 O-GlcNAc transferase (OGT) attaches a GlcNAc moiety on specific substrate proteins using UDP-GlcNAc as the sugar donor.
O-GlcNAc transferase (OGT) attaches a GlcNAc moiety on specific substrate proteins using UDP-GlcNAc as the sugar donor.
Abstract
O-GlcNAc transferase (OGT) attaches a GlcNAc moiety on specific substrate proteins using UDP-GlcNAc as the sugar donor. This modification can alter protein function by regulating cellular signaling and transcription pathways in response to altered nutrient availability and stress. Specific inhibitors of OGT would be valuable tools for biological studies and lead structures for therapeutics. The existing OGT inhibitors are mainly derived from the sugar donor substrate, but poor cell permeability and off-target effects limit their use. Here, we describe our progress on OGT inhibition based on substrate peptides identified by array screening. Subsequently, bisubstrate inhibitors were prepared by conjugating these peptides to uridine in various ways. In parallel, an in silico fragment screening was conducted to obtain small molecules targeting the UDP binding pocket. After evaluation of the initial hits, one of these small molecules was elaborated into a novel OGT hybrid inhibitor, as the replacement of uridine. The novel compounds inhibit OGT activity with IC50 values in the micromolar range.
O-GlcNAc transferase (OGT) is an essential mammalian enzyme involved in the dynamic O-GlcNAcylation of cytosolic and nuclear proteins. Through catalyzing the attachment of N-acetylglucosamine to specific serines and threonines of proteins, OGT is associated with numerous biological processes such as transcription, the cell cycle progression, the stress response and nutrient sensing.1–5 Prior studies have noted the potential of OGT as a therapeutic target not only because it is one of only two enzymes modulating O-GlcNAcylation but also because its expression is correlated with the metabolic status of a cell directly.6–9 In metabolic diseases like cancer and diabetes, increase of various metabolic products like glucose into the cell alters the production of UDP-GlcNAc through the hexosamine biosynthetic pathway. This promotes O-GlcNAcylation since OGT is highly sensitive to intracellular UDP-GlcNAc levels. Elevated O-GlcNAcylation of key signaling molecules and transcription factors can be involved in the development and pathology of diseases.8,10–12 For this reason OGT is emerging as a therapeutic target of current interest. A selective and cell-permeable OGT inhibitor is strongly needed to further decipher its function in biological processes and explore its potential as a therapeutic target.
In recent years, biochemical and crystallographic studies have clearly revealed the mechanistic aspects of OGT's catalytic process. The sugar donor substrate UDP-GlcNAc binds to the active site of the enzyme first, followed by the sugar acceptor substrate.13,14 In this ternary complex, the transition state is approached for glycosyl transfer of the N-acetylglucosamine from the donor onto the specific serine or threonine of the acceptor. In the discovery of OGT inhibitors, a few OGT sugar donor analogues were identified that showed a moderate inhibitory effect.15–17 However, these compounds were not cell permeable, which restricts their use in research. In addition, most of these compounds have a severe off-target effects because of their similarity to the commonly used UDP-GlcNAc.16 Non-substrate inhibitors were also described but off-target or toxicity effects were still noted,18 although recent progress was made with low toxicity inhibitors.19,20
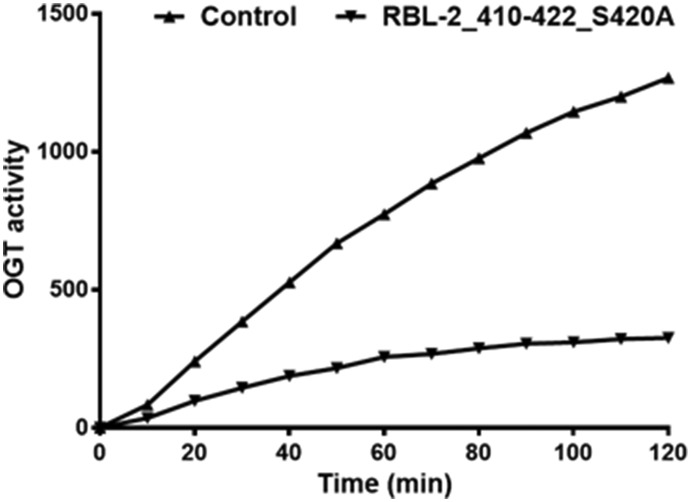
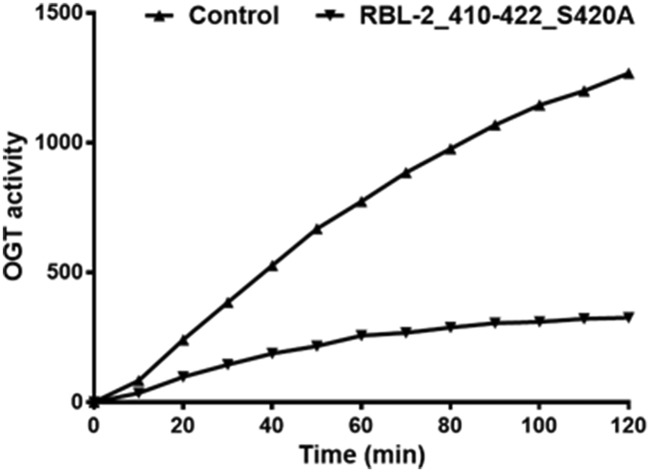
Using a peptide microarray approach (Scheme 1), we recently identified the peptide RBL-2_410–422 as an OGT substrate. This peptide is derived from the RBL-2 protein, which is a key regulator of entry into cell division and may function as a tumor suppressor. The serine that is modified by O-GlcNAc was identified in this peptide.21,22 Interestingly, replacing this serine with an alanine in peptide RBL-2_410–422_S420A resulted in a competitive OGT inhibitor (Fig. 1).
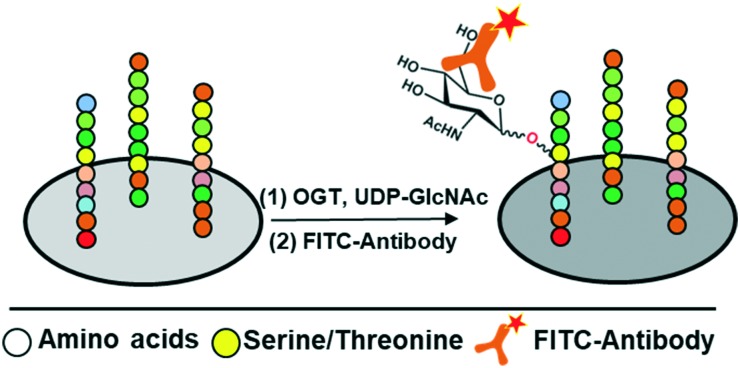
Scheme 1. Schematic depiction of the peptide microarray approach to identify peptides as substrates of OGT. O-GlcNAcylation is detected by a FITC-labelled antibody.
Fig. 1. The RBL-2_410–422_S420A peptide inhibited OGT activity competitively. The effect of RBL-2_410–422_S420A (0.5 mM) on OGT activity was determined using a peptide microarray assay on which RBL-2_410–422 was immobilized. A parallel assay was performed using water as a control. Real time OGT activity was reflected by the fluorescent antibody bound to O-GlcNAcylated RBL-2_410–422.
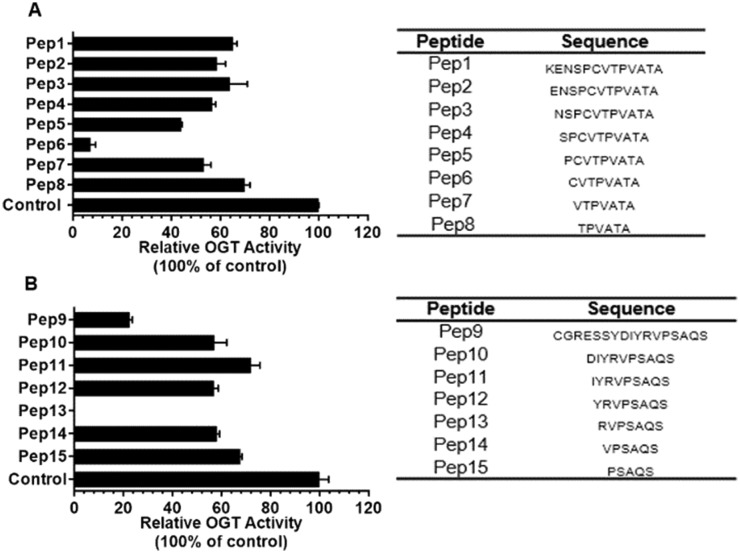
This finding suggests a significant interaction between the peptide substrate and OGT that could form the basis for selective OGT inhibitor development. Another specific substrate peptide was identified among tyrosine kinase substrates and was derived from the tight junction ZO-3 protein: ZO-3_357–371.23 Again replacing the serine at the modification site with an alanine yielded an OGT inhibitor. To identify the minimum structural requirements for OGT inhibition, two series of shorter peptides based on the original sequences were synthesized and they were tested as OGT inhibitors (Fig. 2). In the series of truncated RBL-2_410–422_S420A peptides, surprisingly, an actual potency improvement was observed for a shorter peptide, i.e. octapeptide Pep6 (Fig. 2A). Additional experiments showed that it exhibited an IC50 value of 385 μM.
Fig. 2. A) A series of peptides derived from RBL-2_410–422_S420A (Pep1) were produced with sequences shown in the right table and their inhibitory effect on OGT activity were determined at a concentration of 1 mM using a UDP-Glo assay (see ESI†). B) The same as above but now for ZO-3_357–371_S369A (Pep9). All peptides were synthesized by standard Fmoc SPPS protocols described in the ESI.† .
Similar results were obtained for the ZO-3_357–371_S369A derivatives with the optimal peptide being heptapeptide Pep13 (Fig. 2B) that showed an IC50 value of 193 μM. Besides providing more support for OGT being sequence specific, the data also motivated us to improve potency and selectivity. To this end, the approach was to develop bisubstrate inhibitors by conjugating parts or mimics of the sugar donor with these identified peptide inhibitors.
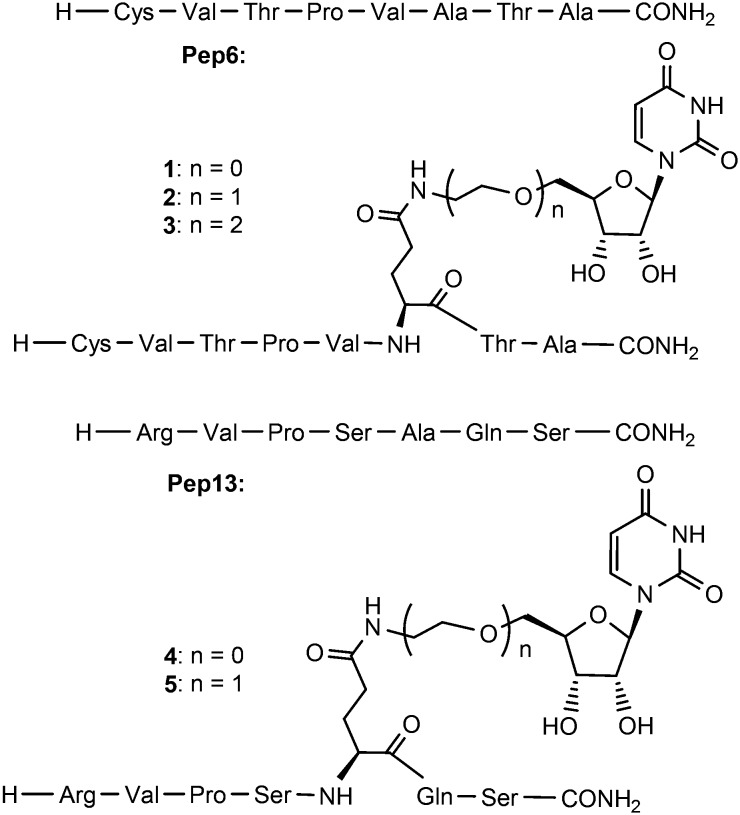
Within this context it is of note that the Kd of UDP-GlcNAc for OGT is higher than that of UDP, indicating that the GlcNAc moiety even hampers the binding.24 Accordingly, we have focused on the UDP alone and its binding site to enhance the peptide affinity. UDP itself as a product of O-GlcNAcylation inhibits OGT in vitro with an IC50 of 1.8 μM, making it the most potent OGT inhibitor to date.17,24 Despite its strong affinity for OGT the charged phosphates preclude cell permeability. Aiming to combine the peptide selectivity with UDP's potency but removing its diphosphate, we prepared uridine–peptide conjugates (compounds 1–5 shown in Fig. 3) with neutral linkers of different length.
Fig. 3. Structure of Pep6, Pep13 and potential bisubstrate inhibitors 1–5.
The abilities of these compounds to inhibit OGT activity were again determined using the UDP-Glo glycosyltransferase assay and compared with those of Pep6 and Pep13. As shown in Table 1, an IC50 of 297 μM was determined for 1 whereas no obvious inhibition was observed for 2 and 3, indicating the importance of spacer length. Similar modifications on Pep13 rendered the compounds inactive, indicating differences in the binding modes of Pep6 and Pep13. Overall the enhancement of the binding due to the introduction of UDP was modest at best. For this reason a search for more potent fragments than uridine was started to be used in the context of a bisubstrate or hybrid inhibitor.
Table 1. OGT inhibition values obtained for Pep6, Pep13 and bisubstrate derivatives a .
| Compound | IC50 [μM] |
| Pep6 | 385 |
| 1 | 297 |
| 2 | >1000 |
| 3 | 493 |
| Pep13 | 193 |
| 4 | >1000 |
| 5 | >1000 |
aIC50 values are reported in μM and are averages obtained from triple independent duplicate analysis of each compound.
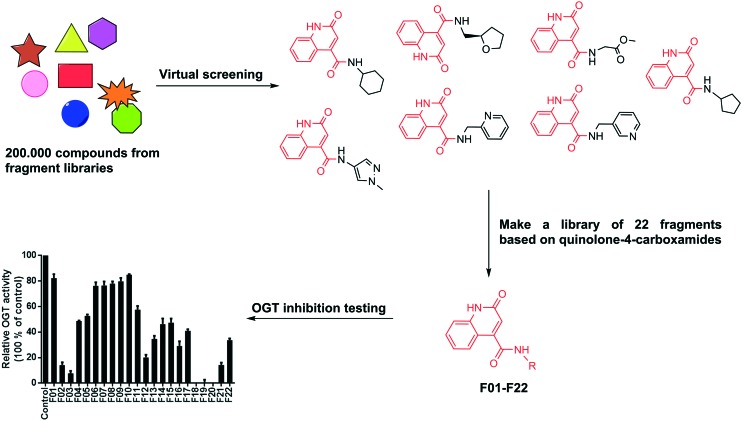
To identify fragments targeting the donor UDP site, we have conducted a structure-based virtual screening in a fragment library containing more than 216 000 molecules using OEDOCKING (OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com)25,26 (see ESI† for details). An analysis of the interactions of UDP in the OGT binding site revealed that the uracil moiety of UDP forms two hydrogen bonds with the Ala896 backbone carbonyl and NH groups (Fig. S1†). To mimic this hydrogen bonding network Ala896 was used as a constraint in virtual screening (docking) experiment and hence only compounds containing proximal hydrogen bond donor and acceptor groups were retrieved as potential hits. Among these virtual hits, seven compounds contained the same scaffolds as they were all quinolone-4-carboxamides. (Fig. 4). A common feature of these molecules is that in the predicted binding mode the quinolone ring is anchored in the uridine binding site of OGT and the additional carboxamides point to the diphosphate binding site.
Fig. 4. Schematic depiction of fragment screening and testing. Fragments screening yielded seven hits with same heterocyclic scaffold. This scaffold was used in the synthesis of 22 new fragments that were tested for OGT inhibition at 1 mM using the UDP-Glo assay.
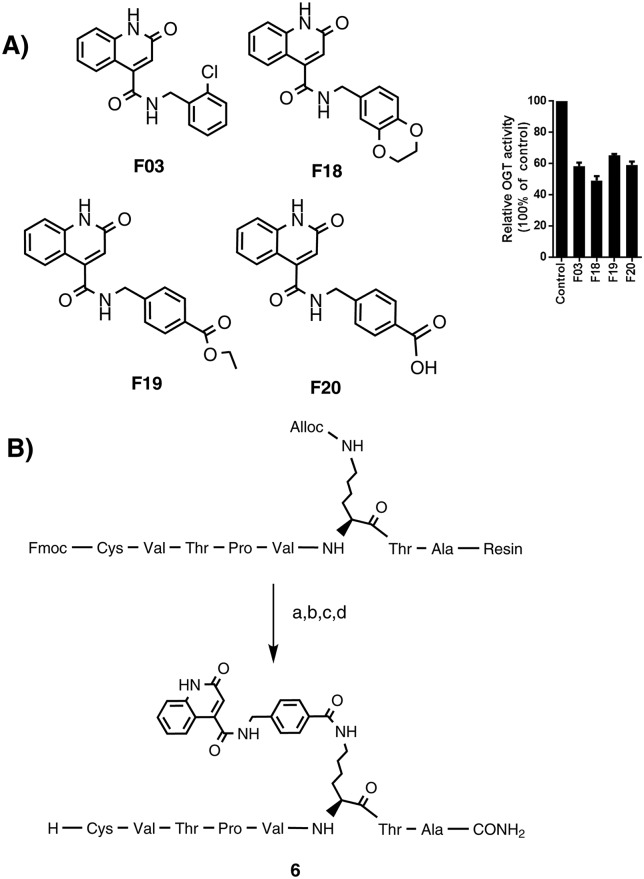
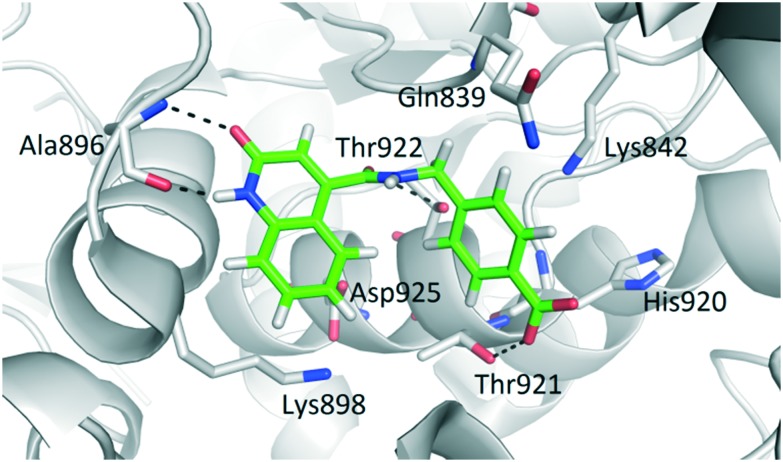
To further explore this finding, a series of 22 fragments (F01–F22) carrying diverse carboxamides (ESI,† Table 2S) was prepared. The synthesis was conducted by coupling 2-hydroxyquinoline-4-carboxylic acid with various amines using EDC/HOBt to effect the coupling. The inhibitory potency of these compounds on OGT activity was measured at a concentration of 1 mM using the UDP-Glo assay and several fragments were found to inhibit OGT activity by 90% under these conditions. The most potent fragments are shown in Fig. 5A, and also their inhibition profile now measured at 200 μM. The docking pose of F20 (structure in Fig. 5) predicted the mentioned two hydrogen bonds with Ala896, which mimic the binding mode of the uracil moiety of UDP (Fig. 6). Additional hydrogen bonds are predicted between the Thr921 side chain and the inhibitor carboxylate group and the Thr922 side chain and the carbonyl group of the carboxamide at position 4 of the quinolone-2-one ring. Moreover, comparing the similar F19 (ethyl ester) and F20 (free acid) suggests that a free carboxylate group is not a prerequisite for binding thus making this a suitable site for conjugation to the peptide.
Fig. 5. A) Top 4 fragments structures and OGT inhibition validation. F03, F18, F19 and F20 were tested for OGT activity inhibition at 0.2 mM using the UDP-Glo assay. B) On resin synthesis of compound 6 as a Pep6–F20 conjugate: a) Pd(PPh3)4, phenylsilane, DCM, rt, 1 h. b) F20, Bop, DiPEA, DMF, 2 h. c) Piperidine/DMF, 1 h. d) TFA/TIPS/EDT/H2O, rt, 3 h.
Fig. 6. Predicted binding mode of OGT inhibitor F20 (green sticks) in the OGT binding site (in grey cartoon, PDB entry: 4N39). The ligand and the neighbouring protein side-chains are shown as stick models, coloured according to the chemical atom type (blue, N; red, O; orange, S; green, Cl). Hydrogen bonds are indicated by black dotted lines.
Considering that the shortest linker was the most effective in 1–3, a lysine was selected for conjugating the fragment, placing the fragment at a similar distance from the peptide compared to the uridine group in 1. This choice also allowed a convenient on-resin synthesis, in which F20 can be covalently linked to the amino group of the Lys side chain. After cleavage and purification, compound 6 was obtained and tested for its ability to inhibit OGT in our assay. An IC50 of 117 μM was determined, which indicated that an improved synergy effect of the two component hybrid inhibitor.
Conclusions
In conclusion, we successfully designed and synthesized several bi-substrate or hybrid OGT inhibitors. In one case bisubstrate inhibitors of OGT inhibitors were reported, in which a UDP moiety was linked to a substrate peptide and yielded promising potencies, although the peptide contribution was negligible.15 Our aim was to enhance/maintain potency while removing the diphosphate group (to enable cell permeability) and enhance the peptidic contribution. Starting from peptide substrates and replacing the modification site serine with an alanine lead to inhibition. Trimming the peptides to 7–8 amino acids actually yielded the more potent inhibitors Pep6 and Pep13. Such an enhancement is unusual as trimming usually leads to reduced potency. Linking uridine to Pep6 in various ways lead to a modest synergy effect but not for Pep13. In addition, fragments were identified to block the sugar donor site of OGT through a combination of virtual screening and rational redesign. Linking a fragment to Pep6 displayed enhanced inhibition showing the promise of this approach for making potent and selective OGT inhibitors. Potency is still a factor of 5 behind a recently discovered natural product inhibitor (L01, IC50 = 22 μM UDP-Glo assay)19 but the lack of diphosphate and the use of a lipophilic fragment in 6 significantly increased the log P and decreased the PSA values over previous bisubstrate inhibitors (see ESI†).
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
Research reported in this article was mainly supported by a scholarship (file No. 201306210049) to HZ from the China Scholarship Council (http://www.csc.edu.cn/) and by the Slovenian Research Agency (Grant No. P1-0208). We thank OpenEye Scientific Software, Santa Fe, NM., for free academic licenses for the use of their software.
Footnotes
†Electronic supplementary information (ESI) available: Analytical methods, synthetic details, MS spectra, biological evaluation and NMR data. See DOI: 10.1039/c8md00115d
References
- Hanover J. A., Krause M. W., Love D. C. Nat. Rev. Mol. Cell Biol. 2012;13:312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- Daou S., Mashtalir N., Hammond-Martel I., Pak H., Yu H., Sui G., Vogel J. L., Kristie T. M., Affar E. B. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2747–2752. doi: 10.1073/pnas.1013822108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkinaree C., Park K., Hart G. W. Biochim. Biophys. Acta, Gen. Subj. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves J. A., Lee A., Yildirir G., Zachara N. E. Cell Stress Chaperones. 2013;18:535–558. doi: 10.1007/s12192-013-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C., Hart G. W. Nat. Rev. Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C., Copeland R. J., Hart G. W. Trends Biochem. Sci. 2010;35:547–555. doi: 10.1016/j.tibs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardini Y., Dehennaut V., Lefebvre T., Issad T. Front. Endocrinol. 2013;4:99. doi: 10.3389/fendo.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood K. R., Hanover J. A. J. Cell Sci. 2014;127:1857–1867. doi: 10.1242/jcs.113233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G. W., Housley M. P., Slawson C. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- de Queiroz R. M., Carvalho E., Dias W. B. Front. Oncol. 2014;4:1–10. doi: 10.3389/fonc.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer C. M., Lynch T. P., Sodi V. L., Falcone J. N., Schwab L. P., Peacock D. L., Vocadlo D. J., Seagroves T. N., Reginato M. J. Mol. Cell. 2014;54:820–831. doi: 10.1016/j.molcel.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus M. B., Jiang J., Gloster T. M., Zandberg W. F., Whitworth G. E., Vocadlo D. J., Walker S. Nat. Chem. Biol. 2012;8:966–968. doi: 10.1038/nchembio.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus M. B., Nam Y., Jiang J., Sliz P., Walker S. Nature. 2011;469:564–569. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodkin V. S., Schimpl M., Gundogdu M., Rafie K., Dorfmueller H. C., Robinson D. A., van Aalten D. M. F. Biochem. J. 2014;457:497–502. doi: 10.1042/BJ20131272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapannone R., Rafie K., van Aalten D. M. F. Biochem. Soc. Trans. 2016;44:88–93. doi: 10.1042/BST20150189. [DOI] [PubMed] [Google Scholar]
- Dorfmueller H. C., Borodkin V. S., Blair D. E., Pathak S., Navratilova I., van Aalten D. M. F. Amino Acids. 2011;40:781–792. doi: 10.1007/s00726-010-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Meoz R. F., Jiang J., Lazarus M. B., Orman M., Janetzko J., Fan C., Duveau D. Y., Tan Z.-W., Thomas C. J., Walker S. ACS Chem. Biol. 2015;10:1392–1397. doi: 10.1021/acschembio.5b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ren Y., Cao Y., Huang H., Wu Q., Li W., Wu S., Zhang J. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-12522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhu J., Zhang L. J. Med. Chem. 2017;60:263–272. doi: 10.1021/acs.jmedchem.6b01237. [DOI] [PubMed] [Google Scholar]
- Shi J., Sharif S., Ruijtenbeek R., Pieters R. J. PLoS One. 2016;11:e0151085. doi: 10.1371/journal.pone.0151085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak S., Alonso J., Schimpl M., Rafie K., Blair D. E., Borodkin V. S., Schüttelkopf A. W., Albarbarawi O., Van Aalten D. M. F. Nat. Struct. Mol. Biol. 2015;22:744–749. doi: 10.1038/nsmb.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Tomašič T., Sharif S., Brouwer A. J., Anderluh M., Ruijtenbeek R., Pieters R. J. FEBS Lett. 2017;591:1872–1883. doi: 10.1002/1873-3468.12708. [DOI] [PubMed] [Google Scholar]
- Schimpl M., Zheng X., Borodkin V. S., Blair D. E., Ferenbach A. T., Schüttelkopf A. W., Navratilova I., Aristotelous T., Albarbarawi O., Robinson D. A., MacNaughtan M. A., Van Aalten D. M. F. Nat. Chem. Biol. 2012;8:969–974. doi: 10.1038/nchembio.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann M. J. Chem. Inf. Model. 2011;51:578–596. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- McGann M. J. Comput.-Aided Mol. Des. 2012;26:897–906. doi: 10.1007/s10822-012-9584-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.