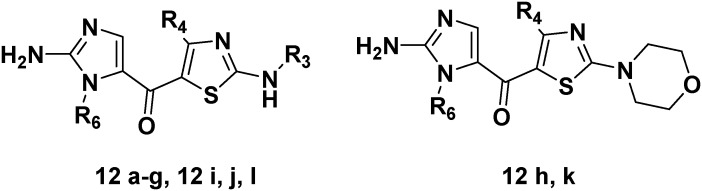
 A combinatorial library of 24 compoundswith 2-amino imidazole & 2-aminoimidazolyl-thiazoles was synthesized.
A combinatorial library of 24 compoundswith 2-amino imidazole & 2-aminoimidazolyl-thiazoles was synthesized.
Abstract
A small-molecule combinatorial library of 24 compounds with 2-aminoimidazole and 2-aminoimidazolyl-thiazole derivatives was synthesized using a 2-chloro trityl resin. The generated compound library was tested against all the human adenosine receptors subtypes. The 2-aminoimidazole derivatives (6a–6l) showed weak to moderate affinity towards the human adenosine receptors. Further modification to 2-aminoimidazolyl-thiazole derivatives (12a–12l) resulted in an improvement of affinity at adenosine A1, A2A and A3 receptor subtypes. Compound 12b was the most potent and selective non-xanthine human adenosine A3 receptor antagonist of this series. A receptor-based modeling study was performed to explore the possible binding mode of these novel 2-aminoimidazole and 2-aminoimidazolyl-thiazole derivatives into human adenosine A1, A2A and A3 receptor subtypes.
Introduction
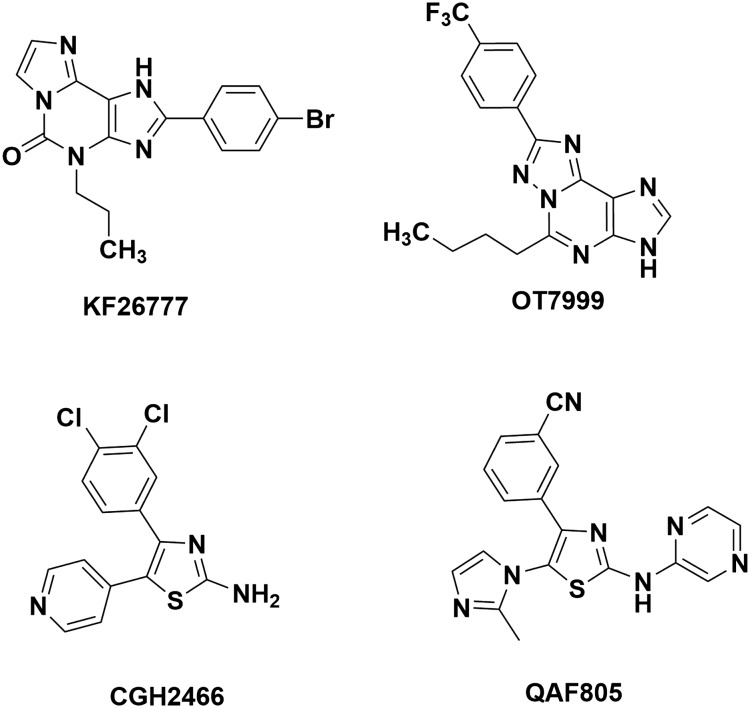
Adenosine is a modulator of multiple physiological processes, including cardiovascular, neurological and respiratory functions. In recent years the search for novel adenosine receptor agonists or antagonists has intensified. Adenosine mediates its effects through specific G-protein-coupled receptors ubiquitously expressed in the body and classified as A1, A2A, A2B and A3 (ARs).1,2 The A3 AR is able to inhibit forskolin- or receptor-mediated cAMP accumulation, to increase phosphatidylinositol-specific phospholipase C and D activity, and to elevate IP3 and intracellular Ca2+ levels.1 Furthermore, the A3 AR subtype has been subject to intensive investigation as a potential therapeutic target in conditions such as inflammation,3 neurodegeneration,4,5 ischaemia,6–8 asthma,9,10 glaucoma11–13 and cancer.14–17 Numerous compounds were synthesized and tested as adenosine receptor antagonists. Many of these compounds are based on a xanthine structure such as caffeine and theophylline which were the first antagonists to be found for adenosine receptors. The problems associated with the xanthine-based compounds, namely, poor receptor selectivity, poor solubility, and low bio-availability, necessitated the search for non-xanthine-like compounds.18 Thus compound KF26777 was found to be a potent and selective adenosine A3 receptor antagonist by Kyowa Hakko Kogyo for the treatment of asthma19 while tricyclic compound OT-7999 is a selective adenosine A3 receptor antagonist reported by Otsuka Pharmaceutical Factory, Inc, investigated as a treatment for glaucoma.11 Further, Novartis developed a series of 2-aminothiazole based compounds out of which CGH2466 was a potent ligand for adenosine A1 and A3 receptors, with no binding activity at the A2A receptor.20 The QAF805 compound was found to be an orally bioavailable dual adenosine A2B/A3 receptor antagonist with therapeutic potential in asthma and COPD21 (Fig. 1).
Fig. 1. Potent and selective adenosine receptor antagonist.
Results and discussion
Chemistry and SAR of aminoimidazole and aminoimidazolyl-thiazole derivatives
Our aim was to develop novel non-xanthine based antagonists for adenosine receptor subtypes. More specifically, our efforts were based on small heterocyles like 2-aminothiazoles and 2-aminoimidazoles derivatives since these five-member heterocycles have been extensively used in the successful design of drugs/drug-like candidates. These rings can pack a relatively large number of polarized bonds in a relatively small molecular space and offer a convenient framework to which various side chains can be attached. Recently, our group has reported a series of 2-aminothiazole derivatives as potent and selective adenosine receptor antagonist.22,23 In fact, we have delineated the minimum structural features for the compounds to be active against the adenosine receptors by substituting 2-aminothiazole scaffold, with aroyl and heteroaryl moiety at the 5 position. In continuation of our findings, we herein report the structure–activity relationships of novel 2-aminoimidazole and 2-aminoimidazolyl-thiazole derivatives with high affinity and selectivity for A3 adenosine receptors. Initially, we synthesized a series of compounds with a 2-aminoimidazole scaffold bearing an aroyl substitution in 5-position and investigated the pharmacological characteristics in radio ligand binding studies at adenosine receptor subtypes with the aim to obtain potent and selective adenosine A3 receptor ligands. However, this series turned out to possess only weak to moderate affinity. Further, the incorporation of a thiazolyl moiety at 5-position of the 2-aminoimidazole scaffold improved the affinity and selectivity towards the adenosine receptor subtypes.
Series 1 – aminoimidazoles
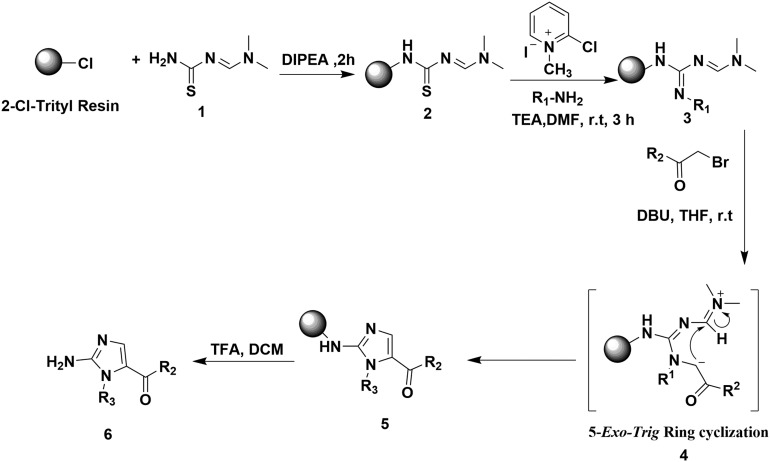
The solid phase synthesis of 2-aminoimidazole library was carried out using the 2-chlorotrityl chloride resin. The resin was treated with amidinothiourea 1 in presence of diisopropylethylamine (DIPEA) to give resin bound amidinothiourea 2. Guanylation of 2 with various arylamines using Mukaiyama's reagent gave amidinoguanidines 3. The amidinoguanidines 3 thus obtained on reaction with a series of α-bromoketones undergo N-alkylation followed by 5-exo-trig cyclization 4 to give resin bound aminoimidazole derivatives 5. Cleavage of 5 from the solid support with trifluoroacetic acid provides 2-aminoimidazoles 6 (Scheme 1).
Scheme 1. Synthesis of 2-aminoimidazole derivatives.
Binding affinities of 2-aminoimidazole derivatives for human adenosine receptors
All the synthesized compounds (6a–6l, Table S1 ESI†) were tested in radioligand binding assays to determine their affinity for the adenosine A1, A2A, and A3 receptor subtypes. The data show that this series of compounds does not bind with measurable affinity to A1and A2B receptors. No measurable interaction with A2B receptors was observed in concentrations up to 30 μM (Table S1†).
However, moderate affinity for A3 receptors (6e–6j). Compound (2-amino-1-(4-(trifluoromethyl)phenyl)-1H-imidazol-5-yl)(4-chlorophenyl) methanone (6g) showed the highest affinity among aminoimidazole series of compounds. (Table S1†). It was the only compound that also bound with moderate affinity to the A2A receptor. The presence of 4-chloro in aryl ketone (R2) and 4-trifluromethyl in N-aryl (R1) substituents in (6g) of 2-aminoimidazole core was found to be important for the affinity towards the A3 adenosine receptor. The presence of a chloro substituent at 2 or 3 positions (6h, 6i) in N-aryl (R1) was also found to be significant to enhance affinity towards adenosine A3 receptor. However, the presence of 4-Me, 4-OMe, 4-SO2Me and 4-pyridyl substituents in aryl ketone (R2) and 4-fluoro, 4-chloro substituents (6a–6e) in N-aryl (R1) found to have deleterious effect on adenosine receptor affinities. The 2-aminoimidazole series (Table S1†) did not show desired affinity as their previously investigated 2-aminothiazole congeners22–24 but shows some good affinity towards the A3 receptor (6e, 6h–j). We hypothesized that replacing the aryl/pyridyl (R2) group with substituted 2-aminothizoles at 5 position of 2-aminoimidazole leading to aminoimidazolyl-thiazoles would be of therapeutic interest and thus synthesized such molecules.
Series 2 – aminoimidazolyl-thiazole
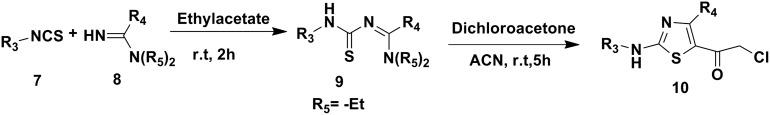
In order to prepare 2-aminoimidazolyl-thiazole derivatives, first step was the synthesis of (2-chloroacetyl) thiazoles (10). Reaction of substituted isothiocynates 7 with amidines 8 was carried out at room temperature to yield amidinothioureas 9 in good to high yields. 9 On further reaction with excess of dichloroacetone gave (2-chloroacetyl) thiazoles 10 in good yields (Scheme 2).
Scheme 2. Preparation of (2-chloroacetyl) thiazoles 10.
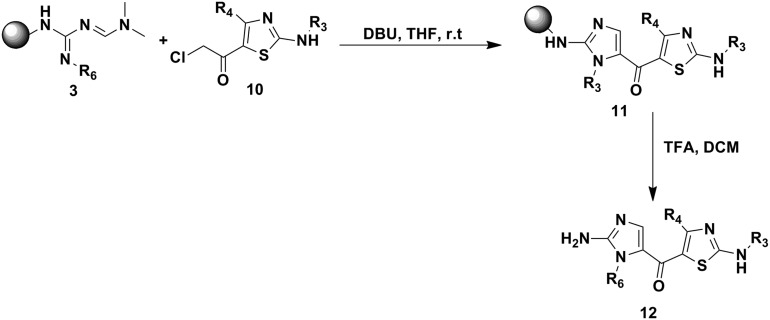
To synthesize the desired compounds 12a–l, reaction of (2-chloroacetyl) thiazoles 10 was carried out with resin bound amidinoguanidine 3 to give 2-aminoimidazolyl-thiazole (Scheme 3).
Scheme 3. Synthesis of 2-aminoimidazolyl-thizole derivatives.
Binding affinities of 2-aminoimidazolyl-thiazole derivatives for human adenosine receptors
The binding affinities for the 2-aminoimidazolyl-thiazole series are summarized in Table 1. In general, the replacement of aryl/pyridyl substituent (R2) (Scheme 1) to thiazole derivatives (Scheme 2) at 5-position of the 2-aminoimidazole scaffold significantly improved the affinity for A1, A2A and A3 adenosine receptors. No measurable interaction with A2B receptors was observed in concentrations up to 10 μM (Table 1). Compounds 12a–c and 12f showed 20–40 fold selectivity toward the A3 receptor. We identified compound 12b as most potent and selective structure in this series with a Ki-value of 0.19 μM at the A3 receptor. Compound 12b is between 40 and 200 fold selective compared to A2A and A1 receptors, respectively.
Table 1. Binding affinities of 2-aminoimidazolyl-thiazole derivatives for human adenosine receptors.
| |||||||
| Compound | R6 | R4 | R3 | Binding experiments Ki (μM) |
Adenylyl cyclase activity Ki (μM) | ||
| hA1 a | hA2A b | hA3 c | hA2B d | ||||
| 12a | 4-OMe–C6H4 | CH3 | C6H5 | 48.6 | 13.4 | 0.574 | >10 |
| (35.4–66.7) | (9.24–19.4) | (0.403–0.816) | |||||
| 12b | 4-F–C6H4 | CH3 | C6H5 | 39.2 | 8.04 | 0.190 | >10 |
| (33.2–46.2) | (6.45–10.0) | (0.154–0.234) | |||||
| 12c | 4-F–C6H4 | Ph | C6H5 | 6.71 | 7.41 | 0.305 | >10 |
| (5.59–8.05) | (4.33–12.7) | (0.268–0.347) | |||||
| 12d | 4-Cl–C6H4 | N(CH3)2 | COC6H4 | 25.4 | 11.8 | 4.32 | >10 |
| (19.8–32.6) | (7.70–18.1) | (3.50–5.33) | |||||
| 12e | 4-CF3–C6H4 | N(CH3)2 | COC6H4 | 19.7 | 16.7 | 4.92 | >10 |
| (18.2–21.3) | (11.9–23.5) | (3.96–6.10) | |||||
| 12f | 4-F–C6H4 | CH3 | 4-Me–C6H4 | 37.5 | 12.4 | 0.487 | >10 |
| (30.1–46.7) | (8.84–17.5) | (0.311–0.76) | |||||
| 12g | 4-OMe–C6H4 | Ph | C6H5 | 3.67 | 8.84 | 0.326 | >10 |
| (2.67–5.10) | (6.64–11.8) | (0.244–0.435) | |||||
| 12h | 4-OMe–C6H4 | Ph | — | >100 | >100 | 13.0 | >10 |
| (10.9–15.5) | |||||||
| 12i | 4-F–C6H4 | CH3 | 4-Cl–C6H4 | 19.8 | 13.0 | 0.871 | >10 |
| (19.1–20.6) | (11.8–14.3) | 0.825–0919 | |||||
| 12j | 4-OMe–C6H4 | CH3 | 4-Cl–C6H4 | 33.3 | 10.3 | 1.60 | >10 |
| (32.0–34.6) | (8.07–13.1) | (1.54–1.65) | |||||
| 12k | 4-Me–C6H4 | Ph | — | >100 | 34.0 | 17.8 | >10 |
| (24.1–48.0) | (15.4–20.6) | ||||||
| 12l | 4-Cl–C6H4 | CH3 | C6H5 | 33.5 | 11.2 | 0.802 | >10 |
| (33.3–33.7) | (8.82–14.2) | (0.673–0.958) | |||||
aDisplacement of specific [3H]CCPA binding at human A1 receptors expressed in CHO cells.
bDisplacement of specific [3H]NECA binding at human A2A receptors expressed in CHO cells.
cDisplacement of specific [3H]HEMADO binding at human A3 receptors expressed in CHO cells.
dInhibition of NECA-stimulated adenylyl cyclase activity at human A2B receptors expressed in CHO cells.
To study the minimum structural requirement of the compounds to be selective for the A3 receptor, N-aryl (R6) substitution of 2-aminoimidazole scaffold was kept constant as Scheme 1 and systematic modification of aminothiazole moiety was carried out. Diverse substituents like phenyl, benzoyl, aryl, morpholine at the amino position of thiazole (R3) and methyl, N,N-dimethyl, phenyl at thiazole (R4) were introduced. All the compounds with phenyl substituent (12a–12c), (12g), (12i), (12l) showed high affinity and selectivity towards the adenosine A3 receptor compared all other subtypes. Further, introduction of 4-chloro (12i–12j) and 4-methyl (12f) substitution to phenyl ring did not show significant improvement in affinity towards A1, A2A and A2B receptors but decreases selectivity towards the A3 receptor. Further, replacement by benzoyl (12d, 12e) and morpholine (12h, 12k) group was also found to significantly decrease selectivity for the A3 receptor.
Molecular modeling study
A structure-based molecular modeling study was conducted to rationalize the compounds' selectivity profile toward human A3 adenosine receptor (AR) over the other ARs subtypes, using a very well consolidated in-house docking pipeline.25–29 The study was focused on human A1, A2A and A3 subtypes, since no binding data were available for A2B. Functional data from adenylyl cyclase experiments suggested that no detectable interaction of the studied compounds occurs with the A2B AR. Docking was performed for all compounds on the three receptors, and the resulting poses were evaluated according to van der Waals and electrostatic interactions. For each ligand one pose was selected by visual inspection, promoting those poses that minimize electrostatic and van der Waals interaction values. Most favorable poses of all compounds on A3, A2A and A1 ARs are shown in Videos SM1–SM3, included in ESI.† With regard to the A1 AR, it was impossible to find a common binding mode for the various ligands, but this was not surprising given the low affinity of the compounds for A1 AR.
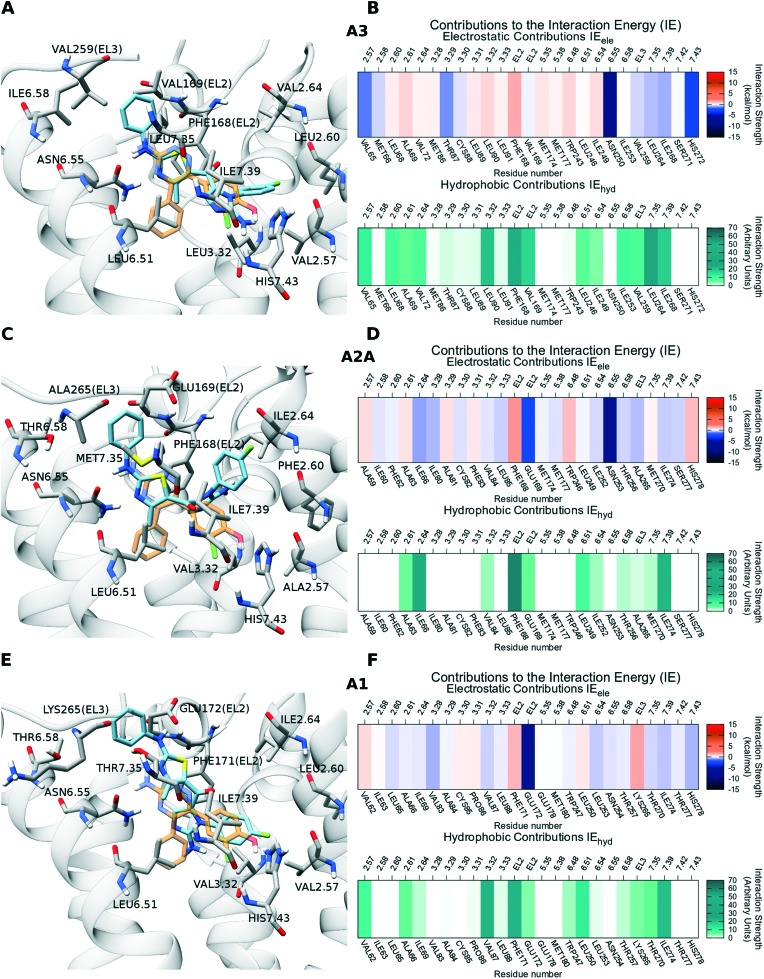
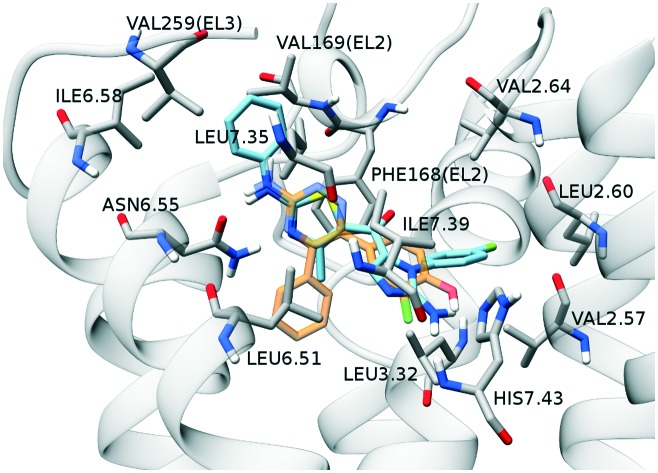
Compound 12b was chosen as representative of the 2-aminoimidazolyl-thiazole series for a detailed description, because of its pronounced selectivity toward A3 (0.19 μM) over A2A (8.0 μM) and A1 (39.2 μM) receptors, as reported in Table 1. The suggested binding mode to the A3 AR (Fig. 2, panel A) shows compound 12b spanning the binding pocket from top to bottom with the 2-phenyl-amino-thiazole portion facing TM6, TM5 and TM7 and the 2-aminoimidazolyl portion pointing toward TM7 and TM2. The compound realizes a double hydrogen bond with Asn6.55, as anticipated by the strong electrostatic contribution of that residue in the IEFs reported in Fig. 2, panel B. The hydrogen bonds engage the endocyclic nitrogen atom of thiazole as acceptor and the exocyclic amine nitrogen as donor. The thiazole ring is stabilized by a π–π stacking interaction with Phe168(EL2). The compound is involved in a series of strong hydrophobic interactions, as reported by IEFs in Fig. 2, panel B, among which Phe168(EL2) emerges as more intense. Phe168(EL2), Val169(EL2), Leu6.51, Ile6.58, Val259(EL3) and Leu7.35 form a hydrophobic cleft which hosts the 1-(4-F-phenyl)-2-aminoimidazolyl scaffold, while Val2.57, Leu2.60, Val2.64, Leu3.32 and Ile7.39 define a pocket that could accommodate the 1-(4-F-phenyl)-2-aminoimidazolyl portion. A similar binding mode and interaction pattern is shared by further compounds with sub-micromolar binding affinity for A3 AR, such as compounds 12c, 12f, 12g, as can be noticed by Video SM1.†
Fig. 2. Panels A–E: Representation of the most favorable docking poses of compound 12b on A3, A2A and A1 adenosine receptors, respectively. The compound is depicted by light-blue sticks, the backbone of the receptor is represented by a transparent light-gray ribbon and most relevant residues are rendered by gray sticks. Helix TM6 is not shown in order to make the visualization of the binding site more clear. The crystallographic conformation of 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol is reported and rendered by orange sticks. For A1 and A3 models, the same ligand is obtained from the superimposition of A2A AR crystallographic structure (PDB ID: ; 3UZC) to the models. Panel B–F: Interaction energy fingerprints (IEFs) of compound 12b on A3, A2A and A1 receptors, respectively. The upper heat map represents IEele fingerprints, with electrostatic interactions strength rendered by blue to red colors for low to high potential values. The lower heat map represents IEhyd fingerprints, with hydrophobic interactions strength rendered by light to dark green colors for low to high potential values.
Compound 12b presents a similar conformation within the binding site of A2A receptor (Fig. 2, panel C). The 2-amino thiazole scaffold is confirmed in its double hydrogen bond interaction pattern with Asn6.55, while the 1-(4-F-phenyl)-2-aminoimidazolyl portion is rotated by almost 90 degrees. For both A2A and A3 receptors, the selected conformation of the compound resembles the one assumed by 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol co-crystallized to A2A AR (PDB ID: ; 3UZC), with the 2-amino-thiazole structure mimicking the 3-amino-trazine scaffold (Fig. 1, panel A and C). The interaction network reported for A2A AR by the IEFs (Fig.2, panel D) confirms many of the interactions encountered for A3: these involve the conserved residues Leu6.51, Ile7.39 and Phe168 (EL2) for hydrophobic interactions, and Asn6.55 for electrostatic ones. However, some hydrophobic interactions lack: positions of Val2.57, Val169(EL2), Ile6.58, Val259(EL3), Leu7.35 are occupied by the less hydrophobic Ala2.57, Glu169(EL2), Thr6.58, Ala265(EL3), Met7.35 on A2A AR, respectively. This may support the drop of binding affinity for A2A in comparison to A3, suggesting that these non-conserved residues may play a role in driving binding selectivity of compound 12b. Indeed, also in A1 AR, Val169(EL2), Ile6.58, Val259(EL3), Leu7.35 are substituted by even more polar residues, Glu172(EL2), Thr6.58, Lys265(EL3) and Thr7.35, implying a general loss of the hydrophobic binding contribution. With regard to A1 AR, among the poses proposed by docking it was impossible to find a conformation similar to the ones just described for the other receptor subtypes. In the selected pose (Fig. 2, panel E), the conformation of the compound within the binding site is driven by the presence of Glu172 on EL2, which engages in a hydrogen bond the exocyclic amine nitrogen atom at position 2 of thiazole.
Conclusions
In conclusion, the present study involves a synthesis of small compound library of 24 2-aminoimidazole and 2-aminoimidazolyl-thiazole derivatives using a 2-chloro trityl resin to explore the SAR. All the compounds were tested for their affinity at A1, A2A, A2B and A3 adenosine receptors. Compounds with 2-aminoimidazole scaffold did not result in good affinity. However, further modification to 2-aminoimidazolyl-thiazoles led to discovery of compound 12b with nanomolar affinity against adenosine A3 receptor and 40 to 200-fold selectivity versus the other adenosine receptor subtypes. The molecular modelling study of compound 12b gave detailed insight into receptor conformations for the lead optimization, which will help to guide further development of the 2-aminoimidazolyl-thiazole series.
Experimental
General remarks
All reagents were purchased from Sigma-Aldrich. All chemicals were reagent grade and used without further purification. The 1H NMR and 13C NMR spectra were recorded on Bruker spectrometer (1H: 300 MHz, 13C: 75 MHz) using TMS as an internal standard. Mass spectra were recorded on Perkin Elmer Sciex API 165.
General procedure for preparation of solid phase synthesis 2-aminoimidazole derivatives (6a–6l)
2-Chlorotrityl resin (1 mmol) was loaded with DMF (10 mL) and stirred. Monosubstituted thiourea 1 and DIPEA were added and the stirring continued for 2 h. Repeated washing of resin with DCM, DMF, THF (4 × 10 ml mmol–1) was carried out in order to remove unbound monocondensed thiourea. Further, guanylation of amidinothiourea with different arylamines was carried out with Mukaiyama's reagent in DMF and stirred for 30 minutes to generate resin bound amidinoguanidine. Excess of reagent was removed by repeated washing of DCM, DMF, THF (4 × 10 ml mmol–1). Cyclization of resin bound amidinoguanidine was carried out by phenacyl bromide in presence of DBU to give resin bound 2-aminoimidazole derivatives. The resin was cleaved using 10% TFA in DCM to give final compound with good to high yield.
(2-Amino-1-(4-fluorophenyl)-1H-imidazol-5-yl)(p-tolyl)methanone (6a)
Yield: 70%, pale yellow solid, mp: 195–197 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.71–7.68 (m, 2H), 7.36–7.32 (m, 3H), 7.28–7.15 (m, 4H), 5.49 (s, 2H), 2.40 (s, 3H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 182.05, 164.0, 160.70, 153.79, 142.44, 140.54, 136.04, 131.71, 128.92, 127.83, 116.79, 116.48, 21.46; C17H14FN3O, exact mass 295.11; LC-ESI-MS/MS: 296.6 [M + 1]+.
(2-Amino-1-(4-fluorophenyl)-1H-imidazol-5-yl)(4-methoxyphenyl)methanone (6b)
Yield: 72%, pale yellow solid, mp: 178–180 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.83–7.77 (m, 2H), 7.39–7.33 (m, 3H), 7.26–7.17 (m, 2H), 6.95–6.92 (m, 2H), 4.94 (s, 2H), 3.87 (s, 3H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 181.38, 164.12, 162.80, 160.81, 153.16, 140.0, 131.41, 130.94, 128.98, 116.91, 116.60, 113.59, 55.42; C17H14FN3O2. Exact mass: 311.11; LC-ESI-MS/MS: 312.2 [M + 1]+.
(2-Amino-1-(4-fluorophenyl)-1H-imidazol-5-yl) (4-chlorophenyl)methanone (6c)
Yield: 78%, pale yellow solid, mp: 230–232 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.74–7.71 (m, 2H), 7.43–7.40 (m, 2H), 7.37–7.33 (m, 3H), 7.24–7.18 (m, 2H), 5.20 (s, 2H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 180.88, 164.21, 160.90, 153.88, 141.19, 138.14, 131.50, 130.10, 129.02, 127.73, 116.96, 116.68; C16H11ClFN3O exact mass: 315.06; LC-ESI-MS/MS: 316.3 [M + 1]+.
(2-Amino-1-(4-chlorophenyl)-1H-imidazol-5-yl) (p-tolyl)methanone (6d)
Yield: 68%, pale yellow solid, mp: 195–197 °C 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.71–7.69 (m, 2H), 7.49–7.45 (m, 2H), 7.36 (s, 1H), 7.31–7.29 (m, 4H), 5.30 (s, 2H), 2.41 (s, 2H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 182.04, 153.43, 142.56, 140.64, 135.96, 134.81, 134.35, 129.92, 128.98, 128.88, 128.33, 127.89, 21.51; C17H14ClN3O exact mass: 311.08; LC-ESI-MS/MS: 312.3 [M + 1]+.
(2-Amino-1-(4-chlorophenyl)-1H-imidazol-5-yl)(pyridin-4-yl)methanone (6e)
Yield: 80%, brown solid, mp: 247 °C; decomposes; 1H NMR (300 MHz, DMSO-d6) δ: 8.77–8.75 (m, 2H), 7.59–7.57 (m, 2H), 7.54–7.50 (m, 2H), 7.34 (s, 1H), 7.33–7.26 (m, 2H), 4.76 (s, 2H); 13C NMR (75 MHz, DMSO-d6) δ: 180.16, 153.85, 150.36, 145.33, 142.63, 135.47, 133.76, 130.19, 128.40, 127.48, 122.20; C15H11ClN4O exact mass: 298.06; LC-ESI-MS/MS m/z 299.4 [M + 1]+.
(2-Amino-1-phenyl-1H-imidazol-5-yl)(4-(methylsulfonyl)phenyl)methanone (6f)
Yield: 62%, pale yellow solid, mp: 192–195 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 8.28–7.79 (m, 2H), 7.88–7.85 (m, 2H), 7.49–7.39 (m, 3H), 7.38–7.21 (m, 3H), 3.35 (s, 3H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 178.47, 155.76, 144.02, 143.51, 142.61, 135.94, 129.21, 129.12, 128.09, 127.22, 127.02, 126.84, 54.90; C17H15N3O3S exact mass: 311.08; LC-ESI-MS/MS m/z 312.3 [M + 1]+.
(2-Amino-1-(4-(trifluoromethyl)phenyl)-1H-imidazol-5-yl)(4-chlorophenyl)methanone (6g)
Yield: 65%, yellow solid, mp: 190–194 °C; 1H NMR NMR (300 MHz, DMSO-d6, ppm) δ: 7.83–7.74 (m, 2H), 7.56–7.49 (m, 2H), 7.41 (s, 1H), 7.37–7.26 (m, 4H), 5.32 (s, 2H); C17H11ClF3N3O, exact mass: 365.05; LC-ESI-MS/MS: 366.3 [M + 1]+.
(2-Amino-1-(2-chlorophenyl)-1H-imidazol-5-yl)(p-tolyl)methanone (6h)
Yield: 64%, pale yellow solid, mp: 163–165 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.81–7.78 (m, 2H), 7.69–7.61 (m, 3H), 7.56–7.41 (m, 4H), 5.51 (s, 2H), 2.41 (s, 3H); C17H14ClN3O, exact mass: 311.08; LC-ESI-MS/MS: 312.3 [M + 1]+.
(2-Amino-1-(3-chlorophenyl)-1H-imidazol-5-yl)(p-tolyl)methanone (6i)
Yield: 78%, white solid, mp: 123–125 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.82–7.79 (m, 2H), 7.71–7.67 (m, 2H), 7.61–7.59 (m, 1H), 7.51–7.45 (m, 4H), 5.49 (s, 2H), 2.45 (s, 3H); C17H14ClN3O exact mass: 311.08; LC-ESI-MS/MS: 312.2 [M + 1]+.
(2-Amino-1-(3-chloro-4-fluorophenyl)-1H-imidazol-5-yl)(4-chlorophenyl)methanone (6j)
Yield: 75%, yellow solid, mp: 220–224 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.81–7.78 (m, 2H), 7.72–7.68 (m, 2H), 7.65–7.58 (m, 1H), 7.57–7.46 (m, 2H), 5.47 (s, 2H); C16H10Cl2FN3O exact mass 349.02; LC-ESI-MS/MS: 350.4 [M + 1]+.
(2-Amino-1-(4-(trifluoromethyl)phenyl)-1H-imidazol-5-yl)(pyridin-4-yl)methanone (6k)
Yield: 64%, brown solid, mp: 229–232 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 8.75–8.69 (m, 2H), 7.78–7.69 (m, 2H), 7.65–7.59 (m, 2H), 7.37 (s, 1H), 7.35–7.22 (m, 2H), 5.15 (s, 2H); C16H11F3N4O exact mass: 332.09; LC-ESI-MS/MS: 333.3 [M + 1]+.
(2-Amino-1-(4-fluorophenyl)-1H-imidazol-5-yl)(pyridin-4-yl)methanone (6l)
Yield: 76%, yellow solid, mp: 236–239 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 8.71–8.65 (m, 2H), 7.71–7.68 (m, 2H), 7.61–7.57 (m, 2H), 7.45 (s, 1H), 6.98–6.87 (m, 2H), 5.27 (s, 2H); C15H11FN4O exact mass: 282.09; LC-ESI-MS/MS: 283.4 [M + 1]+.
General procedure for preparation of solid phase synthesis aminoimidazolyl-thiazole (12a–12l)
2-Chlorotrityl resin (1 mmol) was loaded and vortex it with DMF (10 mL) to swell the resin, the monosubstituted thiourea 1 and DIPEA was added followed by stirring it for about 2 h, then repeated washing of resin with DCM, DMF, THF (4 × 10 ml mmol–1) was given to remove unbound mono condensed thiourea. Further, guanylation of amidinothiourea with various arylamine was carried out with Mukaiyama's reagents in DMF and stirred for 30 minutes to generate resin bound amidinoguanidine. Excess of reagents was removed by repeated washing of DCM, DMF, THF (4 × 10 ml mmol–1). Cyclization of resin bound amidinoguanidine was carried out by (2-chloroacetyl) thiazole derivatives in presence of DBU to give resin bound aminoimidazolyl-thiazole derivatives. The cleavage of resin was done by 10% TFA in DCM to give final compound with good to high yield.
(2-Amino-1-(4-methoxyphenyl)-1H-imidazol-5-yl)(4-methyl-2-(phenylamino)thiazol-5-yl)methanone (12a)
Yield: 64%, brown solid, mp: 235–239 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 10.58 (S, 1H) 7.61–7.58 (d, J = 9, 2H), 7.50 (s, 1H) 7.36–7.31 (m, 2H), 7.19–7.16 (d, J = 9, 2H), 7.02–6.97 (m, 3H), 6.14 (s, 2H), 3.79 (s, 3H), 2.49 (s, 3H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 171.59, 163.90, 158.65, 155.04, 154.90, 140.32, 139.16, 129.12, 128.74, 128.60, 128.36, 122.26, 117.92, 117.73, 114.34, 55.33, 18.19; C21H19N5O2S exact mass: 405.13; LC-ESI-MS/MS: 406.5 [M + 1]+.
(2-Amino-1-(4-fluorophenyl)-1H-imidazol-5-yl)(4-methyl-2-(phenylamino)thiazol-5-yl)methanone (12b)
Yield: 64%, yellow solid, mp: 178–179 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 8.85 (s, 1H), 7.61 (s, 1H) 7.42–7.31 (m, 7H), 7.29–7.11 (m, 3H), 4.84 (s, 2H), 2.50 (s, 3H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 166.26, 164.16, 157.64, 139.37, 137.83, 131.55, 129.69, 128.93, 128.81, 127.92, 124.23, 119.19, 117.48, 117.08, 116.77, 18.26; C20H16FN5OS exact mass: 393.11; LC-ESI-MS/MS: 394.3 [M + 1]+.
(2-Amino-1-(4-fluorophenyl)-1H-imidazol-5-yl)(4-phenyl-2-(phenylamino)thiazol-5-yl)methanone (12c)
Yield: 64%, yellow solid, mp: 230–231 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 10.59 (s, 1H), 7.66–7.64 (d, J = 6, 2H), 7.57–7.55 (d, J = 6, 2H), 7.40–7.34 (m, 5H), 7.32–7.24 (m, 4H), 7.15 (s, 1H), 7.02–6.67 (m, 1H), 6.31 (s, 2H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 171.69, 163.05, 159.88, 155.02, 152.56, 142.40, 140.47, 135.14, 132.07, 129.54, 129.07, 128.30, 128.08, 127.62, 122.14, 119.86, 117.55, 116.22, 115.92; C25H18FN5OS exact mass: 455.12; LC-ESI-MS/MS: 455.6 [M + 1]+.
N-(5-(2-Amino-1-(4-chlorophenyl)-1H-imidazole-5-carbonyl)-4-(dimethylamino)thiazol-2-yl)benzamide (12d)
Yield: 64%, white solid, mp: 180–183 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 12.56 (s, 1H), 9.21 (s, 1H), 8.15–7.74 (m, 5H), 7.65 (s, 1H), 7.31–7.18 (m, 4H), 6.82 (s, 2H), 3.22 (s, 6H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 174.25, 166.15, 163.15, 162.19, 141.57, 140.7, 138.9, 136.96, 134.15, 133.07, 131.91, 129.20, 128.15, 126.57, 122.24, 41.15; C22H19ClN6O2S exact mass: 466.1; LC-ESI-MS/MS : 467.3 [M + 1]+.
N-(5-(2-Amino-1-(4-(trifluoromethyl)phenyl)-1H-imidazole-5-carbonyl)-4(dimethylamino)thiazol-2-yl)benzamide (12e)
Yield: 64%, white solid, mp: 230 °C decomposes; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 12.73 (s, 1H), 9.20 (s, 1H), 7.82 (s, 1H), 8.18–7.78 (m, 5H), 7.81 (s, 1H), 7.31–7.18 (m, 4H), 6.82 (s, 2H), 3.24 (s, 6H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 174.28, 166.20, 163.09, 162.37, 141.61, 140.82, 138.9, 137.01, 134.20, 133.09, 131.94, 129.24, 128.20, 126.59, 122.26, 41.24; C23H19F3N6O2S; exact mass: 500.12; LC-ESI-MS/MS: 501.2 [M + 1]+.
(2-Amino-1-(4-fluorophenyl)-1H-imidazol-5-yl)(4-methyl-2-(p-tolylamino)thiazol-5-yl)methanone (12f)
Yield: 70%, yellow solid, mp: 215–217 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 9.23 (s, 1H), 7.40–7.32 (m, 4H), 7.27–7.12 (m, 4H), 4.19 (s, 2H), 2.50 (s, 3H), 2.34 (s, 3H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 166.21, 164.23, 163.21, 158.01, 140.15, 138.10, 131.61, 129.70, 128.94, 128.82, 127.92, 124.23, 119.20, 117.15, 117.01, 20.34, 18.26; C21H18FN5OS exact mass: 407.12; LC-ESI-MS/MS: 408.2 [M + 1]+.
(2-Amino-1-(4-methoxyphenyl)-1H-imidazol-5-yl)(4-phenyl-2-(phenylamino) thiazol-5-yl)methanone (12g)
Yield: 60%, brown solid, mp: 175–178 decomp. °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 10.61 (s, 1H), 7.67–7.65 (d, J = 6, 2H), 7.60–7.58 (d, J = 6, 2H), 7.49–7.30 (m, 5H), 7.24 (s, 1H), 7.23–7.16 (m, 2H), 7.03–6.95 (m, 3H), 6.20 (s, 2H), 3.79 (s, 3H); 13CNMR (75 MHz, DMSO-d6, ppm) δ: 171.73, 162.99, 158.78, 155.25, 155.07, 152.45, 143.49, 140.99, 140.52, 135.15, 129.47, 129.21, 128.44, 127.88, 127.68, 127.32, 120.45, 116.47, 113.33, 56; C26H21N5O2S exact mass: 467.14; LC-ESI-MS/MS: 468.5 [M + 1]+.
(2-Amino-1-(4-methoxyphenyl)-1H-imidazol-5-yl)(2-morpholino-4-phenylthiazol-5-yl)methanone (12h)
Yield: 62%, white solid, mp: 178–180 °C; purification by column chromatography (ethyl acetate/hexane (3 : 7)) 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.51–7.29 (m, 6H), 7.18–7.01 (m, 4H), 6.11 (s, 2H), 3.81 (s, 3H), 3.74–3.72 (m, 4H), 3.51–3.32 (m, 4H); C24H23N5O3S exact mass 461.15; LC-ESI-MS/MS: 461.5 [M + 1]+.
(2-Amino-1-(4-fluorophenyl)-1H-imidazol-5-yl)(2-(4-chlorophenylamino)-4-methylthiazol-5-yl)methanone (12i)
Yield: 74%, pale yellow solid, mp: 136–138 °C; purification by column chromatography (ethyl acetate/hexane (3 : 7)) 1H NMR (300 MHz, DMSO-d6, ppm) δ: 10.15 (s, 1H), 7.51–7.45 (m, 4H), 7.37 (s, 1H), 7.29–7.14 (m, 4H), 4.81 (s, 2H), 2.45 (s, 3H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 167.62, 165.15, 158.16, 140.47, 139.19, 131.57, 129.72, 128.91, 128.74, 128.15, 124.29, 119.21, 117.45, 117.20, 117.15, 19.15; C20H15ClFN5OS; exact mass: 427.07; LC-ESI-MS/MS: 428.4 [M + 1]+.
(2-Amino-1-(4-methoxyphenyl)-1H-imidazol-5-yl)(2-(4-chlorophenylamino)-4-methylthiazol-5-yl)methanone (12j)
Yield: 70%, white solid, mp: 123–125 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 10.28 (s, 1H), 7.61–7.54 (m, 4H), 7.43 (s, 1H), 7.37–7.29 (m, 4H), 5.07 (s, 2H), 2.93 (s, 3H), 2.45 (s, 3H); C21H18ClN5O2S exact mass: 439.09; LC-ESI-MS/MS: 439.9, 442.1 [M + 1]+.
(2-Amino-1-p-tolyl-1H-imidazol-5-yl)(2-morpholino-4-phenylthiazol-5-yl)methanone (12k)
Yield: 60%, brown solid, mp: 230 °C decomposes; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 7.47–7.25 (m, 6H), 7.20–6.94 (m, 4H), 6.18 (s, 2H), 3.76–3.71 (m, 4H), 3.52–3.35 (m, 4H), 2.56 (s, 3H); C24H23N5O2S; exact mass: 445.16; LC-ESI-MS/MS: 446.3 [M + 1]+.
(2-Amino-1-(4-chlorophenyl)-1H-imidazol-5-yl)(4-methyl-2-(phenylamino)thiazol-5-yl)methanone (12l)
Yield: 65%, pale brown solid, mp: 225–227 °C; 1H NMR (300 MHz, DMSO-d6, ppm) δ: 8.96 (s, 1H), 7.59 (s, 1H), 7.40–7.32 (m, 5H), 7.30–7.15 (m, 4H), 4.86 (s, 2H), 2.56 (s, 3H); 13C NMR (75 MHz, DMSO-d6, ppm) δ: 166.24, 163.20, 159.70, 140.15, 137.91, 131.56, 129.70, 129.01, 128.89, 127.91, 124.98, 120.09, 117.51, 117.07, 116.98, 21.96; C20H16ClN5OS; exact mass: 409.08, LC-ESI-MS/MS: 410.01 [M + 1]+.
Biological assays
For radioligand binding studies (A1, A2A, and A3 receptor) and measurement of adenylyl cyclase activity (A2B) membranes were prepared from CHO cells stably transfected with the respective human adenosine receptor subtype. For details see ESI.†
Abbreviations
- TM
Transmembrane helix
- EL
Extracellular loop
- AR
Adenosine receptor
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors declare no conflict of interests.
Supplementary Material
Acknowledgments
We thank Dr. Manish Nivsarkar, Professor Harish Padh and Professor C. J. Shisoo, Directors, B. V. Patel PERD centre, for their constant encouragement and support. A. N. P. thanks PERD Centre for providing the doctoral fellowship. This work was funded by DST through grant no. SR/S1/OC-83/2009. The authors wish to thank DST for its financial support. The molecular modeling work coordinated by S. M. was carried out with financial support from the University of Padova, Italy, and the Italian Ministry for University and Research (MIUR), Rome, Italy. S. M. is also very grateful to Chemical Computing Group and to OpenEye for the scientific and technical partnership.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7md00643h
References
- Fredholm B. B., IJzerman A. P., Jacobson K. A., Klotz K.-N., Linden J. Pharmacol. Rev. 2001;53:527. [PMC free article] [PubMed] [Google Scholar]
- Fredholm B. B., Arslan G., Halldner L., Kull B., Schulte G., Wasserman W. Naunyn-Schmiedeberg's Arch. Pharmacol. 2000;362:364. doi: 10.1007/s002100000313. [DOI] [PubMed] [Google Scholar]
- Baharav E., Bar-Yehuda S., Madi L., Silberman D., Rath-Wolfson L., Halpren M., Ochaion A., Weinberger A., Fishman P. J. Rheumatol. 2005;32:469. [PubMed] [Google Scholar]
- Jacobson K. A., Kim H. O., Siddiqui S. M., Olah M. E., Stiles G. L., Von Lubitz D. K. J. E. Drugs Future. 1995;20:689. doi: 10.1358/dof.1995.020.07.531583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno Y., Ji X. D., Mawhorter S. D., Koshiba M., Jacobson K. A. Blood. 1996;88:3569. [PMC free article] [PubMed] [Google Scholar]
- Jacobson K. A. Trends Pharmacol. Sci. 1998;19:184. doi: 10.1016/s0165-6147(98)01203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Lubitz D. K. J. E., Lin R. C. S., Popik P., Carter M. F., Jacobson K. A. Eur. J. Pharmacol. 1994;263:59. doi: 10.1016/0014-2999(94)90523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivo J., Zeira E., Galun E., Matot I. Anesthesiology. 2004;101:1153. doi: 10.1097/00000542-200411000-00015. [DOI] [PubMed] [Google Scholar]
- Brown R. A., Spina D., Page C. P. Br. J. Pharmacol. 2008;153:446. doi: 10.1038/bjp.2008.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx D., Ezeamuzie C. I., Nieber K., Szelenyi I. Drug News Perspect. 2001;14:89. [PubMed] [Google Scholar]
- Okamura T., Kurogi Y., Hashimoto K., Sato S., Nishikawa H., Kiryu K., Nagao Y. Bioorg. Med. Chem. Lett. 2004;14:3775. doi: 10.1016/j.bmcl.2004.04.099. [DOI] [PubMed] [Google Scholar]
- Schlotzer-Schrehardt U., Zenkel M., Decking U., Haubs D., Kruse F. E., Junemann A., Coca-Prados M., Naumann G. O. Invest. Ophthalmol. Visual Sci. 2005;46:2023. doi: 10.1167/iovs.04-0915. [DOI] [PubMed] [Google Scholar]
- Avila M. Y., Stone R. A., Civan M. M. Invest. Ophthalmol. Visual Sci. 2002;43:3021. [PubMed] [Google Scholar]
- Gessi S., Varani K., Merighi S., Cattabriga E., Avitabile A., Gavioli R., Fortini C., Leung E., Mac Lennan S., Borea P. A. Mol. Pharmacol. 2004;65:711. doi: 10.1124/mol.65.3.711. [DOI] [PubMed] [Google Scholar]
- Ohana G., Bar-yehuda S., Barer F., Fishman P. J. Cell. Physiol. 2001;186:19. doi: 10.1002/1097-4652(200101)186:1<19::AID-JCP1011>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Baraldi P. G., Borea P. A. Trends Pharm. Sci. 2000;21:456. doi: 10.1016/s0165-6147(00)01581-9. [DOI] [PubMed] [Google Scholar]
- Merighi S., Mirandola P., Varani K., Gessi S., Leung E., Baraldi P. G., Tabrizi M. A., Borea P. A. Pharmacol. Ther. 2003;1001:31. doi: 10.1016/s0163-7258(03)00084-6. [DOI] [PubMed] [Google Scholar]
- Chang L., Brussee J., IJzerman A. P. Chem. Biodiversity. 2004;1:1591. doi: 10.1002/cbdv.200490122. [DOI] [PubMed] [Google Scholar]
- Saki M., Tsumuki H., Nonaka H., Shimada J., Ichimura M. Eur. J. Pharmacol. 2002;444:133. doi: 10.1016/s0014-2999(02)01662-x. [DOI] [PubMed] [Google Scholar]
- Trifilieff A., Keller T. H., Press N. J., Howe T., Gedeck P., Beer D., Walker C. Br. J. Pharmacol. 2005;144:1002. doi: 10.1038/sj.bjp.0706132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press N. J., Keller T. H., Tranter P., Beer D., Jones K., Faessler A., Heng R., Lewis C., Howe T., Gedeck P., Mazzoni L., Fozard J. R. Curr. Top. Med. Chem. 2004;4:863. doi: 10.2174/1568026043451023. [DOI] [PubMed] [Google Scholar]
- Scheiff A. B., Yerande S. G., El-Tayeb A., Li W., Inamdar G. S., Vasu K. K., Sudarsanam V., Muller C. Bioorg. Med. Chem. 2010;18:2195. doi: 10.1016/j.bmc.2010.01.072. [DOI] [PubMed] [Google Scholar]
- Inamdar G. S., Pandya A. N., Thakar H. M., Sudarsanam V., Kachler S., Sabbadin D., Moro S., Klotz K.-N., Vasu K. K. Eur. J. Med. Chem. 2013;63:924. doi: 10.1016/j.ejmech.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Pandya D. H., Sharma J. A., Jalani H. B., Pandya A. N., Sudarsanam V., Kachler S., Klotz K. N., Vasu K. K. Bioorg. Med. Chem. Lett. 2015;25:1306. doi: 10.1016/j.bmcl.2015.01.040. [DOI] [PubMed] [Google Scholar]
- Floris M., Sabbadin D., Medda R., Bulfone A., Moro S. Eur. J. Med. Chem. 2012;58:248–257. doi: 10.1016/j.ejmech.2012.10.022. [DOI] [PubMed] [Google Scholar]
- Floris M., Sabbadin D., Ciancetta A., Medda R., Cuzzolin A., Moro S. In Silico Pharmacol. 2013;20:1–25. doi: 10.1186/2193-9616-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancetta A., Cuzzolin A., Moro S. J. Chem. Inf. Model. 2014;54:2243–2254. doi: 10.1021/ci5002857. [DOI] [PubMed] [Google Scholar]
- Cuzzolin A., Sturlese M., Malvacio I., Ciancetta A., Moro S. Molecules. 2015;20:9977–9993. doi: 10.3390/molecules20069977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancetta A., Sabbadin D., Federico S., Spalluto G., Moro S. Trends Pharmacol. Sci. 2015;36:878–890. doi: 10.1016/j.tips.2015.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.