Abstract
Gastric cancer (GC) is one of the most common malignant tumors with a high mortality rate. Reversing the multi-drug resistance (MDR) of GC offers the potential for significant enhancement of the effect of chemotherapy and improvement of prognosis. Aberrant microRNA expression can attribute to the pathogenesis of GC. However, the effects of microRNA (miR)-195-5p on the MDR of GC cells remains to be fully elucidated. In the present study, the effect of miR-195-5p in regulating the MDR of GC cells was investigated. Reverse transcription quantitative-polymerase chain reaction was used to analyze the levels of miR-195-5p in GC cells. Western blot analysis was performed to analyze the protein levels of ZNF139, P-gp, BCL-2 and MRP1. The chemosensitivity of GC cells was determined by MTT. The results showed that the expression of miR-195-5p was decreased in poorly differentiated GC tissues with a higher chemosensitivity. The overexpression of miR-195-5p promoted the chemosensitivity of GC cells. Bioinformatics analysis indicated that Zing finger 139 (ZNF139) was a target of miR-195-5p. miR-195-5p negatively regulated the expression of ZNF139 by binding to its 3′-untranslated region. The silencing of ZNF139 promoted the chemosensitivity of GC cells, and the downregulation of ZNF139 reversed the effect of miR-195-5p inhibitor on the chemosensitivity of GC cells. In conclusion, miR-195-5p regulated the MDR of GC cells via targeting ZNF139.
Keywords: microRNA-195-5p, Zing finger 139, multi-drug resistance, gastric cancer
Introduction
Gastric cancer (GC) is one of the most common malignant tumors with a high mortality rate. There are no specific clinical symptoms or signs in the early stage of GC. Therefore, the majority of cases of GC are diagnosed at moderate or advanced stages (1). The five-year overall survival rate of GC is <30% in the worldwide (2). There are several factors that influence the prognosis of GC, and its invasion and metastasis. Chemotherapy is an important method in the comprehensive treatment for GC (3). However, the presence of multi-drug resistance (MDR) of GC often leads to the failure of chemotherapy, which is a main contributor to mortality rates in patients with GC (4). Therefore, reversing the MDR of GC offers potential for significant enhancement of the effect of chemotherapy and improvement of prognosis (5). In the present study, the molecular mechanism underlying the MDR of GC, and the correlation between differentiation and MDR in GC were examined. The aim was to provide a therapeutic target for GC.
MicroRNAs are a group of small, non-coding RNAs, which are 21–23 nucleotides in length. MicroRNAs can regulate downstream gene expression as post-transcriptional regulators. Mature microRNAs can bind to the 3′-untranslated region (UTR) of target gene mRNAs and negatively regulate protein expression. It has been reported that microRNAs are involved in several biological processes, including cell proliferation, differentiation and apoptosis, and aberrant microRNA expression can attribute to the pathogenesis of GC. For example, the upregulation of microRNA (miR)-185 can promote GC cell apoptosis via regulating B-cell lymphoma 2 (Bcl-2), survivin and X-linked inhibitor of apoptosis (6). miR-320 acts as a suppressor of GC cell proliferation and metastasis via the Eph receptor A2/Wnt/b-catenin/epithelial-mesenchymal transition signal pathway (7). miR-340 and miR-124 act as tumor-suppressive factors by regulating SLIT-ROBO Rho GTPase activating protein 1 in GC cells (8). miR-155 regulates cell growth and migration by negatively regulating transforming growth factor-β receptor 2 in GC cells (9). miR-204 can negatively regulate Cyclin-dependent kinase regulatory subunit 1, Chemokine (C-X-C motif) ligand 1 and G protein-coupled receptor class C group 5 member A, and suppress the proliferation of GC cells (10). miR-195-5p belongs to the miR-15 family. It has been reported that miR-195-5p is involved in various types of cancer, including thyroid cancer, hepatocellular cancer and breast cancer (11–16). However, the effects of miR-195-5p on the MDR of GC cells remains to be elucidated. In the present study, it was determined that miR-195-5p regulated the MDR of GC cells by targeting Zing finger 139 (ZNF139).
Materials and methods
Gastric adenocarcinoma tissue specimens
A total of 12 tissue samples of gastric adenocarcinoma were collected at the Fourth Hospital of Hebei Medical University (Hebei, China) between April 2014 and 2015. The patients included six male patients and six female patients (age 38–78 years, average age 60 years). Histopathologic features of the tissue samples were determined by hematoxylin and eosin staining, and histological grades of tumor samples were assigned according to the World Health Organization classification criteria (17). All gastric adenocarcinoma tissues were divided into two groups, the well differentiated group and poorly differentiated group. Each group contained six samples. All tissue samples were stored in 4% paraform at −80°C.
Cell culture
The MKN28 GC cell line, a derivative of the MKN74 cell line (18), was used, which was from the Central Laboratory of the Fourth Hospital of Hebei Medical University. All cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Sigma; EMD Millipore, Billerica, MD, USA), 80 U/ml penicillin and 0.1 mg/ml streptomycin (Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a 5% CO2 humidified atmosphere at 37°C.
miR-195-5p mimic and inhibitor transfection
The miR-195-5p mimic and inhibitor were synthesized by GenePharm, Inc. (Sunnyvale, CA, USA). The sequences were as follows: miR-195-5p mimic, sense 5′-UAGCAGCACAGAAAUAUUGGC-3′ and miR-195-5p mimic, antisense 5′-CAAUAUUUCUGUGCUGCUAUU-3′; miR-195-5p inhibitor: 5′-GCCAAUAUUUCUGUGCUGCUA-3′. The miR-195-5p mimic and inhibitor were transfected into cells using Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. At 2 day pre-transfection, the cells were cultured in a 6-well-plate at 1.0×105 cells per well. In brief, 20 ng miRNA oligo was diluted in 50 µl Opti-MEM medium, and 2 µl Lipofectamine™ 2000 reagent was diluted in 50 µl Opti-MEM reduced serum medium. Following gentle mixing and incubation at room temperature for 5 min, the two mixtures were combined and incubated for 20 min. The mixture was added into each well and the cells were incubated at 37°C. The medium was replaced 6 h later and incubated for 44 h prior to harvesting cells.
Quantification of miR-19a levels by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from cells and tissues were isolated using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA (1 µg) was reverse-transcribed into cDNA using the EasyScript First-strand cDNA Synthesis kit (Beijing Transgen Biotech Co., Ltd., Beijing, China). The analysis of miR-195-5p levels was performed by qPCR analysis according the protocol of the SYBR-Green II kit (Takara Biotechnology Co., Ltd., Dalian, China). cDNA (1 µl), 10 µl SYBR Green mixture, 0.8 µl primers (10 µM) and 8.2 µl ddH2O were mixed. The procedures of PCR were described as follow: 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 20 sec. The relative level of miR-195-5p was normalized by U6 small nucleolar RNA, which was used as the housekeeping gene. The gene expression level was calculated using the 2−ΔΔCq method (19). All analyses were performed in triplicate. The primers were designed using Primer Premier 5 (Premier Biosoft International, Palo Alto, CA, USA. The sequences of the RT primers were as follows: miR-195-5p, 5′-TTCCGATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGTT-3′; U6, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATATG-3′. The nucleotide primers used for qPCR were as follows: miR-195-5p, forward, 5′-GCGATAGCAGCACAGAAATA-3′; U6, forward 5′-GCGCGTCGTGAAGCGTTC-3′ and universal reverse, 5′-GTGCAGGGTCCGAGGT-3′.
Western blot analysis
The total protein from tissues and cells was extracted with RIPA (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). The protein concentration was measured using a BCA kit (Thermo Fisher Scientific, Inc.). The proteins (15 µg) from each sample were separated by 10% SDS-PAGE and electrotransferred onto a polyvinylidene fluoride (PVDF) membrane (EMD Millipore). Then membranes were blocked with 5% non-fat milk in TBST at room temperature for 2 h, and then incubated with the following 1:1,000 diluted primary antibodies at 4°C for 12–18 h: ZNF139 (cat. no. ab32124; Abcam, Cambridge, UK), P-glycoprotein (P-gp; cat. no. 13978; Cell Signaling Technology, Inc., Danvers, MA, USA), Multi-drug resistance-associated protein 1 (MRP1; cat. no. 14182; Cell Signaling Technology, Inc.), Bcl-2-associated × protein (Bax; cat. no. 2772; Cell Signaling Technology, Inc.), Bcl-2 (cat. no. 3498; Cell Signaling Technology, Inc.), or GAPDH (cat. no. 5174; Cell Signaling Technology, Inc.). Following washing with TBST five times, the membranes were incubated with 1:10,000 diluted horseradish peroxidase-conjugated secondary antibody (cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature for 2 h. Following washing with TBST five times, the specific bands were detected with an enhanced chemiluminescence detection system (EMD Millipore). GAPDH was used as the housekeeping gene to normalized other genes. The quantification of protein was performed by using ImageJ version 1.42 (National Institutes of Health, Bethesda, MD, USA). The relative protein levels were calculated as the ration of treatment group to control group. The experiments were performed in triplicate.
Luciferase reporter assay
It was predicted that there was a binding site of miR-195-5p on the 3′-UTR of ZNF139 using miRNA target prediction databases, including miRanda (http://www.microrna.org/), TargetScan (http://www.targetscan.org/) and PicTar (http://www.pictar.org/). The fragment of ZNF139 3′-UTR containing the binding site of miR-195-5p was amplified from human gastric adenocarcinoma cells using the following primers: znf139, forward 5′-ATGAGCTCAGAAGAATTTGCCATCAAGCC-3′ (SacI); znf139, reverse 5′-ATTCTAGAGTTCTGATCTCTGGGATGAGGAG-3′ (Xbal). The pmiRGLO vector was purchased from Promega Corporation (Madison, WI, USA). The PCR product was digested with SacI and XbaI endonuclease. The fragment was purified and inserted into the pmiRGLO vector (pmiRGLO-ZNF139). On the day prior to transfection, the MKN28 cells were seeded into a 96-well plate at 5,000 cells/well in 100 µl of DMEM medium. The MKN28 cells were co-transfected with the pmiRGLO vector, pmiRGLO-ZNF139 and a miR-195-5p mimic (or negative control) using Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h. Luciferase activity was assessed by using the Dual-Glo Luciferase assay system (Promega Corp.). All the assays were performed with six repeats.
MTT assay
At 24 h prior to transfection, the MNK28 cells were seeded into a 96-well plate at 1×104 cells/well. At 24 h post-transfection, 20 µl MTT (Sigma; EMD Millipore) assay solution (5 mg/ml) was added into the wells for 4 h at 37°C. The supernatant medium was then discarded, and 150 µl dimethyl sulfoxide was added. The absorbance was measured at 490 nm on a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Chemosensitivity of cancer cells
The MNK28 cells (1×104 per well) were cultured in a 96-well plate. The cells were treated with 50 µM 5-fluorouracil (5-FU; Sigma; EMD Millipore) or 10 µM oxaliplatin (L-OHP; Sigma; EMD Millipore) for 24 h at 37°C. The cell viability was then determined using the MTT assay. The MNK28 cells (8×105 per well) were cultured in a 6-well plate. The miR-195-5p mimic and inhibitor were transfected into MNK28 cells for 48 h, followed by treatment with 50 µM 5-FU or 10 µM L-OHP for 24 h. The cells were harvested and total protein was extracted for western blot analysis.
Statistical analysis
Statistical analysis was performed using the SPSS 13.0 statistical software package (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard error of the mean. The non-parametric Mann-Whitney U test was used to compare two groups. One-way analysis of variance followed by Tukey's post hoc test was used to compare three or more groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-195-5p is decreased in poorly differentiated GC tissues with higher chemosensitivity
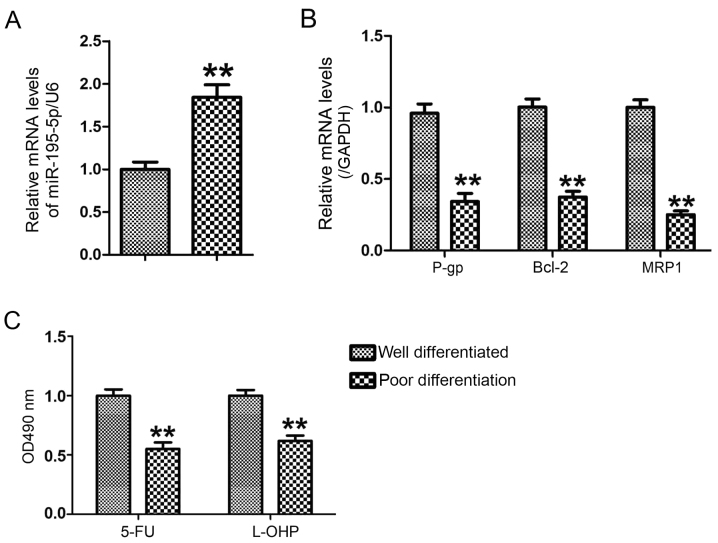
To analyze the changes of miR-195-5p between well differentiated GC tissues and poorly differentiated GC tissues, the levels of miR-195-5p in well differentiated GC tissues (n=6) and poorly differentiated GC tissues (n=6) were measured using RT-qPCR analysis. Compared with the well differentiated GC tissues, the expression of miR-195-5p was significantly upregulated in the poorly differentiated GC tissues (P<0.01; Fig. 1A), accompanied by decreased mRNA levels of P-gp, MRP1 and Bcl-2 (Fig. 1B). The GC cancer cells were separated into well differentiated GC tissues and poorly differentiated GC tissues, respectively. The results of the MTT assay showed that the poorly differentiated GC cells were more sensitive to 5-FU and L-OHP than the well differentiated GC cells (Fig. 1C). This result suggested that decreased miR-195-5p may be associated with the chemosensitivity of GC cells.
Figure 1.
Expression of miR-195-5p is decreased in poorly differentiated gastric cancer tissues with higher chemosensitivity. The levels of (A) miR-195-5p, and (B) p-gp, bcl-2 and mrp1 were analyzed by reverse transcription-quantitative polymerase chain reaction analysis. (C) Chemosensitivities of 5-FU and L-OHP were determined by MTT. **P<0.01, vs. well differentiated group. miR, microRNA; p-gp, p-glycoprotein; bcl-2, B-cell lymphoma 2; mrp1, multi-drug resistance-associated protein 1; 5-FU, 5-fluorouracil; L-OHP, oxaliplatin.
miR-195-5p regulates the MDR of GC cells
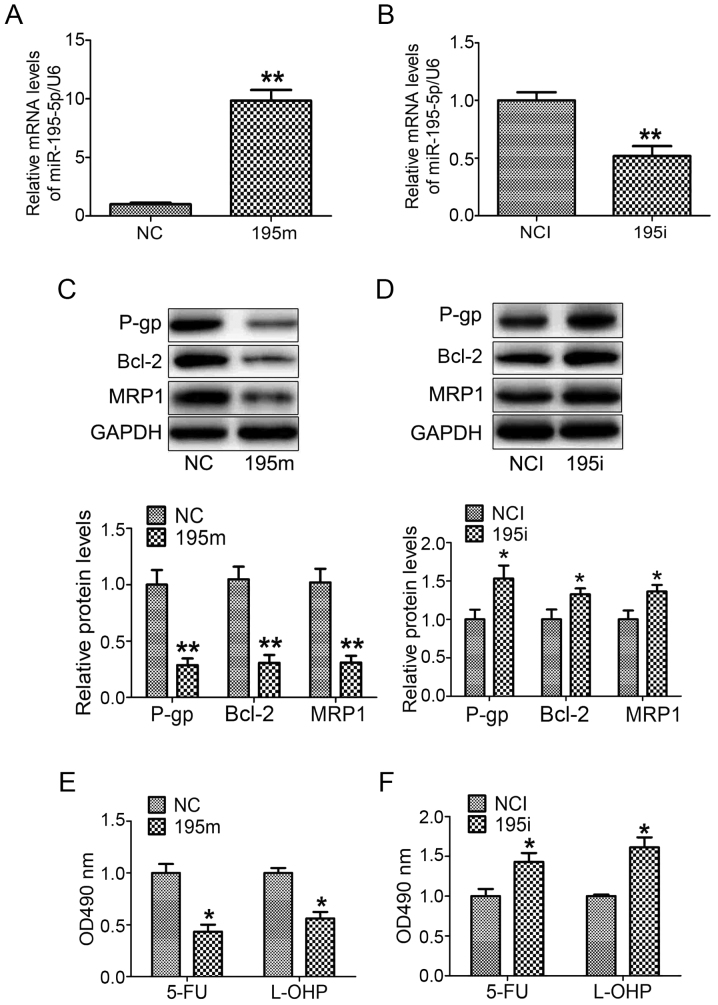
In order to further investigate the impact of miR-195-5p on the MDR of GC cells, miR-195-5p mimic and miR-195-5p inhibitor were transfected into MNK28 cells for 48 h, respectively. The expression of miR-195-5p was increased ~10-fold in the MNK28 cells transfected with miR-195-5p mimic, whereas the level of miR-195-5p was reduced by 60% in the MNK28 cells transfected with the miR-195-5p inhibitor (Fig. 2A and B). In addition, the overexpression of miR-195-5p reduced the protein levels of P-gp, MRP1 and Bcl-2 (Fig. 2C). By contrast, the downregulation of miR-195-5p induced the expression of P-gp, MRP1 and Bcl-2 (Fig. 2D). To determine the effect of miR-195-5p on the chemosensitivity of GC cells, the MKN28 cells were transfected with miR-195-5p mimic and miR-195-5p inhibitor, respectively for 48 h, followed by treatment with 5-FU and L-OHP, respectively, for 24 h. The results of the MTT assay showed that the survival rate of the miR-195-5p mimic-transfected cells was lower than that of the negative control miRNA mimic-transfected cells (Fig. 2E). The survival rate of the miR-195-5p inhibitor cell group was higher than that of the negative control miRNA inhibitor group (Fig. 2F). These results suggested that miR-195-5p regulated the MDR of MNK28 cells.
Figure 2.
miR-195-5p regulates the multi-drug resistance of gastric cancer cells. The levels of miR-195-5p were analyzed in MNK28 cells transfected with (A) miR-195-5p mimic and (B) miR-195-5p inhibitor. The protein levels of P-gp, BCL-2 and MRP1 were measured by western blot analysis in MNK28 cells transfected with (C) miR-195-5p mimic and (D) miR-195-5p inhibitor. The chemosensitivities of 5-FU and L-OHP were determined by MTT in MNK28 cells transfected with (E) miR-195-5p mimic and (F) miR-195-5p inhibitor. *P<0.05 and **P<0.01, vs. control group. miR, microRNA; P-gp, P-glycoprotein; BCL-2, B-cell lymphoma 2; MRP1, Multi-drug resistance-associated protein 1; 5-FU, 5-fluorouracil; L-OHP, oxaliplatin; NC, negative control; 195m, miR-195-5p mimic; NCI, negative control inhibitor; 195i, miR-195-5p inhibitor.
miR-195-5p negatively regulates the expression of ZNF139 by binding to its 3′-UTR
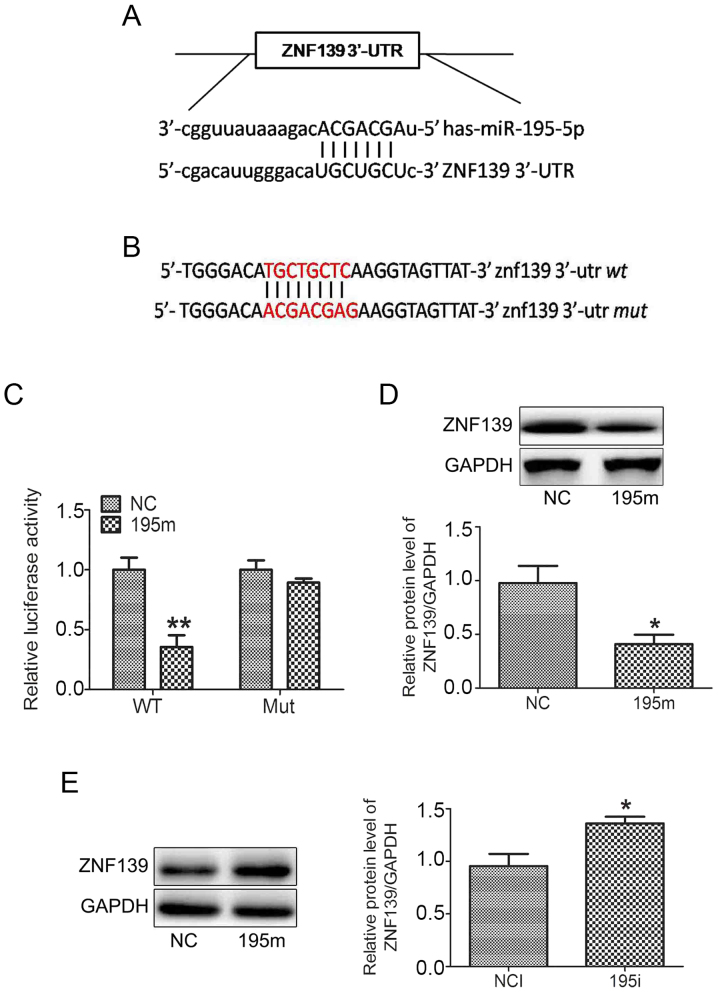
It was predicted that ZNF139 was a target gene of miR-195-5p by the MiRanda, TargetScan and PicTar miRNA databases. There was a binding site of miR-195-5p at the ZNF139 3′-UTR (Fig. 3A). The fragment of ZNF139 3′-UTR was then amplified and inserted into a pmiRLGO vector to construct a recombinant vector. The nucleotides of the binding site were mutated as shown in Fig. 3B. As shown in Fig. 3C, the miR-195-5p mimic significantly decreased the luciferase activity only in the MNK28 cells transfected with pmiRLGO vector containing the wild-type ZNF139 3′-UTR. The miR-195-5p mimic did not affect the luciferase activity in the MNK28 cells transfected with pmiRLGO vector containing the mutated ZNF139 3′-UTR. In addition, transfection with the miR-195-5p mimic decreased the protein expression of ZNF139 in MNK28 cells (Fig. 3D). By contrast, the miR-449a inhibitor upregulated the protein expression of ZNF139 (Fig. 3E). These results suggested that miR-195-5p negatively regulated the expression of ZNF139 by binding to its 3′-UTR.
Figure 3.
miR-195-5p negatively regulates the expression of ZNF139 by binding to its 3′-UTR. (A) Bioinformatics database prediction of the binding site of miR-195-5p on the ZNF139 3′-UTR. (B) Nucleotides of the binding site were mutated. (C) Luciferase assay confirmed that miR-195-5p was able to bind to the 3′-UTR of ZNF139. The protein levels of ZNF139 were determined by western blot analysis in in MNK28 cells transfected with (D) miR-195-5p mimic and (E) miR-195-5p inhibitor. *P<0.05, vs. control group. miR, microRNA; ZNF139, Zing finger 139; 3′-UTR, 3′-untranslated region; WY, wild-type; Mut, mutated; NC, negative control; 195m, miR-195-5p mimic; NCI, negative control inhibitor; 195i, miR-195-5p inhibitor.
Silencing of ZNF139 promotes the chemosensitivity of GC cells
In our previous studies, it was found that ZNF139 was involved in the progression of GC (20,21). In order to determine the effect of ZNF139 on the chemosensitivity of GC cells, three pairs of siRNAs specifically targeting ZNF139 were selected. The results of the western blot analysis showed that only siRNA-3 effectively inhibited the protein expression of ZNF139 (Fig. 4A). Silencing of ZNF139 reduced the protein levels of P-gp, MRP1 and Bcl-2 (Fig. 4B), and the chemosensitivity of MNK28 cells was enhanced via the downregulation of ZNF139 (Fig. 4C).
Figure 4.
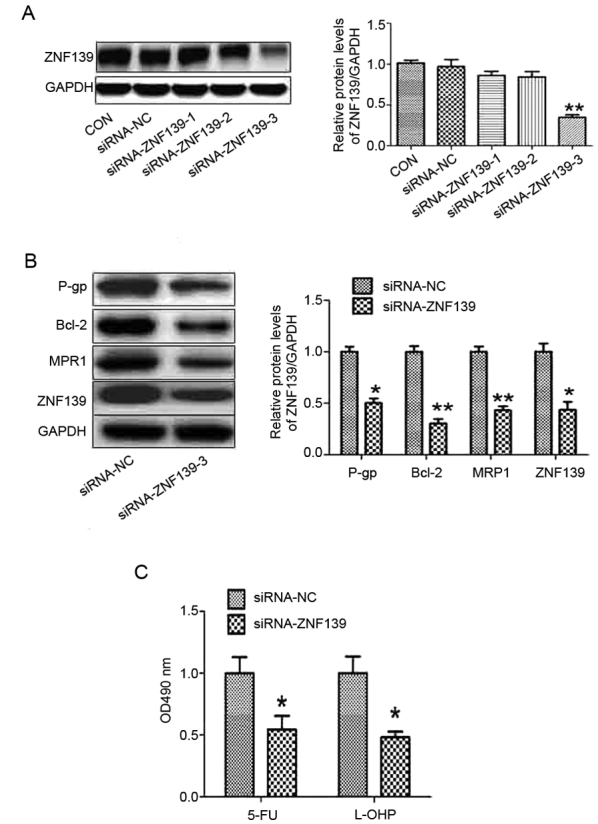
Silencing of ZNF 139 promotes the chemosensitivity of gastric cancer cells. (A) Three pairs of siRNAs targeting ZNF139 were designed. (B) Protein levels of P-gp, BCL-2, MRP1 and ZNF139 were measured using western blot analysis in MNK28 cells transfected with siRNA-ZNF139. (C) Chemosensitivities of 5-FU and L-OHP were determined by MTT in MNK28 cells transfected with siRNA-ZNF139. *P<0.05 and **P<0.01, vs. control group. miR, microRNA; ZNF139, Zing finger 139; p-gp, p-glycoprotein; bcl-2, B-cell lymphoma 2; mrp1, multi-drug resistance-associated protein 1; 5-FU, 5-fluorouracil; L-OHP, oxaliplatin; CON, control; siRNA, small interfering RNA; NC, negative control.
miR-195-5p regulates the MDR of GC cells via targeting ZNF139
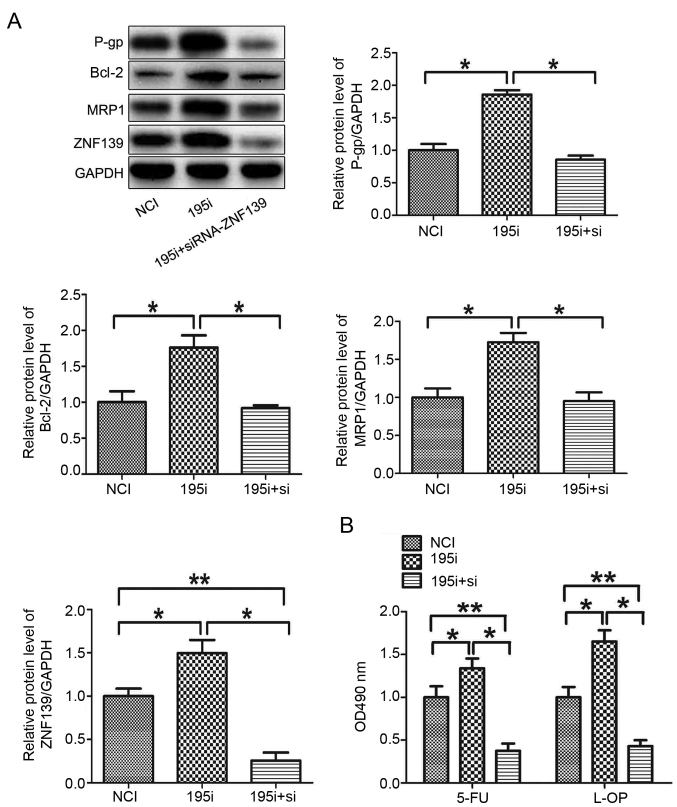
As it was suggested that ZNF139 may regulate the MDR of GC cells, in order to further determine the role of ZNF139 in the miR-195-5p inhibitor-induced MDR of MNK28 cells, siRNA targeting ZNF139 and miR-449a inhibitor was co-transfected into MNK28 cells. The silencing of ZNF139 reversed the effects of the miR-195-5p inhibitor on the protein levels of P-gp, MRP1 and Bcl-2, and the chemosensitivity of the MNK28 cells (Fig. 5A and B). These results demonstrated that miR-195-5p regulated the MDR of MNK28 cells via targeting ZNF139.
Figure 5.
miR-195-5p regulates the multi-drug resistance of gastric cancer cells via targeting ZNF139. (A) Protein levels of P-gp, BCL-2, MRP1 and ZNF139 were measured in MNK28 cells using western blot analysis. (B) Chemosensitivities of 5-FU and L-OHP were determined by MTT in MNK28 cells transfected with siRNA-ZNF139 *P<0.05 and **P<0.01. miR, microRNA; ZNF139, Zing finger 139; P-gp, P-glycoprotein; BCL-2, B-cell lymphoma 2; MRP1, multi-drug resistance-associated protein 1; NCI, negative control inhibitor; 195i, miR-195-5p inhibitor; si, small interfering RNA; 5-FU, 5-fluorouracil; L-OHP, oxaliplatin.
Discussion
GC is one of the most common malignant tumors with a high mortality rate. The majority of patients with GC are diagnosed at an advanced stage. Therefore, chemotherapy is important in the treatment of GC. The MDR of GC cells is a major factor leading to the failure of chemotherapy (22). Therefore, preventing the MDR of GC is beneficial for improving the effect of chemotherapy against GC.
It had been suggested that the degree of tumor differentiation is associated with various biological characteristics, including invasion, adhesion, proliferation, metastasis and drug resistance. Previous studies have shown that the degree of GC tumor differentiation affects their MDR. Consistent with this, the results of the present study showed that poorly differentiated GC cells were more sensitive to 5-FU and L-OHP than well differentiated GC cells. In addition, miR-195-5p was upregulated in poorly differentiated GC cells. These data suggested that miR-195-5p may regulate the MDR of GC cells.
miR-195-5p is a member of the miR-15 family, which is located on chromosome 17. Aberrant expression of miR-195-5p is associated with various types of cancer. For example, miR-195-5p serves as a tumor suppressor in renal cell carcinoma via REGγ-mediated regulation of the wnt/β-catenin pathway (23). The overexpression of miR-195-5p promotes apoptosis, and reduces cell growth, migration and invasion in oral squamous cell carcinoma via targeting tripartite motif-containing 14 (11). The results of the present study revealed that the overexpression of miR-195-5p enhanced chemotherapy sensitivity to 5-FU and L-OHP. The protein levels of P-gp, BCL-2 and MRP1 were decreased in GC cells transfected with the miR-195-5p mimic. P-gp and MRP1 are produced by the mdr1 and mrp1 genes, which regulate tumor MDR. P-gp and MRP1 are cell membrane proteins, which function as an efflux pump to transport chemotherapy drugs out of cells and lead to the MDR of cancer cells (24). BCL-2 induces the MDR of cancer cell by promoting cell proliferation and inhibiting cell apoptosis (25,26).
MicroRNAs are a group of small, non-coding RNAs, which exert their biological function by negatively regulating their target genes. In the present study, bioinformatics databases predicted that ZNF139 was a target gene of miR-195-5p. The results of the luciferase assay confirmed that miR-195-5p was able to directly bind to the 3′-UTR of ZNF139. ZNF139 is a zinc finger protein; the zinc finger protein family function as transcriptional factors to regulate gene expression, which are associated with tumorigenesis, metastasis and drug resistance (27). It has been determined that ZNF139 affects the MDR of GC cells by promoting the expression of P-gp, MRP1 and BCL-2 (20,21). In accordance, the results of the present study indicated that silencing of ZNF139 reversed the effects of miR-195-5p inhibitor on the MDR of GC cells.
In conclusion, the present study demonstrated that miR-195-5p was associated with the degree of differentiation and MDR of GC cells. The upregulation of miR-195-5p negatively regulated the protein level of ZNF139 and promoted the chemosensitivity of GC cells via affecting the protein expression of P-gp, BCL-2 and MRP1.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National Science Foundation of China (grant no. 81072033).
Availability of data and materials
The analysed datasets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HN and YL planned the experiments and wrote the manuscript; JM and JW performed the experiments and analyzed the data. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethical Review committee of the Fourth Hospital of Hebei Medical University. Written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Barra WF, Moreira FC, Pereira Cruz AM, Khayat AS, Calcagno DQ, Carneiro Dos Santos NP, Mascarenhas Junior RW, Thomaz Araújo TM, Ishak G, Demachki S, Rodríguez Burbano RM. GEJ cancers: Gastric or esophageal tumors? searching for the answer according to molecular identity. Oncotarget. 2017;8:104286–104294. doi: 10.18632/oncotarget.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham SC, Kamangar F, Kim MP, Hammoud S, Haque R, Maitra A, Montgomery E, Heitmiller RE, Choti MA, Lillemoe KD, et al. Survival after gastric adenocarcinoma resection: Eighteen-year experience at a single institutuion. J Gastrointest Surg. 2005;9:718–725. doi: 10.1016/j.gassur.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Hwang JH. Understanding gastric cancer risk factors: We need to close the gap. Gut Liver. 2018;12:1–2. doi: 10.5009/gnl17503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng PL, Zhou XY, Yi GD, Chen PF, Wang F, Dong WG. Identification of a novel gene pairs signature in the prognosis of gastric cancer. Cancer Med. 2018;7:344–350. doi: 10.1002/cam4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nienhüser H, Schmidt T. Angiogenesis and anti-angiogenic therapy in gastric cancer. Int J Mol Sci. 2017;19:E43. doi: 10.3390/ijms19010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan L, Tan B, Li Y, Zhao Q, Yuan H, Liu Y, Wang D, Zhang Z. Upregulation of miR185 promotes apoptosis of the human gastric cancer cell line MGC803. Mol Med Rep. 2018;17:3115–3122. doi: 10.3892/mmr.2017.8206. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, He Y, McLeod HL, Xie Y, Xiao D, Hu H, Chen P, Shen L, Zeng S, Yin X, et al. miR-302b inhibits tumorigenesis by targeting EphA2 via Wnt/β-catenin/EMT signaling cascade in gastric cancer. BMC Cancer. 2017;17:886. doi: 10.1186/s12885-017-3875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang T, Zhou Y, Zhang J, Wong CC, Li W, Kwan JSH, Yang R, Chan AKY, Dong Y, Wu F, et al. SRGAP1, a crucial target of miR-340 and miR-124, functions as a potential oncogene in gastric tumorigenesis. Oncogene. 2018;37:1159–117. doi: 10.1038/s41388-017-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu Y, Zhang H, Sun W, Han Y, Li S, Qu Y, Ying G, Ba Y. MiR-155 promotes gastric cancer growth and invasion by negatively regulating transforming growth factor β receptor 2. Cancer Sci. 2018;109:618–62. doi: 10.1111/cas.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrestha S, Yang CD, Hong HC, Chou CH, Tai CS, Chiew MY, Chen WL, Weng SL, Chen CC, Chang YA, et al. : Integrated MicroRNA-mRNA analysis reveals miR-204 inhibits cell proliferation in gastric cancer by targeting CKS1BCXCL1GPRC5A. Int J Mol Sci. 2017;19:E87. doi: 10.3390/ijms19010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T, Ren Y, Liu R, Ma J, Shi Y, Zhang L, Bu R. miR-195-5p suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14. Biomed Res Int. 2017;2017:7378148. doi: 10.1155/2017/7378148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu S, Jing L, Yin XR, Wang MC, Chen YM, Guo Y, Nan KJ, Han LL. MiR-195 suppresses the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by inhibiting YAP. Oncotarget. 2017;8:99757–99771. doi: 10.18632/oncotarget.20909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B, Yan F, Wu C. Overexpressed miR-195 attenuated immune escape of diffuse large B-cell lymphoma by targeting PD-L1. Biomed Pharmacother. 2018;98:95–101. doi: 10.1016/j.biopha.2017.11.146. [DOI] [PubMed] [Google Scholar]
- 14.Hong Z, Zhang R, Qi H. Diagnostic and prognostic relevance of serum miR-195 in pediatric acute myeloid leukemia. Cancer Biomark. 2018;21:269–275. doi: 10.3233/CBM-170327. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Liu J, Wang L, Yang X, Liu X. MicroRNA195 inhibits cell proliferation, migration and invasion in laryngeal squamous cell carcinoma by targeting ROCK1. Mol Med Rep. 2017;16:7154–7162. doi: 10.3892/mmr.2017.7460. [DOI] [PubMed] [Google Scholar]
- 16.Qattan A, Intabli H, Alkhayal W, Eltabache C, Tweigieri T, Amer SB. Robust expression of tumor suppressor miRNA's let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer. 2017;17:799. doi: 10.1186/s12885-017-3776-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fléjou JF. WHO classification of digestive tumors: The fourth edition. Ann Pathol. 2011;31(Suppl 5):S27–S31. doi: 10.1016/j.annpat.2011.08.001. (In French) [DOI] [PubMed] [Google Scholar]
- 18.Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, Macleod RA, Master JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Tan BB, Zhao Q, Fan LQ, Liu Y, Wang D. Regulatory mechanism of ZNF139 in multi-drug resistance of gastric cancer cells. Mol Biol Rep. 2014;41:3603–3610. doi: 10.1007/s11033-014-3224-4. [DOI] [PubMed] [Google Scholar]
- 21.Nie HF, Li Y, Li ZX, Mu JX, Wang JS. Effects of ZNF139 on gastric cancer cells and mice with gastric tumors. Oncol Lett. 2016;12:2550–2554. doi: 10.3892/ol.2016.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu W, Yang Z, Lu N. Molecular targeted therapy for the treatment of gastric cancer. J Exp Clin Cancer Res. 2016;35:1. doi: 10.1186/s13046-015-0276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Wang L, Yao X, Chen H, Xu C, Tong L, Shah A, Huang T, Chen G, Chen J, et al. miR-195-5p is critical in REGγ-mediated regulation of wnt/β-catenin pathway in renal cell carcinoma. Oncotarget. 2017;8:63986–64000. doi: 10.18632/oncotarget.19256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefan K, Schmitt SM, Wiese M. 9-Deazapurines as broad-spectrum inhibitors of the ABC transport proteins P-glycoprotein, multidrug resistance-associated protein 1, and breast cancer resistance protein. J Med Chem. 2017;60:8758–8780. doi: 10.1021/acs.jmedchem.7b00788. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Wang X, Zhao H, Liang B, Du Q. Clusterin confers resistance to TNF-alpha-induced apoptosis in breast cancer cells through NF-kappaB activation and Bcl-2 overexpression. J Chemother. 2012;24:348–357. doi: 10.1179/1973947812Y.0000000049. [DOI] [PubMed] [Google Scholar]
- 26.Tomek M, Akiyama T, Dass CR. Role of Bcl-2 in tumour cell survival and implications for pharmacotherapy. J Pharm Pharmacol. 2012;64:1695–1702. doi: 10.1111/j.2042-7158.2012.01526.x. [DOI] [PubMed] [Google Scholar]
- 27.Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53. doi: 10.1186/s12929-016-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analysed datasets generated during the present study are available from the corresponding author on reasonable request.