, Met357, ILe358, Leu379, and Trp383 of ERα. Therefore, these results indicated that the amino group of PBP-NC10 played an important role in leading the alkyl chain to the hydrophobic groove. Furthermore, the results of the docking analysis suggested that the length of the hydrophobic groove interacting with the long alkyl chain was the best around a decyl group on the amino group of PBP-NC10 (Fig. 6).
Fig. 6. Computational model of the estrogen receptor (ER) α ligand binding domain and PBP-NC10, ICI164,384. The hydrophobic region is colored orange, PBP-NC10 is colored green, and ICI164,384 is colored blue.
Conclusions
We have developed a novel selective estrogen degradation inducer PBP-NC10, which could be useful as a lead structure in the development of compounds with more potent and selective ER degradation-inducing activities. We also revealed that the amino group and optimal length of the long alkyl chain of PBP-NC10 are essential for exerting its ERα-downregulating activity. Furthermore, a docking study was performed and predicted that the interaction between the hydrogen atom on the amino group of PBP-NC10 and the carboxylic acid of Glu351 of ERα leads the long alkyl chain to bind to the hydrophobic groove of ERα.
Experimental section
Chemistry
4-Hydroxytamoxifen was purchased from Fluka. All other reagents and solvents were purchased from Sigma-Aldrich, Wako Pure Chemical and Tokyo Chemical Industry, and were used without purification. Analytical TLC was carried out using Merck silica gel 60 F254 pre-coated plates and visualized using a 254 nm UV lamp, phosphomolybdic acid, p-anisaldehyde or ninhydrin staining. Column chromatography was performed with silica gel (spherical, neutral) purchased from Kanto Chemical. The microwave reactions were carried out using a Biotage Initiator. 1H and 13C NMR spectra were obtained on a Varian AS 400 Mercury spectrometer (400 MHz for 1H and 100 MHz for 13C). FT-IR spectra were recorded on a JASCO FT/IR-4100 spectrometer at 1 cm–1 resolution, with an average of 128 scans used for the solution (CDCl3) method and a 0.1 mm path length for NaCl cells. Chemical shifts are expressed as ppm downfield from a solvent residual peak or internal standard tetramethylsilane (TMS). High-resolution mass spectra were obtained on a Shimadzu IT-TOF MS equipped with an electrospray ionization source.
4-[4-(4-Ethoxy-3-methylphenyl)heptan-4-yl]-2-methylphenol (PBP-C2)
A solution of 321 (95.5 mg, 0.31 mmol) and K2CO3 (47.4 mg, 0.34 mmol) in MeCN (2 mL) was added ethyl iodide (47 mg, 0.34 mmol) and stirred at 60 °C for 6 h. The reaction mixture was acidified with 3% HCl and extracted with Et2O. The organic layer was washed with brine and dried over Na2SO4. After filtration, concentration in vacuo, and purification by silica gel flash column chromatography (AcOEt/hexane = 1/4), PBP-C2 (35.5 mg, 34%) was obtained as colorless oil; IR (CDCl3, cm–1): 3602, 2958, 2932, 2872, 1607, 1503, 1477; 1H NMR (400 MHz, CDCl3): δ 6.90–6.93 (m, 3H), 6.85 (d, J = 8.4 Hz, 1H), 6.67 (d, J = 8.4 Hz, 1H), 6.61 (d, J = 8.4 Hz, 1H), 4.72 (br s, 1H), 3.98 (q, J = 14.0 Hz, 2H), 2.18 (s, 3H), 2.16 (s, 3H), 1.92–1.96 (m, 4H), 1.39 (t, J = 7.2 Hz, 3H), 0.93–0.98 (m, 4H), 0.81–0.85 (m, 6H); 13C NMR (100 MHz, CDCl3): δ 154.9, 151.5, 142.1, 141.3, 130.6, 130.5, 126.8, 126.1, 125.7, 122.7, 114.3, 110.2, 63.7, 48.2, 40.6, 17.6, 16.9, 16.4, 15.3, 15.2; [HR-ESI(+)]: m/z calcd for C23H33O2 [M + H]+: 341.2475, found: 341.2445.
4-[4-(4-n-Butoxy-3-methylphenyl)heptan-4-yl]-2-methylphenol (PBP-C4)
Compound PBP-C4 was prepared from 3 (62.0 mg, 0.20 mmol) using the procedure described for PBP-C2. Yield 16%; colorless oil; IR (CDCl3, cm–1): 3603, 2959, 2933, 2872, 1605, 1503, 1468; 1H NMR (400 MHz, CDCl3): δ 6.89–6.93 (m, 3H), 6.86 (d, J = 8.0 Hz, 1H), 6.67 (d, J = 8.4 Hz, 1H), 6.64 (d, J = 8.4 Hz, 1H), 4.47 (br s, 1H), 3.93 (t, J = 6.2 Hz, 2H), 2.19 (s, 3H), 2.16 (s, 3H), 1.91–1.95 (m, 4H), 1.73–1.79 (m, 2H), 1.48–1.53 (m, 2H), 0.91–0.99 (m, 7H), 0.82–0.85 (m, 6H); 13C NMR (100 MHz, CDCl3): δ 154.8, 151.1, 141.9, 140.8, 130.3, 130.1, 126.6, 125.8, 125.4, 122.3, 114.0, 109.7, 67.4, 47.9, 40.3, 31.6, 19.4, 17.3, 16.5, 16.1, 14.9, 14.0; [HR-ESI(+)]: m/z calcd for C25H37O2 [M + H]+: 369.2788, found: 369.2826.
4-[4-(4-n-Hexyloxy-3-methylphenyl)heptan-4-yl]-2-methylphenol (PBP-C6)
Compound PBP-C6 was prepared from 3 (62.0 mg, 0.20 mmol) using the procedure described for PBP-C2. Yield 47%; colorless oil; IR (CDCl3, cm–1): 3602, 2958, 2932, 2871, 1607, 1503, 1468; 1H NMR (400 MHz, CDCl3): δ 6.89–6.93 (m, 3H), 6.86 (d, J = 8.4 Hz, 1H), 6.66 (d, J = 8.4 Hz, 1H), 6.62 (d, J = 8.4 Hz, 1H), 4.59 (br s, 1H), 3.92 (t, J = 6.6 Hz, 2H), 2.19 (s, 3H), 2.16 (s, 3H), 1.92–1.96 (m, 4H), 1.73–1.80 (m, 2H), 1.43–1.49 (m, 2H), 1.31–1.36 (m, 4H), 0.88–0.98 (m, 7H), 0.84 (t, J = 7.0 Hz, 6H); 13C NMR (100 MHz, CDCl3): δ 154.8, 151.2, 141.9, 140.9, 130.4, 130.2, 126.5, 125.8, 125.5, 122.4, 114.0, 109.8, 67.8, 47.9, 40.4, 31.7, 29.5, 25.9, 22.7, 17.3, 16.6, 16.1, 14.9, 14.1; [HR-ESI(+)]: m/z calcd for C27H41O2 [M + H]+: 397.3101, found: 397.3078.
4-[4-(4-n-Octyloxy-3-methylphenyl)heptan-4-yl]-2-methylphenol (PBP-C8)
Compound PBP-C8 was prepared from 3 (59.1 mg, 0.19 mmol) using the procedure described for PBP-C2. Yield 13%; colorless oil; IR (CDCl3, cm–1): 3602, 2958, 2931, 2871, 1607, 1503, 1468; 1H NMR (400 MHz, CDCl3): δ 6.91–6.93 (m, 3H), 6.85 (d, J = 8.4 Hz, 1H), 6.66 (d, J = 8.4 Hz, 1H), 6.62 (d, J = 8.4 Hz, 1H), 4.81 (br s, 1H), 3.91 (t, J = 6.2 Hz, 2H), 2.19 (s, 3H), 2.16 (s, 3H), 1.91–1.95 (m, 4H), 1.75–1.79 (m, 2H), 1.45–1.46 (m, 2H), 1.30–1.31 (m, 8H), 0.82–0.97 (m, 13H); 13C NMR (100 MHz, CDCl3): δ 155.1, 151.6, 142.2, 141.2, 130.7, 130.5, 126.9, 126.2, 125.8, 122.8, 114.3, 110.1, 68.2, 48.3, 40.7, 32.2, 29.9, 29.8, 29.7, 26.6, 23.1, 17.7, 16.9, 16.5, 15.3, 14.6, 14.5; [HR-ESI(+)]: m/z calcd for C29H45O2 [M + H]+: 425.3414, found: 425.3392.
4-[4-(4-n-Decyloxy-3-methylphenyl)heptan-4-yl]-2-methylphenol (PBP-C10)
Compound PBP-C10 was prepared from 3 (95.3 mg, 0.31 mmol) using the procedure described for PBP-C2. Yield 39%; colorless oil; IR (CDCl3, cm–1): 3602, 2957, 2929, 2871, 1607, 1503, 1468; 1H NMR (400 MHz, CDCl3): δ 6.90–6.93 (m, 3H), 6.85 (d, J = 8.4 Hz, 1H), 6.66 (d, J = 8.4 Hz, 1H), 6.61 (d, J = 8.4 Hz, 1H), 4.69 (br s, 1H), 3.91 (t, J = 6.8 Hz, 2H), 2.18 (s, 3H), 2.16 (s, 3H), 1.92–1.96 (m, 4H), 1.73–1.80 (m, 2H), 1.42–1.48 (m, 2H), 1.23–1.34 (m, 12H), 0.81–0.99 (m, 13H); 13C NMR (100 MHz, CDCl3): δ 154.8, 151.2, 141.9, 140.9, 130.4, 130.2, 126.5, 125.8, 125.5, 122.5, 114.0, 109.8, 67.8, 47.9, 40.4, 32.0, 29.7, 29.6, 29.5, 29.4, 26.2, 22.7, 17.3, 16.6, 16.1, 14.9, 14.2; [HR-ESI(+)]: m/z calcd for C31H49O2 [M + H]+: 453.3727, found: 453.3767.
4-[4-(4-n-Dodecyloxy-3-methylphenyl)heptan-4-yl]-2-methylphenol (PBP-C12)
Compound PBP-C12 was prepared from 3 (92.8 mg, 0.30 mmol) using the procedure described for PBP-C2. Yield 33%; colorless oil; IR (CDCl3, cm–1): 3607, 2927, 2855, 1604, 1505, 1457; 1H NMR (400 MHz, CDCl3): δ 6.89–6.93 (m, 3H), 6.86 (d, J = 8.4 Hz, 1H), 6.66 (d, J = 8.4 Hz, 1H), 6.64 (d, J = 8.4 Hz, 1H), 4.47 (br s, 1H), 3.91 (d, J = 6.4 Hz, 2H), 2.19 (s, 3H), 2.16 (s, 3H), 1.91–1.95 (m, 4H), 1.75–1.79 (m, 2H), 1.44–1.46 (m, 2H), 1.24–1.34 (m, 16H), 0.82–0.98 (m, 13H); 13C NMR (100 MHz, CDCl3): δ 154.8, 151.1, 141.9, 140.8, 130.3, 130.1, 126.6, 125.8, 125.4, 122.3, 114.0, 109.7, 67.7, 47.9, 40.3, 31.9, 29.7, 29.6, 29.5, 29.4, 26.2, 22.7, 17.3, 16.5, 16.1, 14.9, 14.1; [HR-ESI(+)]: m/z calcd for C33H53O2 [M + H]+: 481.4040, found: 481.4002.
4-{4-[4-(2-Chloroethoxy)-3-methylphenyl]heptan-4-yl}-2-methylphenol (4)
To a solution of 3 (525.8 mg, 1.7 mmol) in MeCN (10 mL) was added 1,2-dichloroethane (333 mg, 3.4 mmol) and K2CO3 (705 mg, 5.1 mmol) at room temperature. The mixture was stirred for 48 h at room temperature, brine was added and the whole mixture was extracted with ethyl acetate. The organic layer was dried over Na2SO4, filtered, and then concentrated in vacuo. The residue was purified by column chromatography (ethyl acetate/n-hexane = 1/4) to give 4 (117 mg, 19%) as colorless oil; IR (CDCl3, cm–1): 3602, 2958, 2932, 2071, 1607, 1503, 1457; 1H NMR (400 MHz, CDCl3): δ 6.89–6.95 (m, 3H), 6.83 (d, J = 8.4 Hz, 1H), 6.65 (t, J = 7.8 Hz, 2H), 5.58 (br s, 1H), 4.18 (t, J = 5.8 Hz, 2H), 3.79 (t, J = 5.8 Hz, 2H), 2.18 (s, 6H), 1.92–1.96 (m, 4H), 0.82–0.99 (m, 10H); 13C NMR (100 MHz, CDCl3): δ 153.8, 151.6, 142.2, 141.4, 130.5, 130.3, 126.4, 125.9, 122.8, 114.0, 110.3, 68.1, 48.0, 42.3, 40.3, 31.6, 22.7, 17.3, 16.5, 16.2, 14.9; [HR-ESI(+)]: m/z calcd for C23H32ClO2 [M + H]+: 375.2085, found: 375.2051.
4-(4-{4-[2-(n-Hexylamino)ethoxy]-3-methylphenyl}heptan-4-yl)-2-methylphenol (PBP-NC6)
A solution of 4 (49.0 mg, 0.13 mmol) in MeCN (0.5 mL) was added n-hexylamine (66.0 mg, 0.908 mmol) and stirred at 120 °C for 4 h assisted microwave irradiation. The reaction mixture was concentrated in vacuo and purified by silica gel flash column chromatography (dichloromethane) to give PBP-NC6 (24.2 mg, 0.055 mmol, 42%) as colorless oil. IR (CDCl3, cm–1): 3602, 2958, 2931, 2871, 1606, 1503, 1466; 1H NMR (400 MHz, CDCl3): δ 6.88–6.93 (m, 3H), 6.71 (d, J = 8.0 Hz, 1H), 6.63 (d, J = 8.4 Hz, 1H), 6.55 (d, J = 8.4 Hz, 1H), 4.05 (t, J = 4.4 Hz, 2H), 3.03 (t, J = 4.2 Hz, 2H), 2.73 (t, J = 7.0 Hz, 2H), 2.17 (s, 3H), 2.13 (s, 3H), 1.91–1.94 (m, 4H), 1.54–1.57 (m, 2H), 1.28–1.32 (m, 6H), 0.82–0.95 (m, 13H); 13C NMR (100 MHz, CDCl3): δ 154.7, 152.6, 142.2, 141.3, 130.9, 130.7, 127.0, 126.4, 125.8, 123.5, 114.6, 110.4, 66.9, 50.0, 49.1, 48.4, 40.9, 32.2, 29.9, 27.4, 23.1, 17.8, 17.1, 16.9, 15.4, 14.6; [HR-ESI(+)]: m/z calcd for C29H46NO2 [M + H]+: 440.3523, found: 440.3482.
4-(4-{4-[2-(n-Octylamino)ethoxy]-3-methylphenyl}heptan-4-yl)-2-methylphenol (PBP-NC8)
Compound PBP-NC8 was prepared from 4 (57.0 mg, 0.15 mmol) using the procedure described for PBP-NC6. Yield 34%; colorless oil; IR (CDCl3, cm–1): 3606, 2958, 2930, 2871, 1605, 1504, 1457; 1H NMR (400 MHz, CDCl3): δ 6.89–6.93 (m, 3H), 6.78 (d, J = 8.4 Hz, 1H), 6.65 (d, J = 8.4 Hz, 1H), 6.59 (d, J = 8.4 Hz, 1H), 4.08 (t, J = 4.8 Hz, 2H), 3.06 (t, J = 4.8 Hz, 2H), 2.76 (t, J = 7.6 Hz, 2H), 2.18 (s, 3H), 2.15 (s, 3H), 1.91–1.95 (m, 4H), 1.56–1.60 (m, 2H), 1.26–1.29 (m, 10H), 0.82–0.98 (m, 13H); 13C NMR (100 MHz, CDCl3): δ 154.7, 152.1, 142.2, 141.8, 130.8, 130.7, 127.0, 126.4, 125.8, 123.2, 114.6, 110.5, 66.8, 50.0, 49.0, 48.4, 40.8, 32.3, 29.9, 29.8, 27.7, 23.2, 17.8, 17.1, 16.7, 15.4, 14.6; [HR-ESI(+)]: m/z calcd for C31H50NO2 [M + H]+: 468.3836, found: 468.3815.
4-(4-{4-[2-(n-Decylamino)ethoxy]-3-methylphenyl}heptan-4-yl)-2-methylphenol (PBP-NC10)
Compound PBP-NC10 was prepared from 4 (33 mg, 0.09 mmol) using the procedure described for PBP-NC6. Yield 32%; colorless oil; IR (CDCl3, cm–1): 3603, 2957, 2929, 2856, 1606, 1503, 1466; 1H NMR (400 MHz, CDCl3): δ 6.88–6.93 (m, 3H), 6.74 (d, J = 8.4 Hz, 1H), 6.64 (d, J = 8.4 Hz, 1H), 6.57 (d, J = 8.4 Hz, 1H), 4.06 (br, 2H), 3.04 (br, 2H), 2.73 (t, J = 7.6 Hz, 2H), 2.18 (s, 3H), 2.14 (s, 3H), 1.91–1.94 (m, 4H), 1.54–1.56 (m, 2H), 1.25–1.29 (m, 14H), 0.82–0.95 (m, 13H); 13C NMR (100 MHz, CDCl3): δ 154.7, 152.4, 142.1, 141.4, 130.8, 130.6, 126.9, 126.4, 125.8, 123.4, 114.6, 110.4, 66.9, 50.0, 49.1, 48.4, 40.8, 40.2, 32.4, 30.1, 30.0, 29.8, 27.7, 27.4, 23.2, 17.8, 17.1, 16.8, 15.4, 14.6; [HR-ESI(+)]: m/z calcd for C33H54NO2 [M + H]+: 496.4149, found: 496.4101.
4-(4-{4-[2-(n-Dodecylamino)ethoxy]-3-methylphenyl}heptan-4-yl)-2-methylphenol (PBP-NC12)
Compound PBP-NC12 was prepared from 4 (68.7 mg, 0.183 mmol) using the procedure described for PBP-NC6. Yield 26%; colorless oil; IR (CDCl3, cm–1): 3603, 2957, 2920, 2855, 1605, 1503, 1465; 1H NMR (400 MHz, CDCl3): δ 6.88–6.93 (m, 3H), 6.77 (d, J = 8.4 Hz, 1H), 6.65 (d, J = 8.4 Hz, 1H), 6.59 (d, J = 8.4 Hz, 1H), 4.07 (t, J = 4.8 Hz, 2H), 3.05 (t, J = 5.2 Hz, 2H), 2.75 (t, J = 7.6 Hz, 2H), 2.18 (s, 3H), 2.14 (s, 3H), 1.91–1.95 (m, 4H), 1.53–1.58 (m, 2H), 1.25–1.30 (m, 18H), 0.82–0.98 (m, 13H); 13C NMR (100 MHz, CDCl3): δ 154.2, 151.6, 141.6, 141.3, 130.3, 130.2, 126.5, 125.9, 125.3, 122.7, 114.1, 109.9, 66.4, 49.5, 48.6, 47.9, 40.3, 39.7, 31.9, 29.7, 29.6, 29.5, 29.4, 29.3, 27.2, 26.9, 22.7, 17.3, 16.6, 16.2, 14.9, 14.1; [HR-ESI(+)]: m/z calcd for C35H58NO2 [M + H]+: 524.4462, found: 524.4445.
4-(4-{4-[2-(n-Hexadecylamino)ethoxy]-3-methylphenyl}heptan-4-yl)-2-methylphenol (PBP-NC16)
Compound PBP-NC16 was prepared from 4 (28.1 mg, 0.08 mmol) using the procedure described for PBP-NC6. Yield 34%; colorless oil; IR (CDCl3, cm–1): 3603, 2957, 2927, 2855, 1606, 1503, 1466; 1H NMR (400 MHz, CDCl3): δ 6.88–6.93 (m, 3H), 6.75 (d, J = 8.4 Hz, 1H), 6.63 (d, J = 8.8 Hz, 1H), 6.58 (d, J = 8.4 Hz, 1H), 4.05 (t, J = 4.8 Hz, 2H), 3.04 (t, J = 5.0 Hz, 2H), 2.73 (t, J = 7.4 Hz, 2H), 2.18 (s, 3H), 2.13 (s, 3H), 1.91–1.95 (m, 4H), 1.25–1.28 (m, 28H), 0.82–0.98 (m, 13H); 13C NMR (100 MHz, CDCl3): δ 154.2, 151.7, 141.6, 141.1, 130.3, 130.2, 126.5, 125.9, 125.3, 122.7, 114.1, 109.9, 66.4, 49.5, 48.6, 47.9, 40.3, 31.9, 29.7, 29.6, 29.5, 29.4, 27.2, 22.7, 17.3, 16.6, 16.3, 14.9, 14.2; [HR-ESI(+)]: m/z calcd for C39H66NO2 [M + H]+: 580.5088, found: 580.5077.
4-{4-[4-(2-Aminoethoxy)-3-methylphenyl]heptan-4-yl}-2-methylphenol (PBP-NH2)
To a solution of 3 (614.3 mg, 1.97 mmol) and Cs2CO3 (321.3 mg, 0.99 mmol) in MeCN (5 mL) was added benzyl (2-bromoethyl)carbamate (264.7 mg, 1.03 mmol), and the mixture was stirred at 40 °C for 16 h. The reaction mixture was neutralized with 3% aqueous HCl and extracted with ethyl acetate. The organic layer was washed with brine, dried over Na2SO4, filtered, and then concentrated in vacuo. The residue was dissolved in MeOH (6 mL) and Pd(OH)2 was added (ca. 20 mg). The solution was stirred at room temperature for 16 h under H2 atmosphere. The reaction mixture was filtered through celite and concentrated in vacuo. The residue was purified with silica gel chromatography (AcOEt/n-hexane = 1/4) to afford PBP-NH2 (175 mg, 25%) as colorless oil; IR (CDCl3, cm–1): 3602, 2958, 2932, 2871, 1606, 1503, 1465; 1H NMR (400 MHz, CDCl3): δ 6.91–6.94 (m, 3H), 6.80 (d, J = 8.4 Hz, 1H), 6.66 (d, J = 8.4 Hz, 1H), 6.61 (d, J = 8.4 Hz, 1H), 3.98 (t, J = 5.0 Hz, 2H), 3.11 (t, J = 5.0 Hz, 2H), 2.19 (s, 3H), 2.16 (s, 3H), 1.91–1.95 (m, 4H), 0.91–0.99 (m, 4H), 0.84 (t, J = 7.0 Hz, 6H); 13C NMR (100 MHz, CDCl3): δ 154.3, 151.7, 141.6, 141.2, 130.3, 130.2, 126.5, 125.9, 125.3, 122.7, 114.0, 109.8, 69.4, 47.9, 41.6, 40.3, 17.3, 16.6, 16.3, 14.9; [HR-ESI(+)]: m/z calcd for C23H34NO2 [M + H]+: 356.2584, found: 356.2547.
4-(4-{4-[2-(N,N-Dimethylamino)ethoxy]-3-methylphenyl}heptan-4-yl)-2-methylphenol (PBP-tam)
To a solution of 3 (265.4 mg, 0.85 mmol), K2CO3 (351.6 mg, 2.5 mmol) and NaI (127.2 mg, 0.85 mmol) in DMF (5 mL) was added 2-chlorodimethylethylamine (122.2 mg, 0.85 mmol) and the mixture was stirred at 100 °C for 16 h. The reaction mixture was concentrated in vacuo, and the residue was purified with silica gel chromatography (AcOEt/MeOH = 9/1) to afford PBP-tam (18.8 mg, 0.05 mmol, 6%) as colorless oil; IR (CDCl3, cm–1): 3607, 3020, 2959, 1382, 1212; 1H NMR (400 MHz, CDCl3): δ 6.80–6.92 (m, 3H), 6.81 (d, J = 8.4 Hz, 1H), 6.65 (d, J = 8.4 Hz, 1H), 6.59 (d, J = 8.4 Hz, 1H), 4.05 (t, J = 6.0 Hz, 2H), 2.78 (t, J = 6.0 Hz, 2H), 2.37 (s, 6H), 2.18 (s, 3H), 2.13 (s, 3H), 1.91–1.95 (m, 4H), 0.93–0.98 (m, 4H), 0.83 (t, J = 6.8 Hz, 6H); 13C NMR (100 MHz, CDCl3): δ 154.8, 151.1, 141.9, 140.8, 130.3, 130.1, 126.6, 125.8, 125.4, 122.3, 114.0, 109.7, 67.4, 47.9, 40.3, 31.6, 19.4, 17.3, 16.5, 16.1, 14.9, 14.0; [HR-ESI(+)]: m/z calcd for C25H38NO2 [M + H]+: 384.2897, found: 384.2898.
Biology
Cell culture
Human breast carcinoma MCF-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 100 μg mL–1 kanamycin with 10 μg ml–1 insulin (Sigma, St. Louis, MO, USA). The cells were treated with various concentrations of the compounds for 6 h. Whole-cell lysates were analyzed using western blotting with the indicated antibodies.
Western blotting
Cells were lysed with SDS lysis buffer (0.1 M Tris-HCl at pH 8.0, 10% glycerol, 1% SDS) and boiled for 10 min. The protein concentration was measured by the BCA method (Pierce, Rockford, IL, USA) and the lysates containing an equal amount of protein were separated by SDS-PAGE, transferred to PVDF membranes (Millipore, Darmstadt, Germany) for western blotting using the appropriate antibodies. The immunoreactive proteins were visualized using the Immobilon Western chemiluminescent HRP substrate (Millipore) or Clarity Western ECL substrate (Bio-Rad, Hercules, CA, USA), and light emission was quantified with a LAS-3000 lumino-image analyzer (Fuji, Tokyo, Japan). The antibodies used in this study were anti-ERα rabbit monoclonal antibody (mAb) (Cell Signaling Technology, Danvers, MA, USA; 8644), anti-β-actin mouse mAb (Sigma; A5316), anti-AR rabbit mAb antibody (Cell Signaling Technology; 5153), anti-AhR rabbit mAb antibody (Cell Signaling Technology; 13790), anti-VDR rabbit mAb antibody (Cell Signaling Technology; 12550), and anti-Lamin B goat pAb (Santa Cruz, Dallas, TX, USA; sc-6216).
Fluorescence polarization assay
Test compounds were dissolved in DMSO to prepare a stock solution (10 mM). Fluorescence polarization-based competition binding assays were conducted to determine the relative binding affinity of compounds for ERα using commercially available kits (P2698, Life Technologies) according to the manufacturer's instructions. Fluorescence polarization signals (mP values) were then measured using an EnVision multiple plate reader (Perkin Elmer) with a 470 nm excitation filter and a 535 nm emission filter. The fraction of compound bound to ERα was correlated with the mP value and plotted against values of competitor concentrations.
Reporter gene assay
ER-antagonistic activities of the compounds were evaluated by means of reporter gene assay using a Gal4-human ERα reporter system.18 A ligand binding domain (LBD) of human ERα was inserted into the pCMX-GAL4 vector to obtain pCMX-GAL4-hER (pCMX-flag vector to make pCMX-ERα). A GAL4-responsive MH100 (USA)x4-tk-LUK reporter was used. Human embryonic kidney (HEK 293) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) without phenol red containing 10% fetal bovine serum (FBS) and antibiotic–antimycotic (Nacalai) at 37 °C in a humidified atmosphere of 5% CO2 in air. Transfections were performed by the calcium phosphate coprecipitation method. Test compounds were added at 8 h after transfection. Cells were harvested 16–20 h after the treatment, and luciferase and β-galactosidase activities were assayed using a luminometer and a microplate reader. DNA cotransfection experiments were performed with 50 ng of reporter plasmid, 15–20 ng of pCMX-β-galactosidase, 10–15 ng of each receptor expression plasmid and pGEM carrier DNA to make a total of 150 ng of DNA per well in a 96-well plate. Luciferase data were normalized to an internal β-galactosidase control, and the reported values are means of triplicate assays. Antagonist activity was measured in the presence of 0.3 nM 17β-estradiol.
Supplementary Material
Acknowledgments
This work was supported in part by a grant for the Research on Development of New Drugs from Japan Agency for Medical Research and Development (AMED; M. N.) and a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (T. M.).
Footnotes
†The authors declare no competing interests.
‡Electronic supplementary information (ESI) available. See DOI: 10.1039/c6md00553e
References
- (a) Ogawa S., Eng V., Taylor J., Lubahn B. D., Korach S. K., Pfaff W. D. Endocrinologia. 1998;139:5070. doi: 10.1210/endo.139.12.6357. [DOI] [PubMed] [Google Scholar]; (b) Imai Y., Youn M. Y., Kondoh S., Nakamura T., Kouzmenko T., Matsumoto T., Takeda I., Takaoka K., Kato S. Ann. N. Y. Acad. Sci. 2009;1173:E31. doi: 10.1111/j.1749-6632.2009.04954.x. [DOI] [PubMed] [Google Scholar]
- Hall J. M., McDonnell D. P. Mol. Interventions. 2005;5:343. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- (a) Kuiper G. G., Enmark E., Pelto-Huikko M., Nilsson S., Gustafsson J. A. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5925. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Thomas C., Gusrafsson J. Nat. Rev. Cancer. 2011;11:597. doi: 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- (a) Walter P., Green S., Greene G., Krust A., Bornert J. M., Jeltsch J. M., Staub A., Jensen E., Scrase G., Waterfield M. Proc. Natl. Acad. Sci. U. S. A. 1985;82:7889. doi: 10.1073/pnas.82.23.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Holst F., Stahl P. R., Ruiz C., Hellwinkel O., Jehan Z., Wendland M., Lebeau A., Terracciano L., Al-Kuraya K., Jänicke F., Sauter G., Simon R. Nat. Genet. 2007;39:655. doi: 10.1038/ng2006. [DOI] [PubMed] [Google Scholar]
- Jordan V. C. Br. J. Pharmacol. 2006;147:269. doi: 10.1038/sj.bjp.0706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grese T. A., Cho S., Finley D. R., Godfrey A. G., Jones C. D., Lugar C. W., Martin M. J., Matsumoto K., Pennington L. D., Winter M. A., Adrian M. D., Cole H. W., Magee D. E., Phillips D. L., Rowley E. R., Short L. L., Glasebrook A. L., Bryant H. U. J. Med. Chem. 1997;40:146. doi: 10.1021/jm9606352. [DOI] [PubMed] [Google Scholar]
- Howell A., Cuzick J., Baum M., Buzdar A., Dowsett M., Forbes J. F., Hoctin-Boes G., Houghton J., Locker G. Y., Tobias J. S. Lancet. 2005;365:60. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- (a) Johnson M. D., Zuo H., Lee K. H., Trebley J. P., Rae J. M., Weatherman R. V., Desta Z., Flockhart D. A., Skaar T. C. Breast Cancer Res. Treat. 2004;85:151. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]; (b) Maximov P. Y., Myers C. B., Curpan R. F., Lewis-Wambi J. S., Jordan V. C. J. Med. Chem. 2010;53:3273. doi: 10.1021/jm901907u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneekloth Jr. J. S., Fonseca F. N., Koldobskiy M., Mandal A., Deshaies R., Sakamoto K., Crews C. M. J. Am. Chem. Soc. 2004;126:3748. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Ishikawa M., Naito M., Hashimoto Y. J. Am. Chem. Soc. 2010;132:5820–5826. doi: 10.1021/ja100691p. [DOI] [PubMed] [Google Scholar]
- Gustafson J. L., Neklesa T. K., Cox C. S., Roth A. G., Buckley D. L., Tae H. S., Sundberg T. B., Stagg D. B., Hines J., McDonnell D. P., Norris J. D., Crews C. M. Angew. Chem., Int. Ed. 2015;54:9659. doi: 10.1002/anie.201503720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Wakeling A. E., Dukes M., Bowler J. Cancer Res. 1991;51:3867. [PubMed] [Google Scholar]; (b) Yeh W. L., Shioda K., Coser K. R., Rivizzigno D., McSweeney K. R., Shioda T. PLoS One. 2013;8:e60889. doi: 10.1371/journal.pone.0060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike A. C., Brzozowski A. M., Walton J., Hubbard R. E., Thorsell A. G., Li Y. L., Gustafsson J. A., Carlquist M. Structure. 2001;9:145. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- Shoda T., Kato M., Harada R., Fujisato T., Okuhira K., Demizu Y., Inoue H., Naito M., Kurihara M. Bioorg. Med. Chem. 2015;23:3091. doi: 10.1016/j.bmc.2015.05.002. [DOI] [PubMed] [Google Scholar]
- (a) McCague R., Jarman M., Leung O. T., Foster A. B., Leclercq G., Stoessel S. J. Steroid Biochem. 1988;31:545. doi: 10.1016/0022-4731(88)90005-2. [DOI] [PubMed] [Google Scholar]; (b) Katzenellenbogen B. S., Norman M. J., Eckert R. L., Peltz S. W., Mangel W. F. Cancer Res. 1984;44:112. [PubMed] [Google Scholar]
- (a) Maruyama K., Noguchi-Yachide T., Sugita K., Hashimoto Y., Ishikawa M. Bioorg. Med. Chem. Lett. 2010;20:6661. doi: 10.1016/j.bmcl.2010.09.011. [DOI] [PubMed] [Google Scholar]; (b) Kainuma M., Kasuga J., Hosoda S., Wakabayashi K., Tanatani A., Nagasawa K., Miyachi H., Makishima M., Hashimoto Y. Bioorg. Med. Chem. Lett. 2006;16:3213. doi: 10.1016/j.bmcl.2006.03.075. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Nakamura M., Tomoshige S., Sugita K., Makishima M., Hashimoto Y., Ishikawa M. Bioorg. Med. Chem. Lett. 2013;23:4031. doi: 10.1016/j.bmcl.2013.05.067. [DOI] [PubMed] [Google Scholar]
- Noguchi-Yachide T., Sugita K., Hashimoto Y. Heterocycles. 2011;83:2137. [Google Scholar]
- Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. Cell. 1998;95:927. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.