Abstract
To survive, cells need to avoid excessive volume change that jeopardizes structural integrity and stability of the intracellular milieu. Searching for the molecular identity of volume-regulated anion channel (VRAC) has yielded multiple potential candidates, but none has been confirmed. Recently, it is reported that leucine-rich repeat-containing 8A (LRRC8A) is a main molecular determinant of VRAC current. The biological functions of LRRC8 family proteins are poorly understood, particularly in cancer. In the present study, we investigated LRRC8A in the most common cancers of the digestive system. LRRC8A proteins were found to be abundantly expressed in the esophagus, stomach, duodenum, colon, rectum, liver and pancreas. LRRC8A was elevated in 60% of colorectal cancer patient tissues, which was higher than that in patients with cancer of the esophagus, stomach, duodenum, liver and pancreas. Colon cancer patients with high- expressed LRRC8A had a survival time of 54.9±5.5 months, shorter than that of patients with low-expressed LRRC8A (77.1±3.7). Moreover, survival time (52.6±7.3 months) of patients with metastases in the lymph nodes was shorter than that of patients without positive lymph nodes (72.2±3.6); patients with positive lymph nodes and an elevated LRRC8A expression had the highest mortality rate (~80%). These rates were not observed in rectal cancer. After LRRC8A protein was knocked down in colon cancer HCT116 cells, VRAC currents, migration and tumorigenesis in nude mice were significantly inhibited. In conclusion, we propose that LRRC8A could be a novel prognostic biomarker for colon cancer patient survival, and that the elevated expression of LRRC8A may enhance cancer cell growth and metastasis, and worsen the outcome of patients.
Keywords: chloride channel, volume-regulated anion channel, leucine-rich repeat-containing 8A, colon cancer, biomarker
Introduction
The digestive system is one of the systems that is most likely to be disordered, including infection, inflammation, atypical hyperplasia, metaplasia and carcinogenesis. According to the World Cancer Report 2014, cancers of the digestive system were the most commonly diagnosed cancers and the most common cause of cancer-related deaths in 2012 (1). Increased attention should be paid to the diagnosis, therapy and prognosis of these cancer patients to prolong survival time and improve their quality of life.
For cell survival, it is essential for cells to avoid excessive change in cell volume, which jeopardizes structural integrity and the stability of the intracellular milieu (2). Cell volume regulation plays pivotal roles in all types of cellular functions, such as epithelial transport, metabolism, excitation, hormone release, cell migration, cell proliferation and cell death (2–4).
Channel proteins on the cell membrane govern the movements of water and electrolytes for the regulation of cell volume. Although it has been confirmed that channel proteins play important roles in a wide variety of physiological and/or pathophysiological processes, the molecular entity for the volume-regulated anion channel (VRAC) is not entirely confirmed. The search for the molecular identity of VRAC has been long and has yielded multiple potential candidates, all of which eventually turned out to have properties not fully compatible with those of VRAC (5). Recently, 2 groups have independently reported that the leucine-rich repeat-containing 8A (LRRC8A) is a main molecular determinant of VRAC current (6,7), which has been immediately confirmed in different cells or tissue by other groups (8–11). However, in human retinal pigment epithelium cells, bestrophin 1, but not LRRC8A is essential for volume regulation (12). This idea concerning a non-essential contribution of LRRC8A to volume regulation has also been supported by others (13,14). These findings indicate that the molecular determinants of VRAC may be more complex and that VRAC could be formed by a cell type-specific or tissue-specific subunit component.
Given its importance in cancer cell proliferation, apoptosis, migration and/or invasion, and drug resistance, VARC has been considered to be a promising target for cancer diagnosis, prognosis and therapy (15–17). However, the biological functions of the LRRC8 family proteins remain poorly understood. LRRC8A was isolated and identified in a girl with congenital agammaglobulinemia (18), indicating that it may be involved in B- and T-cell development (19–21). In vascular smooth muscle cells, it was found that LRRC8A takes part in inflammation via supporting tumor necrosis factor-α-induced superoxide production (22). In human ovarian (A2780) and alveolar (A549) cancer cells, a reduced expression of LRRC8A may contribute to the acquisition of cisplatin resistance (23,24). The contribution of LRRC8A varies and is largely dependent on the cell type; thus, the precise role of LRRC8A warrants further investigation.
In the present study, we first disclosed that the high expression of LRRC8A may be correlated with the short survival time of colon cancer patients by enhancing the growth and metastasis of cancer cells.
Materials and methods
Cell culture, transfection and infection
Human colon cancer cells (HCT116) were routinely grown in Roswell Park Memorial Institute (RPMI)-1640 culture medium with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 µg/ml streptomycin in an atmosphere with 100% humidity, 5% CO2, and 95% O2 at 37°C. The cells were trypsinized and subcultured every 2 days.
The small interfering RNAs (siRNAs) were chemically synthesized and labeled with or without FAM carboxyfluorescein (Shanghai GenePharma Co., Ltd., Shanghai, China). The sense strand of LRRC8A siRNA was 5′-ACCAAGCUCAUCGUCCUCAACtt-3′; and, the sense strand of the negative control siRNA was 5′-UUCUCCGAACGUGUCACGUtt-3′. The short hairpin RNA (shRNA) was designed according to the sequence of siRNA, inserted into the pGLVH1 lentivirus shRNA vector containing the GFP gene, and then packaged as infectious lentivirus by the Shanghai GenePharma Co. The sequence of LRRC8A shRNA was, 5′-CCGGACCAAGCTCATCGTCCTCAACCTCGAGGTTGAGGACGATGAGCTTGGTTTTTTG-3′ (12).
For siRNA transfection, HCT116 cells were cultured in RPMI-1640 medium without antibiotics in 12-well culture plates for 24 h and reached 30–40% confluence. The cells were transfected with 100 nM siRNA in the presence of Invitrogen™ Lipofectamine 2000 (2.5 µl in 1,000 µl medium; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in serum and antibiotic-free culture medium for 6 h, and were incubated in normal RPMI-1640 medium for 72 h. For lentivirus infection, when cells reached 70% confluence, the lentivirus particles [multiplicity of infection (MOI=5)] were diluted and added into the culture medium; after a 12-h incubation, the cells were incubated in normal medium and subcultured for 1 week. The transfection efficiencies were detected using flow cytometry.
Animal feeding and tumor cell implantation
The animal experimental protocol was approved by the Laboratory Animal Administration Committee of Xi'an Jiaotong University and was carried out in accordance with the Guidelines for Animal Experimentation of Xi'an Jiaotong University and the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication no. 85–23, revised 2011).
Male BALB/c nude mice, 4 weeks of age, were provided by the Laboratory Animal Center of Xi'an Jiaotong University and maintained under a 12-h dark/light cycle with ad libitum self-feeding in specific pathogen-free conditions (55% humidity and 22°C). Every nude mouse was subcutaneously injected with the scramble and LRRC8A shRNA lentivirus-infected HCT116 cells (1×106) at the armpits of the right and left forelimb, respectively. The initial body weight of 10 mice was 14.7±0.3 g and then measured once a week; after 3 weeks, the mice were anesthetized with 1.5% isoflurane inhalation, and then the tumorigenesis (indicated by green fluorescence) was detected using the in vivo non-invasive small animal molecular imaging system (Xenogen; Caliper Life Sciences, Inc., Hopkinton, MA, USA); according to the IACUC guidelines for mice, tumor cannot reach the maximum allowable size (diameter, 1.5 cm; area, 1.8 cm2; and volume 1.8 cm3). Next, the solid tissues of the neoplasms were dissected and weighed; the tissues were fixed in 10% formalin solution and embedded with paraffin.
Tissue microarray, immunostaining, and hematoxylin and eosin staining
Tissue microarrays were commercial and purchased from the Shanghai Outdo Biotech Co., Ltd. (Shanghai, China) (HDgS-C120PT-01, HOrg-C120PG-02, HDgS-C140PT-01, HRec-Ade180Sur-01 and HCol-Ade180Sur-02); ethics approval for research using human tissue was obtained from the Ethics Committee in Taizhou Hospital, Zhejiang Province, and included a waiver for consent. Patients involved in HRec-Ade180Sur-01 underwent surgery during the period from July 2006 to August 2007, and then were followed up in August 2013. Patients involved in HCol-Ade180Sur-02 underwent surgery during the period from May 2007 to April 2008, and were followed up in September 2014. The tissues or tissue microarrays were immunolabeled using the UltraSensitive™ SP system (KIT-9710; Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) and stained with hematoxylin and eosin (H&E; C0105; Beyotime Institute of Biotechnology, Haimen, China) following the manufacturer's instructions; the dilution of primary antibodies for LRRC8A (cat. no. ab157489; Abcam, Cambridge, MA, USA) and CD31 (cat. no. ab134168; Abcam) was 1:100. Images were captured under the digital pathology whole slide scanners. Using Aperio ImageScope software (Leica Microsystems, Inc., Buffalo Grove, IL, USA), parenchyma in cancer and cancer adjacent tissues were depicted, and the intensity and distribution of the immunostaining reaction (LRRC8A) were analyzed.
Solutions and chloride current recording
The isotonic bath solution contained (in mM) the following: 70 NaCl, 0.5 MgCl2, 2 CaCl2, 10 N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid (HEPES), and 140 D-mannitol. The hypotonic bath solution (47% hypotonic, compared with the isotonic solution) was obtained by omitting the D-mannitol from the isotonic solution. The pipette solution contained (in mM) 70 N-methyl-d-glucamine chloride (NMDG-Cl), 1.2 MgCl2, 10 HEPES, 1 ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 140 D-mannitol and 2 ATP. The osmolarity of solutions was detected with an automatic cryoscopic osmometer (Osmomat 030; Gonotec, Berlin, Germany). The pH of the bath and pipette solutions was adjusted to 7.40 and 7.25, respectively.
Coverslips with cells were put in a bath chamber, and whole-cell chloride currents were recorded with 4–6 MΩ pipettes and an EPC-7 patch clamp amplifier (HEKA Electronik, Lambrecht, Germany) at 20–24°C. The membrane potential was held at the Cl− equilibrium potential (0 mV) and stepped to the 200 msec-pulses of ±80, ±40 and 0 mV in sequence and repeatedly, with a 4-sec interval between pulses. Currents were measured at 10 msec after the onset of each voltage step. Electrophysiological signals were recorded with a sampling rate of 3 kHz, filtered in 2 steps (filter 1, 10 kHz; filter 2, 3 kHz), and transferred to a computer via a 1401 interface (CED; Cambridge Electronic Design Ltd., Cambridge, UK). Data were collected and analyzed using the EPC software package (CED; Cambridge Electronic Design Ltd.).
Western blotting
HCT116 cells or tumor tissues were lysed using radio-immunoprecipitation assay (RIPA) lysis buffer. The collected supernatant was quantified using bicinchoninic acid (BCA) protein assay (cat. no. 23227; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and boiled with the loading buffer for 5 min to denature the proteins. The proteins were loaded onto sodium dodecylsulfate-polyacrylamide gel electrophoresis (12% SDS-PAGE) gels, separated and transferred onto a polyvinylidene difluoride (PVDF) membrane. The PVDF membrane was blocked with 3% non-fat dry milk for 1 h at 37°C, soaked in TBS-Tween-20 (TBST) solutions containing the primary antibodies (1:1,000 for LRRC8A; cat. no. ab157489; Abcam) overnight at 4°C, rinsed with TBS to wash the unbound primary antibodies away, and exposed to TBST solutions containing peroxidase-labeled secondary antibodies (1:5,000; cat. no. ab6721; Abcam) for 1 h at 37°C. After washing the unbound second antibodies away, the chemiluminescent method was used to detect the expression of LRRC8A (cat. no. 34079; Thermo Fisher Scientific, Inc.). Images were obtained and analyzed by the ChemiDoc™ XRS+ system with Image Lab™ software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell migration
The cell migratory potential was assessed by ex vivo wound-scratch experiments. HCT116 cells were seeded in 24-well culture plates and allowed to reach 80–90% confluence at 1 week after lentivirus infection. The monolayer cells were scratched with a 200-µl pipette tip. The cells were incubated in the medium containing 10 ng/ml epidemic (EGF; E9644; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 48 h. The images were captured under an inverted fluorescence microscope (Nikon Eclipse Ti-U) and the widths of wounds were measured by the Nikon NIS-Elements Basic Research software (both from Nikon Corp., Tokyo, Japan).
Apoptosis detection
TdT-mediated dUTP nick end labeling (TUNEL) was used to detect cell apoptosis in the neoplasm tissue. The Colorimetric TUNEL Apoptosis Assay kit (C1091; Beyotime Institute of Biotechnology) was used according to the manufacturer's protocol. The images were captured under an upright Olympus BX51 microscope with a DP71 digital camera (Olympus Corp., Tokyo, Japan).
Statistical analysis
Values are expressed as the mean ± standard error. The differences among the multiple groups were assessed by one-way analysis of variance; all pairwise multiple comparison procedures were tested by Holm-Sidak method. The differences in 2 groups were analyzed by the Mann-Whitney U test or the Student's t-test. The survival curves were plotted according to the Kaplan-Meier method and checked by the log-rank test. The comparisons of ratios in different groups were analyzed by the Chi-square test. All tests were computed using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to be indicative of statistical significance.
Results
LRRC8A proteins are widely expressed in the digestive system
VRAC is widely distributed in mammals. LRRC8A was recently found to be a main molecular determinant of VRAC (6,7), which has been confirmed to be expressed in the lymphocytes, vascular smooth muscle cells, astrocytes and several cancer cell lines. Yet, there is little knowledge concerning the expression of LRRC8A in the human digestive system. Thus, the distribution of LRRC8A was observed first.
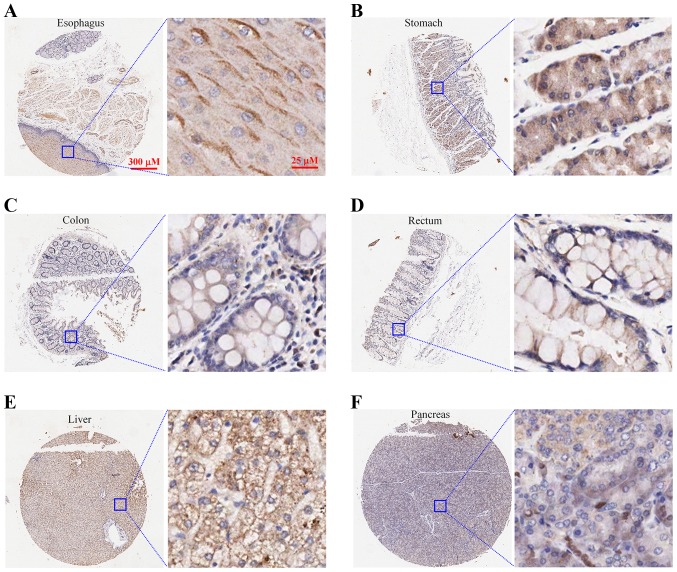
The expression of LRRC8A was immunolabeled in the normal human tissues including the esophagus (n=3), stomach (n=6), colon (n=5), rectum (n=2), liver (n=2) and pancreas (n=2). In the stratified squamous epithelium of the esophagus (Fig. 1A) and the columnar epithelium and glands of the stomach (Fig. 1B), colon (Fig. 1C), and rectum (Fig. 1D), LRRC8A proteins were abundantly expressed and primarily located on the cell membrane. In hepatocytes, most of the LRRC8A proteins were distributed on the cell membrane (Fig. 1E); in islet and acinar cells of the pancreas, LRRC8A was weakly immunolabeled and located in the cell membrane and cytoplasm (Fig. 1F).
Figure 1.
Expression of LRRC8A proteins in the main organs of the human digestive system. (A-D) LRRC8A proteins were abundantly expressed in the stratified squamous epithelium of the esophagus and the columnar epithelium and glands of the stomach, colon and rectum, which were primarily located on the cell membrane. (E and F) LRRC8A proteins were distributed in the liver and pancreas. LRRC8A, leucine-rich repeat-containing 8A.
LRRC8A may be a novel prognostic biomarker for survival in colon cancer patients
Cancer is a leading cause of death worldwide, and the most common causes of cancer-related deaths are cancers of the lung, liver, stomach, colorectum, breast and esophagus (1). To investigate the roles of LRRC8A in cancers of the digestive system, we analyzed the expression of LRRC8A in primary cancer and cancer adjacent tissues of 277 patients with cancers of the esophagus, stomach, duodenum, colon, rectum, liver and pancreas.
As illustrated in Table I, the expression of LRRC8A was significantly higher in nearly 60% of the colorectal cancer patients (51/90 colon cancer; 52/90 rectal cancer), which was significantly more than that in patients with cancer of the esophagus (12.5%, 3/24), stomach (13.6%, 3/22), duodenum (11.1%, 1/9), liver (4.8%, 1/21) and pancreas (19.0%, 4/21) (P<0.05).
Table I.
Percentages of patients with elevated expression of LRRC8A among 277 patients with cancer of the esophagus, stomach, duodenum, colon, rectum, liver and pancreas.
| Cancer tissue | No. of patients | No. of patients with higher LRRC8A expression in cancer than adjacent tissue | Percentage (%) |
|---|---|---|---|
| Esophagus | 24 | 3 | 12.5b |
| Stomach | 22 | 3 | 13.6b |
| Duodenum | 9 | 1 | 11.1a |
| Colon | 90 | 51 | 56.7 |
| Rectum | 90 | 52 | 57.8 |
| Liver | 21 | 1 | 4.8b |
| Pancreas | 21 | 4 | 19.0b |
P<0.05
P<0.01; vs. colon or rectum; Chi-square test. LRRC8A, leucine-rich repeat-containing 8A.
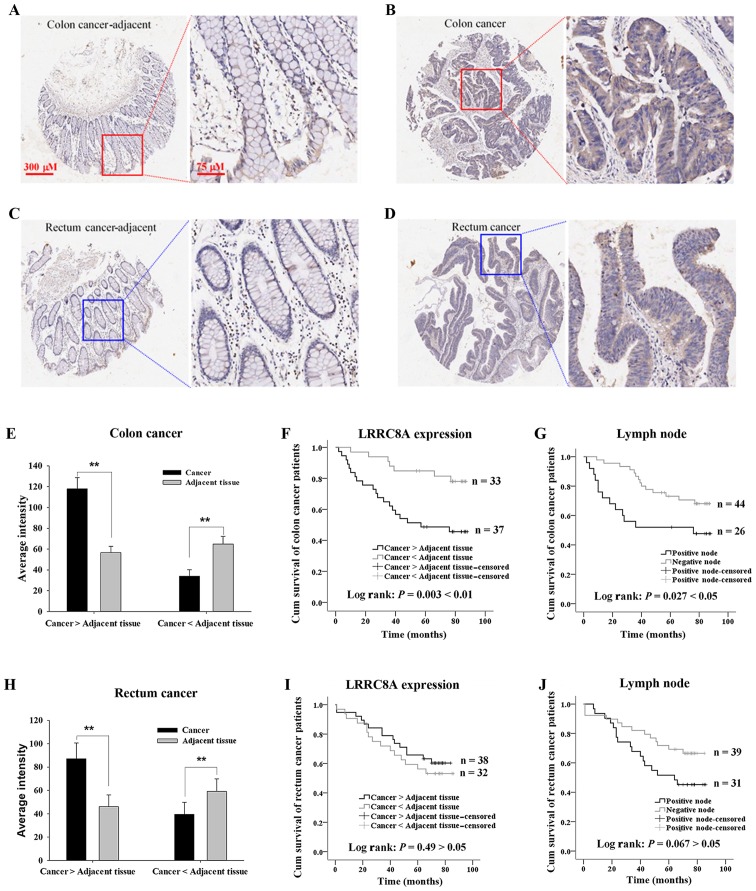
In colorectal cancer patients, LRRC8A expression in the primary cancer tissue was higher than that in adjacent tissue (Fig. 2A-D, E and H). In the cancer tissue, it was hard to distinguish the membrane from the cell body due to the compact growth of the cancer cells; however, it was obviously observed that LRRC8A proteins were distributed in the cytoplasm and on the cell membrane. We further analyzed the impact of LRRC8A expression in the cancer tissues on cancer-related survival time in patients with colon cancer (n=70) and patients with rectal cancer (n=70); there was no significant difference in the main clinical characteristics between the elevated and low expressed LRRC8A groups (Table II). As shown in Fig. 2F, the colon cancer patients with elevated expression of LRRC8A had poorer survival (P=0.003), of which the survival time (54.9±5.5 months; n=37 was significantly shorter than that of patients with the lower LRRC8A expression (77.1±3.7 months; n=33. There was no significant difference between the ages of these 2 groups: 68.0±1.9 and 64.4±1.6 years, respectively, for elevated and low LRRC8A groups (P>0.05). Conversely, there was no significant difference in survival time in rectal cancer patients between the elevated and low expressed LRRC8A groups; the mean ages of patients are 62.7±2.0 and 65.6±2.2 years, respectively (P>0.05, Fig. 2I).
Figure 2.
Effects of LRRC8A expression and metastases in lymph nodes on the survival time of colorectal cancer patients. (A and B) Expression of LRRC8A proteins in colon cancer adjacent and cancer tissues, respectively. (C and D) LRRC8A proteins in rectal cancer adjacent tissue and cancer tissue, respectively. (E and H) Average intensity of LRRC8A protein in the cancer tissue and the corresponding adjacent tissue of colorectal cancer patients with elevated and low expressed LRRC8A. (F) Colon cancer patients with elevated expression of LRRC8A in cancer tissue had poorer survival than patients with low expression of LRRC8A. (G) Colon cancer patients with metastases in the lymph nodes (node positive) had a shorter survival time than that of patients without metastases in the lymph nodes. (I and J) Graphs respectively demonstrate the influences of high expression of LRRC8A and positive lymph nodes on the survival of patients with rectal cancer. **P<0.01. LRRC8A, leucine-rich repeat-containing 8A.
Table II.
Main clinical characteristics of the colorectal cancer patients according to LRRC8A expression.
| Colon cancer | Rectal cancer | |||||
|---|---|---|---|---|---|---|
| LRRC8A expression | LRRC8A expression | |||||
| Variables | Cancer>adjacent tissue n (%) | Cancer<adjacent tissue n (%) | P-value | Cancer>adjacent tissue n (%) | Cancer<adjacent tissue n (%) | P-value |
| Sex | ||||||
| Male | 20 (54.1) | 15 (45.5) | 0.473 | 29 (76.3) | 18 (56.2) | 0.075 |
| Female | 17 (45.9) | 18 (54.5) | 9 (23.7) | 14 (43.8) | ||
| Age (years) | ||||||
| <60 | 8 (21.6) | 5 (15.2) | 0.487 | 16 (42.1) | 10 (31.2) | 0.349 |
| ≥60 | 29 (78.4) | 28 (84.8) | 22 (57.9) | 22 (68.8) | ||
| Differentiation | ||||||
| Well (I) | 6 (16.2) | 8 (24.2) | 0.687 | 3 (7.9) | 4 (12.5) | 0.594 |
| Moderate (II) | 28 (75.7) | 23 (69.7) | 23 (60.5) | 21 (65.6) | ||
| Poor (III) | 3 (8.1) | 2 (6.1) | 12 (31.6) | 7 (21.9) | ||
| T stage | ||||||
| Tis | 0 (0) | 0 (0) | 0.058 | 1 (2.6) | 0 (0) | 0.418 |
| T1 | 0 (0) | 0 (0) | 0 (0) | 1 (3.1) | ||
| T2 | 0 (0) | 5 (15.1) | 3 (7.9) | 7 (21.9) | ||
| T3 | 33 (89.2) | 22 (66.7) | 31 (81.6) | 22 (68.8) | ||
| T4a | 3 (8.1) | 4 (12.1) | 2 (5.3) | 1 (3.1) | ||
| T4b | 1 (2.7) | 2 (6.1) | 1 (2.6) | 1 (3.1) | ||
| N stage | ||||||
| N0 | 22 (59.5) | 22 (66.7) | 0.536 | 23 (60.5) | 16 (50) | 0.222 |
| N1a | 5 (13.5) | 4 (12.1) | 4 (10.5) | 7 (21.9) | ||
| N1b | 6 (16.2) | 3 (9.1) | 4 (10.5) | 7 (21.9) | ||
| N1c | 1 (2.7) | 0 (0) | 0 (0) | 0 (0) | ||
| N2a | 3 (8.1) | 2 (6.1) | 5 (13.2) | 2 (6.2) | ||
| N2b | 0 (0) | 2 (6.1) | 2 (5.3) | 0 (0) | ||
| Metastasis | ||||||
| M0 | 37 (100) | 33 (100) | 38 (100) | 32 (100) | ||
| M1 | 0 | 0 | 0 | 0 | ||
| Clinical stage | ||||||
| 0 | 0 (0) | 0 (0) | 0.150 | 1 (2.6) | 0 (0) | 0.106 |
| I | 0 (0) | 5 (15.1) | 1 (2.6) | 7 (21.9) | ||
| IIA | 19 (51.4) | 12 (36.4) | 19 (50) | 9 (28.1) | ||
| IIB | 1 (2.7) | 3 (9.1) | 1 (2.6) | 0 (0) | ||
| IIC | 2 (5.4) | 2 (6.1) | 0 (0) | 0 (0) | ||
| IIIA | 1 (2.7) | 0 (0) | 2 (5.3) | 2 (6.2) | ||
| IIIB | 13 (35.1) | 9 (27.3) | 10 (26.3) | 12 (37.5) | ||
| IIIC | 1 (2.7) | 2 (6.1) | 4 (10.5) | 2 (6.2) | ||
| IV | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
LRRC8A, leucine-rich repeat-containing 8A.
The lymph nodes were assessed for metastasis, which may help to determine cancer therapy and prognosis. The impact of metastases in lymph nodes on cancer-related survival was analyzed. The colon cancer patients with metastases in the lymph nodes (node positive) had a shorter survival time (52.6±7.3 months) than that of patients without metastases in the lymph nodes (72.2±3.6 months, P<0.05; Fig. 2G). However, there was no significant difference in survival time of the rectal cancer patients with and without metastases in lymph nodes (Fig. 2J). Notably high or low expression of LRRC8A could occur in patients with or without the positive lymph nodes. The colon cancer patients who had positive lymph nodes and high expression of LRRC8A had the highest mortality rate, which was 80% (Table III).
Table III.
Survival of colorectal cancer patients evaluated by combining of metastases in the lymph nodes and LRRC8A expression.
| Cancer | Lymph node status | LRRC8A expression | Percentage of deaths (%) | No. of deaths | No. of surviving patients | Chi-square test |
|---|---|---|---|---|---|---|
| Colon | Positive | Cancer>adjacent tissue | 80.0 | 12 | 3 | P=0.007 <0.01 |
| Cancer<adjacent tissue | 27.3 | 3 | 8 | |||
| Negative | Cancer>adjacent tissue | 36.4 | 8 | 14 | P=0.176 >0.05 | |
| Cancer<adjacent tissue | 18.2 | 4 | 18 | |||
| Rectum | Positive | Cancer>adjacent tissue | 53.3 | 8 | 7 | P=0.870 >0.05 |
| Cancer<adjacent tissue | 56.3 | 9 | 7 | |||
| Negative | Cancer>adjacent tissue | 30.4 | 7 | 16 | P=0.217 >0.05 | |
| Cancer<adjacent tissue | 50.0 | 8 | 8 |
LRRC8A, leucine-rich repeat-containing 8A.
These results suggest that LRRC8A may be used as a novel prognostic biomarker for survival in colon cancer patients.
Knockdown of LRRC8A inhibits EGF-induced migration of HCT116 cells
The previous results suggest that high expression of LRRC8A is associated with a shorter survival time in colon cancer patients who have positive lymph nodes. This implies that LRRC8A proteins may facilitate the migration of colon cancer cells. We therefore assessed the effect of LRRC8A on cell migratory potential using ex vivo wound-scratch experiments in human colon cancer HCT116 cells.
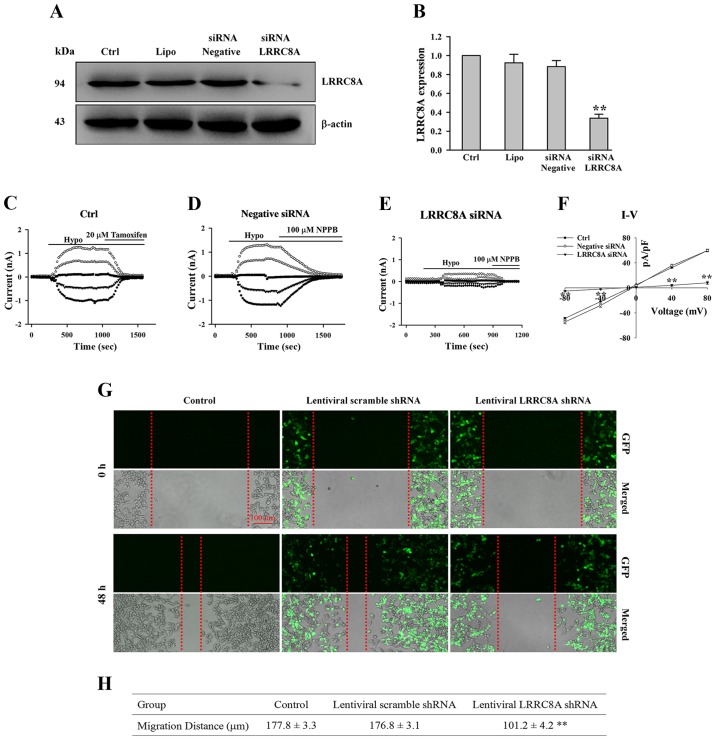
As illustrated in Fig. 3A and B, after HCT116 cells were transfected with LRRC8A siRNA for 72 h, the expression of LRRC8A protein was substantially reduced (n=3 experiments, P<0.01 vs. all control groups). A current was induced by a hypotonic solution in HCT116 cells, which could be almost inhibited by chloride channel blockers (20 µM tamoxifen and 100 µM NPPB; Fig. 3C and D). The hypotonicity-activated chloride current was clearly attenuated by the treatment of LRRC8A siRNA; mean current density in the LRRC8A siRNA group was 8.1±2.6 at + 80 mV, which is significantly lower than that in the negative siRNA group (59.7±2.3 at + 80 mV; n=6–8 cells, P<0.01, Fig. 3C-F). These data suggest that LRRC8A was the main determinant of VRAC current in HCT116 cells, which was consistent with a previous study (6), and that this specific siRNA for LRRC8A could efficiently interfere with the expression of LRRC8A protein.
Figure 3.
Downregulation of LRRC8A inhibits the EGF-induced migration of human colon cancer HCT116 cells. (A and B) Expression of LRRC8A protein was significantly decreased after HCT116 cells were transfected with LRRC8A siRNA for 72 h. (C-E) The typical time courses of hypotonicity-activated currents in non-treated (Ctrl), negative siRNA-treated, and LRRC8A siRNA-treated cells, respectively; currents induced by the hypotonic solutions could be inhibited by 20 µM tamoxifen and 100 µM NPPB chloride channel blockers, respectively. (F) The I–V relationships of chloride currents in the hypotonic solutions. (G) Images of EGF-induced cell migration captured under an inverted fluorescence microscope. The 2 upper rows are the images indicating that the wounds were scratched on the monolayer cells, referred to as 0 h. The two bottom rows present the migration of cells incubated in medium containing 10 ng/ml EGF for 48 h (referred to as 48 h; GFP, green fluorescent protein). (H) The migration distances of cells in the control, lentiviral scramble shRNA and lentiviral LRRC8A shRNA groups. n=3 experiments; **P<0.01 vs. control. LRRC8A, leucine-rich repeat-containing 8A.
According to the sequence of this specific siRNA, the LRRC8A shRNA vector was constructed and packaged as an infectious lentivirus, which was used to interfere with LRRC8A expression. One week after the lentivirus infection, wounds on the monolayer cells were scratched and observed to repair for 48 h. As shown in Fig. 3G and H, in the control and lentiviral scramble shRNA groups, the wounds were mostly recovered by cells migrating from the wound edges; the average distances of migration in the control and scramble shRNA groups were 177.8±3.3 and 176.8±3.1 µm, respectively. In the LRRC8A sRNA group, an obvious gap remained in the wound; the migration distance was 101.2±4.2 µm, significantly shorter than that in the control and scramble shRNA groups (n=3 experiments, P<0.01).
These results suggest that LRRC8A may have an important role in modulating the migration of colon cancer HCT116 cells, and that high expression of LRRC8A may facilitate the metastasis of colon cancer in patients.
Downregulation of LRRC8A inhibits the tumorigenesis of HCT116 cells in the nude mouse
Cancerous cells have strong capabilities of adaptation and proliferation, which always are required for cells to metastasize and survive in regions distant from the primary locus. Therefore, the role of LRRC8A in cancer cell growth was investigated via an in vivo method.
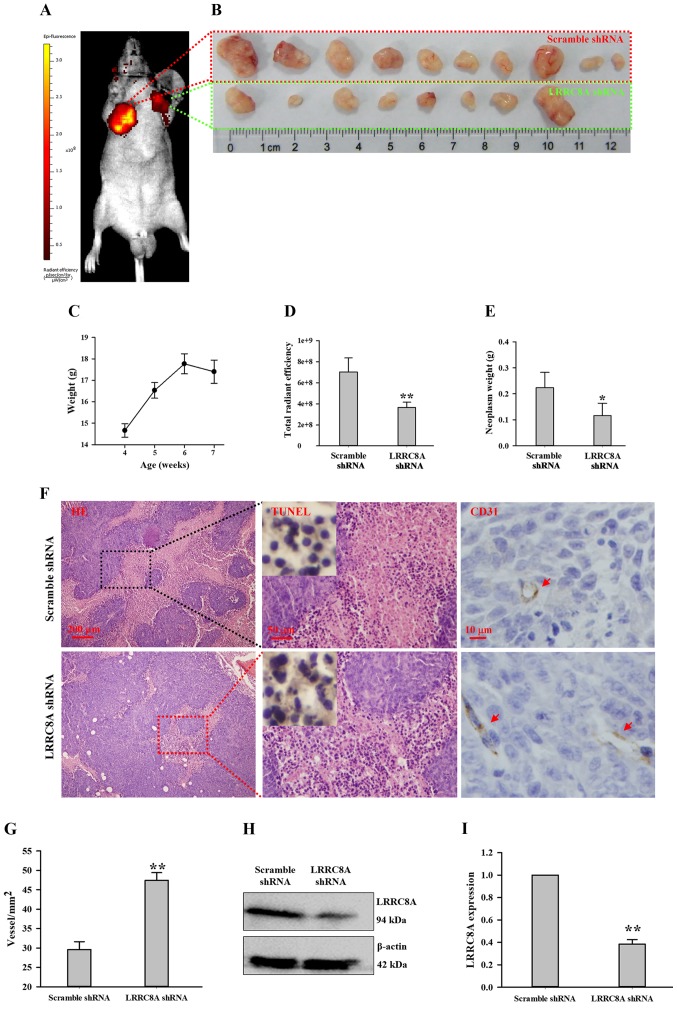
The scramble and LRRC8A shRNA lentivirus-infected HCT116 cells were subcutaneously injected at the armpits (right and left forelimb, respectively) of the nude mice. After 1 week, a very small nodule was observed at the armpit of the right forelimb; the nude mice were fed for another 2 weeks, and the nodules gradually became large. When body weights began to decrease at the third week (Fig. 4C), the total radiant efficiency of GFP indicating neoplasm growth was captured by the in vivo non-invasive small animal molecular imaging system. As illustrated in Fig. 4A and D, the intensity of radiance at the right forelimb armpit was much higher than that at the left forelimb armpit (n=10, P<0.01), suggesting downregulation of LRRC8A may inhibit HCT116 cell growth in the nude mouse. This finding was further verified by dissecting and weighing these nodules. In the scramble shRNA lentivirus group, the neoplasm always had a larger volume compared with the paired cancer tissue (Fig. 4B); the average weight of neoplasms in the scramble shRNA group was more than that in the LRRC8A shRNA group (Fig. 4E; n=8–10, P<0.05). In addition, there were two mice at whose left forelimb armpits no radiance was detected; also, there were no solid tissues to be found and dissected (Fig. 4B).
Figure 4.
Downregulation of LRRC8A inhibits the tumorigenesis of HCT116 cells in the nude mouse. (A) Images of HCT116 cell tumorigenesis captured by the in vivo non-invasive small animal molecular imaging system; the scramble and LRRC8A shRNA lentivirus-infected HCT116 cells were subcutaneously implanted at the nude mouse armpits of the right and left forelimb, respectively. (B) Sizes of cancer nodules dissected from the armpits of the right and left forelimbs after mice were fed for 3 weeks. (C) Mean body weights of mice during a consecutive 3 weeks after HCT116 cells were injected. (D and E) Radiant efficiencies and weights of cancer nodules in the scramble and LRRC8A shRNA groups, respectively. (F) Cancer nodule sections stained with H&E, TUNEL and CD31 in the scramble and LRRC8A shRNA groups; red arrows indicate the vessels. (G) Statistical analysis of vessel density. (H and I) Expression of LRRC8A protein in cancer nodules originating from the scramble or LRRC8A shRNA lentivirus-infected HCT116 cells. *P<0.05; **P<0.01. LRRC8A, leucine-rich repeat-containing 8A; H&E, hematoxylin and eosin.
The tissue slices of the neoplasms were stained with H&E. As shown in Fig. 4F, in solid tumor being formed with HCT116 cells, the organizational structures were compact and unorganized; the cell nuclei were large, atypical and deeply stained by hematoxylin (blue); the cytoplasm were lightly labeled with eosin (pink). In the scramble shRNA group, there appeared a large area of necrosis-like cells, having characteristics of karyopyknosis, karyolysis and intensely eosinophilic cytoplasm. The negative TUNEL staining suggested that most of these cells were not apoptotic cells, by which these necrotic cells were further confirmed. In the LRRC8A shRNA group, necrosis appeared, but not widely.
A wide range of necrosis occurred in the tissue slices of the scramble shRNA group; one possible mechanism may have been that the speed of angiogenesis did not meet the requirement of nutrition for HCT116 cell proliferation. In the LRRC8A shRNA group, the speed of angiogenesis may have met the requirement of nutrition for cell growth, which may have been inhibited by the downregulation of LRRC8A proteins (Fig. 4H and I). Thus, vessels in these cancer tissues were detected by CD31, a marker of endothelial cells. As illustrated in Fig. 4F and G, the vessel density of cancer tissue in the LRRC8A shRNA group was higher than that in the scramble shRNA group (P<0.01), which may relatively supply more nutrition for cells.
Discussion
The cancer burden is growing at an alarming pace, and implementation of efficient prevention strategies to curb the disease are urgently needed.
In the growth/proliferation, migration, and/or invasion of cancer cells, abnormal expression and/or activity of a number of ion channels have been confirmed to be involved, such as voltage-gated K+, Na+, Ca2+ and TRP channels and epithelial Na+/degenerin family of ion channels (15). It was proposed that ion channels may become potential targets for the diagnosis, therapy and prognosis of cancer (25–28).
It is well known that volume-regulated anion channel (VRAC) plays key roles in all types of cellular functions; however there have always been controversies concerning the molecular entity for VRAC. ClC-3, a member of the ClC superfamily of voltage-gated chloride channels, is widely expressed and hypothesized to be VRAC. As previously reported by us and others, the cell cycle-dependent expression and distribution of ClC-3 is closely related to cell proliferation, migration and apoptosis, particularly in cancerous cells (15,16,29–33). This suggests that ClC-3 may be a promising target of anticancer drugs or a novel prognostic biomarker for survival in tumor patients.
In 2003, a novel gene, leucine-rich repeat-containing 8 (LRRC8), was isolated from a girl with congenital agammaglobulinemia in peripheral blood, which may be required for B-cell development (18). One year later, the authors identified another 4 LRRC8-like genes, named TA-LRRP, AD158, LRRC5 and FLJ23420 (20). All 5 genes, designated as the LRRC8 family, encode proteins consisting of 17 extracellular leucine-rich repeats (LRRs), all of which have 4 transmembrane helices except for FLJ23420 (20,34); later, these genes were called LRRC8A-E. The LRRC8 family was nearly forgotten, and few investigations were reported in the following 10 years. The recent discovery of LRRC8A as an essential component of VRAC (6,7) profoundly motivated us to investigate the physiological and pathophysiological functions of LRRC8 proteins, including cell volume regulation, inflammation, T-cell development, small molecule transporting, neurotransmitter release and anticancer drug resistance (11,14,19,21–23,35,36). There is little knowledge concerning the biological functions of LRRC8A in cancers, particularly cancers of the digestive system. We therefore investigated the roles of LRRC8A in cancers of the digestive system.
As VRAC is widely distributed in the tissues of mammals, LRRC8A proteins were found to be widely and abundantly expressed in the tissues of the digestive system. Our previous study indicated that there were >50% of patients with gastric adenocarcinoma, colorectal adenocarcinoma, and esophageal squamous cell carcinoma who had high-expressed ClC-3 proteins (16). Among 277 cancer patients (including cancers of the esophagus, stomach, duodenum, colon, rectum, liver and pancreas), we found that patients with elevated expression of LRRC8A proteins in the primary cancer tissue >50% occurred only in colorectal cancer patients (colon cancer, 56.7%, and rectal cancer, 57.8%; Table I). It has been previously reported that reduced expression of LRRC8A contributes to the acquisition of cisplatin resistance in human ovarian and alveolar cancer cells (23,24), suggesting that patients with high LRRC8A expression may benefit from platinum drug treatment. Although colorectal patients are not usually treated with platinum drugs, it may be a clinically relevant question to investigate whether patients with high LRRC8A expression could be treated with platinum agents. Moreover, in cancers of the esophagus, stomach, duodenum, liver and pancreas, whether patients with low LRRC8A expression obtain benefits from the combined application of platinum agents and targeted upregulation of LRRC8A may be an important strategy to explore.
In cancer patients, elevated expression of ClC-3 proteins was closely associated with poor survival (16). Thus, the effects of high-expressed LRRC8A on cancer-related survival in cancer patients of the colon (n=70) and rectum (n=70) were respectively analyzed. Surprisingly, it was found that high-expressed LRRC8A was related to shorter survival time in colon cancer patients, but not in rectal cancer patients. The high-expressed LRRC8A had no relationship with survival time in rectal cancer patients, which could be explained by the following mechanisms. i) Carcinoma in the colon and rectum may originate from tumor-initiating cells with various directions of differentiation, which may decide the features of colon and rectal adenocarcinoma. ii) Difference in post-translational modification for LRRC8A may exist in colon and rectal cancer, such as phosphorylation-dephosphorylation, acetylation-deacetylation and oxidation-reduction. iii) Difference in the proteins interacting with LRRC8A may exist in colon and rectal carcinoma, which remain unclarified to help us better understand why high expression of LRRC8A has no association with survival time in rectal cancer patients. Furthermore, we observed that colon cancer patients who had positive lymph node status and high expression of LRRC8A had the highest mortality rate, ~80%. These data disclose that colon cancer patients may be prone to high expression of LRRC8A, which is related with poor survival. Therefore, we propose that LRRC8A may be used as a novel prognostic biomarker to evaluate survival in colon cancer patients. Due to the small sample size in this study, this potential prognostic biomarker may be confirmed by a multicenter study.
LRRC8A may become a potential target by which to cure or curb colon cancer in patients with high expression of LRRC8A proteins. This hypothesis was supported by our following findings. Human colon cancer HCT116 cells with downregulated LRRC8A were used to assay cell migration ex vivo and cell growth in vivo, respectively. The high expression of LRRC8A may be associated with the poorest survival in colon cancer patients who had positive lymph nodes, which implied that LRRC8A proteins may contribute to colon cancer metastasis via facilitating cell migration. The EGF-induced migration in HCT116 cells with downregulated LRRC8A proteins was significantly inhibited (Fig. 3), suggesting that LRRC8A may play an important role in cell migration. The in vivo experiment of tumorigenesis in nude mice demonstrated that HCT116 cells with downregulated LRRC8A proteins grew slowly and formed smaller tumor nodules (Fig. 4), suggesting that LRRC8A may be closely related with cell growth. LRRC8A, as one of the molecular determinants for VRAC, is responsible for cell volume regulation in HCT116 cells. During migration and proliferation, the dysfunction of cell volume regulation may be one of the possible mechanisms with which the downregulation of LRRC8A inhibits migration and growth. Although the detailed molecular mechanisms of LRRC8A in modulating cell migration and growth are poorly understood, the novel findings herein may provide a promising strategy to curb the development of colon cancer by inhibiting the function or expression of LRRC8A protein. It should be further investigated whether LRRC8A could be a novel and potential target for curing patients with colon cancer.
Although the molecular determinant for VRAC is still being debated and the biological function of LRRC8A is still not completely understood, we present an important and interesting finding: poor survival in more than half of colon cancer patients may be closely related with the high expression of LRRC8A protein, which facilitates cell migration and growth. In conclusion, LRRC8A may be used as a novel prognostic biomarker for survival and a potential target for therapy in colon cancer patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (no. 81402312) and the China Postdoctoral Science Foundation (no. 2014M552462).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
HZ and JS conceived and designed the study. ZD, DZ, HL, LZ and JN performed the experiments. HZ wrote the manuscript. WZ, RF, LF and JHY edited the manuscript and were also involved in the conception of the study. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The animal experimental protocol was approved by the Laboratory Animal Administration Committee of Xi'an Jiaotong University and was carried out in accordance with the Guidelines for Animal Experimentation of Xi'an Jiaotong University and the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication no. 85–23, revised 2011). Ethics approval for research using human tissue was obtained from the Ethics Committee in Taizhou Hospital, Zhejiang Province, and included a waiver for consent. The Outdo Biotech Co., Ltd. obtained the ethical approval from the Taizhou Hospital for research using human tissue. This institution used tumor tissues of Taizhou Hospital to make microarray; we purchased these microarrays for our research, which be approved by Ethics Committee of Health Science Center, Xi'an Jiaotong University.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing interests.
References
- 1.McGuire S. World cancer report 2014. Geneva, Switzerland: World Health Organization, International agency for research on cancer, WHO press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lang F, Busch GL, Ritter M, Völkl H, Waldegger S, Gulbins E, Häussinger D. Functional significance of cell volume regulatory mechanisms. Physiolo Rev. 1998;78:247–306. doi: 10.1152/physrev.1998.78.1.247. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol Rev. 2009;89:193–277. doi: 10.1152/physrev.00037.2007. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen SF, Hoffmann EK, Novak I. Cell volume regulation in epithelial physiology and cancer. Front Physiol. 2013;4:233. doi: 10.3389/fphys.2013.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen SF, Klausen TK, Nilius B. The identification of a volume-regulated anion channel: An amazing Odyssey. Acta Physiol (Oxf) 2015;213:868–881. doi: 10.1111/apha.12450. [DOI] [PubMed] [Google Scholar]
- 6.Voss FK, Ullrich F, Münch J, Lazarow K, Lutter D, Mah N, Andrade-Navarro MA, von Kries JP, Stauber T, Jentsch TJ. Identification of LRRC8 heteromers as an essential component of the volume-regulated anion channel VRAC. Science. 2014;344:634–638. doi: 10.1126/science.1252826. [DOI] [PubMed] [Google Scholar]
- 7.Qiu Z, Dubin AE, Mathur J, Tu B, Reddy K, Miraglia LJ, Reinhardt J, Orth AP, Patapoutian A. SWELL1, a plasma membrane protein, is an essential component of volume-regulated anion channel. Cell. 2014;157:447–458. doi: 10.1016/j.cell.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada T, Wondergem R, Morrison R, Yin VP, Strange K. Leucine-rich repeat containing protein LRRC8A is essential for swelling-activated Cl-currents and embryonic development in zebrafish. Physiological Rep. 2016;4:e12940. doi: 10.14814/phy2.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaitán-Peñas H, Gradogna A, Laparra-Cuervo L, Solsona C, Fernández-Dueñas V, Barrallo-Gimeno A, Ciruela F, Lakadamyali M, Pusch M, Estévez R. Investigation of LRRC8-mediated volume-regulated anion currents in xenopus oocytes. Biophys J. 2016;111:1429–1443. doi: 10.1016/j.bpj.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syeda R, Qiu Z, Dubin AE, Murthy SE, Florendo MN, Mason DE, Mathur J, Cahalan SM, Peters EC, Montal M, Patapoutian A. LRRC8 proteins form volume-regulated anion channels that sense ionic strength. Cell. 2016;164:499–511. doi: 10.1016/j.cell.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyzinski-Garcia MC, Rudkouskaya A, Mongin AA. LRRC8A protein is indispensable for swelling-activated and ATP-induced release of excitatory amino acids in rat astrocytes. J Physiol. 2014;592:4855–4862. doi: 10.1113/jphysiol.2014.278887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milenkovic A, Brandl C, Milenkovic VM, Jendryke T, Sirianant L, Wanitchakool P, Zimmermann S, Reiff CM, Horling F, Schrewe H, et al. Bestrophin 1 is indispensable for volume regulation in human retinal pigment epithelium cells. Proc Natl Acad Sci USA. 2015;112:E2630–E2639. doi: 10.1073/pnas.1418840112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirianant L, Wanitchakool P, Ousingsawat J, Benedetto R, Zormpa A, Cabrita I, Schreiber R, Kunzelmann K. Non-essential contribution of LRRC8A to volume regulation. Pflugers Arch. 2016;468:805–816. doi: 10.1007/s00424-016-1789-6. [DOI] [PubMed] [Google Scholar]
- 14.Okada T, Islam MR, Tsiferova NA, Okada Y, Sabirov RZ. Specific and essential but not sufficient roles of LRRC8A in the activity of volume-sensitive outwardly rectifying anion channel (VSOR) Channels. 2017;11:109–120. doi: 10.1080/19336950.2016.1247133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Li H, Yang L, Deng Z, Luo H, Ye D, Bai Z, Zhu L, Ye W, Wang L, Chen L. The ClC-3 chloride channel associated with microtubules is a target of paclitaxel in its induced-apoptosis. Sci Rep. 2013;3:2615. doi: 10.1038/srep02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu B, Jin X, Min L, Li Q, Deng L, Wu H, Lin G, Chen L, Zhang H, Li C, et al. Chloride channel-3 promotes tumor metastasis by regulating membrane ruffling and is associated with poor survival. Oncotarget. 2015;6:2434–2450. doi: 10.18632/oncotarget.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Deng Z, Yang L, Luo H, Liu S, Li Y, Wei Y, Peng S, Zhu L, Wang L, Chen L. The AQP-3 water channel is a pivotal modulator of glycerol-induced chloride channel activation in nasopharyngeal carcinoma cells. Int J Biochem Cell Biol. 2016;72:89–99. doi: 10.1016/j.biocel.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Sawada A, Takihara Y, Kim JY, Matsuda-Hashii Y, Tokimasa S, Fujisaki H, Kubota K, Endo H, Onodera T, Ohta H, et al. A congenital mutation of the novel gene LRRC8 causes agammaglobulinemia in humans. J Clin Invest. 2003;112:1707–1713. doi: 10.1172/JCI18937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zúñiga-Pflücker JC. New role identified for LRR-containing proteins in T cell development. J Exp Med. 2014;211:746–747. doi: 10.1084/jem.2115insight4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubota K, Kim JY, Sawada A, Tokimasa S, Fujisaki H, Matsuda-Hashii Y, Ozono K, Hara J. LRRC8 involved in B cell development belongs to a novel family of leucine-rich repeat proteins. FEBS Lett. 2004;564:147–152. doi: 10.1016/S0014-5793(04)00332-1. [DOI] [PubMed] [Google Scholar]
- 21.Kumar L, Chou J, Yee CS, Borzutzky A, Vollmann EH, von Andrian UH, Park SY, Hollander G, Manis JP, Poliani PL, Geha RS. Leucine-rich repeat containing 8A (LRRC8A) is essential for T lymphocyte development and function. J Exp Med. 2014;211:929–942. doi: 10.1084/jem.20131379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi H, Ettinger N, Rohrbough J, Dikalova A, Nguyen HN, Lamb FS. LRRC8A channels support TNFalpha-induced superoxide production by Nox1 which is required for receptor endocytosis. Free Radic Biol Med. 2016;101:413–423. doi: 10.1016/j.freeradbiomed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sørensen BH, Nielsen D, Thorsteinsdottir UA, Hoffmann EK, Lambert IH. Downregulation of LRRC8A protects human ovarian and alveolar carcinoma cells against Cisplatin-induced expression of p53, MDM2, p21Waf1/Cip1, and Caspase-9/-3 activation. Am J Physiol Cell Physiol. 2016;310:C857–C873. doi: 10.1152/ajpcell.00256.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sørensen BH, Dam CS, Stürup S, Lambert IH. Dual role of LRRC8A-containing transporters on cisplatin resistance in human ovarian cancer cells. J Inorg Biochem. 2016;160:287–295. doi: 10.1016/j.jinorgbio.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Xiong ZG. Ion channels as targets for cancer therapy. Int J Physiol Pathophysiol Pharmacol. 2011;3:156–166. [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser SP, Pardo LA. Ion channels: Functional expression and therapeutic potential in cancer. Colloquium on ion channels and cancer. EMBO Rep. 2008;9:512–515. doi: 10.1038/embor.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arcangeli A, Pillozzi S, Becchetti A. Targeting ion channels in leukemias: A new challenge for treatment. Curr Med Chem. 2012;19:683–696. doi: 10.2174/092986712798992093. [DOI] [PubMed] [Google Scholar]
- 28.Arcangeli A, Crociani O, Lastraioli E, Masi A, Pillozzi S, Becchetti A. Targeting ion channels in cancer: A novel frontier in antineoplastic therapy. Curr Med Chem. 2009;16:66–93. doi: 10.2174/092986709787002835. [DOI] [PubMed] [Google Scholar]
- 29.Mao J, Li X, Chen W, Xu B, Zhang H, Li H, Wang L, Jin X, Zhu J, Lin G, et al. Cell cycle-dependent subcellular distribution of ClC-3 in HeLa cells. Histochem Cell Biol. 2012;137:763–776. doi: 10.1007/s00418-012-0937-0. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Zhu L, Zuo W, Luo H, Mao J, Ye D, Li Y, Liu S, Wei Y, Ye W, et al. The ClC-3 chloride channel protein is a downstream target of cyclin D1 in nasopharyngeal carcinoma cells. Int J Biochem Cell Biol. 2013;45:672–683. doi: 10.1016/j.biocel.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Li H, Liu E, Guang Y, Yang L, Mao J, Zhu L, Chen L, Wang L. The AQP-3 water channel and the ClC-3 chloride channel coordinate the hypotonicity-induced swelling volume in nasopharyngeal carcinoma cells. Int J Biochem Cell Biol. 2014;57:96–107. doi: 10.1016/j.biocel.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Ye D, Luo H, Lai Z, Zou L, Zhu L, Mao J, Jacob T, Ye W, Wang L, Chen L. ClC-3 chloride channel proteins regulate the cell cycle by up-regulating cyclin D1-CDK4/6 through suppressing p21/p27 expression in nasopharyngeal carcinoma cells. Sci Rep. 2016;6:30276. doi: 10.1038/srep30276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang LW, Chen LX, Jacob T. ClC-3 expression in the cell cycle of nasopharyngeal carcinoma cells. Acta Physiol Sin. 2004;56:230–236. [PubMed] [Google Scholar]
- 34.Smits G, Kajava AV. LRRC8 extracellular domain is composed of 17 leucine-rich repeats. Mol Immunol. 2004;41:561–562. doi: 10.1016/j.molimm.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Lee CC, Freinkman E, Sabatini DM, Ploegh HL. The protein synthesis inhibitor blasticidin s enters mammalian cells via leucine-rich repeat-containing protein 8D. J Biol Chem. 2014;289:17124–17131. doi: 10.1074/jbc.M114.571257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Planells-Cases R, Lutter D, Guyader C, Gerhards NM, Ullrich F, Elger DA, Kucukosmanoglu A, Xu G, Voss FK, Reincke SM, et al. Subunit composition of VRAC channels determines substrate specificity and cellular resistance to Pt-based anticancer drugs. EMBO J. 2015;34:2993–3008. doi: 10.15252/embj.201592409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.