 Screening of a natural product library identified several selective cystathionine β-synthase inhibitors, which suppressed the proliferation of HT29 cancer cells.
Screening of a natural product library identified several selective cystathionine β-synthase inhibitors, which suppressed the proliferation of HT29 cancer cells.
Abstract
A high-throughput assay was developed to identify inhibitors of cystathionine β-synthase (CBS), which is one of three key enzymes involved in H2S biosynthesis. Screening of 6491 natural compounds identified several selective CBS inhibitors, which suppressed the proliferation of HT29 cancer cells, with IC50 values of less than 10 μM.
Cystathionine β-synthase (CBS) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme that catalyzes the condensation of serine and homocysteine to generate cystathionine and plays a key role in sulfur metabolism. CBS deficiency is the single most common cause of homocystinuria and leads to a number of clinical and pathological abnormalities in the central nervous, cardiovascular, skeletal and ocular systems.1 Additionally, CBS is also one of the critical enzymes that catalyze the condensation of homocysteine and cysteine to produce hydrogen sulfide (H2S), a recently recognized endogenous gasotransmitter that mediates diverse biological functions.2 Recently published studies indicated that in ovarian and colorectal cancers, a substantially high level of human cystathionine β-synthase (hCBS) expression plays a key role in promoting the proliferation and migration of cancer cells.3 Consequently, the development of potent and selective CBS inhibitors would be of great value in studying the CBS-mediated H2S signaling pathway and treating CBS-related diseases.
Although there has been some progress in the development of CBS inhibitors, only a few inhibitors are available.4 Currently, aminooxyacetic acid (AOAA) and hydroxylamine (HA) are the only two small molecular compounds that have widely been used to inhibit hCBS activity in vitro and in vivo. However, both AOAA and HA have poor selectivity for hCBS over the other PLP-dependent enzymes, including H2S-producing cystathionine γ-lyase (CSE), and poor potency, as a considerably higher dose of AOAA or HA (1–10 mM) is required to inhibit hCBS activity under physiological conditions.5 Thus far, only a few potent and selective hCBS inhibitors have been identified through high-throughput screening assays.4
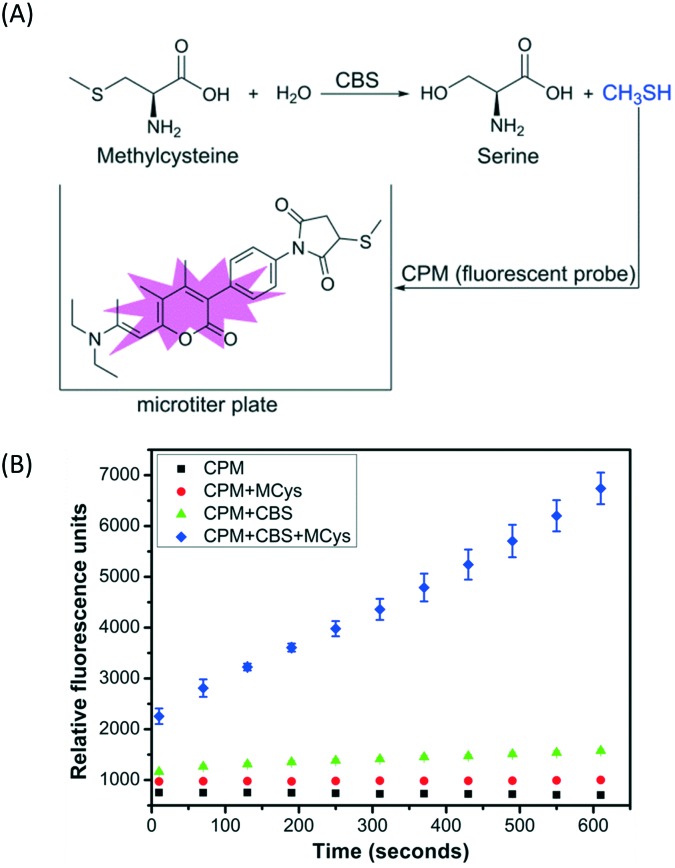
A simple, sensitive and continuous enzyme activity assay was required to establish a high-throughput screening method for monitoring CBS activity. We have previously shown that CBS can catalyze the decomposition of methylcysteine, a thiol-free substrate analog of cysteine, to produce methanethiol (CH3SH).6 Therefore, CH3SH production catalyzed by CBS can be continuously monitored with the commercial fluorescent thiol probe 7-diethylamino-3-(4-maleimidophenyl)-4-methylcoumarin (CPM) using microplate readers, without any interference from the enzyme assay buffer (Fig. 1A). The production of CH3SH from methylcysteine catalyzed by full-length, wild-type hCBS was monitored using the CPM fluorescent probe at physiological pH 7.4 to verify the reliability of the screening method. As indicated in Fig. 1B, the produced CH3SH reacted with the fluorescent probe CPM and generated a concomitant increase in fluorescence intensity as the incubation time increased. Moreover, this assay for monitoring the CH3SH-producing activity of CBS is sensitive and takes much less time (<10 min) compared to the other high-throughput assays reported previously.4
Fig. 1. Illustration of the high-throughput assays for cystathionine β-synthase (CBS). (A) Methanethiol (CH3SH) generation catalyzed by CBS using methylcysteine (Mcys) as a substrate was determined with the CPM fluorescent thiol probe. (B) The CBS activity was monitored using 10 mM methylcysteine as the substrate in the presence of a 15 μM CPM fluorescent probe and 2 μg of hCBS in a 200 μL reaction mixture containing 50 mM HEPES, pH 7.4. The fluorescence of the reaction mixture at 460 nm (λex = 400 nm) was monitored for 600 seconds. CPM = 7-diethylamino-3-(4-maleimidophenyl)-4-methylcoumarin.
Based on the initial success of monitoring the CH3SH-producing activity of CBS with the CPM probe using methylcysteine as a substrate, we wondered whether CSE, another PLP-dependent enzyme that is responsible for the biogenesis of H2S in vivo, would catalyze the degradation of methylcysteine to produce CH3SH that can be readily monitored in the presence of CPM. Indeed, as shown in Fig. S1,‡ we were able to monitor human cystathionine γ-lyase (hCSE) activity using methylcysteine as the substrate. The Michaelis constants (Km) of hCBS and hCSE were measured by varying the concentrations of methylcysteine to determine the appropriate concentration of substrate. As indicated in Fig. S2,‡ the Km values of hCBS and hCSE for methylcysteine were 5.22 mM and 1.98 mM, respectively, which are similar to the previously reported values obtained for hCBS (6.8 mM)2d and hCSE (2.75 mM) using cysteine as the substrate.7 According to the measured Km values, 10 mM methylcysteine was chosen for the subsequent hCBS and hCSE assays.
We next investigated the effect of AOAA, a general inhibitor of PLP-dependent enzymes, on the CH3SH-producing activities of hCBS and hCSE. As shown in Fig. S3A and B,‡ AOAA showed a dose–response curve with an IC50 value of 1 μM for hCBS and 0.57 μM for hCSE in 50 mM HEPES, pH 7.4. We used a standard assay in which cysteine and homocysteine were employed as substrates to measure the H2S-producing activity of the two enzymes in the presence of 0.4 mM lead nitrate to compare the effects of AOAA on the hCBS and hCSE activities in the different enzyme assays.8 As shown in Fig. S3C and D,‡ AOAA in 50 mM HEPES (pH 7.4) inhibited the activities of both hCBS and hCSE, with IC50 values of ∼4 μM, which were close to the previously reported values using the methylene blue assay to determine the H2S-producing enzyme activity.5 These results indicated that in contrast with the standard H2S-producing assay, AOAA was slightly more potent in inhibiting hCBS and hCSE using the CH3SH-producing assay conditions. Additionally, in these studies, the full-length, wild-type hCBS was employed instead of a truncated version of hCBS, which does not contain the regulatory domain and does not exist in vivo.9
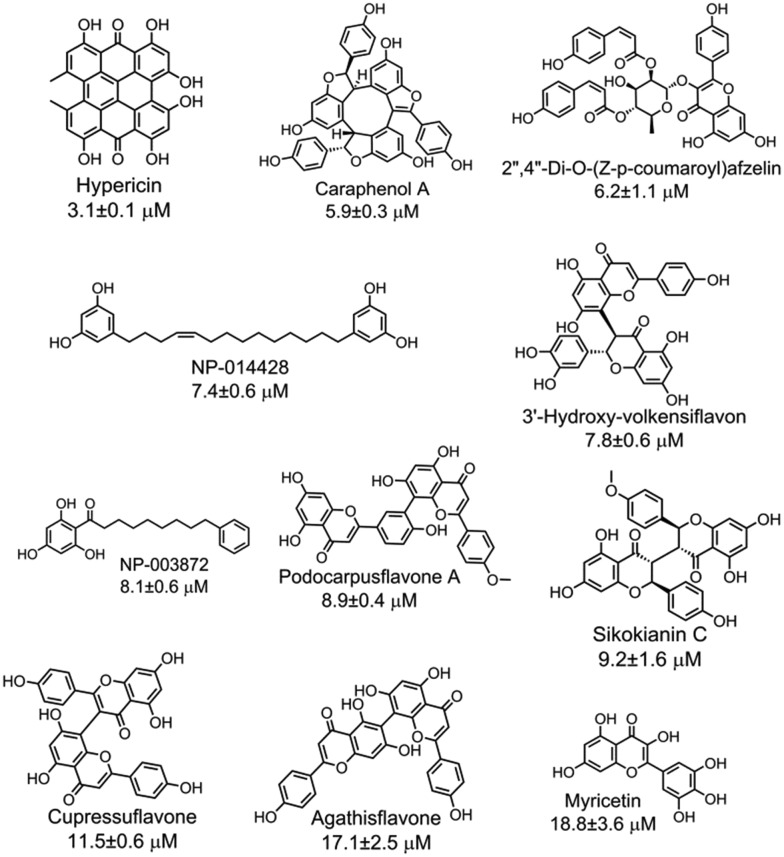
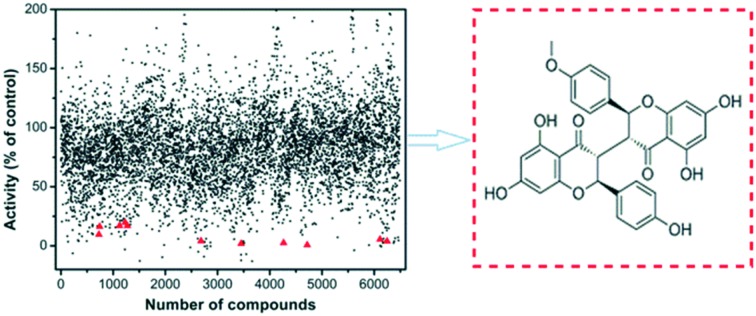
With the purpose of identifying potent and selective CBS inhibitors, we screened 6491 compounds from a natural product library. The library contains compounds isolated from plants and microorganisms with a wide range of chemical diversity and biological activities. All the compounds were screened at a concentration of 100 μM, and then the compounds that were capable of decreasing the CBS activity by more than 80% were accepted for further investigation (Fig. S4‡). The average Z′ value of the screening assay was 0.79 using 100 μM AOAA as a positive control (ESI‡). The secondary screening at a 50 μM concentration of the compounds confirmed that 25 compounds inhibited the enzyme activity by more than 50%. As methylcysteine is not a natural substrate of CBS in vivo, the lead acetate assay using cysteine and homocysteine as substrates was employed to measure the CBS activity.8 The dose-dependent curve of each selective hit was measured to obtain the IC50 values for inhibition of hCBS activity, thus resulting in the identification of 11 compounds with IC50 values of less than 20 μM (Fig. S5‡). The chemical structure of each validated hit and its IC50 value are shown in Fig. 2. In addition, for each compound shown in Fig. 2, a series of control experiments were carried out to ensure that the hits did not react with either the CPM probe or CH3SH to an appreciable extent (ESI‡). These compounds could be categorized into four structural classes (Table S1‡). Hypericin, a photosensitizer in the photodynamic therapy of cancer isolated from medicinal herb Hypericum perforatum or St. John's wort,10 was the most potent inhibitor of hCBS, with an IC50 value of ∼3 μM. Interestingly, seven out of the eleven hits are categorized as flavonoid compounds, five of which are biflavonoids.
Fig. 2. Eleven validated hits from the natural product library that inhibited the hCBS activity with IC50 values of less than 20 μM.
The effect of each hit on hCSE activity was measured using the H2S-producing assay at physiological pH 7.4 to investigate the selectivity of the eleven hits on H2S production. As a result, two compounds (caraphenol A and myricetin) showed similar inhibitory activity against both hCSE and hCBS, three compounds (hypericin, NP-014428, and NP-003872) showed a more than ∼10-fold higher selectivity for hCBS than for hCSE, and the six remaining compounds did not inhibit hCSE activity, even at high concentrations (IC50 > 400 μM). Interestingly, these six compounds that showed selectivity for hCBS fall into the category of flavonoids, of which five compounds are biflavonoids. In addition, to determine the mode of action of the 11 hits, the reversibility of their inhibitory effect was measured by ultrafiltering the enzyme-plus-inhibitor solutions. The results indicated that these 11 inhibitors reversibly bind to hCBS (Fig. S6‡).
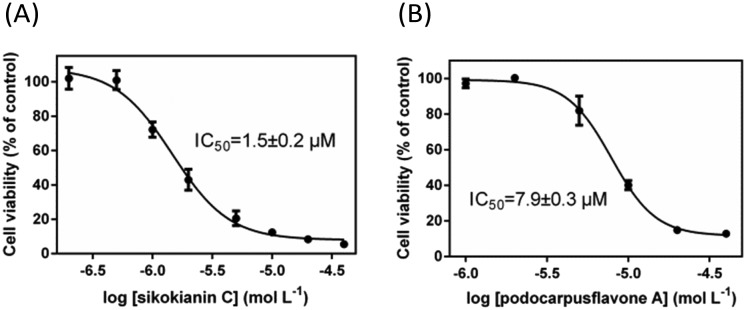
We next investigated whether these six selective hCBS inhibitors would inhibit the proliferation of HT29 colon cancer cells, which express high levels of CBS compared with the normal colon mucosal epithelial cells, NCM356 (Fig. S7‡).3b First, the effect of varying the concentration of AOAA, a commonly used pharmacological CBS inhibitor, on the proliferation of HT29 cells was determined. As indicated in Fig. S8,‡ in contrast with the IC50 values in the low micromolar range determined with purified hCBS,5 a high concentration of AOAA was required to inhibit the proliferation of HT29 cells, with an IC50 value of ∼140 μM. Subsequently, the dose–response curve of each selective inhibitor was measured to obtain the IC50 values for inhibiting the proliferation of HT29 cells. Two compounds, 3′-hydroxy-volkensiflavon and cupressuflavone, showed a weak inhibition of HT29 cells, with IC50 values of ≥50 μM. The other four inhibitors had IC50 values ranging between 1.5 and 13.5 μM, which were far less than the IC50 value of AOAA (Fig. 3 and Fig. S9‡). In particular, sikokianin C, which was isolated from the medicinal herb Stellera chamaejasme,11 had the most potent cytotoxic activity against HT29 cells, with an IC50 value of 1.5 μM (Fig. 3A). Collectively, we identified six compounds that selectively inhibited hCBS activity and did not affect hCSE by screening a natural product library, among which four compounds efficiently inhibited the proliferation of HT29 human colon cancer cells.
Fig. 3. Effects of two selective hCBS inhibitors, sikokianin C (A) and podocarpusflavone A (B), on the viability of HT29 colon cancer cells. The data represented the cell viability of the experimental group compared with the control group and are presented as the means ± SD (n = 6).
To determine whether the two selective inhibitors, sikokianin C and podocarpusflavone A, targeted specifically the CBS protein in mammalian cells, the dose–response curve of each inhibitor was measured to obtain the IC50 values for inhibiting the proliferation of NCM356 cells (which overexpress CSE, but only very low levels of CBS3b). As indicated in Fig. S10,‡ the IC50 values of sikokianin C and podocarpusflavone A against NCM356 cells were 19 and 36 μM respectively, and increased by ∼10 and ∼5 times compared to the IC50 values determined in HT29 cells. Expectedly, the IC50 values of AOAA against NCM356 cells also increased to ∼800 μM compared to ∼140 μM determined in HT29 cells (Fig. S11‡). These results indicated that the CBS protein could be targeted by these two inhibitors in mammalian cells. It should be noted, however, that the inhibitors probably have other target(s) existing in mammalian cells besides the CBS protein. Further work is needed to determine the possible mechanism in these inhibitors' anti-proliferative actions.
In conclusion, a simple, efficient, sensitive and continuous assay was developed to monitor the activity of CBS and CSE using a fluorescent thiol probe. Using this assay, several potent and selective hCBS inhibitors, derived from medicinal herbs, have been identified, and these inhibitors should be of great value in understanding the biological functions of hCBS and the hCBS-mediated H2S signaling pathway. Moreover, this CH3SH-producing assay could readily be adapted to future high-throughput screening of more potent and selective CBS or CSE inhibitors.
This work was supported by the National Natural Science Foundation of China (Grant Number: 31471086), the Fundamental Research Funds for the Central Universities of China (Grant Number: 3102015ZY084) and the Seed Foundation of Innovation and Creation for Graduate Students in Northwestern Polytechnical University (Grant Number: Z2016197). We would like to thank the National Compound Resource Center for providing compound library.
Supplementary Material
Footnotes
†The authors declare no competing interests.
‡Electronic supplementary information (ESI) available. See DOI: 10.1039/c6md00493h
References
- (a) Kraus J., Janosik M., Kozich V., Mandell R., Shih V., Sperandeo M., Sebastio G., de Franchis R., Andria G., Kluijtmans L., Blom H., Boers G., Gordon R., Kamoun P., Tsai M., Kruger W., Koch H., Ohura T., Gaustadnes M. Hum. Mutat. 1999;13:362. doi: 10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]; (b) Miles E., Kraus J. J. Biol. Chem. 2004;279:29871. doi: 10.1074/jbc.R400005200. [DOI] [PubMed] [Google Scholar]
- (a) Kabil O., Banerjee R. Antioxid. Redox Signaling. 2014;20:770. doi: 10.1089/ars.2013.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Whiteman M., Le Trionnaire S., Chopra M., Fox B., Whatmore J. Clin. Sci. 2011;121:459. doi: 10.1042/CS20110267. [DOI] [PubMed] [Google Scholar]; (c) Kimura H. Antioxid. Redox Signaling. 2010;12:1111. doi: 10.1089/ars.2009.2919. [DOI] [PubMed] [Google Scholar]; (d) Singh S., Padovani D., Leslie R., Chiku T., Banerjee R. J. Biol. Chem. 2009;284:22457. doi: 10.1074/jbc.M109.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Hellmich M., Coletta C., Chao C., Szabo C. Antioxid. Redox Signaling. 2015;22:424. doi: 10.1089/ars.2014.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Szabo C., Coletta C., Chao C., Módis K., Szczesny B., Papapetropoulos A., Hellmich M. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12474. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bhattacharyya S., Saha S., Giri K., Lanza I., Nair K., Jennings N., Rodriguez-Aguayo C., Lopez-Berestein G., Basal E., Weaver A., Visscher D., Cliby W., Sood A., Bhattacharya R., Mukherjee P. PLoS One. 2013;8:e79167. doi: 10.1371/journal.pone.0079167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Thorson M., Majtan T., Kraus J., Barrios A. Angew. Chem., Int. Ed. 2013;52:4641. doi: 10.1002/anie.201300841. [DOI] [PubMed] [Google Scholar]; (b) Zhou Y., Yu J., Lei X., Wu J., Niu Q., Zhang Y., Liu H., Christen P., Gehring H., Wu F. Chem. Commun. 2013;49:11782. doi: 10.1039/c3cc46719h. [DOI] [PubMed] [Google Scholar]; (c) McCune C., Chan S., Beio M., Shen W., Chung W., Szczesniak L., Chai C., Koh S., Wong P., Berkowitz D. ACS Cent. Sci. 2016;2:242. doi: 10.1021/acscentsci.6b00019. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Thorson M., Van Wagoner R., Harper M., Ireland C., Majtan T., Kraus J., Barrios A. Bioorg. Med. Chem. Lett. 2015;25:1064. doi: 10.1016/j.bmcl.2015.01.013. [DOI] [PubMed] [Google Scholar]; (e) Druzhyna N., Szczesny B., Olah G., Módis K., Asimakopoulou A., Pavlidou A., Szoleczky P., Gerö D., Yanagi K., Törö G., López-García I., Myrianthopoulos V., Mikros E., Zatarain J., Chao C., Papapetropoulos A., Hellmich M., Szabo C. Pharmacol. Res. 2016;113:18. doi: 10.1016/j.phrs.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimakopoulou A., Panopoulos P., Chasapis C., Coletta C., Zhou Z., Cirino G., Giannis A., Szabo C., Spyroulias G., Papapetropoulos A. Br. J. Pharmacol. 2013;169:922. doi: 10.1111/bph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W., Yadav P., Adamec J., Banerjee R. Antioxid. Redox Signaling. 2015;22:350. doi: 10.1089/ars.2014.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Collins R., Huang S., Holmberg-Schiavone L., Anand G., Tan C., van-den-Berg S., Deng L., Moore P., Karlberg T., Sivaraman J. J. Biol. Chem. 2009;284:3076. doi: 10.1074/jbc.M805459200. [DOI] [PubMed] [Google Scholar]
- Chiku T., Padovani D., Zhu W., Singh S., Vitvitsky V., Banerjee R. J. Biol. Chem. 2009;284:11601. doi: 10.1074/jbc.M808026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank N., Kent J., Meier M., Kraus J. Arch. Biochem. Biophys. 2008;470:64. doi: 10.1016/j.abb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Kleemann B., Loos B., Scriba T., Lang D., Davids L. PLoS One. 2014;9:e103762. doi: 10.1371/journal.pone.0103762. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lin S., Lei K., Du W., Yang L., Shi H., Gao Y., Yin P., Liang X., Liu J. Int. J. Biochem. Cell. Biol. 2016;71:24. doi: 10.1016/j.biocel.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Wang Z., Cheng M., Zhang X., Hong Z., Gao M., Kan X., Li Q., Wang Y., Zhu X., Xiao H. Fitoterapia. 2014;99:334. doi: 10.1016/j.fitote.2014.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.