Abstract
The abnormal expression of homeobox A5 (HOXA5) has been observed in breast and colon cancer; however, the clinical significance of HOXA5 in gastric cancer (GC) is not yet clear. In this study, we found that HOXA5 expression was decreased in GC tissues at the mRNA and protein level compared with paracancerous tissues using reverse transcription-quantitative PCR (RT-qPCR) and western blot analysis, respectively. Immunohistochemistry and Kaplan-Meier survival analysis confirmed that the underexpression of HOXA5 was associated with GC progression and indicated a poor prognosis of patients with GC. Given that proliferation-related genes may be potential target genes of HOXA5, we performed a series of experiments in vitro to examine the effects of HOXA5 on the proliferation of GC cells. A CCK-8 assay, colony formation assay and flow cytometry revealed that HOXA5 inhibited the abnormal proliferation of GC cells, and this finding was further supported by a 5-ethynyl-2′-deoxyuridine (EdU) assay. Further mechanistic investigation clarified that HOXA5 promoted the protein expression of p21 and inhibited the protein expression of c-Myc and Ki67. Additionally, the use of nude mouse models also verified that HOXA5 suppressed the proliferation of GC cells in vivo. Collectively, the findings of this study demonstrate that HOXA5 acts as a tumor suppressor gene during the development and progresion of GC, possibly functioning by inhibiting the abnormal proliferation of cancer cells.
Keywords: homeobox A5, gastric cancer, prognosis, proliferation
Introduction
Gastric cancer (GC) is the second most common malignant tumor in terms of morbidity and mortality. There are 900,000 new cases of GC and 700,000-related deaths worldwide each year (1). However, no biomarkers for GC are routinely used to predict its occurrence and prognosis.
Homeobox A5 (HOXA5) is located on chromosome 7, has a total length of 2,319 bp and is a member of the HOX family. Its encoded protein, HOXA5, contains 403 amino acids. HOXA5 is a transcription factor that is involved in regulating human embryonic development and adult stem cell differentiation (2,3). The role of HOXA5 in cancer progression has been gradually clarified. It has been demonstrated that HOXA5 inhibits the activity of the Wingless (Wnt) pathway through the regulation of β-catenin inhibitory proteins and forms a negative feedback loop with Wnt signals to inhibit the stemness characteristics of colorectal cancer cells (4). Retinoic acid (RA) is a promising anticancer drug, and it has been found that the enforced overexpression of HOXA5 induces the apoptosis and differentiation of breast cancer cells. Further mechanistic research has revealed that the RA response element upstream of the HOXA5 promoter combines with RA receptor β (RARβ), thus mediating the anticancer effects of RA (5). F-actin reorganization and filopodia formation are known to promote cell movement, and HOXA5 in lung cancer cells has been shown to decrease the level of myosin-X proteins expressed on filopodia tips and to reduce the number of pseudopods, thereby preventing cancer cell metastases. Survival analysis has indicated that HOXA5 expression is associated with a better overall and disease-free survival of patients with non-small cell lung cancer expressing wild-type EGFR (6). These studies have suggested that HOXA5 may be a broad-spectrum tumor suppressor gene with the potential for wide-ranging significance and applications.
A recent study found that HOXA5 was significantly differentially methylated between GC tissues and adjacent non-cancer tissues (7). However, to the best of our knowledge, no information is available to date regarding the expression of HOXA5 and its function in human GC. Thus, in this study, in order to explore the vital role of HOXA5 in the tumorigenesis and progression of GC, the expression of HOXA5 in 81 patients with GC was detected by immunohistochemistry, and the association between HOXA5 expression and the clinicopathological characteristics of the patients with GC was analyzed. Additionally, the potential value of HOXA5 as a prognostic indicator was assessed in patients with GC. Finally, the effects of HOXA5 on cell proliferation were investigated in vitro and in vivo.
Materials and methods
Patients and specimens
A total of 81 pairs of GC tissues and matched paracancerous tissues (≥6 cm away from the tumor) for immunohistochemistry and reverse transcription-quantitative PCR (RT-qPCR) were collected from patients with GC after surgical resection at the First Affiliated Hospital of Chongqing Medical University from January to October, 2009. Due to the long storage time, only 72 in 81 pairs of tissue samples were qualified for RT-qPCR. All patients had complete pathological parameters and follow-up information, including 53 males and 28 females with an average age of 64.1 years (range, 42–83 years). TNM staging was as follows: 5 cases of stage I, 23 cases of stage II, 48 cases of stage III and 5 cases of stage IV. In total, 30 pairs of GC and matched paracancerous specimens for western blot analysis were collected from patients with GC admitted to the First Affiliated Hospital of Chongqing Medical University from July to August, 2016. The patient cohort comprised 18 male and 12 female patients with an average age of 58.3 years (range, 31–83 years). All patients underwent total or subtotal gastrectomy and did not receive radiotherapy and chemotherapy prior to surgery. The use of human tissue samples and experimental protocols were approved by the Medical Ethics Review Committee of the First Affiliated Hospital of Chongqing Medical University and written informed consent was obtained from all patients.
Cells and lentiviral transduction
The human GC cell line, SGC7901, was obtained from the Chinese Academy of Sciences Shanghai Cell Bank (Shanghai, China) and cultured in RPM-1640 (HyClone, Shanghai, China) supplemented with 10% fetal bovine serum (HyClone) in an incubator with 5% CO2 at 37°C. The overexpression vector pLV-HOXA5-GFP-puro, the control vector pLV-GFP-puro and polybrene were obtained from Hanbio Technology (Shanghai, China). The SGC7901 cells were seeded at 1×105/well in 6-well plates (1 ml/well). After 10 h, lentiviruses were added into the plates at a MOI of 40. Polybrene was added at a final concentration of 5 µg/ml to each well. The culture media were replaced after 15 h. After 72 h, the cells were examined to determine the transduction efficiency under a fluorescence microscope (Olympus, Tokyo, Japan) and puromycin (Beyotime, Shanghai, China) was then added at a final concentration of 2 µg/ml to each well. After selection for 2 weeks, puromycin was removed and the transduced cells were used in further experiments.
Antibodies and reagents
Rabbit anti-human HOXA5 (Cat. #ab82645), p21 (Cat. #ab227443) and c-Myc (Cat. #ab32072) antibodies were purchased from Abcam (Cambridge, UK). Rabbit anti-human Ki67 (Cat. #AP19895b) antibody was purchased from Abgent (San Diego, CA, USA). Rabbit anti-human GAPDH (Cat. #10494-1-AP), cyclin D1 (Cat. #12363-1-AP) and cyclin E (Cat. #11554-1-AP) antibodies and horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (Cat. #SA00001-15) were purchased from Proteintech (Wuhan, China). Immunohistochemistry-related reagents, western blot analysis-related reagents, CCK-8 reagents and DAPI were purchased from Beyotime (Shanghai, China). All reagents used in RT-qPCR were purchased from Takara (Dalian, China). The 5-ethynyl-2′-deoxyuridine (EdU) proliferation assay kit was obtained from Ribobio (Guangzhou, China). All primers used for RT-qPCR were obtained from Sangon Biotech (Shanghai, China).
RT-qPCR
RT-qPCR was used to assess the differences in HOXA5 mRNA expression levels between the GC tissues and adjacent paracancerous tissues. Total RNA was extracted using TRIzol reagent (Takara, Dalian, China) and reverse transcribed into cDNA using the reverse transcription kit from Takara. The reverse transcription conditions were as follows: 37°C, 15 min; 85°C, 5 sec. Two-step RT-qPCR was performed using a SYBR-Green assay on a CFX96 PCR machine (Bio-Rad, USA) according to the instruction. The PCR conditions were as follows: Pre-denaturation at 95°C for 30 sec; 45 cycles of denaturation at 95°C for 5 sec and annealing at 60°C for 30 sec, extension at 65°C for 1 min. The data were analyzed using the ΔΔCq method (8). The primers used are shown in Table I.
Table I.
The sequences of the PCR primers used in this study.
| Gene | Primer |
|---|---|
| GAPDH | F: 5′-CTTTGGTATCGTGGAAGGACTC-3′ |
| R: 5′-GTAGAGGCAGGGATGATGTTCT-3′ | |
| HOXA5 | F: 5′-AGCCACAAATCAAGGACACA-3′ |
| R: 5′-GCTCGCTCACGGAACTATG-3′ | |
| E-cadherin | F: 5′-TGGCTTCCCTCTTTCATCTCC-3′ |
| R: 5′-TCATAGTTCCGCTCTGTCTTTGG-3′ | |
| N-cadherin | F: 5′-CGTGAAGGTTTGCCAGTGTGA-3′ |
| R: 5′-CCTGGCGTTCTTTATCCCG-3′ | |
| Vimentin | F: 5′-TCAATGTTAAGATGGCCCTTG-3′ |
| R: 5′-TGAGTGGGTATCAACCAGAGG-3′ | |
| CD44 | F: 5′-TTACTCTGCTGCGTTGTCATTG-3′ |
| R: 5′-ACAACACCACGCCCAGAGGA-3′ | |
| EpCAM | F: 5′-CGCAGCTCAGGAAGAATGTGT-3′ |
| R: 5′-AGCCATTCATTTCTGCCTTCAT-3′ | |
| Lgr5 | F: 5′-GAGCTGCCTTCCAACCTCAG-3′ |
| R: 5′-CCCGCAAGACGTAACTCCTC-3′ | |
| CCND1 | F: 5′-ATGTTCGTGGCCTCTAAGATGA-3′ |
| R: 5′-CAGGTTCCACTTGAGCTTGTTC-3′ | |
| CCNB1 | F: 5′-AATAAGGCGAAGATCAACATGGC-3′ |
| R: 5′-TTTGTTACCAATGTCCCCAAGAG-3′ | |
| Ki67 | F: 5′-ATCGTCCCAGGTGGAAGAGTT-3′ |
| R: 5′-ATAGTAACCAGGCGTCTCGTGG-3′ | |
| MMP2 | F: 5′-GACATACATCTTTGCTGGAGAC-3′ |
| R: 5′-TTCAGGTAATAGGCACCCTT-3′ | |
| MMP7 | F: 5′-CGGATGGTAAGCAGTCTAGGG-3′ |
| R: 5′-AGGTTGGATACATCACTGCATTAG-3′ | |
| VEGFA | F: 5′-CACACAGGATGGCTTGAAGA-3′ |
| R: 5′-AGGGCAGAATCATCACGAAG-3′ | |
| VEGFR2 | F: 5′-CCAGATGACAACCAGACGGA-3′ |
| R: 5′-AGCCTTCAGATGCCACAGACTC-3′ | |
| EGFR | F: 5′-AGGCACGAGTAACAAGCTCAC-3′ |
| R: 5′-ATGAGGACATAACCAGCCACC-3′ | |
| ABCC1 | F: 5′-GTGATGGCCATGAAGACCAAGA-3′ |
| R: 5′-GCCAGCTCCCAGGCATAAAG-3′ | |
| ABCG2 | F: 5′-TCTCTTCTTCCTGACGACCAA-3′ |
| R: 5′-AAACCACACTCTGACCTGCTG-3′ | |
| Caspase-3 | F: 5′-TACCAGTGGAGGCCGACTTC-3′ |
| R: 5′-GCACAAAGCGACTGGATGAAC-3′ | |
| BCL2 | F: 5′-CTGCACCTGACGCCCTTCACC-3′ |
| R: 5′-CACATGACCCCACCGAACTCAAAGA-3′ | |
| BECN1 | F: 5′-GGCTGAGAGACTGGATCAGG-3′ |
| R: 5′-CTGCGTCTGGGCATAACG-3′ | |
| p21 | F: 5′-AGCGGAACAAGGAGTCAG-3′ |
| R: 5′-CGTTAGTGCCAGGAAAGAC-3′ | |
| p53 | F: 5′-GCGTAAACGCTTCGAGATGTT-3′ |
| R: 5′-TTTTTATGGCGGGAAGTAGACTG-3′ | |
| C-Myc | F: 5′-ACACCCGAGCAAGGACGCGA-3′ |
| R: 5′-CGCGGGAGGCTGCTGGTTTTC-3′ | |
| β-catenin | F: 5′-GAGTGCTGAAGGTGCTATCTGTC-3′ |
| R: 5′-CTGAACAAGAGTCCCAAGGAGA-3′ |
F, forward; R, reverse.
Western blot analysis
The cancer tissues and matched paracancerous tissues were lysed using RIPA lysis buffer, and the protein concentration was measured using the BCA assay after the lysates were harvested by centrifugation (12,000 rpm) at 4°C. Subsequently, 20 µg of protein samples were separated by electrophoresis on a 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes. The membranes were incubated overnight at 4°C with HOXA5 antibody (1:1,000), p21 antibody (1:1,000), p53 antibody (1:1,000), cyclin D1 antibody (1:2,000), cyclin E antibody (1:500), c-Myc antibody (1:1,000) and Ki67 antibody (1:1,000) following blocking of the non-specific binding sites for 2 h in 5% non-fat milk. After washing with TBS-T, membranes were incubated with secondary antibody (1:2,000) at 37°C for 2 h. GAPDH was used as a loading control. An enhanced chemiluminescence kit (ECL) was used for visualizing immunobands and the signal intensity of each band was measured by Fusion software (Vilber Lourmat, Paris, France) to calculate protein levels.
Immunohistochemistry
Tissues were fixed by formalin and embedded in paraffin blocks, and a series of sections (4-mm-thick) were prepared. The tissue antigens were repaired by sodium citrate buffer (pH 6.0). The sections were incubated with rabbit anti-human HOXA5 antibody at 4°C overnight after non-specific binding was blocked with goat serum. The scoring criteria were as follows: 5 fields were randomly selected from each section under a light microscope (Olympus), and the percentage of positively stained cells to total cells was calculated. The sections were scored as follows: 0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. Additionally, the staining intensity was scored as follows: 0, no staining; 1, light yellow; 2, brownish yellow; and 3, brown. The total score was the sum of staining range and intensity, and was classified as low expression (≤3 points) or high expression (≥4 points).
CCK-8 and colony-forming assays
For the CCK-8 assay, the cells were seeded at 2×103/well in 96-well plates (200 µl/well). After cell adherence, 20 µl CCK-8 reagent were added to each well on days 0, 1, 2 and 3. Following incubation at 37°C for 1.5 h, the OD value at 450 nm was measured using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA), and the growth curve was then plotted. For the colony formation assay, the cells were seeded at 500/well in 6-well plates (1 ml/well) and cultured in complete medium. The culture medium was replaced every 4 days. After 15 days, the cells were fixed with 4% paraformaldehyde and stained with crystal violet (Beyotime, Shanghai, China) at room temperature for 10 min. Images were obtained to calculate the number of clones.
Flow cytometry
At least 5×105 cells were suspended and fixed with 70% ethanol for 2 h. Propidium iodide (IP) was used to stain the cells and the cell cycle distribution was detected using a flow cytometer (BD Biosciences, San Jose, CA, USA).
5-ethynyl-2′-deoxyuridine (EdU) assay
The cells were seeded at 2×103/well in 96-well plates (200 µl/well) and were treated with 50 µM EdU for 2 h at 37°C. After washing with PBS and fixing with 4% paraformaldehyde for 30 min, the cells were permeabilized with 0.5% Triton X-100 for 5 min. The cells were then incubated with 100 µl 1× Apollo reaction cocktail for 30 min. DAPI was then used to stain the nuclei for 15 min. Images were obtained under a fluorescence microscope (Olympus) to calculate the proliferation rate.
Nude mouse models
A total of 12 nude mice aged 4 weeks old (female, weighing 13 to 16 g) were obtained from the Animal Experimental Center of Chongqing Medical University and were maintained at 24±2°C under a 40–70% humidity with free access to food and water. The experiment was divided into SGC7901-NC group and SGC7901-HOXA5 group, with 6 mice in each group. When mice were 5 weeks old, 2×106 cells suspended in 100 µl PBS were injected subcutaneously into the left flank. The tumor volume was monitored by measuring diameters and calculated as LxS2/2 (L indicates the long side and S indicates the short side) every 3 days. After 3 weeks, the nude mice were sacrificed by ether anesthesia combined with carbon dioxide asphyxiation. The flow rate of the gas was 20% of the chamber volume per minute and the death of the mice was judged according to breathing, heartbeat, nerve reflex and muscle relaxation after 5 min. The tumor tissues were stained with hematoxylin and eosin (H&E) for pathological confirmation. The animal experimental protocols were approved by the Medical Ethics Review Committee of the First Affiliated Hospital of Chongqing Medical University.
GEPIA analysis
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn), an online cancer microarray database, was used to analyze the effect of HOXA5 on the overall survival of GC patients (9). Patients were divided into high expression group and low expression group according to the median expression level of HOXA5 mRNA (Normalized by GAPDH) and the overall survival was assessed by a Kaplan-Meier survival plot.
Statistical analysis
All experiments were repeated 3 times and the data were analyzed using SPSS 19.0 software. Measurement data are expressed as the means ± standard deviation (SD). Comparisons were made between two groups using the Student's t-test. Wilcoxon signed-rank tests were used to assess the differences in HOXA5 mRNA expression levels between the GC tissues and adjacent paracancerous tissues of 72 patients. The correlation between HOXA5 protein expression and each clinicopathological parameter was examined using χ2 tests. Survival curves for different expression levels of HOXA5 were plotted according to the Kaplan-Meier method, and prognostic significance of all factors was calculated by the log-rank test. Significant parameters (P<0.05) in the univariate model were further evaluated using multivariate Cox regression, after which the independence and hazard ratio (HR) of each prognostic factor could be calculated. Differences were considered statistically significant at a P-value <0.05. The correlation between the expression of HOXA5 mRNA and 23 genes related to malignant biological behavior in 30 GC tissues was determined using Spearman's correlation analysis.
Results
Decreased expression of HOXA5 in human GC tissues
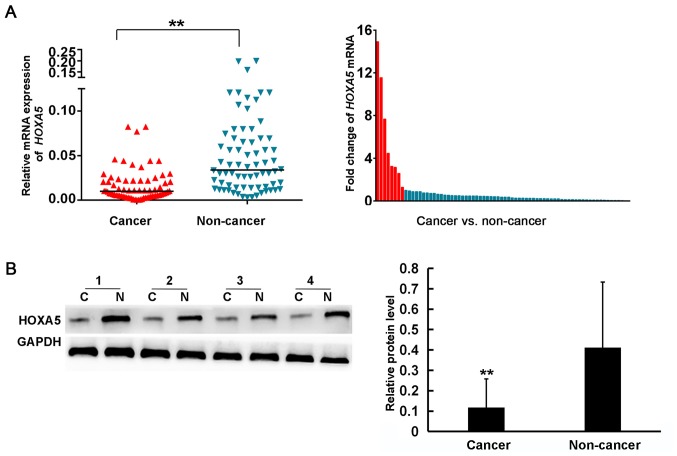
To examine the role of HOXA5 in GC, RT-qPCR was used to detect the mRNA expression of HOXA5 in 72 pairs of GC issues and adjacent paracancerous tissues. The results demonstrated that 63 patients (87.5%) exhibited a lower mRNA expression of HOXA5 in GC tissues compared to the matched non-cancer tissues (Fig. 1A). The average mRNA expression level of HOXA5 in the cancer tissues was 3.23-fold lower than that in the adjacent non-cancer tissues. Subsequently, western blot analysis was performed to determine whether HOXA5 was also downregulated at the protein level in 30 GC patients. As shown in Fig. 1B, the average protein expression level of HOXA5 in the GC tissues was 3.73-fold lower than that in the non-cancer tissues (0.11±0.14 vs. 0.41±0.32, respectively). These results suggested that HOXA5 expression was decreased in GC.
Figure 1.
Homeobox A5 (HOXA5) expression is downregulated in gastric cancer (GC) tissues. (A) The mRNA expression levels of HOXA5 in GC and adjacent non-cancer tissues of 72 patients were detected by RT-qPCR. GAPDH was used as an endogenous control (left panel), **P<0.01; The fold change is expressed as the ratio of HOXA5 mRNA expression in GC tissue to the expression of non-cancer tissues (right panel). (B) The protein expression of HOXA5 in GC tissues and adjacent non-cancer tissues was detected by western blot analysis.
HOXA5 acts as a tumor suppressor gene in GC
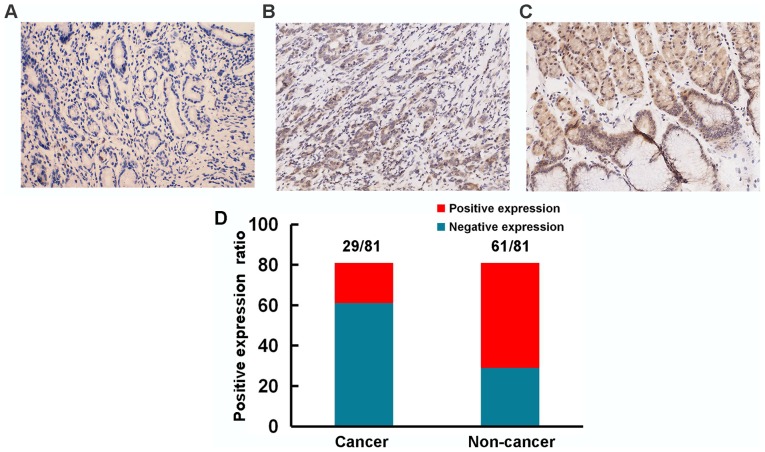
To further illuminate the role of HOXA5 in the progression of GC, we used immunohistochemistry to detect the expression of HOXA5 in samples from 81 patients with GC and analyzed the association between HOXA5 and clinical parameters. We found that HOXA5 staining was mainly located in the nucleus and cytoplasm (Fig. 2). Compared with a 75.3% positive expression of HOXA5 (61/81) in the adjacent non-cancer tissues, the positive expression in the GC tissues was only 35.8% (29/81).
Figure 2.
Representative immunohistochemical images showing the expression of homeobox A5 (HOXA5) in paraffin-embedded sections (magnification, ×200). (A) A gastric cancer sample without HOXA5 expression. (B) A gastric cancer sample with a high expression level of HOXA5. (C) A non-cancer sample with a high expression level of HOXA5. (D) The positive HOXA5 staining ratio in GC samples and adjacent non-cancer samples.
The GC specimens were then divided into a high expression group and a low expression group, according to the immunohistochemistry score for HOXA5 expression. As shown in Table II, a decreased HOXA5 expression was significantly associated with tumor size and histological grade. These results thus suggest that HOXA5 acted as a tumor suppressor gene in the progression of GC.
Table II.
Association between the expression of HOXA5 and clinical parameters of patients with gastric cancer (GC).
| HOXA5 | |||||
| Index | Case | + | − | χ2 | P-value |
|---|---|---|---|---|---|
| Sex | 0.974 | 0.324 | |||
| Male | 53 | 21 | 32 | ||
| Female | 28 | 8 | 20 | ||
| Age (years) | 0.822 | 0.365 | |||
| <60 | 31 | 13 | 18 | ||
| ≥60 | 50 | 16 | 34 | ||
| Histological grade | 7.814 | 0.005b | |||
| Well-differentiated | 39 | 20 | 19 | ||
| Poorly-differentiated | 42 | 9 | 33 | ||
| Tumor size (cm) | 10.65 | 0.001b | |||
| <5 | 39 | 21 | 18 | ||
| ≥5 | 42 | 8 | 34 | ||
| TNM stage | 2.486 | 0.115a | |||
| 1+2 | 38 | 17 | 21 | ||
| 3+4 | 43 | 12 | 31 | ||
| Lymph node involvement | 0.510 | 0.475 | |||
| No | 24 | 10 | 14 | ||
| Yes | 57 | 19 | 38 | ||
| Vascular invasion | 2.523 | 0.112 | |||
| Absent | 16 | 3 | 13 | ||
| Present | 65 | 26 | 39 | ||
| Distant metastasis | 0.008 | 0.928 | |||
| No | 78 | 28 | 50 | ||
| Yes | 3 | 1 | 2 | ||
P<0.05
P<0.01; Chi-square test.
Decreased expression of HOXA5 protein as a prognostic marker for GC
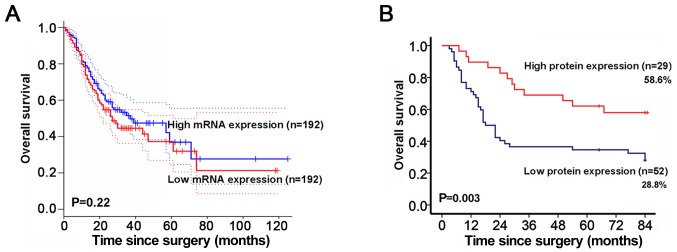
To investigate the prognostic value of HOXA5 for GC, survival analysis from GEPIA database was obtained. HOXA5 mRNA was not associated with the overall survival of patients with GC (Fig. 3A). Given that HOXA5 functions as a transcription factor, we further assessed the association between HOXA5 protein expression and survival according to the immunohistochemistry score. It is worth noting that the 75% survival duration of the patients in the low protein expression group was 10 months, which was significantly shorter than that of 30 months for the patients in the high protein expression group. Patients with a low protein expression level of HOXA5 had an overall survival rate of 28.8% in comparison to 58.6% for patients with a high protein expression level (Fig. 3B).
Figure 3.
Decreased expression of homeobox A5 (HOXA5) protein indicates a poor prognosis of patients with gastric cancer (GC). (A) The overall survival of patients with GC was compared between the patients with a high HOXA5 mRNA expression and those with a low HOXA5 mRNA expression by Kaplan-Meier survival curve analysis. (B) The overall survival of patients with GC was estimated by Kaplan-Meier survival curve analysis and compared between the patients with high HOXA5 protein expression and those with low HOXA5 protein expression using the log-rank test.
According to a univariate analysis of the prognostic factors of GC, the overall survival was found to be significantly associated with each of the following: Histological grade (P=0.031), tumor size (P<0.001), lymph node involvement (P<0.001), TNM stage (P<0.001), distant metastasis (P<0.001), and decreased HOXA5 expression (P=0.003). To investigate whether a decreased HOXA5 is an independent prognostic marker in patients with GC, a multivariate Cox regression analysis was performed. We found that tumor size, lymph node involvement, TNM stage, distant metastasis and a decreased HOXA5 expression were significantly associated with overall survival after controlling for other prognostic factors in the multivariate Cox regression analysis (Table III). These data suggested that a decreased HOXA5 expression was an independent risk factor for the prognosis of GC and could be a prognostic marker for patients with GC.
Table III.
Kaplan-Meier univariate survival analysis and multivariate Cox regression analysis of prognostic factors in gastric cancer (GC) for overall survival.
| Univariate | Multivariate analysis | |||
|---|---|---|---|---|
| Variables | P-value | HR | 95% CI | P-value |
| Sex | 0.307 | |||
| Age (years) | 0.943 | |||
| Histological grade | 0.031a | 0.683 | 0.379–1.234 | 0.206 |
| Tumor size | <0.001b | 2.697 | 1.309–5.558 | 0.007b |
| TNM stage | <0.001b | 3.083 | 1.474–6.447 | 0.003b |
| Lymph node | <0.001b | 3.787 | 1.308–10.962 | 0.014a |
| Vascular invasion | 0.284 | 0.714 | 0.334–1.526 | 0.385 |
| Distant metastasis | <0.001b | 9.263 | 2.068–41.502 | 0.004b |
| HOXA5 expression | 0.003b | 0.429 | 0.202–0.912 | 0.028a |
P<0.05
P<0.01; log rank test and cox regression.
Identification of potential target genes of HOXA5
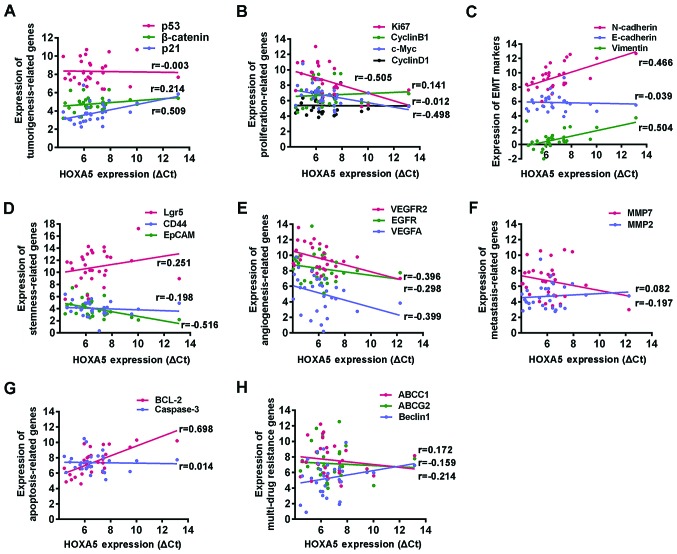
The progression of GC involves a series of cellular biological events, including the loss of control of cell proliferation, an enhanced angiogenesis, the acquisition of epithelial-mesenchymal transition (EMT) and stemness, and sustained invasion and drug resistance. Our data suggested that HOXA5 inhibited the progression of GC. In order to further explore the underlying mechanisms, we used RT-qPCR to detect the expression of the following genes in 30 GC tissues, including tumorigenesis-related genes (p21, p53 and β-catenin), proliferation-related genes (cyclin B1, cyclin D1, c-Myc and Ki67), angiogenesis-related genes [vascular endothelial growth factor (VEGF)A, VEGFR2 and epidermal growth factor (EGFR)], metastasis-related genes [matrix metalloproteinase (MMP2) and (MMP7)], apoptosis-related genes (BCL2 and caspase-3), EMT markers (vimentin, N-cadherin and E-cadherin), stemness markers (Lgr5, CD44 and EpCAM) and multi-drug resistance genes (ABCC1, ABCG2 and Beclin1) (10–15). A Spearman's correlation model was used to examine the correlation between each of these genes and HOXA5 mRNA expression. We found that HOXA5 mRNA expression positively correlated with the tumor suppressor gene, p21, and negatively correlated with the proliferation-related genes, c-Myc and Ki67, the angiogenesis genes, VEGFA and VEGAFR2, and the stemness marker, EpCAM. No correlation was observed with the other genes examined. Surprisingly, although HOXA5 exhibited a role in inhibiting the progression of GC, its mRNA expression positively correlated with the mesenchymal factors, vimentin and N-cadherin, which indicated the acquisition of EMT; it also positively correlated with the anti-apoptotic molecule, BCL2, which is known to play an important role in apoptosis resistance (Fig. 4 and Table IV).
Figure 4.
Correlation between homeobox A5 (HOXA5) and cancer malignant behavior-related genes. HOXA5 and the mRNA expression of 23 typical malignant behavior-related genes were detected using RT-qPCR in 30 GC tissues. GAPDH was used as a loading control and the relative expression levels were expressed by ΔΔCq. Spearman's correlation analysis was used to determine the correlation HOXA5 and (A) tumorigenesis-related genes, (B) proliferation-related genes, (C) EMT markers, (D) stemness markers, (E) angiogenesis-related genes, (F) metastasis-related genes, (G) apoptosis-related genes, and (H) multi-drug resistance genes.
Table IV.
Spearman's correlation analysis of the correlation between the expression of HOXA5 and malignant behavior- related genes.
| Spearman's correlation analysis | ||
|---|---|---|
| Variables | r value | P-value |
| Carcinogenesis-related genes | ||
| p21 | 0.509 | <0.01b |
| p53 | −0.003 | 0.987 |
| β-catenin | 0.214 | 0.256 |
| Proliferation-related genes | ||
| CCNB1 | 0.141 | 0.456 |
| CCND1 | −0.012 | 0.951 |
| Ki67 | −0.505 | <0.01a |
| C-Myc | −0.498 | <0.01a |
| EMT markers | ||
| E-cadherin | −0.039 | 0.836 |
| N-cadherin | 0.466 | 0.01a |
| Vimentin | 0.504 | <0.01a |
| Stemness markers | ||
| Lgr5 | 0.251 | 0.181 |
| CD44 | −0.198 | 0.295 |
| EpCAM | −0.516 | <0.01a |
| Angiogenesis-related genes | ||
| VEGFA | −0.399 | 0.029b |
| VEGFR2 | −0.396 | 0.03b |
| EGFR | −0.298 | 0.109 |
| Metastasis-related genes | ||
| MMP2 | 0.082 | 0.668 |
| MMP7 | −0.197 | 0.296 |
| Apoptosis-related genes | ||
| BCL2 | 0.698 | <0.01a |
| Caspase-3 | −0.014 | 0.941 |
| Multi-drug resistance genes | ||
| ABCC1 | −0.214 | 0.256 |
| ABCG2 | −0.159 | 0.401 |
| Beclin1 | 0.172 | 0.364 |
P<0.05
P<0.01; Spearman's correlation analysis.
Enforced overexpression of HOXA5 suppresses the aberrant proliferation of GC cells
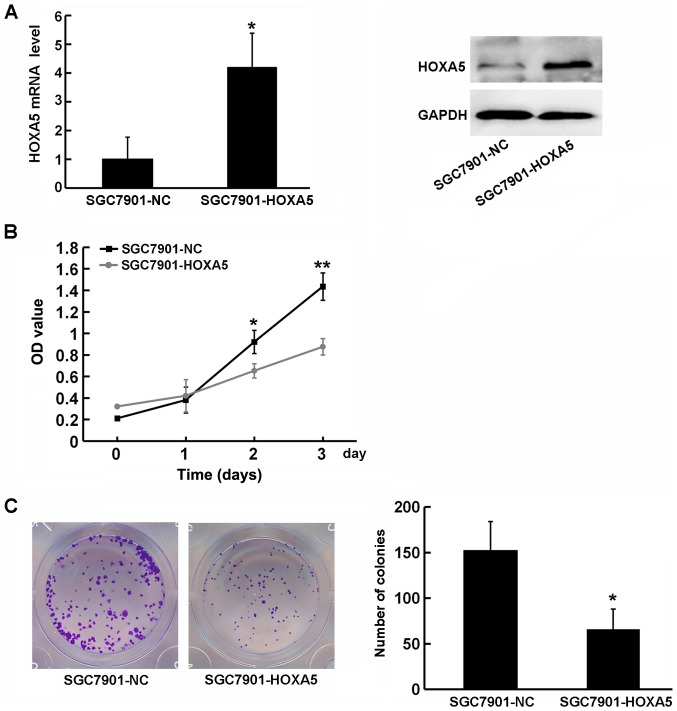
The immunohistochemistry results revealed that HOXA5 expression negatively correlated with the size of the tumor. Moreover, correlation analysis revealed that Ki67, c-Myc and p21 (these 3 genes are involved in the regulation of tumor proliferation) may be potential target genes of HOXA5. This indicated that HOXA5 may exert its anticancer effects through the negative regulation of cancer cell proliferation. To examine the function of HOXA5 in the progression of GC, we used HOXA5 overexpression vectors to transfect GC SGC7901 cells, which express a low level of HOXA5, and detected its effect on cell proliferation using a CCK-8 assay (Fig. 5A). The growth curve indicated that HOXA5-overexpressing cells (SGC7901-HOXA5) exhibited a significantly decreased proliferation compared with the negative control cells (SGC7901-NC) (Fig. 5B). Similarly, a colony formation assay revealed that HOXA5 overexpression resulted in an impairment of SGC7901 colony formation (Fig. 5C). These results indicated that the upregulation of HOXA5 in GC cells was beneficial in inhibiting cell growth, and that HOXA5 suppressed the proliferation of GC cells.
Figure 5.
Homeobox A5 (HOXA5) attenuates the aberrant proliferation of gastric cancer (GC) cells. (A) SGC7901 cells were transduced with lentiviruses containing HOXA5 overexpression plasmids. After 72 h, the overexpression efficiency was confirmed by RT-qPCR (left panel) and western blot analysis (right panel). (B) The growth curves of SGC7901-NC and SGC7901-HOXA5 cells were plotted according to the CCK-8 assay. (C) A colony formation assay was used to investigate the colon formation ability of SGC7901-NC and SGC7901-HOXA5 cells.
Enforced overexpression of HOXA5 decelerates the G1-S phase transition of GC cells
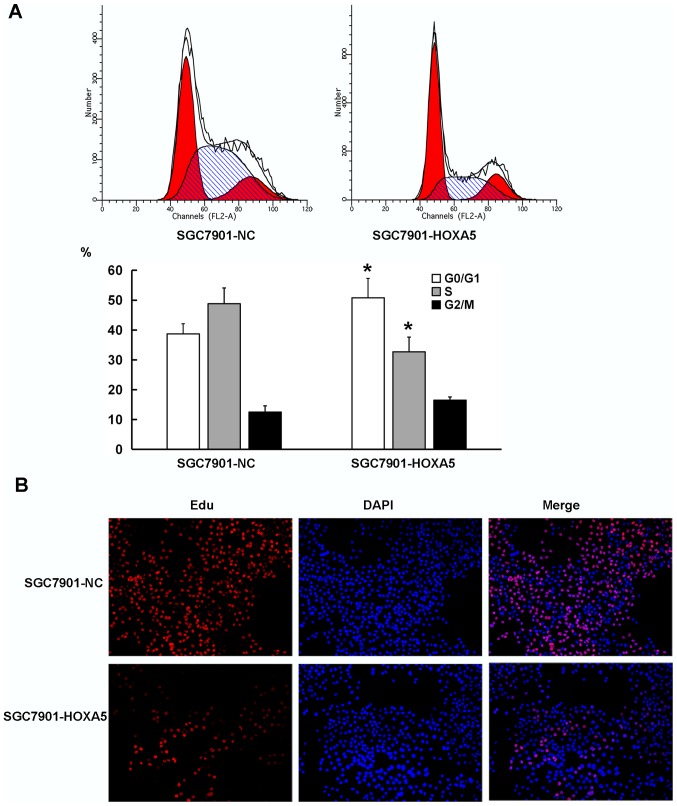
Cell proliferation depends on the proper progression of the cell cycle, particularly in the G1-S phase transition. As shown in Fig. 6A, the results from flow cytometry revealed that compared with the SGC7901-NC cells, the SGC7901-HOXA5 cells exhibited a significantly increased percentage of cells in the G0/G1 phase (38.7% vs. 50.8%), but an decreased percentage of cells in the S phase (48.6% vs. 32.7%). We then employed an EdU staining assay to detect the cell proliferation rate of the cells in the SGC7901-NC and SGC7901-HOXA5 groups. Fluorescence microscopy revealed a lower proliferative activity in the SGC7901-HOXA5 group (19.2%) than that noted in the SGC7901-NC group (55.7%), which was consistent with the flow cytometry data (Fig. 6B). These results indicated that the enforced overexpression of HOXA5 decelerated the G1-S transition of the GC cells.
Figure 6.
Effect of homeobox A5 (HOXA5) on cell cycle progression in gastric cancer (GC) cells. (A) The cell cycle distributions of SGC7901-NC and SGC7901-HOXA5 cells were evaluated by flow cytometry. Representative images of cell cycle distribution (high) and cell cycle analysis (low). (B) EdU staining was performed to determine the cell proliferation rate of SGC7901-NC and SGC7901-HOXA5 cells (magnification, ×200). The proliferation rate was calculated using EdU-stained cell (red) number compared to the total cell number (DAPI-stained, blue). *P<0.05.
HOXA5 regulates the expression of cell cycle-related proteins
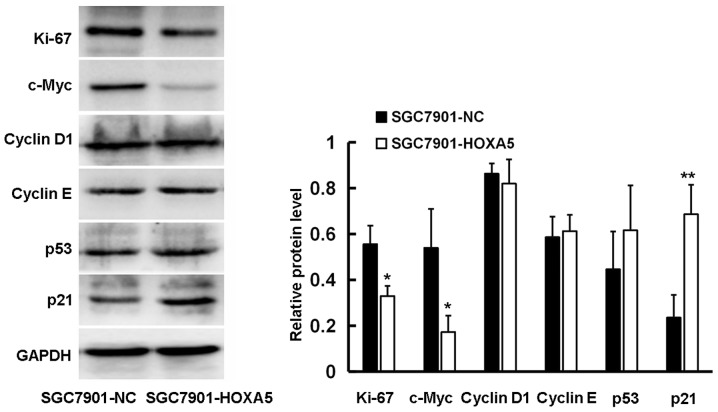
Our data suggested that HOXA5 decelerated the cell cycle, thus inhibiting the proliferation of GC cells. In order to further clarify the underlying mechanisms, we used western blot analysis to detect the effect of HOXA5 on a series of cell cycle-related proteins. We found that HOXA5 promoted the expression of p21 and inhibited the expression of c-Myc and Ki67 at the protein level, although it had no marked effect on the expression of p53, cyclin D1 and cyclin E (Fig. 7). This may be the underlying mechanism through which HOXA5 exerts its inhibitory effect on GC cell proliferation.
Figure 7.
Homeobox A5 (HOXA5) regulation of cell cycle-related proteins. The protein expression levels of p21, p53, cyclin D1, cyclin E, c-Myc and Ki67 in SGC7901-NC and SGC7901-HOXA5 cells were detected by western blot analysis. GAPDH was used as a loading control. *P<0.05, **P<0.01.
HOXA5 suppresses the proliferation and promotes the differentiation of GC cells in vivo
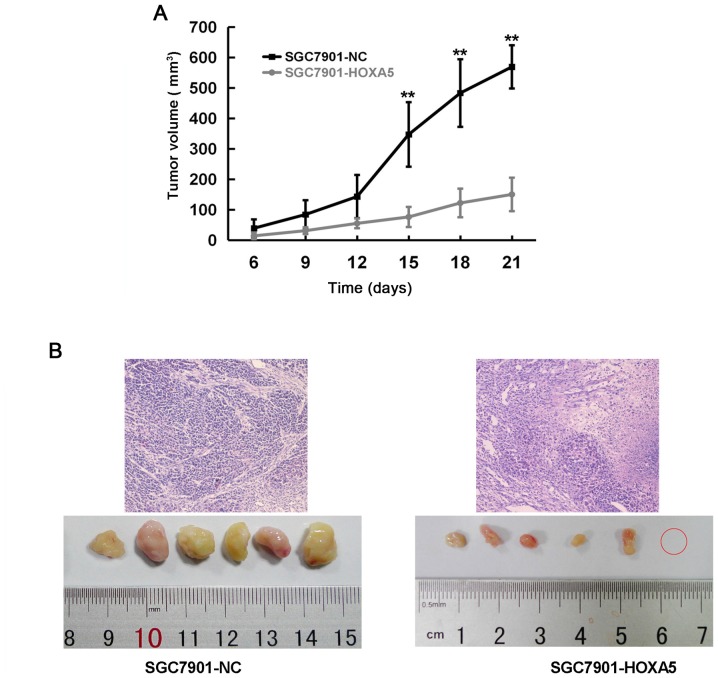
Xenograft experiments in vivo are the gold standard to determine cell proliferation. Thus, we injected SGC7901-NC or SGC7901-HOXA5 cells subcutaneously into nude mice to observe their different effects on proliferation. As shown in the tumor growth curve, the SGC7901-HOXA5 group began to display a smaller tumor volume than that of the SGC7901-NC group from day 15 (Fig. 8A). In addition, H&E staining revealed that compared with the NC group, the tumors in the SGC7901-HOXA5 group were loosely arranged, with more mesenchymal and inflammatory cell infiltration and fewer abnormal cell mitoses. The overexpression of HOXA5 promoted the tumor cells to enter a more differentiated state (Fig. 8B). These results suggested that HOXA5 suppressed the proliferation and promoted the differentiation of GC cells in vivo.
Figure 8.
In vivo assessment of homeobox A5 (HOXA5) on the proliferation of gastric cancer (GC) cells. (A) Cells (2×106) were subcutaneously injected into the left flank of nude mice, with 6 mice in each group. The tumor growth curve was plotted on the basis of the tumor volume monitored every 3 days. (B) After 3 weeks, the mice were sacrificed and the tumor tissues were stained for hematoxylin and eosin (H&E) for pathology confirmation. **P<0.01.
Discussion
In the present study, we demonstrated that HOXA5 was downregulated in GC and that the underexpression of HOXA5 was associated with the progression of GC. In addition, we also demonstrated that the underexpression of HOXA5 was an independent prognostic factor for patients with GC. Furthermore, we revealed that HOXA5 may exert its anticancer effects through the inhibition of the abnormal proliferation of GC cells.
GC is the second most common malignant tumor worldwide and is associated with a poor prognosis. Despite the existence of comprehensive treatments based on surgery and chemotherapy, the 5-year survival rate of patients with GC has not improved significantly. It is thus of great clinical significance to find an effective molecular target for the early diagnosis and treatment of GC.
HOXA5 is a member of the homeobox gene family, and it encodes the 29 kDa HOXA5 protein, which functions as a critical master regulatory factor in controlling embryonic development and adult stem cell differentiation. Recently, accumulating evidence has revealed that the human HOXA5 protein limits the aggressiveness of breast cancer and colon cancer (4,5). To clarify the associatoin between HOXA5 and GC, we detected the expression of HOXA5 using RT-qPCR and western blot analysis in 30 patients with GC and found that HOXA5 expression was significantly lower in GC tissues than that in adjacent non-cancer tissues. This result was consistent with a previous finding, in that HOXA5 was hypermethylated in GC tissues (7). A recent study reported that HOXA5 was a marker of a good prognosis in patients with colon cancer (4), although another study demonstrated that the overexpression of HOXA5 was associated with a poor prognosis in non-small cell lung cancer (16). In this study, we found that a decreased HOXA5 expression in GC tissues was associated with larger tumors that were poorly differentiated and a higher TNM stage.
Although that the mRNA level of HOXA5 was not directly associated with the prognosis of patients with GC, patients with a low HOXA5 protein expression had a significantly shorter overall survival than patients with a high expression. One possible explanation for this is that HOXA5 is a transcription factor which functions as a protein and there may be post-transcriptional modifications during the process of translation of HOXA5 mRNA. Further multivariate analysis confirmed that a decreased HOXA5 expression, together with tumor size, lymph node involvement, distant metastasis and TNM stage, were independent prognostic factors in patients with GC. Therefore, our results support the notion that a decreased HOXA5 expression promotes the progression of GC and is an indicator of a poor prognosis of patients with GC.
As is widely known, cancer progression involves a series of cellular biological events, including the loss of control of cell proliferation, angiogenesis, EMT, stemness, invasion and drug resistance (17–20). In this study, we found that the mRNA expression of HOXA5 positively correlated with the broad spectrum tumor suppressor gene, p21. Although it has been shown that HOXA5 promotes apoptosis by the transcriptional regulation of p53 in breast cancer cells expressing wild-type p53 (21), our study did not show an association of HOXA5 with p53. In addition, HOXA5 was found to negatively correlate with the proliferation-related genes, c-Myc and Ki67, the angiogenesis-related genes, VEGFA and VEGAFR2, and the stemness marker EpCAM, suggesting that HOXA5 may suppress GC progression by inhibiting abnormal proliferation, angiogenesis and the acquisition of stemness. These results are consistent with previous findings on breast cancer and colon cancer (4,5). It is worth noting that HOXA5 exhibited a significant positive correlation with the mesenchymal factors, N-cadherin and vimentin, and the anti-apoptotic molecule, BCL2. It is well known that the upregulation of mesenchymal molecules can induce cytoskeletal remodeling, polarity loss and an increased migratory capacity, which are early markers of metastasis in epithelium-derived tumors (22). However, taking into account that HOXA5 is expressed in the mesenchyme and regulates organ patterning through signal pathways such as the hedgehog (Hh) and transforming growth factor-β (TGF-β) pathways in the processes of respiratory and digestive tract development, it is not difficult to envision that HOXA5 also increases N-cadherin and vimentin expression in GC (23). Although HOXA5 may increase BCL2 expression according to our data, the association between HOXA5 and apoptosis remains to be further verified given the presence of multiple complex signaling pathways in the regulation of apoptosis. Our results suggested that HOXA5 may inhibit the proliferation, angiogenesis and the stemness of GC through mediators or signaling pathways. Despite possibly facilitating EMT, HOXA5 exerted an inhibitory effect on the progression of GC.
Previous studies have reported that HOXA5 inhibits the proliferation of breast and lung cancer cells (24,25). Moreover, in this study, the results from immunohistochemistry also indicated that HOXA5 may exert its anticancer effect through the negative regulation of cancer cell proliferation. To further confirm the role of HOXA5 in GC cell proliferation, we overexpressed HOXA5 by the transduction of lentiviral vectors in SGC7901 cells. As was suspected, the enforced overexpression of HOXA5 significantly inhibited the proliferation of GC cells, as shown by CCK-8 and colony formation assays. The results from flow cytometry and EdU assay revealed that HOXA5 overexpression induced arrested cell cycle at the G1 phase. The G1-S transition is regulated by a series of factors (26,27).
In this study, we demonstrated that HOXA5 upregulated p21 and downregulated c-Myc and Ki67, although the expression of two important G1 phase proteins, cyclin D1 and cyclin E, was not altered. The cyclin-dependent kinase inhibitor, p21, inhibits the activity of the cyclin D1-CDK4 and cyclin E-CDK2 complexes and prevents the cells from entering the S phase from the G1 phase, which is the most important pathway to block the G1-S transition (28,29). By contrast, the proliferation-related proteins, c-Myc and Ki67, contribute to the progression of the cell cycle. It is worth noting that the correlation model in tissues revealed that the correlation between these three genes and HOXA5 was unimpressive. Our function assays in cells demonstrated that the overexpression of HOXA5 can cause significant p21 upregulation and Ki67, c-Myc downregulation. One possible explanation is that HOXA5 is a type of transcription factor and can function through downstream signaling pathways (27), or direct transcriptional activation or inhibition of target genes (30) in the form of a protein. Once HOXA5 protein in the GC cells was upregulated, the downstream target genes revealed obvious changes. Therefore, we hypothesized that HOXA5 suppressed the aberrant proliferation of GC cells through the regulation of p21, c-Myc, and Ki67. This may be the mechanism through which HOXA5 exerts its anti-tumor effects on GC. Finally, nude mouse models also verified that HOXA5 suppressed the proliferation of GC cells in vivo.
In conclusion, in this study, we demonstrated that HOXA5 was a tumor suppressor gene and was decreased in GC. Its underexpression may be used as a direct prognostic indicator of a negative outcome. Our research may provide an opportunity for developing a novel therapeutic target as well as a prognostic marker in GC.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Key Clinical Specialties Construction Program of China [grant no. (2012)649].
Availability of data and materials
All data generated or analyzed during this study are included in this published article and are freely available to any researchers.
Authors' contributions
ZW conceived and designed the experiments; XP, LZ and AC performed the experiments and analyzed the data; and XP wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The use of human tissue samples and experimental protocols were approved by the Medical Ethics Review Committee of the First Affiliated Hospital of Chongqing Medical University and written informed consent was obtained from all patients. The animal experimental protocols were approved by the Medical Ethics Review Committee of the First Affiliated Hospital of Chongqing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Jiang H, Shah S, Hilt DC. Organization, sequence, and expression of the murine S100 beta gene. Transcriptional regulation by cell type-specific cis-acting regulatory elements. J Biol Chem. 1993;268:20502–20511. [PubMed] [Google Scholar]
- 3.Boucherat O, Montaron S, Bérubé-Simard FA, Aubin J, Philippidou P, Wellik DM, Dasen JS, Jeannotte L. Partial functional redundancy between Hoxa5 and Hoxb5 paralog genes during lung morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2013;304:L817–L830. doi: 10.1152/ajplung.00006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ordóñez-Morán P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 counteracts stem cell traits by inhibiting wnt signaling in colorectal cancer. Cancer Cell. 2015;28:815–829. doi: 10.1016/j.ccell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Zhang H, Lee J, Liang X, Wu X, Zhu T, Lo PK, Zhang X, Sukumar S. HOXA5 acts directly downstream of retinoic acid receptor beta and contributes to retinoic acid-induced apoptosis and growth inhibition. Cancer Res. 2007;67:8007–8013. doi: 10.1158/0008-5472.CAN-07-1405. [DOI] [PubMed] [Google Scholar]
- 6.Wang CC, Su KY, Chen HY, Chang SY, Shen CF, Hsieh CH, Hong QS, Chiang CC, Chang GC, Yu SL, Chen JJ. HOXA5 inhibits metastasis via regulating cytoskeletal remodelling and associates with prolonged survival in non-small-cell lung carcinoma. PLoS One. 2015;10:e124191. doi: 10.1371/journal.pone.0124191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh M, Liem N, Vaithilingam A, Lim PL, Sapari NS, Elahi E, Mok ZY, Cheng CL, Yan B, Pang B, et al. DNA methylation subgroups and the CpG island methylator phenotype in gastric cancer: A comprehensive profiling approach. BMC Gastroenterol. 2014;14:55. doi: 10.1186/1471-230X-14-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 9.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng QY, Hu ZX, Song XL, Pan HW. Aberrant expression of genes and proteins in pterygium and their implications in the pathogenesis. Int J Ophthalmol. 2017;10:973–981. doi: 10.18240/ijo.2017.06.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marhaba R, Klingbeil P, Nuebel T, Nazarenko I, Buechler MW, Zoeller M. CD44 and EpCAM: Cancer-initiating cell markers. Curr Mol Med. 2008;8:784–804. doi: 10.2174/156652408786733667. [DOI] [PubMed] [Google Scholar]
- 12.Simon T, Gagliano T, Giamas G. Direct effects of anti-angiogenic therapies on tumor cells: VEGF signaling. Trends Mol Med. 2017;23:282–292. doi: 10.1016/j.molmed.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Pan K, Wang P, Cao Z, Wang W, Wang S, Hu N, Xue J, Li H, Jiang W, et al. HBP1-mediated regulation of p21 protein through the Mdm2/p53 and TCF4/EZH2 pathways and its impact on cell senescence and tumorigenesis. J Biol Chem. 2016;291:12688–12705. doi: 10.1074/jbc.M116.714147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta A, Kaur CD, Jangdey M, Saraf S. Matrix metalloproteinase enzymes and their naturally derived inhibitors: Novel targets in photocarcinoma therapy. Ageing Res Rev. 2014;13:65–74. doi: 10.1016/j.arr.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Spitzwieser M, Pirker C, Koblmüller B, Pfeiler G, Hacker S, Berger W, Heffeter P, Cichna-Markl M. Promoter methylation patterns of ABCB1, ABCC1 and ABCG2 in human cancer cell lines, multidrug-resistant cell models and tumor, tumor-adjacent and tumor-distant tissues from breast cancer patients. Oncotarget. 2016;7:73347–73369. doi: 10.18632/oncotarget.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang ML, Nie FQ, Sun M, Xia R, Xie M, Lu KH, Li W. HOXA5 indicates poor prognosis and suppresses cell proliferation by regulating p21 expression in non small cell lung cancer. Tumour Biol. 2015;36:3521–3531. doi: 10.1007/s13277-014-2988-4. [DOI] [PubMed] [Google Scholar]
- 17.Ilson DH. Angiogenesis in gastric cancer: Hitting the target? Lancet. 2014;383:4–6. doi: 10.1016/S0140-6736(13)61892-9. [DOI] [PubMed] [Google Scholar]
- 18.Mikhail S, Albanese C, Pishvaian MJ. Cyclin-dependent kinase inhibitors and the treatment of gastrointestinal cancers. Am J Pathol. 2015;185:1185–1197. doi: 10.1016/j.ajpath.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okimoto RA, Breitenbuecher F, Olivas VR, Wu W, Gini B, Hofree M, Asthana S, Hrustanovic G, Flanagan J, Tulpule A, et al. Inactivation of Capicua drives cancer metastasis. Nat Genet. 2017;49:87–96. doi: 10.1038/ng.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang B, Chen Q, Cao Y, Ma X, Yin C, Jia Y, Zang A, Fan W. LGR5 is a gastric cancer stem cell marker associated with stemness and the EMT signature genes NANOG, NANOGP8, PRRX1, TWIST1, and BMI1. PLoS One. 2016;11:e168904. doi: 10.1371/journal.pone.0168904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 22.Nieto MA. Context-specific roles of EMT programmes in cancer cell dissemination. Nat Cell Biol. 2017;19:416–418. doi: 10.1038/ncb3520. [DOI] [PubMed] [Google Scholar]
- 23.Aubin J, Déry U, Lemieux M, Chailler P, Jeannotte L. Stomach regional specification requires Hoxa5-driven mesenchymal-epithelial signaling. Development. 2002;129:4075–4087. doi: 10.1242/dev.129.17.4075. [DOI] [PubMed] [Google Scholar]
- 24.Teo WW, Merino VF, Cho S, Korangath P, Liang X, Wu RC, Neumann NM, Ewald AJ, Sukumar S. HOXA5 determines cell fate transition and impedes tumor initiation and progression in breast cancer through regulation of E-cadherin and CD24. Oncogene. 2016;35:5539–5551. doi: 10.1038/onc.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Xu L, Jiang L. MiR-1271 promotes non-small-cell lung cancer cell proliferation and invasion via targeting HOXA5. Biochem Biophys Res Commun. 2015;458:714–719. doi: 10.1016/j.bbrc.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 26.Yang HW, Chung M, Kudo T, Meyer T. Competing memories of mitogen and p53 signalling control cell-cycle entry. Nature. 2017;549:404–408. doi: 10.1038/nature23880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratti S, Ramazzotti G, Faenza I, Fiume R, Mongiorgi S, Billi AM, McCubrey JA, Suh PG, Manzoli L, Cocco L, Follo MY. Nuclear inositide signaling and cell cycle. Adv Biol Regul. 2018;67:1–6. doi: 10.1016/j.jbior.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Waldman T. The inaugural use of gene editing for the study of tumor suppressor pathways in human cells-p21WAF1/CIP1. Cancer Res. 2016;76:4598–4601. doi: 10.1158/0008-5472.CAN-16-1972. [DOI] [PubMed] [Google Scholar]
- 29.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 30.Feng F, Ren Q, Wu S, Saeed M, Sun C. Hoxa5 increases mitochondrial apoptosis by inhibiting Akt/mTORC1/S6K1 pathway in mice white adipocytes. Oncotarget. 2017;8:95332–95345. doi: 10.18632/oncotarget.20521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and are freely available to any researchers.