Abstract
TRIM58 is a member of the tripartite motif protein (TRIM) family of E3 ubiquitin ligases. Aberrant gene methylation of TRIM58 has been reported in liver and lung cancer and indicates a poor patient prognosis. However, the expression level and functional role of TRIM58 in colorectal cancer (CRC) have yet to be elucidated. In the present study, we found that TRIM58 expression was significantly suppressed in human CRC and was inversely correlated with CRC progression. Additionally, overall survival was significantly reduced in patients with low TRIM58 expression in CRC tumors. In vitro studies demonstrated that ectopic TRIM58 overexpression strongly inhibited CRC cell invasion but had minimal effects on cell proliferation, colonization and migration. Furthermore, TRIM58 suppression enhanced the expression of epithelial-to-mesenchymal transition (EMT) and matrix metalloproteinase (MMP) genes. Thus, our findings suggest that TRIM58 is a potential prognostic marker of CRC and functions as a tumor-suppressor gene via inhibition of cancer cell invasion through EMT and MMP activation.
Keywords: TRIM58, suppressor gene, colorectal cancer, invasion, EMT
Introduction
Colorectal cancer (CRC) is one of the leading contributors to cancer-related mortality and morbidity in China (1). The morphological features and molecular subtypes of colorectal tumors have been studied extensively and include microsatellite instability, the CpG island methylation phenotype and chromosomal instability. Furthermore, typical molecular patterns have been identified to distinguish different CRC subtypes derived from distinctive locations. For example, BRAFV600E occurs more frequently in serrated CRC, while tubular CRC has an increased rate of the KRAS mutation (2,3). Moreover, several molecular markers are also used in targeted therapy. For example, the epidermal growth factor receptor (EGFR) is a target of panitumumab, a fully human monoclonal antibody specific to EGFR used to treat EGFR-expressing metastatic CRC with disease progression (4). Similarly, bevacizumab, which targets VEGF, is used in metastatic CRC with standard chemotherapy treatment (5). However, a more detailed network of the molecular pathways deregulated in CRC is needed. Studying these molecular alterations can help further elucidate the mechanisms underlying CRC and improve clinical care.
The ubiquitin-proteasome system controls protein homeostasis of almost the entire proteome. Ubiquitin-mediated proteolysis employs an enzymatic cascade that primarily involves E1 activating enzymes, E2 conjugating enzymes and E3 ubiquitin ligases. Deregulation of ubiquitin ligase has been shown to play crucial roles in many pathological activities. Tripartite motif proteins (TRIMs) represent one of the largest families of E3 ubiquitin ligases. TRIM58, a TRIM protein containing a RING motif, a B box type 1 and 2, and a coiled-coil domain in its N-terminal region, was identified as an E3 ubiquitin ligase and was first shown to regulate terminal erythropoiesis (6). TRIM58 was reported to be involved in a variety of cancers, including hepatocellular carcinoma and lung cancer. Hypermethylation of TRIM58 and downregulation of its mRNA expression were associated with tumorigenesis of hepatocellular carcinoma and worse prognosis of patients after hepatectomy (7). Similarly, aberrant inactivation of TRIM58 caused by CpG island hypermethylation promoted early carcinogenesis of early-stage lung adenocarcinoma regardless of smoking status (8). However, the association between TRIM58 expression and CRC prognosis and the role of TRIM58 in CRC have not yet been determined.
Herein, we studied the potential role of TRIM58 in colorectal carcinogenesis. Instead of measuring gene promoter methylation, we directly evaluated the expression of TRIM58 and its clinical significance in a cohort of patients with CRC. Furthermore, we investigated the effects of TRIM58 on CRC progression and the underlying molecular mechanisms. Our data suggest that both mRNA and protein levels of TRIM58 are commonly suppressed in CRC tissues and cell lines. In vitro studies showed that ectopic TRIM58 expression specifically influenced cell invasion, while it had minimal effects on cell proliferation and migration. In addition, TRIM58 expression modulated the activation of epithelial-to-mesenchymal transition (EMT) and the matrix metalloproteinase (MMP) genes. This study indicates that TRIM58 is a potential biomarker of CRC prognosis; it acts as a tumor suppressor and has a specific role in the regulation of cancer cell invasion in CRC.
Materials and methods
Patients and sample collection
This study was approved by the Institutional Review Board of the Sixth Affiliated Hospital, Sun Yat-sen University. All samples were collected with the patients' written informed consent and approval from the Institutional Review Board.
Fresh frozen paired samples (n=48 for mRNA assay with 30 males vs. 18 females, and n=30 for protein assay containing 25 males vs. 5 females) of primary CRC and adjacent normal colon tissue (2 cm from the tumor border), ranging from stage I to stage IV, were collected from the Department of Surgery at the Sixth Affiliated Hospital of Sun Yat-sen University during the period of October 2010 to July 2015. Age of all patients with TRIM58 mRNA detected ranged from 30 to 71 years old, while patients whose samples were used for TRIM58 protein detection varied from 32 to 88 years old.
Clinical tissue samples were all confirmed histopathologically and stored in Invitrogen™ RNAlater solution (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and RIPA lysis buffer (Thermo Fisher Scientific, Inc.) containing PMSF (Beijing Dingguo Biotechnology, Co., Ltd., Beijing, China) and PPI (Beijing Dingguo Biotechnology) at −80°C until extraction.
Cell culture
Eleven human colorectal cell lines, HCT8, KM12, Caco-2, DLD-1, HCT116, LoVo, HT-29, SW480, SW620, RKO and HCT15, and 293 cells with SV40-T antigen (293T) were used. Caco-2, DLD-1, HCT15, HCT116, HT-29, HCT8 and KM12 cells were cultured in RPMI-1640 medium with 10% fetal bovine serum (FBS). SW480, SW620 and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS. RKO cells were cultured in EMEM with 10% FBS, and LoVo cells were cultured in F-16 with 10% FBS. All cells were incubated at 37°C with 5% CO2.
Transient transfection and establishment of stable clone cells
The TRIM58 construct vectors (TRIM58, pTRIM58-IRES2-EGFP; Empty, H316) or TRIM58 siRNA (siT58#1, GGAG GGAGCTCTTAAGGAA; siT58#2, AAAUUUCAUUCUA CAAUGUCA) were used to transiently transfect HCT8 and KM12 cells using Invitrogen Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, Inc.). Briefly, 4 µg of vectors or 5 µl of siRNA was mixed with 4 µl of transfection reagent. The transfection protocol was performed according to the manufacturer's instructions, and transfection was confirmed by RT-qPCR and western blot analysis. For stable transfected cell selection, after 48 h of transfection, the cells were seeded onto fresh media with 10% FBS (Gibco; Thermo Fisher Scientific) and 1 mg/ml G418 (Geneticin; Thermo Fisher Scientific, Inc.). Resistant clones were selected for 7 days and passaged.
Tissue microarray (TMA) construction and immunohistochemistry (IHC)
A paraffin-embedded tissue microarray and related clinicopathological parameters were obtained from the Sixth Affiliated Hospital of Sun Yat-sen University. Paraffin-embedded tissues were cut into 4-µm thick sections, deparaffinized in xylene, rehydrated through a graded alcohol series, and heat treated for 30 min in citrate buffer (pH 6.0; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) for antigen retrieval. Endogenous peroxidase activity was blocked for 10 min with reagent from an IHC kit (cat. no. SP9000; ZSGB-Bio, Beijing, China). Samples were then blocked with 5% blocking buffer for 1 h, followed by incubation overnight with primary anti-TRIM58 antibody (HPA023637, rabbit anti-human, 1:200; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 4°C. The next day, the sections were incubated for 1 h with secondary antibody provided by the kit (cat. no. SP9000; ZSGB-Bio) after 3 washes with PBST, 10 min per wash, and then were stained by DAB kit (cat. no. ZLT-9017; ZSGB-Bio). Finally, the sections were observed under a light microscope (Olympus BX53; Olympus Corp., Tokyo, Japan). The immunostaining results were evaluated by two independent pathologists blinded to the patient status to avoid possible bias.
The staining intensity was scored as: 0 points, negative; 4 points, weak intensity; 8 points, moderate intensity; or 12 points, strong intensity. Staining density was scored separately based on the percentage of cells stained as follows: 0 points, 0%; 4 points, 1–25%; 8 points, 26–50%; 10 points, 51–75%; or 12 points, >75%. The final score was calculated as the sum of the intensity score and the density score. Final scores ≥8 points were considered high expression, and final scores <8 points were considered low expression.
RNA isolation and RT-qPCR
RNA was extracted from CRC cell lines, human CRC tissues and normal tissues with Invitrogen TRIzol reagent (Thermo Fisher Scientific, Inc.). cDNAs were synthesized from extracted RNA (1 µg/20 µl) with a ReverTra Ace-α RT-PCR kit (Toyobo Co., Ltd., Osaka, Japan), and real-time quantitative PCR amplification of TRIM58 and β-actin was performed with 1 µl of reverse cDNA from CRC cell lines, human CRC tissues and normal tissues per well in a LightCycler 488 system (Roche Diagnostics, Indianapolis, IN, USA). The gene encoding β-actin was used as the reference gene, and the ∆∆Cq method was used to analyze the results (9). The final results are presented as fold changes in the charts/graphs. [TRIM58 (forward) 5′-GCGGGATCCAGCTTTACAT-3′ and (reverse) 5′-GGCTGGAAGCAGAGAACATC-3′; β-actin (forward) 5′-TTGTTACAGGAAGTCCCTTGCC-3′ and (reverse) 5′- ATGCTATCACCTCCCCTGTGTG-3′. The thermocycling conditions were as follows: precubation at 95°C for 10 min; amplification at 95°C for 10 sec, 60°C for 10 sec, 72°C for 10 sec, 45 cycles; melting at 95°C for 10 sec, 65°C for 60 sec and 97°C for 1 sec.
Western blotting
Cells were washed twice with PBS and lysed with RIPA lysis buffer (Thermo Fisher Scientific, Inc.) containing PMSF (Beijing Dingguo Biotechnology) and PPI (Beijing Dingguo Biotechnology). The protein concentration was determined with a bicinchoninic acid (BCA) protein assay kit (Beijing Dingguo Biotechnology). Equal amounts of protein (~20 µg) were separated using 8, 10 or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, depending on the molecular weight of the protein, and were then transferred to 0.22-µm polyvinylidene difluoride filter (PVDF) membranes. After that, the transferred membranes were blocked in TBST buffer containing 5% non-fat milk or bovine serum albumin (BSA) for 1 h and then incubated overnight at 4°C with primary antibodies. Next, the membranes were washed three times with TBST for at least 30 min and probed with HRP-linked secondary antibodies for 1.5 h at room temperature. Protein bands were visualized with a chemiluminescence detection kit (Thermo Fisher Scientific, Inc.) after three washes with TBST buffer and semi-quantified analysis was carried out using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
The primary antibodies and concentrations used were as follows: anti-TRIM58 (dilution 1:1,000; cat. no. H6-NBP1-88608; Novus Biologicals LLC, Littleton, CO, USA) and anti-GAPDH (dilution 1:1,000; cat. no. 60004–1-Ig; Proteintech Group, Rosemont, IL, USA). The following primary rabbit anti-human polyclonal antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA): anti-β-catenin (dilution 1:1,000; cat. no. 8480), anti-E-cadherin (dilution 1:1,000; cat. no. 3195), anti-N-cadherin (dilution 1:500; cat. no. 13116), anti-Snail (dilution 1:500; cat. no. 3879), anti-Slug (dilution 1:500; cat. no. 9585), anti-MMP-2 (dilution 1:250; cat. no. 13132S) and anti-MMP-9 (dilution 1:250; cat. no. 13667S). The secondary antibodies were anti-rabbit-IgG-HRP and anti-mouse-IgG-HRP (dilution 1:10,000; cat. nos. 7074S and 7076).
Proliferation assay using an IncuCyte imaging system
First, 2×104 cells were seeded in 96-well plates with RPMI-1640 plus 10% FBS. The plate was then placed into and incubated in an IncuCyte Essens Bioscience incubator (Essens Bioscience, Birmingham, UK). Live cell images were collected every 4 h. Proliferation rates based on cell confluence were determined by live cell imaging using IncuCyte software (Essens Bioscience).
Colony formation assay
First, 2,000 cells were seeded in 6-well plates with RPMI-1640 plus 10% FBS and incubated at 37°C with 5% CO2 for 7 to 10 days. After colony formation, cells were fixed with 4% PFA at room temperature for 30 min and stained with 0.01% crystal violet solution for 1 min. Data were collected after the plate was completely dry.
Wound-healing assays with the IncuCyte time-lapse imaging system
After transfection with the TRIM58 vector or TRIM58 siRNA, cells were seeded in 6-well plates and incubated until 100% confluence was reached. The cell layer was scratched with a 10-µl pipette tip (Rainin; Mettler-Toledo AG, Greifensee, Switzerland) and washed with PBS three times. The plate was placed in the IncuCyte Essens Bioscience incubator (Essens Bioscience) and cultured in fresh serum-free DMEM containing the TRIM58 vector or TRIM58 siRNA transfection mix for 24–72 h. Live cell images were collected every 4 h. Migration distance was calculated using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Invasion assay
Cell invasion was evaluated with 8-µm Transwell filters (Corning Inc., Corning, NY, USA). The upper chambers of the Transwell filters were coated with Matrigel (Corning Inc.) and incubated at 37°C for at least 30 min. Then, 4×104 cells were seeded on the Matrigel-coated filter with RPMI-1640 plus 1% FBS, and RPMI-1640 containing 10% FBS was added to the lower chamber. After 24 or 36 h, non-migrating cells were carefully removed from the upper chamber with a cotton swab, and the filters were fixed and stained with 0.01% crystal violet solution for 1 min. Invading cells were photographed, and the invaded cell proportion was determined by measuring the OD570 nm value of the crystal violet eluted with ethanol.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). All data are presented as the mean ± SEM. Comparisons to determine significant differences between two experimental groups were made using two-tailed t-tests or Chi-square test, and one-way ANOVA with Dunnett's post hoc test or Bonferroni post hoc test was conducted to determine significant differences among three or more groups. Potential risk factors of OS were evaluated by univariate analysis and multivariate analysis using Cox proportional hazards model. Kaplan-Meier survival curves were determined by log-rank test. Significance was set at P<0.05.
Validation of TRIM58 expression based on the Oncomine database
We utilized the Oncomine database (http://www.oncomine.org) to validate the expression of TRIM58 in patients. The differential expression analysis was directly performed using Oncomine online analysis tools (10).
Results
Trim58 expression is suppressed in CRC tissues and cell lines
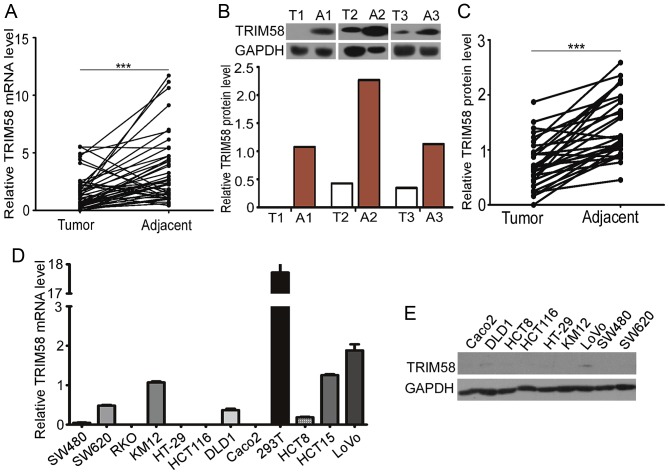
To examine TRIM58 expression in clinical CRC samples, we evaluated the TRIM58 mRNA and protein levels in 48 tumor tissues with matched adjacent normal samples from CRC patients. Trim58 expression was significantly lower in human CRC than in non-tumorous tissues (Fig. 1A). Western blot analysis further confirmed that expression of TRIM58 protein was lower in 30 pairs of human CRC tissues than the level in the matched adjacent normal tissues (Fig. 1B and C).
Figure 1.
Trim58 expression is suppressed in colon cancer tissues and cell lines. (A) The relative mRNA level of TRIM58 was significantly lower in human colorectal cancer (CRC) tissues (n=48) than in adjacent colorectal tissues (n=48) from patients admitted to the hospital from 2011 to 2014. ***P<0.001, two-tailed Student's t-test. (B) Representative western blot for TRIM58 in human CRC and adjacent colorectal tissues. T, tumor tissue; A, adjacent colorectal tissue. (C) Statistical analysis of western blot results showed that TRIM58 protein levels were significantly decreased in human CRC tissues (n=30) compared with adjacent colorectal tissues (n=30) from patients admitted to the hospital from 2011 to 2014. ***P<0.001, two-tailed Student's t-test. (D) TRIM58 expression level in 11 human CRC cell lines was analyzed by RT-PCR. (E) TRIM58 expression level in 9 human CRC cell lines was analyzed by western blot analysis. TRIM, tripartite motif protein; CRC, colorectal cancer.
Having shown downregulation of TRIM58 in human CRC tissues, we further investigated the expression of TRIM58 in human CRC cell lines. We examined the mRNA levels of TRIM58 in 11 human CRC cell lines and protein levels in 9 of the cell lines (Fig. 1D and E). The TRIM58 mRNA levels in CRC cell lines were generally low compared with that in the 293T cell line, and TRIM58 protein was expressed at low levels in most of the CRC cell lines examined. Taken together, these results indicate that TRIM58 expression was suppressed in both CRC tissues and cell lines.
Low expression of TRIM58 is correlated with poor prognosis in CRC patients
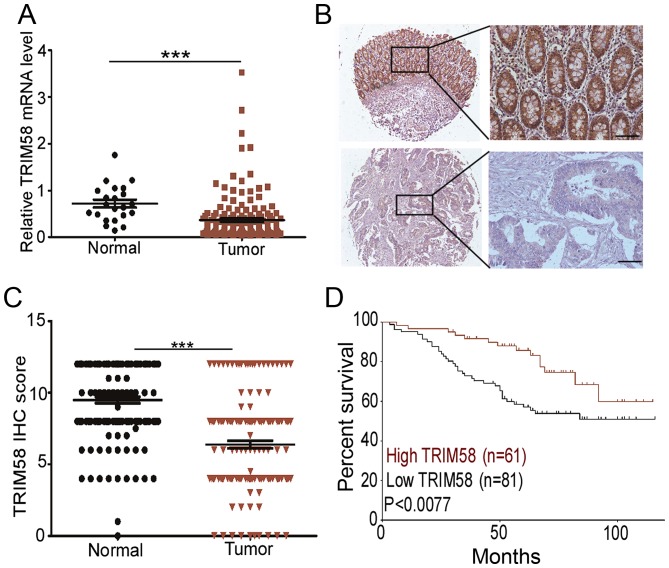
To determine the clinical implications of TRIM58 in CRC, we evaluated the relationship between TRIM58 expression level and pathological features of the disease. First, we analyzed TRIM58 mRNA expression with the Oncomine database using RNA sequencing data of CRC patients from The Cancer Genome Atlas (TCGA). The results revealed that the TRIM58 mRNA level was lower in cancer tissues than in normal tissues (Fig. 2A). To further validate the suppressed expression of TRIM58 in CRC, we recruited a cohort of 152 patients for TMA analysis. The IHC staining confirmed that TRIM58 was primarily localized in the cytoplasm (Fig 2B) in the tissues. IHC scores of TRIM58 revealed that in 68.57% (96/140) of the paired samples, TRIM58 expression was downregulated in CRC compared to normal tissues (Fig. 2C)
Figure 2.
Low expression of TRIM58 is correlated with poor prognosis in CRC patients. (A) TRIM58 expression in CRC tissues (n=161) was prominently inhibited compared with that in normal tissues (n=22) in a TCGA cohort. ***P<0.001, two-tailed Student's t-test. (B) Representative IHC staining for TRIM58 in human CRC cancer (lower panels) and adjacent normal colon tissues (upper panels). Scale bar, 100 µm. (C) Scatterplot of TRIM58 IHC scores in CRC and normal tissues. TRIM58 expression in a tissue microarray of 140 paired samples of CRC and normal colon tissues of patients was assessed by IHC. The receiver operating characteristic curve was used to define the cutoff. ***P<0.001, two-tailed Student's t-test. (D) Kaplan-Meier survival curves of overall survival duration based on TRIM58 expression in the CRC tissues of the testing cohorts. Survival curves show that patients with high TRIM58 expression survived significantly longer than those with low TRIM58 expression. P<0.0077, log-rank test. TRIM, tripartite motif protein; CRC, colorectal cancer; TCGA, The Cancer Genome Atlas.
Since TRIM58 expression was significantly decreased in CRC, we next investigated whether TRIM58 expression was associated with the clinical characteristics of CRC patients. Kaplan-Meier analysis showed that a low level of TRIM58 was correlated with the poor overall survival in the cohorts (P<0.0077, Fig. 2D). Clinicopathological characteristic analysis revealed that low expression was positively correlated with various clinical stages in the cohort (P<0.05, Table I), in particular T status (P<0.004, Table I). Furthermore, univariate Cox regression analysis revealed that TRIM58 expression was an independent prognostic factor for poor survival (P<0.05, Table II).
Table I.
Association between expression of TRIM58 and clinicopathological characteristics of the CRC patients.
| TRIM58 | |||
|---|---|---|---|
| Variables | Low expression n (%) | High expression n (%) | P-value |
| Sex | 0.803 | ||
| Male | 44 (54.3) | 40 (56.3) | |
| Female | 37 (45.7) | 31 (43.7) | |
| Median age (years) | 0.324 | ||
| <60 | 43 (53.1) | 32 (45.1) | |
| ≥60 | 38 (46.9) | 39 (54.9) | |
| Histological grade | 0.428 | ||
| G1 | 5 (8.2) | 5 (11.6) | |
| G2 | 55 (90.2) | 38 (88.4) | |
| G3 | 1 (1.6) | 0 (0.00) | |
| pT status | 0.004b | ||
| T1 | 2 (2.5) | 3 (4.2) | |
| T2 | 9 (11.1) | 18 (25.4) | |
| T3 | 63 (77.8) | 49 (69.0) | |
| T4 | 7 (8.6) | 1 (1.4) | |
| pN status | 0.055 | ||
| N0 | 41 (51.2) | 50 (70.4) | |
| N1 | 31 (38.8) | 17 (23.9) | |
| N2 | 8 (10%) | 4 (5.6%) | |
| pM status | 0.556 | ||
| M0 | 77 (96.3) | 69 (97.2) | |
| M1 | 3 (3.8) | 2 (2.8) | |
| Clinical stage (Duke's) | 0.025a | ||
| I | 10 (12.8) | 16 (22.5) | |
| II | 30 (38.5) | 33 (46.5) | |
| III | 38 (48.7) | 21 (29.6) | |
| IV | 0 (0.0) | 1 (1.4) | |
| Neo chemotherapy (%) | 0.17 | ||
| No | 6 (7.4) | 1(1.4) | |
| Yes | 75 (92.6) | 70 (98.6) | |
| Recurrence-metastasis | 0.025a | ||
| No | 40 (58.0) | 46 (76.7) | |
| Yes | 29 (42.0) | 14 (23.3) | |
All data are expressed as the number of patients (%). P-values were calculated using SPSS 22 using a Chi-square test. P-values <0.05 were considered to indicate statistical significance.
P<0.01
P<0.05. CRC, colorectal cancer.
Table II.
Univariate and multivariate analyses of different prognostic parameters of the CRC patients in the testing cohort (n=152).
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Parameters | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex (male vs. female) | 1.1 (0.6–1.8) | 0.781 | 1.1 (0.6–2.0) | 0.689 |
| Age (<60 vs. ≥60 years) | 1.1 (0.6–1.8) | 0.759 | 1.2 (0.7–2.1) | 0.421 |
| Neo-chemotherapy (no vs. yes) | 2.4 (1.0–6.0) | 0.06 | 1.1 (0.3–3.8) | 0.831 |
| Histological grade (G1 or G2 vs. G3 or G4) | 0.4 (0.2–0.6) | <0.001 | 0.5 (0.9–2.2) | 0.335 |
| pT status (T1 or T2 vs. T3 or T4) | 0.2 (0.05–0.5) | 0.002 | 0.2 (0.6–0.7) | 0.012 |
| pN status (N0 vs. N1or N2) | 0.3 (0.2–0.6) | <0.001 | 1.0 (0.2–4.7) | 0.951 |
| pM status (M0 vs. M1) | 0.2 (0.09–0.6) | 0.001 | 0.3 (0.1–0.9) | 0.024 |
| TRIM58 expression (low vs. high) | 1.8 (1.0–3.0) | 0.044a | 1.2 (0.7–2.2) | 0.562 |
Hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated using univariate or multivariate Cox proportional hazards regression in SPSS 22. P-values were calculated using univariate or multivariate Cox proportional hazards regression in SPSS 16.0. P-values <0.05 were considered to indicate statistical significance.
P<0.05.
In conclusion, these results indicated that low expression of TRIM58 is correlated with the poor prognosis of CRC patients, indicating that TRIM58 expression may play an important role in the tumorigenesis of CRC.
TRIM58 overexpression has no significant influence on the proliferation, colonization or migration of human CRC cell lines
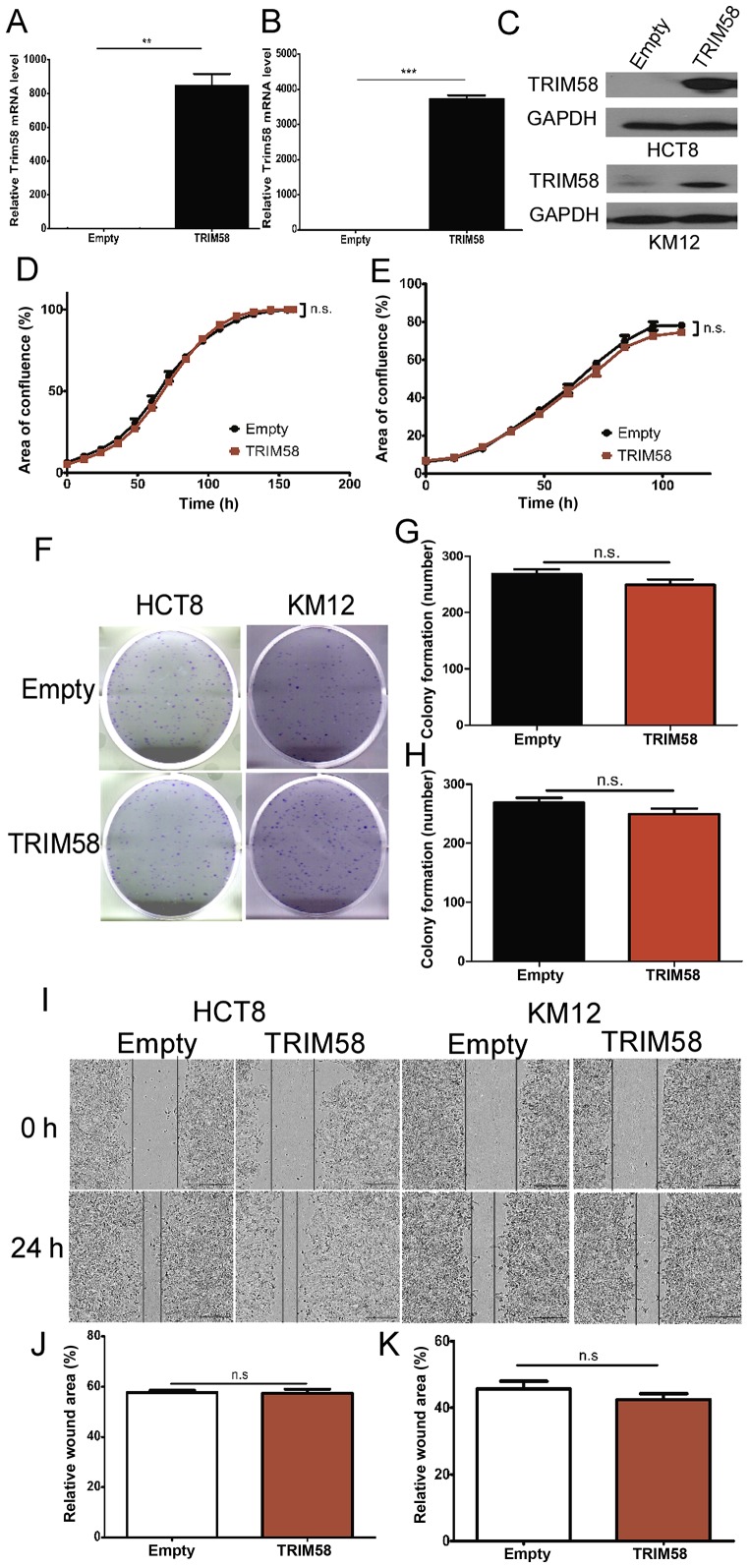
Our results clearly indicated that high TRIM58 expression was associated with the TNM stage, suggesting the role of TRIM58 in cell proliferation, cell colonization and cell motility. Therefore, to identify the specific roles in which TRIM58 modulates tumorigenesis, we randomly selected two CRC cell lines (HCT8 and KM12) with low expression of TRIM58 (Fig. 1D and E) and transfected the cells with a TRIM58 overexpression vector (TRIM58, pTRIM58-IRES2-EGFP) or an empty vector as a negative control (Empty, H316). The efficacy of TRIM58 overexpression was examined by RT-PCR and western blot analyses. The results showed that HCT8 and KM12 cells efficiently overexpressed TRIM58 (Fig. 3A-C).
Figure 3.
TRIM58 overexpression has no significant influence on the proliferation, colonization or migration of human colorectal cancer cell lines. (A-C) The overexpression of TRIM58 mRNA (A: HCT8 cells and B: KM12 cells) and (C) protein in CRC cell lines was confirmed by RT-PCR and western blot analysis.**: n=3, P<0.001 and ***: n=3, P<0.0001, two-tailed Student's t-test. (D and E) Proliferation assay (D: HCT8 cells and E: KM12 cells) conducted using IncuCyte. Cells were transfected with the TRIM58 overexpression vector on day 1 and day 3. n.s. indicates not significant, n=3, P>0.05, two-tailed Student's t-test. (F-H) Images (F) and statistical analysis (G: HCT8 cells and H: KM12 cells) of colony formation assays indicated that TRIM58 overexpression had no significant influence on cell colonization. n.s. indicates not significant, n=3, P>0.05, two-tailed Student's t-test. (I-K) Images of wound-healing assays (I) and statistical analysis (J: HCT8 cells and K: KM12 cells) were performed to show that TRIM58 did not inhibit cell migration. n.s. indicates not significant, n=3, P>0.05, two-tailed Student's t-test. Scale bar, 100 µm. TRIM, tripartite motif protein; CRC, colorectal cancer.
Next, we determined whether TRIM58 overexpression inhibits CRC cell proliferation. To this end, cellular proliferation was assessed using IncuCyte time-lapse imaging. We observed that TRIM58 overexpression barely inhibited the proliferation of HCT8 and KM12 cells during the observation time (Fig. 3D and E). To further investigate the role of TRIM58 in CRC progression, we analyzed the cell colony formation ability of HCT8 and KM12 cells overexpressing TRIM58. Our results demonstrated that TRIM58 overexpression had little influence on CRC cell colonization (Fig. 3F-H). Similarly, wound-healing assays showed that TRIM58 overexpression did not affect the migration of the HCT8 and KM12 cells (Fig. 3I-K). These data suggested that TRIM58 inhibits tumor progression via other processes.
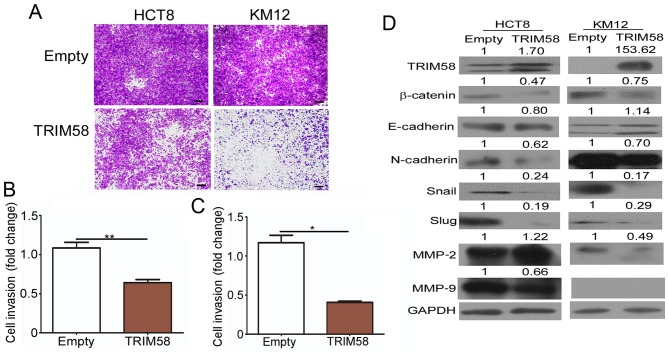
Overexpression of TRIM58 specifically reduces invasion of CRC cells and inhibits the expression of related genes
Evidence has shown that local and distant invasion is highly associated with cancer malignancy (11). We thus conducted invasion assays to determine whether high TRIM58 expression negatively impacts CRC development by interfering with the invasion of CRC cells. As shown in Fig. 4A-C, a significant reduction in the number of invading HCT8 and KM12 CRC cells was observed following transient overexpression of TRIM58 in these cells in the invasion assays. Since the EMT program is a prominent regulator of invasion, and MMPs have been shown to play an active role in tumor invasion and metastasis (12), we detected the TRIM58-mediated changes in EMT proteins and MMP expression (Fig. 4D). Western blot results showed that TRIM58 overexpression downregulated the levels of most mesenchymal proteins, including N-cadherin, Snail and Slug, in both HCT8 and KM12 cells, while it upregulated the epithelial protein E-cadherin in KM12 cells. Furthermore, β-catenin, a key regulator of EMT, was downregulated when TRIM58 was overexpressed in both tested cell lines. Moreover, we also determined the MMP2 and MMP9 levels and found that the MMP2 level was decreased in KM12 cells and MMP9 was reduced in the HCT8 cells under TRIM58 overexpression. The above results suggested that TRIM58 plays an important role in inhibiting the invasive ability of CRC cells during cancer progression and might be regulated by Wnt-β-catenin-mediated regulation of EMT signaling and MMP expression.
Figure 4.
Overexpression of TRIM58 specifically reduces invasion in CRC cells and inhibits expression of related genes. (A) Invasion assays indicated that TRIM58 overexpression inhibits cell invasion. Cells were transiently transfected with control vector or TRIM58 expression vector before they were seeded on the Transwell filters. All the results were collected after 36 h. Scale bar, 100 µm. (B and C) Statistical analysis of the number of invasive HCT8 (B) and KM12 (C) cells. The number of invasive HCT8 and KM12 cells treated with empty or TRIM58 expression vectors was quantified. **: n=3, P<0.01 and *: n=3, P<0.05, two-tailed Student's t-test. (D) Western blotting confirmed that TRIM58 attenuated the expression of EMT-associated genes and MMPs. TRIM, tripartite motif protein; CRC, colorectal cancer; MMPs, matrix metalloproteinases.
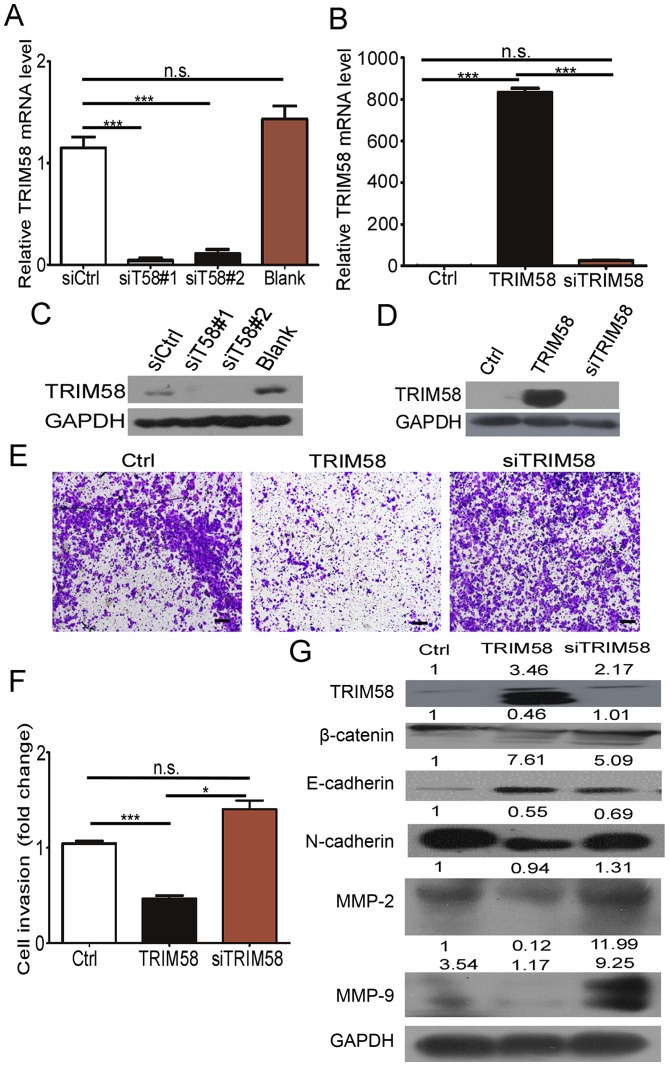
Silencing of TRIM58 overexpression reverses CRC cell invasion and related protein levels
To further determine the effect of TRIM58 on cell invasion, we neutralized TRIM58 overexpression by introducing siRNAs into stable TRIM58-overexpressing cells. First, we tested the efficiency of the TRIM58 siRNAs by RT-PCR and western blotting. The results (Fig. 5A and C) showed that siT58#1 and siT58#2 effectively reduced the mRNA and protein levels of TRIM58 in SW620 cells. We then mixed siT58#1 and siT58#2 and transfected the mixture into the TRIM58-overexpressing HCT8 cells (termed siTRIM58), while HCT8 cells were transfected with empty vector and siCtrl as the control (termed Ctrl), and stable TRIM58-expressing HCT8 cells were transfected with siCtrl (termed TRIM58) (Fig. 5B and D). The results showed that the invasive ability recovered and was slightly stronger than that in the Ctrl-HCT8 cells after knocking down TRIM58 in TRIM58-overexpressing HCT8 cells (siTRIM58) compared to TRIM58-overexpressing HCT8 cells transfected with the control siRNA (TRIM58) (Fig. 5E and F). Additionally, expression of mesenchymal proteins (N-cadherin, Snail, Slug), MMP2 and MMP9 were prominently increased and E-cadherin was reduced after silencing of TRIM58 in HCT8 overexpression cells, which was similar to the results in the Ctrl HCT8 cells (Fig. 5G). The above results further demonstrated that TRIM58 induced CRC cell progression by enhancing the invasion modulated by EMT signaling and MMPs.
Figure 5.
Silencing of TRIM58 overexpression reversed invasive ability in the CRC cells and related protein level. (A and C) siRNA silencing of TRIM58 mRNA and protein in CRC cell lines was confirmed by RT-PCR and western blot analysis. Blank, cells treated with ddH2O as a blank control. n.s. indicates not significant, n=3, P>0.05; ***: n=3, P<0.0001, one-way ANOVA with Dunnett's post hoc test (A). (B and D) Knockdown of TRIM58 mRNA and protein in stable TRIM58-overexpressing CRC cell lines was confirmed by RT-PCR and western blot analysis. n.s. indicates not significant, n=3, P>0.05; ***: n=3, P<0.0001, one-way ANOVA with Bonferroni post hoc test (B). (E) Invasion assays indicated that the cell invasion inhibition of TRIM58 overexpression was neutralized after introducing TRIM58 siRNA into the stable TRIM58-overexpressing HCT8 cells, and the cell invasion was similar to that in Ctrl cells with empty vector that were transfected with siCtrl. All the results were collected after 24 h. Scale bar, 100 µm. (F) Statistical analysis of the number of invasive HCT8 cells. The number of invasive HCT8 cells treated with empty or TIRIM58 expression vectors was quantified. **: n=3, P<0.01, and *: n=3, P<0.05, one-way ANOVA with Bonferroni post hoc test. (G) Western blot analysis and semi-quantative statistical results for EMT-related proteins and MMPs. n.s. indicates not significant, n=3, P>0.05. TRIM, tripartite motif protein; CRC, colorectal cancer; MMPs, matrix metalloproteinases.
Discussion
Discovery of potential biomarkers for early tumor detection is now a major concern of researchers. Various aberrant genes have been reported and translated into clinical diagnostic applications.
The TRIM family is a large protein family comprising more than 70 members. The majority of TRIM proteins are believed to be E3 ubiquitin ligases because of their RING domain, which frequently displays E3 ligase activity. Indeed, several TRIM proteins have been shown to modify target proteins with ubiquitination and to be associated with many biological processes, and abnormal changes in their abundance or activity are commonly involved in pathological conditions, including cancer (13,14). Accumulated reports have shown that TRIM proteins participate in tumorigenesis and progression of breast cancer, liver cancer, gastric cancer, osteosarcoma and skin cancer (15–18). For example, TRIM31 indirectly overactivates the mTORC pathway by ubiquitinating the TSC1-TSC2 complex to promote hepatocellular carcinoma progression (15).
However, the pro-tumorigenesis TRIM family has primarily been studied for their oncogenic functions, and less attention has been paid to tumor-suppressor gene functions. TRIM58 was recently shown to act as a tumor-suppressor gene and to play a crucial role in the tumorigenesis of different cancers (7,8). Accordingly, our data also indicated that TRIM58 downregulation in CRC was significantly correlated with poor clinicopathological characteristics, especially T status. We therefore hypothesized that TRIM58 might be a tumor suppressor whose aberrant expression plays a crucial role in CRC tumorigenesis and progression. To confirm this hypothesis, we performed a series of in vitro assays, including proliferation, colony formation, migration and invasion assays, to investigate the role of TRIM58 in CRC cell growth and motility, which are the crucial factors leading to tumor malignancy. Surprisingly, our results showed that TRIM58 overexpression only inhibited cell invasion, which is likely associated with EMT and MMP regulation, but not other malignant behaviors, such as cell proliferation, colony formation and migration.
These results not only support our view that TRIM58 may function as a tumor suppressor in CRC, expanding upon its previously reported role in other cancers, but also, interestingly, demonstrated a distinct function of TRIM58. Aberrant TRIM58 expression only influenced the CRC cell invasion capacity, in contrast with the results of other studies showing that TRIM58 suppression enhances proliferation, colony formation, migration and invasion in lung and liver tumors. The functional differences among these tumor types is likely due to phenotypic distinctions and genomic pattern variations in tumors derived from diverse origins (19,20). Furthermore, our results revealed that although TRIM58 overexpression inhibited CRC cell invasion, it had little effect on CRC cell motility in the wound healing assays. This is likely because cell motility or directional movement is one of several invasion processes that TRIM58 has little or no effect on in the CRC cells used in this study (21). We also tested the expression of EMT genes that have been confirmed as important cell invasion regulators. Consistent with the TRIM58-induced changes in cell invasion, Snail and Slug, the key transcriptional genes that control cell-cell adhesion, cell shape, degradation of E-cadherin protein and basement membrane integrity during EMT, were downregulated when TRIM58 was overexpressed. Furthermore, the expression level of the mesenchymal protein N-cadherin was reduced when TRIM58 was overexpressed and rescued after silencing of TRIM58 by siRNA, which is opposite to the trend observed for E-cadherin, an epithelial cell gene involved in EMT. These findings indicate that TRIM58 regulates EMT and thus influences CRC invasion.
Further investigation indicated that the expression of β-catenin, a crucial downstream effector of canonical Wnt-β-catenin signaling during EMT (16), is negatively regulated by TRIM58 and that TRIM58 might affect EMT by modulating Wnt-β-catenin signaling. However, further studies are needed.
Collectively, we suggest that TRIM58 is a tumor suppressor that inhibits tumorigenesis and progression by regulating EMT via Wnt-β-catenin signaling, and TRIM58 suppression may be a marker for early CRC detection.
Acknowledgements
We thank for the support by the National Key Clinical Discipline.
Glossary
Abbreviations
- TRIM
tripartite motif protein
- CRC
colorectal cancer
- EMT
epithelial-mesenchymal transition
- OS
overall survival
- MMP
matrix metalloproteinase
- TCGA
The Cancer Genome Atlas
Funding
This work was financially supported by the National Natural Science Foundation (grant no. 81573078 to LW), the Natural Science Foundation of Guangdong Province (grant no. 2017A030313805 to DC and 2016A030311021 to LW), the Science and Technology Program of Guangzhou City (grant no. 201604020174 to DC), and the Science and Technology Planning Project of Tianhe District of Guangzhou (201504KW044 to HW).
Availability of data and materials
The datasets and materials used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
DC conceived the project, designed the experiments and edited the manuscript. LW edited the manuscript and provided overall support. XZ performed the experiments, analyzed the results, conducted statistical analysis of data and wrote the manuscript. HW assessed the IHC microarray scores and performed RT-qPCR and western blotting of the tissue samples. ML collected the clinical samples, counted the HCT8 and KM12 cell colonies, assessed the IHC microarray scores and edited the manuscript. JC provided the tissue microarray. YL, QL and ZY helped culture cells and in vitro assay. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The study was approved by the Human Medical Ethics Committee of Sun Yat-Sen University (SYSU). Informed consent was obtained from all patients enrolled in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest to disclose.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Fessler E, Medema JP. Colorectal cancer subtypes: Developmental origin and microenvironmental regulation. Trends Cancer. 2016;2:505–518. doi: 10.1016/j.trecan.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Shia J, Schultz N, Kuk D, Vakiani E, Middha S, Segal NH, Hechtman JF, Berger MF, Stadler ZK, Weiser MR, et al. Morphological characterization of colorectal cancers in The Cancer Genome Atlas reveals distinct morphology-molecular associations: Clinical and biological implications. Mod Pathol. 2017;30:599–609. doi: 10.1038/modpathol.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson TB, Ranganathan A, Grothey A. Randomized phase III trial results of panitumumab, a fully human anti-epidermal growth factor receptor monoclonal antibody, in metastatic colorectal cancer. Clin Colorectal Cancer. 2006;6:29–31. doi: 10.3816/CCC.2006.n.01. [DOI] [PubMed] [Google Scholar]
- 5.Los M, Roodhart JML, Voest EE. Target practice: Lessons from phase III trials with bevacizumab and vatalanib in the treatment of advanced colorectal cancer. Oncologist. 2007;12:443–450. doi: 10.1634/theoncologist.12-4-443. [DOI] [PubMed] [Google Scholar]
- 6.Thom CS, Traxler EA, Khandros E, Nickas JM, Zhou OY, Lazarus JE, Silva AP, Prabhu D, Yao Y, Aribeana C, et al. Trim58 degrades Dynein and regulates terminal erythropoiesis. Dev Cell. 2014;30:688–700. doi: 10.1016/j.devcel.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu X, Huang Y, Zhou Y, Zheng F. Aberrant methylation of TRIM58 in hepatocellular carcinoma and its potential clinical implication. Oncol Rep. 2016;36:811–818. doi: 10.3892/or.2016.4871. [DOI] [PubMed] [Google Scholar]
- 8.Kajiura K, Masuda K, Naruto T, Kohmoto T, Watabnabe M, Tsuboi M, Takizawa H, Kondo K, Tangoku A, Imoto I. Frequent silencing of the candidate tumor suppressor TRIM58 by promoter methylation in early-stage lung adenocarcinoma. Oncotarget. 2017;8:2890–2905. doi: 10.18632/oncotarget.13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Dufour A, Overall CM. Missing the target: Matrix metalloproteinase antitargets in inflammation and cancer. Trends Pharmacol Sci. 2013;34:233–242. doi: 10.1016/j.tips.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. BioEssays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 14.Elabd S, Meroni G, Blattner C. TRIMming p53's anticancer activity. Oncogene. 2016;35:5577–5584. doi: 10.1038/onc.2016.33. [DOI] [PubMed] [Google Scholar]
- 15.Guo P, Ma X, Zhao W, Huai W, Li T, Qiu Y, Zhang Y, Han L. TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1-TSC2 complex. Oncogene. 2018;37:478–488. doi: 10.1038/onc.2017.349. [DOI] [PubMed] [Google Scholar]
- 16.Kawabata H, Azuma K, Ikeda K, Sugitani I, Kinowaki K, Fujii T, Osaki A, Saeki T, Horie-Inoue K, Inoue S. TRIM44 is a poor prognostic factor for breast cancer patients as a modulator of NF-κB signaling. Int J Mol Sci. 2017;18:E1931. doi: 10.3390/ijms18091931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G, Guo Y, Xu D, Wang Y, Shen Y, Wang F, Lv Y, Song F, Jiang D, Zhang Y, et al. TRIM14 regulates cell proliferation and invasion in osteosarcoma via promotion of the AKT signaling pathway. Sci Rep. 2017;7:42411. doi: 10.1038/srep42411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn EJ, Albor A, Liu Y, El-Hizawi S, Vanderbeek GE, Babcock M, Bowden GT, Hennings H, Lozano G, Weinberg WC, et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25:157–167. doi: 10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- 19.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22:1342–1350. doi: 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 21.Friedl P, Alexander S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and materials used during the present study are available from the corresponding author upon reasonable request.