Abstract
Background
Antibody-mediated rejection is a major cause of premature graft loss in kidney transplantation. Multiple scoring systems are available to assess the HLA mismatch between donors and recipients at the molecular level; however, their correlation with the development of de novo donor-specific antibody (dnDSA) has not been compared in recipients on active immunosuppression.
Methods
HLA-DRβ1/3/4/5/DQα1β1 molecular mismatch was determined using eplet analysis, amino acid mismatch, and electrostatic mismatch for 596 renal transplant recipients and correlated with HLA-DR/DQ dnDSA development. The molecular mismatch scores were evaluated in multivariate models of posttransplant dnDSA-free survival.
Results
Eplet mismatch correlated with amino acid mismatch and electrostatic mismatch (R2 = 0.85-0.96). HLA-DR dnDSA-free survival correlated with HLA-DR eplet mismatch (hazards ratio [HR], 2.50 per 10 eplets mismatched; P < 0.0001), amino acid mismatch (HR, 1.49 per 10 amino acids mismatched; P < 0.0001), and electrostatic mismatch (HR, 1.23 per 10 units mismatched; P < 0.0001). HLA-DQ dnDSA-free survival correlated with HLA-DQ eplet mismatch (HR, 1.98 per 10 eplets mismatched; P < 0.0001), amino acid mismatch (HR, 1.24 per 10 amino acids mismatched; P < 0.0001), and electrostatic mismatch (HR, 1.14 per 10 units mismatched; P < 0.0001). All 3 methods were significant multivariate correlates of dnDSA development after adjustment for recipient age, baseline immunosuppression, and nonadherence.
Conclusions
HLA molecular mismatch represents a precise method of alloimmune risk assessment for renal transplant patients. The method used to determine the molecular mismatch is likely to be driven by familiarity and ease of use as highly correlated results are produced by each method.
In order to assess alloimmune risk in kidney transplant recipiennts, the authors compare 3 methods evaluating HLA molecular mismatch (HLA-DG eplet, amino acid and electrostatic) which all correlate with adjusted dnDSA development. Supplemental digital content is available in the text.
Antibody-mediated rejection is a major cause of allograft dysfunction and allograft loss in kidney transplantation.1-3 Improvements in HLA histocompatibility assessment and HLA antibody screening methods have made it possible to avoid transplanting across known donor-specific antibodies (DSA), however, twenty to thirty percent of recipients develop de novo DSA (dnDSA) after 5 to 10 years of follow-up.4 Therapy for late antibody-mediated rejection is limited; therefore, strategies to minimize dnDSA development through more precise HLA mismatch evaluation and through appropriate immunosuppression management are paramount.5,6
Advances in genetics and protein modelling have made it possible to compare donor-recipient HLA mismatch at a molecular level. Traditional HLA mismatch quantification is constrained by a limited range of possible values (0, 1, or 2 per locus) at the whole antigen level. However, assessment of HLA mismatch at the molecular level enables quantification of the degree of mismatch and in turn immunogenicity between donor-recipient HLA improving the precision of immunological risk assessment with dnDSA development as the immune response readout. One such approach, based on enumerating all mismatched amino acid sequence polymorphisms on donor HLA and scoring them according to their physicochemical properties, has been shown to be independently associated with dnDSA development after graft failure.7 An extension of this work, to assess the impact of donor sequence polymorphisms on the HLA tertiary structure, suggested that surface exposed antibody epitopes have unique electrostatic potential profiles that help explain HLA cross-reactive antigen groups.8 A different approach, namely, HLA matchmaker, identifies small patches of surface exposed mismatched amino acids named “eplets” on each HLA molecule, which are hypothesized to drive DSA specificity.9 The quantity of mismatched eplets between donor and recipient alleles has been shown to correlate with dnDSA development, rejection, chronic glomerulopathy, and graft loss.6,10–12 Whether one of these methods is a superior correlate for dnDSA development in the setting of active immunosuppression has not been determined.
The purpose of this analysis was to compare the eplet mismatch (EpMM), amino acid mismatch (AAMM), and electrostatic mismatch (ESM) computational methodologies in a large cohort of well characterized renal transplant recipients for their correlation with the development of dnDSA posttransplant. Unique to this consecutive patient cohort is the strict exclusion of preexisting HLA DSA, the availability of high-resolution donor and recipient HLA typing, immunosuppression adherence, serial sera obtained posttransplantation to characterize the timing of dnDSA onset, and long-term graft outcomes.
MATERIALS AND METHODS
Study Population
Approval was obtained from the institutional review board (H2011: 211) and was in adherence with the declaration of Helsinki. 654 adult and pediatric consecutive renal transplants between January 1999 and January 2015 were considered for inclusion. Patients with primary nonfunction (n = 16), or pretransplant DSA (n = 42) were excluded, leaving 596 recipients (adult n = 541, pediatric n = 55) for analysis. Recipients who moved (n = 21) or died with a functioning graft (n = 82) were censored at last follow-up. Standard maintenance immunosuppression consisted of a calcineurin inhibitor (tacrolimus (86%) or cyclosporine (14%)), mycophenolate mofetil, and prednisone. Induction therapy with thymoglobulin (16%) or basiliximab (19%) was used in 35% of patients.
HLA Typing and Molecular Mismatch Identification
High-resolution (4-digit) class II HLA typing (HLA-DRβ1/3/4/5 and HLA-DQα1/β1) was performed using sequence-specific oligonucleotide probes or sequence-specific primer technology (LABType HD SSO, Micro SSP; One Lambda, Canoga Park, CA). HLAMatchmaker software (HLA DRDQDP matching version 2.0) was used to define Class II EpMM between donors and recipients. The AAMM score and ESM score for mismatched donor-recipient HLA combinations were determined using the Cambridge HLA Immunogenicity algorithm, as described previously.7 For a given patient, when more than a single HLA mismatch was present within a locus, individual scores for each HLA mismatch were added to represent an overall immunogenicity score.
Antibody Monitoring
Posttransplant serum samples were collected and stored at 0, 1, 2, 3, 6, 12, 18, and 24 months, then yearly, or at the time of biopsy for graft dysfunction, as routine clinical practice in our program since 1990. Since 2007 posttransplant surveillance for dnDSA was instituted for all renal transplant patients. DSA screening was performed using FlowPRA beads representing HLA-A, -B, -Cw, -DR, -DQ, and -DP antigens (One Lambda). If the screening assay was positive, determination of HLA antibody specificities was performed using FlowPRA single antigen class I and II beads (One Lambda) and analyzed according to the manufacture’s recommendations. HLA antibody specificities were validated using LABScreen single antigen beads (One Lambda) using a threshold mean fluorescence intensity ≥ 500 (mean fluorescence intensity ≥1000 initially or on a subsequent sample in 98% of cases).
Pretransplant all patients had remote and immediate pretransplant sera screened by FlowPRA and if positive evaluated by FlowPRA single antigen beads. Even if the FlowPRA screen was negative, patient sera were still evaluated by FlowPRA single antigen beads if there was elevated risk of sensitization (eg, pregnancy, history of transfusion). To rule out a DSA pretransplant, the mismatched donor antigens had to be represented on the single antigen beads. If donor-specific antibodies were absent pretransplant, as determined by solid phase assays and a negative flow crossmatch, and became detectable posttransplant they were classified as dnDSA. Patients with dnDSA had banked posttransplant serum tested to determine the approximate timing of dnDSA onset by FlowPRA single antigen beads. All patients continue to be prospectively tested for dnDSA according to the serum collection schedule outlined above to detect new dnDSA or to assess the persistence of existing dnDSA.
Statistics
Comparisons between baseline variables and clinical outcomes were done using Student’s t-test for parametric continuous variables and Wilcoxon-rank test for nonparametric data. Chi-squared or Fisher’s exact tests were used to test categorical variables. Comparisons across multiple groups were done using Kruskal-Wallis test for nonparametric data and analysis of variance for parametric variables. Survival analysis was done by the Kaplan-Meier method using the log-rank test for significance. Cox proportional hazards model was used to evaluate correlates of dnDSA-free survival. Akaike information criterion (AIC) was calculated with Cox models to allow model comparisons within specific cohorts. The ability of the models to correctly classify subjects for their actual outcomes (dnDSA development) was examined using time-dependent receiver operator characteristic curves and area under the curve (AUC) statistics. Variables for multivariate regression were selected on the basis of bivariate screening, with P values of 0.2 or less used to identify candidates for inclusion in the final model. The proportional hazard assumption was not violated (assessed by both Schoenfeld residuals and Harrell’s rho). Co-linearity was assessed and all variance inflation factors were less than 3. Statistical software used was R version 3.0.1 and JMP (version 12.2).
RESULTS
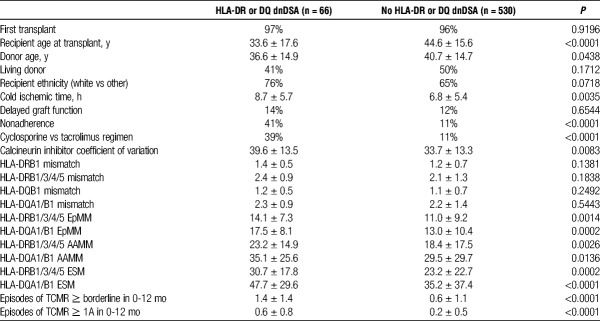
This consecutive cohort (n = 596) represented a low immunological risk group (96% first transplant, <10% with calculated panel-reactive antibody >80%) by conventional criteria. Median follow-up was 87 months (range, 18-210). HLA-DR or DQ dnDSA developed in 66 (11%) recipients at a median of 55 months (range, 6-170) posttransplant. At the time of dnDSA development 15 (23%) of 66 had HLA-DR dnDSA alone, 37 (56%) of 66 had HLA-DQ dnDSA alone, and 14 (21%) of 66 had both HLA-DR and DQ dnDSA. Significant correlates with Class II dnDSA were younger recipient and donor ages, Class II HLA-DR and DQ EpMM, class II HLA-DR and DQ AAMM, class II HLA-DR, and DQ ESM, greater cold ischemic time, calcineurin inhibitor regimen (cyclosporine vs tacrolimus), immunosuppression nonadherence, calcineurin inhibitor coefficient of variation, and T-cell mediated rejection in the first year (Table 1).
TABLE 1.
Recipient characteristics
Correlates of dnDSA-Free Survival
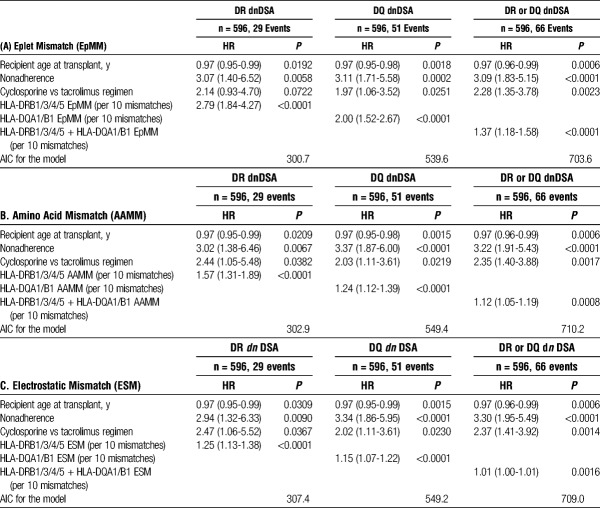
The median number of HLA-DRβ1/3/4/5 EpMM, AAMM, and EMS were 10 (range, 0-41), 15 (range, 0-82), and 22 (range, 0-147). HLA-DRβ1/3/4/5 EpMM (hazard ratio [HR], 2.50 per 10 mismatches; 95% confidence interval [CI], 1.71-3.64; P < 0.0001), AAMM (HR, 1.49; 95% CI, 1.25-1.76; P < 0.0001), and EMS (HR, 1.23; 95% CI, 1.11-1.35; P < 0.0001) were each significant univariate correlates of HLA-DR dnDSA-free survival posttransplant (Table S1, SDC, http://links.lww.com/TP/B531).
The median number of HLA-DQα1β1 EpMM, AAMM, and EMS was 13 (range, 0-42), 18 (range, 0-97), 24 (range, 0-164). HLA-DQ α1β1 EpMM (HR, 1.98 per 10 mismatches; 95% CI, 1.53-2.58; P < 0.0001), AAMM (HR, 1.24; 95% CI, 1.12-1.37, P < 0.0001), and EMS (HR, 1.14; 95% CI, 1.07-1.21; P < 0.0001) were each significant correlates of HLA-DQ dnDSA-free survival posttransplant (Table S1, SDC, http://links.lww.com/TP/B531). Receiver operating characteristic curve analysis showed that all molecular mismatch methods had similar AUC scores (0.71 to 0.74) (Figure S1, SDC, http://links.lww.com/TP/B531).
There were strong correlations between intra-locus molecular mismatch scores (Figure 1). HLA-DRβ1/3/4/5 EpMM correlated with HLA-DRβ1 AAMM (R2 = 0.96), and EMS (R2 = 0.85). HLA-DQα1β1 EpMM correlated with HLA- DQα1β1 AAMM (R2 = 0.95), and EMS (R2 = 0.90).
FIGURE 1.
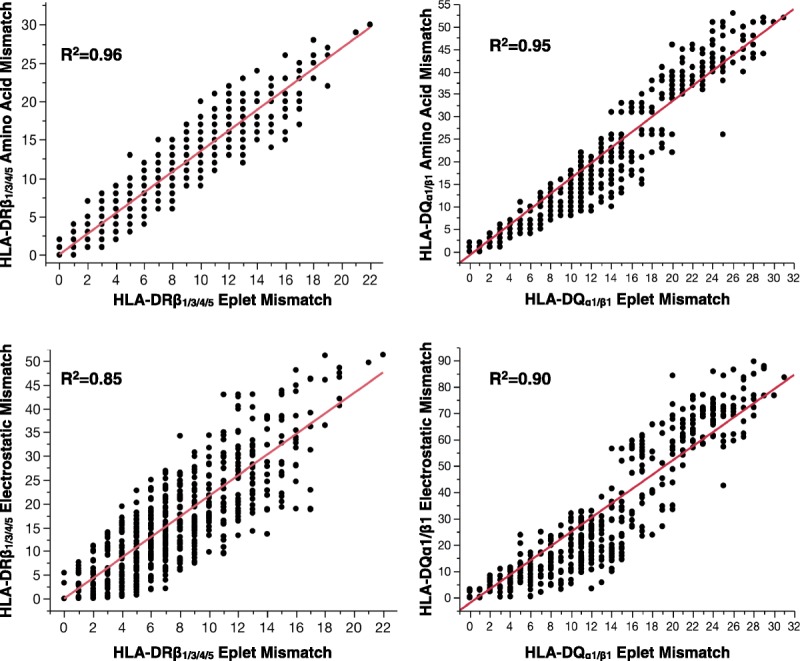
There was a strong correlation between EpMM and AAMM (top row) and ESM (bottom row) scores at the HLA-DRβ1/3/4/5, HLA-DQ α1β1 loci.
Multivariate Models
In multivariate analyses, each of the molecular mismatch scores were independent correlates of dnDSA development after adjustment for younger recipient age, cyclosporine versus tacrolimus, and nonadherence (Table 2). AIC (a measure of the relative quality of multivariate statistical models) were similar among the molecular mismatch scores examined.
TABLE 2.
Multivariate correlates of dnDSA development
DISCUSSION
Current assessment of donor-recipient histocompatibility and of the risk of humoral alloresponses after kidney transplantation is based on simple enumeration of HLA antigenic differences at individual class I and II loci without consideration of the relative immunogenicity of donor HLA mismatches according to the recipient HLA type. In the present study, we examined 3 different approaches for assessment of HLA class II immunogenicity ranging from simply enumerating the number of AAMMs between donor and recipient HLA (AAMM), to counting the number of polymorphic surface accessible amino acid residues at discontinuous positions of donor HLA that cluster together to form a potential epitope (EpMM), to assessing the physicochemical disparity between the side chains of mismatched amino acids of donor and recipient HLA (EMS). Our study is the first to compare the capacity of these approaches to assess the risk of dnDSA development in a cohort of renal transplant patients on active immunosuppressive therapy and where the timing of dnDSA development posttransplant was monitored prospectively. The principal finding was that assessment of donor HLA immunogenicity based on AAMM, EpMM or EMS is superior to that of conventional HLA mismatch grade for assessing the risk of dnDSA development after kidney transplantation. We did not demonstrate an advantage in using one approach over another and, in this patient cohort, each method provided equivalent assessment of immunological risk associated with donor HLA class II mismatches.
Development of dnDSA after kidney transplantation is associated with rejection, accelerated estimated glomerular filtration rate decline, and graft loss.1,13 Currently, no therapies have been proven effective to eliminate dnDSA after its development nor prevent progression of allograft dysfunction, therefore, prevention of dnDSA is of paramount importance.5 Nonadherence with immunosuppression, younger recipient age, cyclosporine-based immunosuppression regimens, early T cell–mediated rejection and HLA mismatch have been established as independent correlates of dnDSA development.6,10,14–16 Molecular assessment of HLA immunogenicity has gained the interest of the transplant community due to its ability to outperform traditional whole molecule mismatch as a correlate of dnDSA development, transplant glomerulopathy, and graft survival.6,10,11 Humoral responses after kidney transplantation are frequently directed against donor HLA class II alloantigens, and our study suggests that assessment of HLA-DR and -DQ immunogenicity based on mismatched eplets produces similar results compared to simply enumerating the number of amino acid polymorphisms between donor and recipient HLA molecules. This is not surprising given that AMS and EpMS both reflect differences in donor-recipient amino acid sequence and a strong correlation between the 2 scoring systems has been demonstrated in this and other studies.7 EMS integrates information on the number of mismatched amino acids and the differences in electrostatic charge of their side chains, and it is, therefore, correlated to the AAMM score. Previous studies suggested that consideration of the electrostatic charge of amino acid polymorphisms on donor HLA-A and -B alloantigens might provide useful information regarding their immunogenic potential,7,17 but this and other studies do not support an advantage in using EMS, over EpMM and AAMM, for assessing the risk of DSA responses against HLA class II mismatches.7,18 Larger studies and assessment of HLA electrostatic properties at the tertiary level are warranted to further explore the relationship between donor-recipient HLA physicochemical differences and humoral alloresponses after transplantation.
HLAMatchmaker defines eplets by considering each polymorphic amino acid at or near the surface of the molecule and then asks the question what other polymorphic amino acids are nearby (3 Å radius).9 This small patch of polymorphic amino acids is known as an eplet, and its specific name is derived from the amino acids involved (ie, 52PQ). By comparison, the AAMM software developed by Kosmoliaptsis et al17 aligns the amino acid sequence of donor and recipient HLA alleles and counts the number of mismatched amino acids irrespective of their position in three-dimensional space (as no advantage was previously demonstrated by exclusion of surface inaccessible polymorphisms). The physicochemical approach compares the isoelectric points of mismatched amino acids between donor and recipient alleles, and the differences are summed to represent an overall ESM score. The strong correlations between EpMM, AAMM, and ESM (Figure 1) are expected given that each scoring system examines a similar, but not identical, list of polymorphic amino acids (nonsurface exposed residues are excluded in HLAMatchmaker).
Using the AIC to compare EpMM, AAMM, and EMS in multivariate models of dnDSA development (Table 2) revealed a small advantage to the HLAMatchmaker model. However, all 3 molecular mismatch methods had similar discrimination measures (AUC) and have been shown to outperform traditional HLA antigen matching in this and in previous reports, and a clinically meaningful difference of using one method over another to correlate with dnDSA development in clinical practice is doubtful. Due to the relatively small sample size and the associated risk of type II error, risk quantification should be interpreted with caution, and should be validated in a larger independent cohort. We acknowledge that this analysis focused on class II dnDSA because, in our cohort, dnDSA against donor HLA Class I mismatches alone was infrequent (2% of cohort) and only 1 patient in the entire cohort suffered allograft failure after developing isolated class I dnDSA. HLA-DPα1β1 dnDSA development was tracked in this cohort (data not shown), however, was too infrequent for meaningful analysis. Development of mature humoral alloimmunity is dependent upon T cell help through linked recognition of HLA derived peptides presented in the context of recipient HLA class II molecules. Recent reports suggest that the presentation of allopeptides by HLA-DR correlate with dnDSA development.19 Although early in development, this may be a promising area for future research. Forthcoming studies should also explore the immunogenicity of individual donor HLA, as determined by molecular mismatch methods, and the risk of de novo HLA-specific antibody development as a time-dependent variable accounting for the effect of relevant confounders.
In conclusion, HLA molecular mismatch methods enable precise assessment of alloimmune risk associated with renal transplantation. Donor HLA AAMM, ESM and EpMM were each significant multivariate correlates of dnDSA development. Relevant studies in larger independent cohorts are warranted but, at present, the use of one method over the other is likely to be driven by familiarity and ease of use as highly correlated results are produced by each method.
Supplementary Material
Footnotes
C.W. received funding by a Research Manitoba operating grant. P.N. is funded by the Canadian Institutes for Health Research and salary support from the Flynn Family Chair in Renal Transplantation. V.K. was supported by an Evelyn Trust Grant and an NIHR PostDoctoral Fellowship (PDF-2016-09-065).
The research was funded in part in part by the National Institute for Health Research Blood and Transplant Research Unit (NIHR BTRU) in Organ Donation and Transplantation at the University of Cambridge in collaboration with Newcastle University and in partnership with NHS Blood and Transplant (NHSBT). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, the Department of Health or NHSBT.
The authors declare no conflicts of interest.
C.W. and V.K. are co-first authors.
C.W. and V.K. authored the article, conceived of the research idea and conducted the analysis. P.W. and C.T. collectively conceived of the research program, provided input into design, and the analysis plan. All coauthors provided review and revisions to the manuscript and ultimately approved the final version for submission and publication.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Wiebe C, Gibson IW, Blydt-Hansen TD, et al. Rates and determinants of progression to graft failure in kidney allograft recipients with de novo donor-specific antibody. Am J Transplant. 2015;15:2921–2930. [DOI] [PubMed] [Google Scholar]
- 2.El-Zoghby ZM, Stegall MD, Lager DJ. Identifying specific causes of kidney allograft loss. Am J Transplant. 2009;9:527–535. [DOI] [PubMed] [Google Scholar]
- 3.Terasaki PI. Humoral theory of transplantation. Am J Transplant. 2003;3:665–673. [DOI] [PubMed] [Google Scholar]
- 4.Wiebe C, Nickerson P. Posttransplant monitoring of de novo human leukocyte antigen donor-specific antibodies in kidney transplantation. Curr Opin Organ Transplant. 2013;18:470–477. [DOI] [PubMed] [Google Scholar]
- 5.Archdeacon P, Chan M, Neuland C, et al. Summary of FDA antibody-mediated rejection workshop. Am J Transplant. 2011;11:896–906. [DOI] [PubMed] [Google Scholar]
- 6.Wiebe C, Rush DN, Nevins TE, et al. Class II eplet mismatch modulates tacrolimus trough levels required to prevent donor-specific antibody development. J Am Soc Nephrol. 2017;28:3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosmoliaptsis V, Mallon DH, Chen Y, et al. Alloantibody responses after renal transplant failure can be better predicted by donor-recipient HLA amino acid sequence and physicochemical disparities than conventional HLA matching. Am J Transplant. 2016;16:2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mallon DH, Bradley JA, Winn PJ, et al. Three-dimensional structural modelling and calculation of electrostatic potentials of HLA bw4 and bw6 epitopes to explain the molecular basis for alloantibody binding: toward predicting HLA antigenicity and immunogenicity. Transplantation. 2015;99:385–390. [DOI] [PubMed] [Google Scholar]
- 9.Duquesnoy RJ, Askar M. HLAMatchmaker: a molecularly based algorithm for histocompatibility determination. V. Eplet matching for HLA-DR, HLA-DQ, and HLA-DP. Hum Immunol. 2007;68:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiebe C, Pochinco D, Blydt-Hansen TD, et al. Class II HLA epitope matching—a strategy to minimize de novo donor-specific antibody development and improve outcomes. Am J Transplant. 2013;13:3114–3122. [DOI] [PubMed] [Google Scholar]
- 11.Sapir-Pichhadze R, Tinckam K, Quach K, et al. HLA-DR and -DQ eplet mismatches and transplant glomerulopathy: a nested case-control study. Am J Transplant. 2015;15:137–148. [DOI] [PubMed] [Google Scholar]
- 12.Wiebe C, Nickerson P. Strategic use of epitope matching to improve outcomes. Transplantation. 2016;100:2048–2052. [DOI] [PubMed] [Google Scholar]
- 13.Everly MJ, Rebellato LM, Haisch CE, et al. Incidence and impact of de novo donor-specific alloantibody in primary renal allografts. Transplantation. 2013;95:410–417. [DOI] [PubMed] [Google Scholar]
- 14.Wiebe C, Gareau AJ, Pochinco D, et al. Evaluation of C1q status and titer of de novo donor-specific antibodies as predictors of allograft survival. Am J Transplant. 2017;17:703–711. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto T, Watarai Y, Takeda A, et al. De novo anti-hla DSA characteristics and subclinical antibody-mediated kidney allograft injury. Transplantation. 2016;100:2194–2202. [DOI] [PubMed] [Google Scholar]
- 16.Wiebe C, Gibson IW, Blydt-Hansen TD, et al. Evolution and clinical pathologic correlations of de novo donor-specific HLA antibody post kidney transplant. Am J Transplant. 2012;12:1157–1167. [DOI] [PubMed] [Google Scholar]
- 17.Kosmoliaptsis V, Chaudhry AN, Sharples LD, et al. Predicting HLA class I alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation. 2009;88:791–798. [DOI] [PubMed] [Google Scholar]
- 18.Kosmoliaptsis V, Sharples LD, Chaudhry AN, et al. Predicting HLA class II alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation. 2011;91:183–190. [DOI] [PubMed] [Google Scholar]
- 19.Lachmann N, Niemann M, Reinke P, et al. Donor-recipient matching based on predicted indirectly recognizable HLA epitopes independently predicts the incidence of de novo donor-specific HLA antibodies following renal transplantation. Am J Transplant. 2017;17:3076–3086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.