 By probing the quinone substrate binding site of mitochondrial complex I with a focused set of quinazoline-based compounds, we identified EVP4593 as a novel, highly potent and specific inhibitor of mitochondrial complex I.
By probing the quinone substrate binding site of mitochondrial complex I with a focused set of quinazoline-based compounds, we identified EVP4593 as a novel, highly potent and specific inhibitor of mitochondrial complex I.
Abstract
By probing the quinone substrate binding site of mitochondrial complex I with a focused set of quinazoline-based compounds, we identified substitution patterns as being critical for the observed inhibition. The structure activity relationship study also resulted in the discovery of the quinazoline 4-N-[2-(4-phenoxyphenyl)ethyl]quinazoline-4,6-diamine (EVP4593) as a highly potent inhibitor of the multisubunit membrane protein. EVP4593 specifically and effectively reduces the mitochondrial complex I-dependent respiration with no effect on the respiratory chain complexes II–IV. Similar to established Q-site inhibitors, EVP4593 elicits the release of reactive oxygen species at the flavin site of mitochondrial complex I. Recently, EVP4593 was nominated as a lead compound for the treatment of Huntingtons disease. Our results challenge the postulated primary mode-of-action of EVP4593 as an inhibitor of NF-κB pathway activation and/or store-operated calcium influx.
1. Introduction
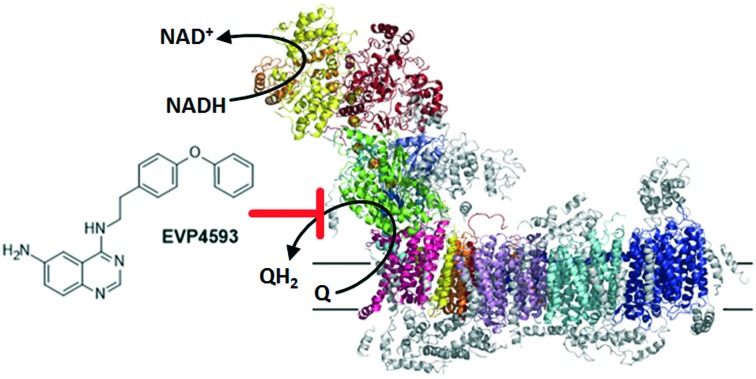
With a mass of almost 1 MDa and more than 40 subunits NADH–ubiquinone oxidoreductase (complex I) is the largest multisubunit membrane protein complex of the mitochondrial respiratory chain.1–3 Complex I contributes to the formation of the proton motive force required for ATP synthesis by coupling the two electron transfer reaction from NADH to ubiquinone with the translocation of four protons across the inner mitochondrial membrane. Dysfunction of complex I is linked to tumour progression4 and with several neuromuscular and neurodegenerative disorders including Parkinson's and Alzheimer's disease.5 Therefore, there is a continuous effort to understand the molecular organisation and function of this multisubunit membrane protein complex. To date, several structures of complex I have been reported.6–14 The common overall architecture of complex I comprises a membrane arm and a peripheral arm. The latter harbours all redox-active cofactors and the catalytic sites for NADH oxidation and ubiquinone reduction. Several lines of evidence indicate that proton translocation is triggered and driven by the redox chemistry of ubiquinone.3 However, the proton pump elements in the membrane arm are spatially separated and the mechanism of long range energy transfer is yet poorly understood.
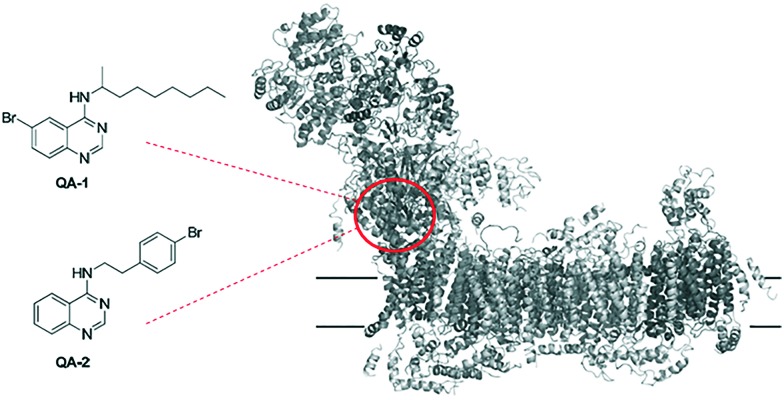
Inhibitors acting at the ubiquinone reduction site of complex I are in the focus of biomedical research because they were shown to induce acute Parkinson's disease15 but might also have a potential to be developed into new antitumor agents4,16 by targeting the hypoxia-inducible factor-1 (HIF-1) pathway.17,18 Moreover, inhibitors have proven to be useful tools to investigate the quinone binding sites of quinone converting enzymes.19–23 For example, quinazoline type inhibitors were reported as potent inhibitors of complex I with concentrations for half-maximal inhibition (IC50) values in the nanomolar range.24–27 Recently, we have designed and synthesized two brominated derivatives, 6-bromo-N-(nonan-2-yl)-4-quinazolinylamine (QA1) and N-[2-(4-bromophenyl)ethyl]-4-quinazolinylamine (QA2) (Fig. 1).8 These inhibitors were used to explore the ubiquinone binding site in the 3.6 to 3.9 Å X-ray structure of Yarrowia lipolytica, a yeast genetic model system to study the eukaryotic complex I. Inhibitor binding was modelled based on bromine signals in anomalous Fourier electron density maps (Fig. 1).8
Fig. 1. Constitution of reported quinazolines QA-1 and QA-2 used for structural elucidation of quinone substrate binding site in mitochondrial complex I (PDB code ; 4WZ7).8.
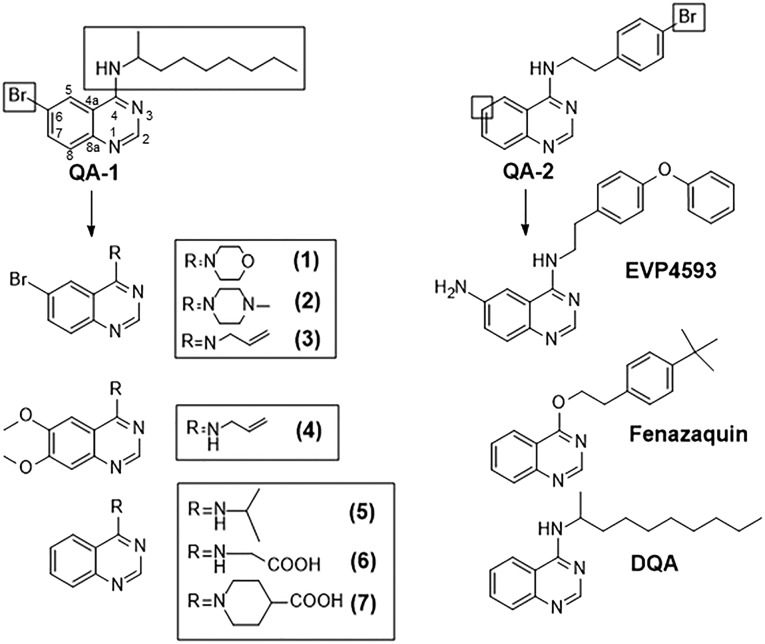
To gain further insight into the quinone-binding pocket, we followed an approach known as “Structure–Activity-relationship (SAR)-by-catalogue”. Several analogues of QA-1 and QA-2 were used to explore the quinone-binding pocket of mitochondrial complex I. In total, ten quinazoline-based compounds were selected for this study and tested for their inhibition of complex I (Fig. 2). The SAR study indicated that substitutions of the quinazoline ring and the structure of the side chain are critical for inhibitory potency. Compound EVP4593, which was originally discovered as an inhibitor of NF-κB activation28,29 and recently nominated as a lead structure for the treatment of Huntington's disease30 displayed in our assays significant complex I inhibition, comparable to the well-known complex I inhibitors decyl-quinazolineamine (DQA) and fenazaquin.
Fig. 2. Constitution of selected quinazolines used in this SAR study. Boxes highlight the site of modification of QA-1 and QA-2 quinazolines.
2. Results and discussion
A set of QA-1 and QA-2 analogues were selected for this study. The rationale behind the selection is highlighted in Fig. 2. First, compounds were selected with modifications at the N4-alkyl side chain of QA-1 (compounds 1–3). Next, compound 4 with modification in the adjacent aromatic ring (C6 and C7) was selected. Finally, compounds 5–7 were chosen, lacking the bromine substitution at C6 position and bearing different N-substitutions. Additionally, quinazoline-based compounds with reported bioactivity, EVP4593, fenazaquin and DQA were also included. EVP4593 was reported to be a high-affinity partial antagonist of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway28,29 by acting as an inhibitor of the store-operated calcium (SOC) entry pathway.31 It appears that EVP4593 targets heteromeric channels containing TRPC1 in neurons of patients with Huntington disease.30EVP4593 was also shown to exhibit neuroprotective effects from glutamate toxicity in nanomolar concentrations.32 Fenazaquin is a pesticide, reported to act through the inhibition of complex I.33DQA is a well-known complex I inhibitor,27 which was included as a positive control in the enzymatic functional assay.
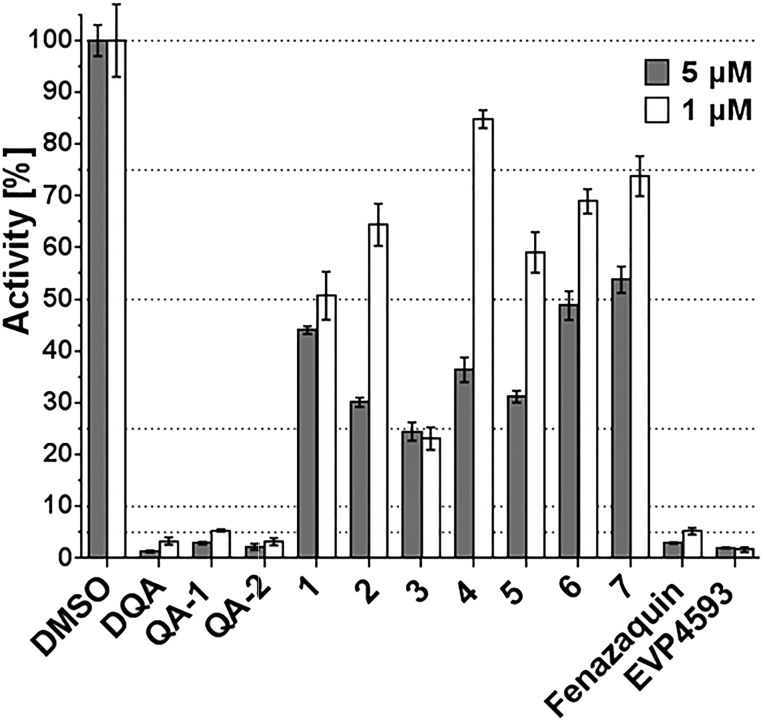
All compounds were tested in replicates against isolated mitochondrial membranes from the Y. lipolytica, which were prepared according to published protocols.25 Quinazolines were pre-incubated with the mitochondrial membranes and the dNADH : quinone catalytic activity was determined by monitoring the rate of absorbance at 340 nm due to dNADH oxidation (ε340–400nm = 6.22 mM–1 cm–1) after the addition of DBQ (decylubiquinone; an artificial quinone substrate) (see ESI,‡ Fig. S1).26 All compounds were tested at a concentration of 1 μM and 5 μM (Fig. 3). A concentration dependent behaviour of inhibition was observed for all tested derivatives, expect compound 3 (see ESI,‡ Fig. S2), confirming the observed inhibitory effect of the primary active molecules.
Fig. 3. Inhibition performance of quinazoline derivatives on the enzymatic reaction of complex I from Yarrawia lipolytica. Analogues were tested at 5 μM and 1 μM in replicates.
In agreement with our previous findings,8QA-1, QA-2 and the parent compound DQA displayed strong inhibition of complex I. In comparison, compound QA-1 was slightly less potent, indicating that the bromine substituent at the C-6 position weakens the inhibitory potential. The same holds true for replacing bromine by methoxy-groups at C-6/C-7 positions as shown for compound 4. This compound showed further decrease in inhibition compared to compound 3. Either shortening the alkyl-side chain of DQA or introducing polar substituents, as demonstrated by compounds 5, 6 and 7, resulted in a dramatic decrease in inhibition. It can also be concluded that larger substituents at N-4, such as in compounds 1, 2 and 7, were less tolerated compared to smaller substituents (e.g. in compound 3). In contrast, the bromo-substituent in QA-2 can be replaced by a tert-butyl group without changes in inhibition, as shown for fenazaquin. This holds also true for the NH isosteric replacement by an oxygen group.
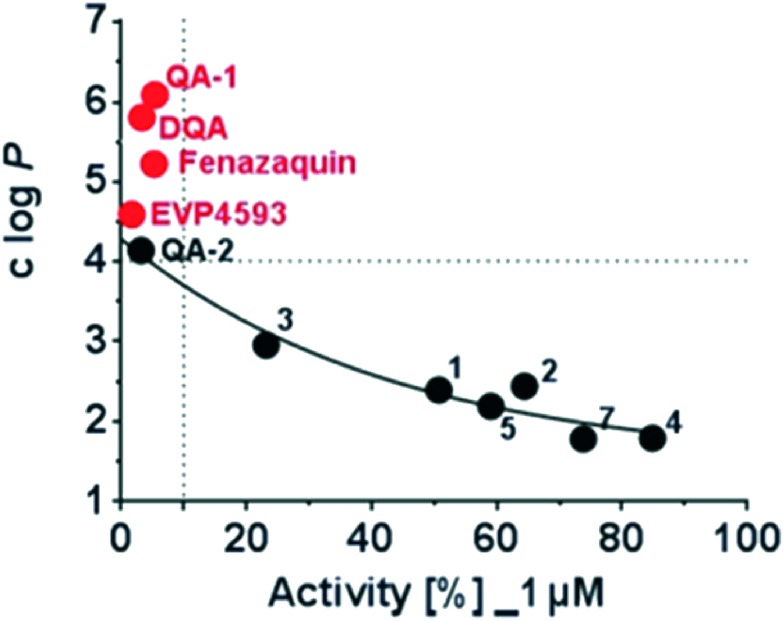
Interestingly, EVP4593 revealed strong inhibition of mitochondrial complex I. The amino-function was well tolerated and the hydrophobic phenoxy-substituent significantly enhanced the inhibition. The observed SAR data confirm the observation, that the putative quinone substrate access path in mitochondrial complex I is largely lipophilic.8,9,12 This finding was illustrated by plotting the calculated log P values, a measure of molecular lipophilicity of the tested compound against their respective inhibitory activities at 1 μM (Fig. 4). A clear correlation was found between clog P values and the observed inhibitory activity. The enzymatic activity decreases with increasing lipophilicity (clog P values) of tested molecules. This trend is observed up to a clog P value of 4 and all compounds above this value were highly potent. According to this plot, there is a threshold of a minimum lipophilicity of clog P = 4 for small molecules to enter the mitochondrial complex I via the hydrophobic substrate channel and to efficiently inhibit its enzymatic activity.
Fig. 4. Correlation of the calculated logarithm of partition coefficient (clog P) as a measure of compound lipophilicity and the enzymatic activity of mitochondrial complex I in the presence of 1 μM compound.
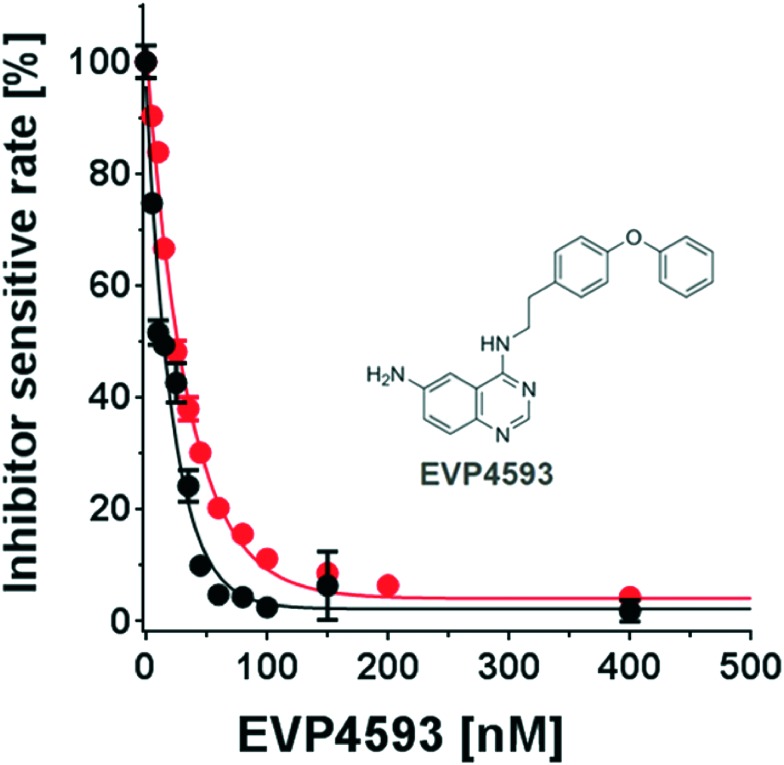
The hit molecule, EVP4593 was further analysed in a series of concentration-dependent experiments revealing an IC50 value of 25 ± 4 nM for the mitochondrial complex I from Y. lipolytica. The inhibitory activity of EVP4593 was also investigated against complex I in membrane preparations from Bos taurus heart mitochondria with similar outcome (IC50 = 14 ± 2 nM; see Fig. 5).
Fig. 5. Concentration-response analysis of EVP4593. The inhibitory efficiency of EVP4593 was determined by titrating the inhibitor in presence of mitochondrial complex I of Y. lipolytica (red circles; IC50 = 25 ± 4 nM) and of Bos taurus (black circles; IC50 = 14 ± 2 nM).
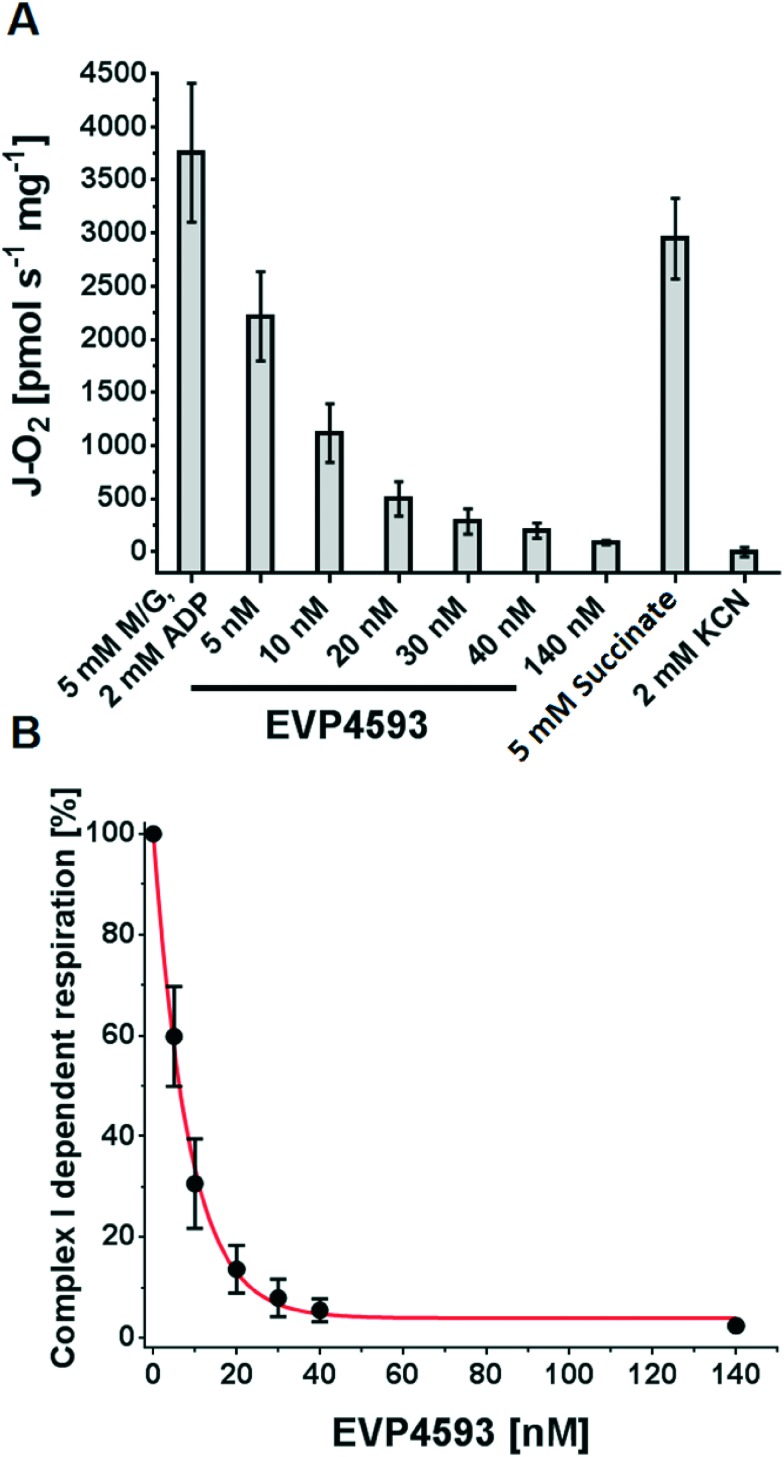
Next, orthogonal techniques were applied to confirm the inhibitory activity of EVP4593 on mitochondrial complex I and to gain more detailed information about the mechanism of inhibition. Therefore, high-resolution respirometry experiments were performed with intact rat heart mitochondria (RHM). RHM were prepared as described34 and the inhibitory effect of EVP4593 on the complex I-dependent respiration was measured at 37 °C using an Oxygraph-2k. After ADP-stimulated “state 3” oxygen consumption with malate and glutamate was stable, aliquots of EVP4593 were added stepwise and the respective respiration was determined. As a result EVP4593 was found to inhibit the complex I-dependent “state 3” respiration (Fig. 6A) with a Ki value of 6.3 nM (Fig. 6B).
Fig. 6. Inhibition of complex I-dependent respiration by EVP4593. (A) Intact rat heart mitochondria were fuelled by 5 mM malate/5 mM glutamate in presence of 2 mM ADP and subsequently inhibited by increasing concentrations of EVP4593. Finally, 5 mM succinate and 2 mM potassium cyanide (KCN) were added to demonstrate specificity for complex I. (B) Analysing the effect of EVP4593 on complex I-dependent respiration revealed a Ki of 6.3 nM.
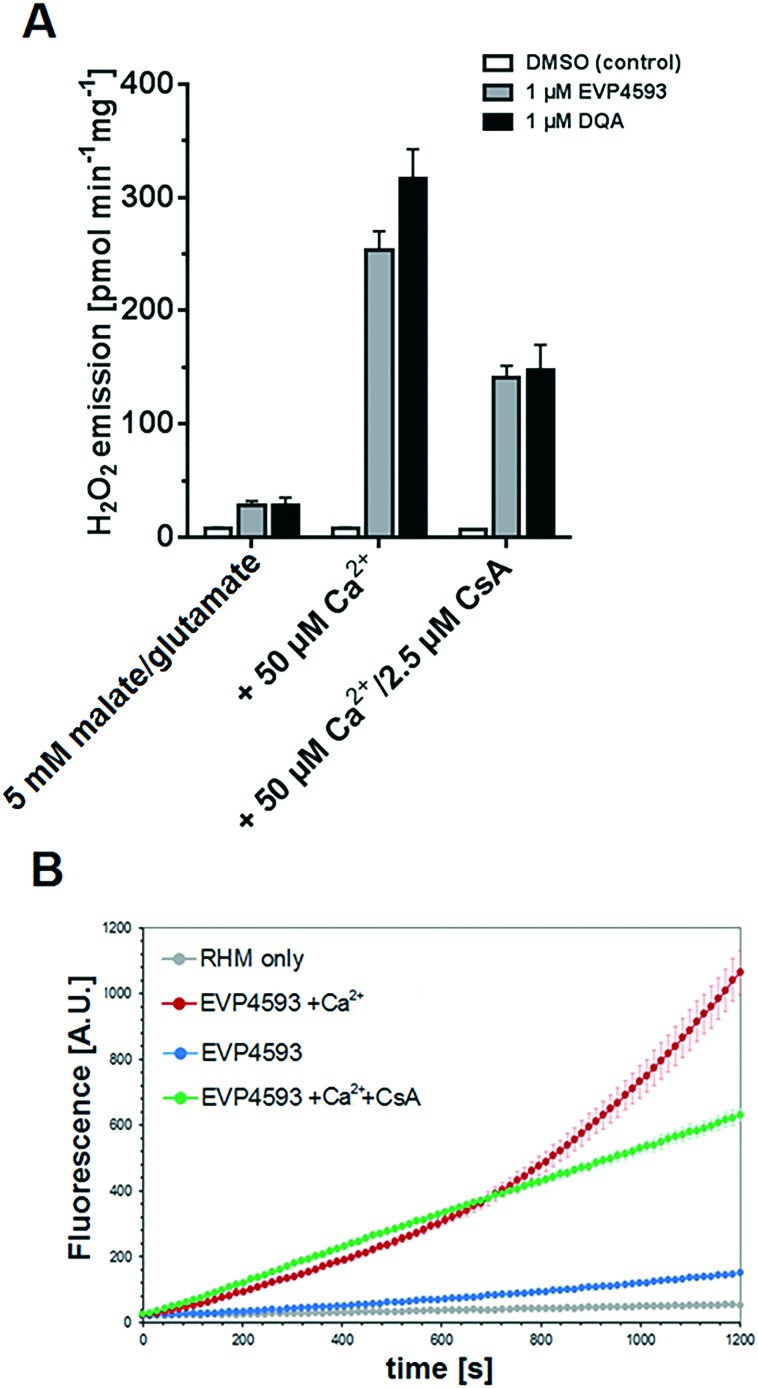
The specificity of EVP4593 for complex I was further supported by the observation that a subsequent addition of succinate restored respiration (Fig. 6A), thereby excluding respiratory chain complexes II–V and mitochondrial substrate transporters as targets of EVP4593. Q-site inhibitors of complex I can induce reactive oxygen species (ROS, i.e. superoxide, O2˙–, and hydrogen peroxide, H2O2) generation at flavin-containing sites of complex I and NAD+-dependent substrate dehydrogenases of the tricarboxylic acid (TCA) cycle.35 To investigate the ability of EVP4593 to induce mitochondrial hydrogen peroxide emission the Ampliflu-Red™/HRP assay was performed at 37 °C as described.36 In malate/glutamate fueled RHM, EVP4593 induced a comparable H2O2 emission as DQA (Fig. 7A). For both, the rate was increased in presence of 50 μM Ca2+ (Fig. 6A) which can be explained by Ca2+-mediated stimulation of the 2-oxoglutarate dehydrogenase that has been identified as a major mitochondrial source for ROS in presence of complex I Q-site inhibitors.37 In addition, the kinetics of EVP4593/Ca2+-induced mitochondrial H2O2 emission revealed an increase over time (Fig. 7B).
Fig. 7. EVP4593 induces H2O2 emission and mitochondrial permeability transition (mPT) in rat heart mitochondria. (A) Equimolar amounts (1 μM) of EVP4593 and DQA had similar effects on the mean H2O2 emission over a period of 20 min. (B) Kinetics course of H2O2 emission (mean values of triplicates and standard deviations of one representative preparation are shown). In presence of 50 μM Ca2+, the EVP4593 induced H2O2 emission was dynamically increased over time, implicating mPT as supported by the suppressing effect of cyclosporine A (CsA).
It is a well-known phenomenon that ROS – caused by complex I inhibition – induce mitochondrial permeability transition (mPT) in presence of Ca2+ leading to deprivation of essential antioxidant defense components (e.g. NADPH and glutathione (GSH)), thereby increasing the rate of ROS (H2O2) emission.38,39 This mechanism is supported by the delaying effect of the well-established mPT inhibitor cyclosporine A (CsA) (Fig. 7B).
In summary, a “SAR-by-catalogue” study was conducted by testing quinazoline-based analogues of previously identified inhibitors. The structure–activity-relationship provided a better understanding of the key parameters for quinone binding. The study led to the identification of EVP4593 as a novel and specific inhibitor of mitochondrial complex I. Our findings have implications for further development and application of EVP4593 as a tool compound to study NF-κB activation and the SOC pathway and challenge the previously reported view on its mechanism of action. Based on our presented data, we postulate that the primary target for EVP4593 is mitochondrial complex I and all the reported effects on NF-κB and SOC are downstream effects arising from inhibition of mitochondrial complex I.
Supplementary Material
Acknowledgments
This work was supported by the German Research Foundation (ZI 552/4-1 to VZ) and the Excellence Initiative of the German Federal and State Governments (EXC 115 to VZ). Excellent technical assistance by Andrea Duchene and Karin Siegmund is gratefully acknowledged. The authors thank Dr. Günter Fritz for helpful discussions and critical reading of this manuscript.
Footnotes
†The authors declare no competing interests.
‡Electronic supplementary information (ESI) available. See DOI: 10.1039/c6md00655h
References
- Brandt U. Annu. Rev. Biochem. 2006;75:69. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- Hirst J. Annu. Rev. Biochem. 2013;82:551. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- Wirth C., Brandt U., Hunte C., Zickermann V. Biochim. Biophys. Acta. 2016;1859:902. doi: 10.1016/j.bbabio.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Vatrinet R., Iommarini L., Kurelac I., De Luise M., Gasparre G., Porcelli A. M. Int. J. Biochem. Cell Biol. 2015;63:41. doi: 10.1016/j.biocel.2015.01.017. [DOI] [PubMed] [Google Scholar]
- Breuera M. E., Koopmana W. J., Koeneb S., Nootebooma M., Rodenburga R. J., Willemsa P. H., Smeitinka J. A. M. Neurobiol. Dis. 2013;51:27. doi: 10.1016/j.nbd.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Sazanov L. A., Hinchliffe P. Science. 2006;311:1430. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- Hunte C., Zickermann V., Brandt U. Science. 2010;329:448. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- Zickermann V., Wirth C., Nasiri H., Siegmund K., Schwalbe H., Hunte C., Brandt U. Science. 2015;347:44. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- Baradaran R., Berrisford J. M., Minhas G. S., Sazanov L. A. Nature. 2013;494:443. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar K. R., Zhu J., Hirst J. Nature. 2014;515:80. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov R. G., Baradaran R., Sazanov L. A. Nature. 2010;465:441. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- Zhu J., Vinothkumar K. R., Hirst J. Nature. 2016;536:354. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorczuk K., Letts J. A., Degliesposti G., Kaszuba K., Skehel M., Sazanov L. A. Nature. 2016;538:406. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts J. A., Fiedorczuk K. F., Sazanov L. A. Nature. 2016;537:644. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- Betarbet R., Sherer T. B., MacKenzie G., Garcia-Osuna M., Panov A. V., Greenamyre J. T. Nat. Neurosci. 2000;3:1301. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. L. J. Nat. Prod. 2008;71:1311. doi: 10.1021/np800191t. [DOI] [PubMed] [Google Scholar]
- Morgan J. B., Mahdi F., Liu Y., Coothankandaswamy V., Jekabsons M. B., Johnson T. A., Sashidhara K. V., Crews P., Nagle D. G., Zhou Y. D. Bioorg. Med. Chem. 2010;18:5988. doi: 10.1016/j.bmc.2010.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus P., Heisler I., Unterschemmann K., Haerter M., Beck H., Greschat S., Ehrmann A., Summer H., Flamme I., Oehme F., Thierauch K., Michels M., Hess-Stumpp H., Ziegelbauer K. Cancer Med. 2013;2:611. doi: 10.1002/cam4.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiri H. R., Madej M. G., Panisch R., Lafontaine M., Bats J. W., Lancaster C. R., Schwalbe H. J. Med. Chem. 2013;56:9530. doi: 10.1021/jm400978u. [DOI] [PubMed] [Google Scholar]
- Palsdottir H., Lojero C. G., Trumpower B. L., Hunte C. J. Biol. Chem. 2003;278:31303. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- Murai M., Murakami S., Ito T., Miyoshi H. Biochemistry. 2015;54:2739. doi: 10.1021/acs.biochem.5b00187. [DOI] [PubMed] [Google Scholar]
- Bridges H. R., Jones A. Y. J., Pollak M. N., Hirst J. Biochem. J. 2014;462:475. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendel U., Tocilescu M. A., Kerscher S., Brandt U. Biochim. Biophys. Acta. 2008;1777:660. doi: 10.1016/j.bbabio.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Murai M., Mashimo Y., Hirst J., Miyoshi H. Biochemistry. 2011;50:6901. doi: 10.1021/bi200883c. [DOI] [PubMed] [Google Scholar]
- Kerscher S., Okun J. G., Brandt U. J. Cell Sci. 1999;112:2347. doi: 10.1242/jcs.112.14.2347. [DOI] [PubMed] [Google Scholar]
- Tocilesc M. A., Fende U., Zwicke K., Kerscher S., Brandt U. J. Biol. Chem. 2007;282:29514. doi: 10.1074/jbc.M704519200. [DOI] [PubMed] [Google Scholar]
- Okun J. G., Lümmen P., Brandt U. J. Biol. Chem. 1999;274:2625. doi: 10.1074/jbc.274.5.2625. [DOI] [PubMed] [Google Scholar]
- Tobe M., Isobe Y., Tomizawa H., Nagasaki T., Takahashi H., Fukazawa T., Hayashi H. Bioorg. Med. Chem. 2003;11:383. doi: 10.1016/s0968-0896(02)00440-6. [DOI] [PubMed] [Google Scholar]
- Tobe M., Isobe Y., Tomizawa H., Nagasaki T., Takahashi H., Fukazawa T., Hayashi H. Bioorg. Med. Chem. 2003;11:3869. doi: 10.1016/s0968-0896(03)00438-3. [DOI] [PubMed] [Google Scholar]
- Wu J., Ryskamp D. A., Liang X., Egorova P., Zakharova O., Hung G., Bezprozvanny I. J. Neurosci. 2016;36:125. doi: 10.1523/JNEUROSCI.1038-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Kim J. H., Roh E. J., Ko M. J., Jung J. E., Kim H. J. J. Biol. Chem. 2006;281:12722. doi: 10.1074/jbc.M601018200. [DOI] [PubMed] [Google Scholar]
- Wu J., Shih H. P., Vigont V., Hrdlicka L., Diggins L., Singh C., Mahoney M., Chesworth R., Shapiro G., Zimina O., Chen X., Wu Q., Glushankova L., Ahlijanian M., Koenig G., Mozhayeva G. N., Kaznacheyeva E., Bezprozvanny I. Chem. Biol. 2011;18:777. doi: 10.1016/j.chembiol.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lümmen P. Biochim. Biophys. Acta. 1998;1364:287–296. doi: 10.1016/s0005-2728(98)00034-6. [DOI] [PubMed] [Google Scholar]
- Dröse S., Brandt U., Hanley P. J. J. Biol. Chem. 2006;281:23733. doi: 10.1074/jbc.M602570200. [DOI] [PubMed] [Google Scholar]
- Dröse S., Stepanova A., Galkin A. Biochim. Biophys. Acta. 2016;1857:946. doi: 10.1016/j.bbabio.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke E., Zwicker K., Dröse S. Arch. Biochem. Biophys. 2015;580:75. doi: 10.1016/j.abb.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Quinlan C. L., Goncalves R. L., Hey-Mogensen M., Yadava N., Bunik V. I., Brand M. D. J. Biol. Chem. 2014;289:8312. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votyakova T. V., Reynolds I. J. J. Neurochem. 2005;93:526–537. doi: 10.1111/j.1471-4159.2005.03042.x. [DOI] [PubMed] [Google Scholar]
- Bonke E., Siebels I., Zwicker K., Dröse S. Free Radical Biol. Med. 2016;99:43. doi: 10.1016/j.freeradbiomed.2016.07.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.