 Molecular dynamics has been demonstrated to be crucial for unveiling otherwise hidden binding sites especially for the undruggable target challenge.
Molecular dynamics has been demonstrated to be crucial for unveiling otherwise hidden binding sites especially for the undruggable target challenge.
Abstract
Molecular dynamics (MD) has become increasingly popular due to the development of hardware and software solutions and the improvement in algorithms, which allowed researchers to scale up calculations in order to speed them up. MD simulations are usually used to address protein folding issues or protein–ligand complex stability through energy profile analysis over time. In recent years, the development of new tools able to deeply explore a potential energy surface (PES) has allowed researchers to focus on the dynamic nature of the binding recognition process and binding-induced protein conformational changes. Moreover, modern approaches have been demonstrated to be effective and reliable in calculating some kinetic and thermodynamic parameters behind the host–guest recognition process. Starting from all of these considerations, several efforts have been made in order to integrate MD within the virtual screening process in drug discovery. Knowledge retrieved from MD can, in fact, be exploited as a starting point to build pharmacophores or docking constraints in the early stage of the screening campaign as well as to define key features, in order to unravel hidden binding modes and help the optimisation of the molecular structure of a lead compound. Based on these outcomes, researchers are nowadays using MD as an invaluable tool to discover and target previously considered undruggable binding sites, including protein–protein interactions and allosteric sites on a protein surface. As a matter of fact, the use of MD has been recognised as vital to the discovery of selective protein–protein interaction modulators. The use of a dynamic overview on how the host–guest recognition occurs and of the relative conformational modifications induced allows researchers to optimise small molecules and small peptides capable of tightly interacting within the cleft between two proteins. In this review, we aim to present the most recent applications of MD as an integrated tool to be used in the rational design of small molecules or small peptides able to modulate undruggable targets, such as allosteric sites and protein–protein interactions.
Introduction
The rational design of new selective chemical entities represents without any doubt the most important issue in medicinal chemistry, which is often referred to as rational drug design or simply rational design. In silico rational drug design is based on an early hypothesis of a desired effect due to the modulation of a specific biological target that is structurally complementary to the designed molecule. This process is then characterized by the search of molecules that are directed to a specific target whose biological role within the pathology is known.1 The main goal of a drug discovery process is the identification of a small molecule responsible for the modulation of a specific target. The hardest challenge for the medicinal chemist involved in this process is therefore the quest for the optimal affinity, selectivity, and stability of the designed molecules.2
If on the one hand the first step of rational drug design is target validation, which is intended to elucidate its real and clear involvement in the biochemical process associated with the pathology, on the other hand there is a non-negligible aspect to be considered for the target: its druggability. This term has been extensively adopted to explain, in different contexts, some properties of proteins, ligands or genes. In medicinal chemistry, it refers to the target capability to bind to a small molecule for the modulation of its activity,3 and sometimes the term druggability is substituted by the synonyms “ligandability” or “bindability”.4,5 In the past years, there has been a huge rise in interest in undruggable sites to be deeply studied in order to find a strategic way to target them effectively. In 2017, a very interesting perspective on undruggable sites was published in Nature Reviews, reporting the experts' opinions on undruggable sites of targets involved in cancer.6 In this work, Dang, Premkumar, Shokat and Soucek explain their own point of view on the definition of undruggable sites. The most curious aspect of this work is the different points of view presented by the different authors on the same issue. Dang mainly focuses on protein–protein interaction (PPI) as one of the main undruggable targets because of the lack of well-defined and deep pockets.7,8 Reddy states that the use of the term undruggable is fairly exaggerated and it would be more correct to define those sites as difficult to drug. Actually, analysing the most commonly studied undruggable sites, during those years several small molecules have been identified as binders, with several candidates reaching the clinical trials (e.g. Bcl-2 family members and transcription factors such as STAT3, MDM2 and others). Another important concept concerning undruggable sites is expressed by Shokat in the same manuscript. He points out that there are two main aspects that must be considered in order to classify a binding site as really undruggable. One is related to the chemical intractability of the target and the other one focuses on the need to have sufficient data demonstrating the clinical meaningfulness of a modulator towards that target. Laura Soucek, also contributes to the same topic expressing an interesting differentiation between those targets that have not been yet “drugged” because of structural difficulties and those difficult to drug because of not being disease-specific, for example, in normal and cancer cells. In the latter case, the intervention could produce severe side effects in normal cells expressing the same target. In recent years, several methods to assess target druggability have been created, with the most used mainly based on protein surface descriptors such as curvature and lipophilicity. These features are considered to be fundamental for the recognition and consequent binding of small molecules.9–11 In the past years, massive advancements have been completed in the basic understanding of the biological properties and biochemical role of undruggable sites, thanks especially to the increase in available structural insights provided both by X-ray crystallography and nuclear magnetic resonance (NMR). Nevertheless, working on undruggable targets is not always possible using only experimental techniques because of difficulties related to time, costs and inappropriate methods available. For example, the use of X-ray structures provides a static picture of the protein, without any kind of information about how the target can structurally evolve in the presence of a modulator. Meanwhile, NMR presents some other limitations related to the target location, size and characteristics (it is not always possible to examine the whole protein–ligand complex with NMR techniques).12 Opportunely, at the same time, computational methods reveal their usefulness, offering a valid and supportive alternative to classical experimental methods.13–17
Recently, the use of computational methods in drug discovery and development has expanded in popularity and implementation, thanks to the good and reliable results obtained. The implementation of computational methods within the drug design process, commonly called computer-aided drug design (CADD), gained its main success, in the early years of use, due to its capability to increase the hit rate of novel drug compounds when compared to the classical high-throughput screening (HTS) approach. The main application of these methods is still the rapid use of virtual screening campaigns to cut the time-to-market for the discovery of new chemical entities. For this purpose, the approach mostly used is the structure-based one, where the three-dimensional structure of the biological target, obtained through methods such as X-ray crystallography, NMR spectroscopy or homology modelling, is used to evaluate the binding capability of a small molecule library.18,19
In the past years, advances in software and hardware performance allowed researchers to adopt molecular dynamics (MD) with great success20 to address drug discovery issues such as protein–ligand interaction stability,21–24 binding kinetics25–29 and the binding process.30–32 The understanding of molecular motions is basically the main issue related to molecular recognition and represents the evolution of the old idea of “Lock-and-key model” where a frozen receptor can house a small molecule without mutating its conformation.33 The dynamic nature of the receptor has been, in fact, largely demonstrated and conformational changes have been related to ligand binding.34,35 This dynamicity of protein conformation, which is related to the binding process of small molecules, has represented the key to unveiling some strategies to target undruggable sites.
In light of these considerations, different approaches have been developed based on MD with the aim of exploring protein flexibility and discovering otherwise accessible hidden binding sites. One of the first methods to unveil undruggable sites through classical MD simulations was published by Seco et al.36 In their work, an explicitly restrained MD simulation was applied in order to evaluate the binding propensities of a probe interacting with the protein surface. Starting from the molecular trajectory, free energy calculations were performed to assess the molecular recognition process.37 More recently, a similar approach has been adopted by Bakan et al.38 to demonstrate that the approach of small molecules to proteins produces global and local structural rearrangements of the protein that represent the starting point for increasing target druggability.38 Nevertheless, some computational techniques adopted for such kind of study do not always reproduce the native milieu in which proteins are normally plunged into the cells. It is actually important to maintain the maximum reliable conditions in order to avoid protein denaturation or wrong folding changes.39 Indeed, the dynamic nature of proteins is particularly crucial for some targets exhibiting active, inactive and intermediate configuration alternation (e.g. GPCRs) and it should be taken into consideration when evaluating the binding mode of small molecules, as demonstrated by Ferruz et al. in their recent publication.40 In their work, the authors used a combination of MD and Markov state models (MSMs) to analyse the binding mode and interaction pattern of a dopamine D3 receptor antagonist (PF-4363467). The use of aggregated MD through MSMs allowed researchers to unveil an otherwise hidden cryptic pocket, created by the positional shift of the highly conserved residue F3466.52. The discovery of this cryptic pocket and the pose of the ligand observed by researchers could not be observed using a canonical static docking approach.
The aim of this review is to present how MD applications have been used in recent years to treat undruggable sites, in order to unravel targetable pockets using small molecules. Particularly, we have focused on the application of MD to two specific undruggable sites of main interest for medicinal chemists: allosteric sites and protein–protein interactions (PPI). The next sections of this review will be dedicated to each of these two issues. In each part of the work, we will present the different approaches adopted by researchers in order to deepen the exploration of binding sites and the design of selective modulators.
Allosteric sites
Protein functions are strictly correlated to their flexibility and conformational transition causing cavity shape modifications and exposure. This dynamic process is fundamental for the recognition of chemical or biological guests useful to a particular biological process. As a matter of fact, interest in biochemistry has always been focused on the conformational changes of proteins related to their biological role with specific regard to the possible cooperative functionalisation between different positions on the protein surface. Such a phenomenon is known as allostery and typically occurs when the protein binding process to a guest molecule transmits some conformational changes to some other different proximal or distal sites on the protein surface.41,42 Some recent observations unveiled that allostery can be facilitated by dynamic and intrinsically disordered proteins, offering new insights into the understanding of allosteric functional regulation.43 The structural rearrangements associated with protein activity modulation can be referred to as small side chains conformational changes as well as important modifications, within the quaternary structure, of spatial protein motif distribution. Following the classical model of allostery proposed in the past years, the real definition should refer to the latter case.44 Molecular modeling, particularly molecular simulations, can indeed help in the understanding of such functional motif rearrangement and movements within the protein. Considering the wide collection of molecular simulations approaches, it is imperative to underline that not every model is appropriate to completely explore the protein conformational space. Some classical simulations can in fact be limited to the exploration of only certain energy landscape portions because of their sampling capabilities.44 When using MD, it is difficult to correctly and widely sample the useful landscape for structure transition and it becomes essential to use biased MD techniques45–47 or other modified approaches such as supervised MD.48 Using biased methods allow researchers to explore a wide landscape of protein motions focusing on the energetic aspect of the allostery phenomenon. Moreover, the use of classical MD simulations is not always capable of providing useful information about conformational changes, unless long simulations are used,49 but in the latter case it must also be considered that the longer the simulation is, the higher the approximation becomes.50 Practically, the best way to catch all the useful conformational sampling information should be the use of enhanced sampling techniques.51,52 One possible alternative to long simulations could be the use of multiple shorter simulations which are then analysed using Markov state models to catch all the quantitative parameters to analyse.53–55 In the latter, all the processes must be monitored well in order to explore the structure and rebuild the whole process. In this section, we will present the most recent approaches based on MD used to deeply study the allosteric regulation of protein functions related to their biological role. We will hereby present the most recent advances grouped based on the approach used (classical MD or biased MD).
Use of molecular probes in molecular dynamics for novel allosteric binding site discovery
Some of the studies reported below present noteworthy approaches to study the structural evolution of allosteric binding site cavities and conformational shifting in proteins when targeted with probe molecules, which can act as modification inducers.
One of the first assays in this field was published by Bakan et al. in 2012.38 In their work, the authors introduced the definition of druggability as something related to the affinity of small molecules towards binding sites available on the protein surface. The use of molecular probes designed on approved drug scaffolds allowed researchers to effectively evaluate the binding affinity and druggability for some challenging targets, especially for some hidden allosteric sites. The use of approved drugs with known experimental binding data constituted the method validation to check if theoretical binding affinities were correctly estimated. In this work, two main aspects were deeply studied, firstly the analysis of the putative binding mode for several probe ligands towards different proteins, and secondly the consequent identification of the most druggable sites. Starting from collected data as geometry and energy parameters of the recognition and binding process, researchers could correctly evaluate the binding affinities. This methodology has been applied to several targets such as protein tyrosine phosphatase 1B (PTP1B), lymphocyte function-associated antigen 1, vertebrate kinesin-5 (Eg5), and p38 mitogen-activated protein kinase (MAPK). One of the most interesting aspects sharpened by this method is the possibility to unveil putative interaction spots on the protein surface, which are otherwise hidden. Remarkably, authors used probes with different physicochemical properties to harvest consistently more reliable predictions, which are not biased in favor of a specific chemotype or physicochemical profile of ligand, therefore widening the type of putative binding sites that can be “gathered” within the protein. The use of mixed probes highlighted some interesting aspects of the binding process for the analysed targets, especially on Eg5 and p38 MAPK. On the other hand, using charged probes allowed the authors to discover some important ligand features useful for binding phosphatase PTP1B catalytic sites.
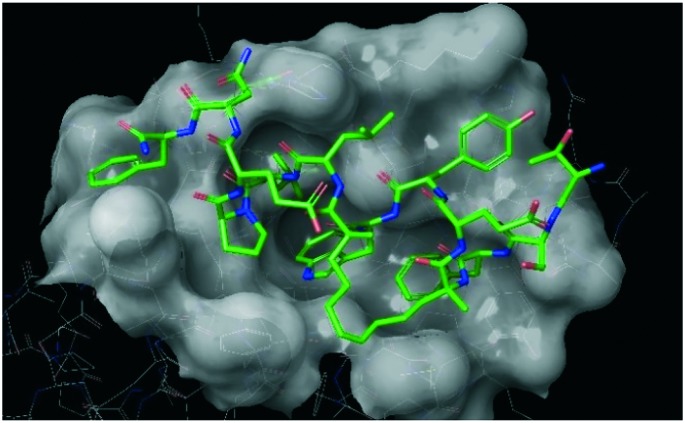
Benzene probes have also been used in a similar manner by Tan et al. in a very recent work.56 In their study, the authors charted putative oncoprotein MDM2 binding sites using a ligand-mapping molecular dynamics (LMMD) simulation technique. In particular, this method allowed the discovery of two new sites both in the N-terminal domain of the protein. The first one, situated between Tyr100 and Tyr104, was mapped in both apo and holo forms of the protein, revealing an interaction region which in the X-ray crystal structure seemed inaccessible to any ligand. This region had already been demonstrated to be essential for the nutlin binding at the p53 cleft.57 The second presumed site, in a region nearby the p53-Pro27 interaction site, was only identified during the simulation of the apo protein. In the holo protein simulation, this region resulted bound to the C-terminal of the p53 binding partner. The efficacy of the adopted method was then demonstrated by biophysical assays that proved the real existence of one of the predicted binding sites close to the consensus p53-binding cleft in MDM2, underlining how molecular simulations can be invaluable for the rational design of new drugs. Based on the simulation outcomes, a series of hydrocarbon stapled peptides were proposed to target the binding sites identified during simulations (Fig. 1). The use of these peptides improved the activity of already known MDM2 ligands because of a cooperative action of the newly discovered allosteric site with the canonical orthosteric site. Moreover, the structural knowledge about these new binding sites opened up novel strategies of peptide optimisation, which can improve the binding mode of other ligands targeted for the MDM2/p53 interaction.
Fig. 1. A stapled peptide (green chain) in complex with MDM2 protein (PDB ID: 4UE1).
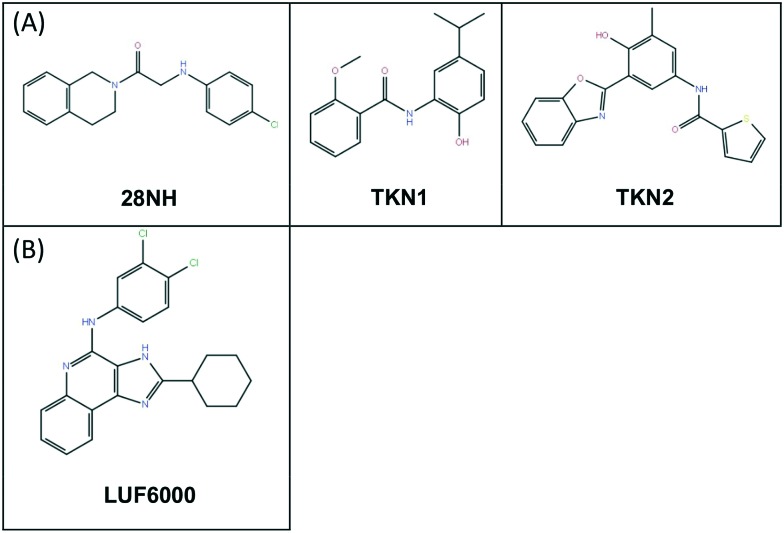
A noteworthy application of molecular probes has been similarly presented by Luo et al.58 In their work, the authors focused on a challenging target such as the two-pore domain potassium channel (K2P). This is a main actor in membrane potential maintenance presenting a unique structural feature of an extracellular cap formed by the E1 and E2 helices, which is not observed in other ion channels. Luo et al. adopted different computational chemistry techniques, mutagenesis, and electrophysiology experiments in order to characterise the binding mode of N-(4-cholorphenyl)-N-(2-(3,4-dihydrosioquinolin-2(1H)-yl)-2-oxoethyl) methanesulfonamide (TKDC), used as probe, to study the process of binding to the extracellular cap of the K2P channel, an allosteric and difficult site to be targeted. The study underlined important differences in the different binding modes of ligands on the different potassium channels (TREK, TRAAK etc.). The first stage of the computational approach was conducted using molecular docking on the previously identified putative binding site. Following the docking outcomes, some mutagenesis experiments within the binding site were conducted in order to confirm the residues responsible for the binding mode retrieved from docking experiments in the different potassium channel models. Starting from the binding mode hypothesis, a chemical probe derived from TKDC was used for molecular dynamics to explore the extracellular binding site of the different channel models and further validate the proposed binding mode previously observed. These outcomes have then been exploited to supervise virtual screening of other molecules and find new allosteric inhibitors of K2P (Fig. 2A).
Fig. 2. (A) Allosteric inhibitors of K2P-28NH, allosteric inhibitor of TREK channels; TKN1 and 2, allosteric inhibitors of TRAAK. (B) Positive allosteric modulator, LUF6000.
Biased molecular dynamics for the design of allosteric modulators
In the past years, several MD applications have been used to better understand the role of allosteric sites within the proteins. One of the most attractive approaches is based on supervised molecular dynamics (SuMD). In this technique, ligand–receptor recognition is well-investigated in a relatively short time period (ns). The method relies on the use of a specific algorithm capable of focusing on the binding process between the protein and the ligand, speeding up the recognition trajectory. Such an approach allows the investigation of some crucial aspects of the binding event with special focus on the meta-binding sites or allosteric sites.59 In their work, Deganutti et al. applied SuMD to a GPCR A3 adenosine receptor to study the effect of a positive allosteric modulator (LUF6000, Fig. 2B).48 This work represents one of the first applications of MD for studying the allosteric recognition mechanism. Indeed, it effectively revealed its importance in the elucidation of what was experimentally observed, opening up two different hypotheses of binding and subsequent regulation of LUF6000 on the A3 adenosine receptor. In their experiment, the authors placed the allosteric modulator about 60 Å away from the orthosteric binding site occupied by the natural agonist adenosine.
The binding process pathway obtained from the simulation showed two possible ways through which the ligand may act on the receptor. In the first hypothesis, LUF6000 produced conformational changes inside the protein, empowering the adenosine to strengthen interactions previously formed in the binding pocket. The other mechanism proposed was the formation of a ternary complex, LUF6000-receptor-adenosine, where the role of LUF6000 was to act as a sort of cap stabilising the binding of the other two interacting partners. Another biased MD technique widely used for the discovery of allosteric modulators is MetaDynamics (MetaD). The great intuition behind this method relies on the use of an additive potential applied to the system analysed in order to overcome some energy barriers of the potential energy surface (PES), allowing the complete exploration of the energy landscape in the protein conformation shift.
MetaD was applied by Laio and Parrinello for the first time and it is usually adopted for molecular simulations in order to expand the exploration of protein conformational changes.60 As a matter of fact, the use of this technique is particularly indicated for allosteric modulation studies because of its capability to deeply explore all the conformational changes in a target. In 2015, Grazioso et al. applied MetaD together with essential dynamics to the Alpha7 nicotinic receptor to give a mechanistic hypothesis of the allosteric modulation.61 In detail, the application of these techniques allowed researchers to explore the free energy landscapes related to the open and closed states of the protein loop C. In this study, the effect of different modulators (including an agonist, a positive allosteric modulator, and a newly reported ago-allosteric modulator) on the conformational change of the protein was investigated. Every ligand considered showed a unique particular free energy profile, and most importantly, the possible interaction between the orthosteric site in the loop C and M helices within the protein structure. This specific interaction was evidenced by the ago-allosteric modulator GAT107. In fact, when bound to the allosteric binding site, GAT107 induced a loop C rearrangement typical of a full agonist, thus providing a possible explication of the experimentally demonstrated ago-allosteric properties of GAT107. The results obtained from the computational approach were in perfect agreement with those observed in the experimental assays and represented an outstanding advancement in the nicotinic receptor biology knowledge.
In 2017, Gomez-Gutierrez et al. used accelerated MD for 6 μs to identify putative allosteric modulators for p38α MAP kinase.62 The method63 is based on an enhanced-sampling algorithm for sampling the conformational space by reducing energy barriers, thus modifying the potential energy profile. At a fixed, defined energy level, the algorithm does not affect energy-profile points above this zone, while it rises up wells that are below the fixed threshold energy level. As a result, the energy profile barriers are reduced, allowing wider exploration of the entire potential energy surface that is otherwise not easily scouted. In their work, Gomez-Gutierrez et al. allowed p38α MAP kinase to undergo a 6 μs accelerated dynamics, collecting all the structures during the trajectory and clustering all the ensembles. Clusters were created using principal component analysis (PCA) first and Cluster Analysis after, in order to collect the most representative structures during the simulation. The collected structures represented the starting point for hot spot analysis conducted on FTMap. Through this approach, it was possible to confirm all the canonical well-known sites of the protein such as the DFG pocket, lipid binding pocket, DEF site and others. Moreover, this study unveiled new allosteric binding sites named NP1 and NP10. These two contact areas in particular caused protein structural rearrangement involving elements responsible for the protein activation (e.g. the activation loop, the catalytic loop, the glycine-rich loop and others).
Classical molecular dynamics for the design of allosteric modulators
Classical MD has been widely applied for studying allosteric modulation of biological targets. One interesting application of molecular simulation to comprehend the structural mechanism associated with signalling pathways of the lymphocyte function has been recently published by Abdullahi et al.64 In their work, the authors applied molecular modeling to examine lymphocyte function activation, through allosteric shape shifting of the lymphocyte-associated antigen, when it is bound to a specific modulator (ICAM binding enhancer-667 – IBE-667). During the simulations, several parameters were collected in order to deeply study and understand the succession of events leading to the active conformation of protein. Particular attention was given to variations in residual distances, dihedral angles, and triCα angles to evaluate how the structural rearrangement was related to these variations. The conformational change between the inactive and active states of the target was characterised by significant fluctuation in residue positions and in the energy stability of the complex. The shape shifting was strictly accompanied by an α7 helix movement driven by a metal-ion dependent adhesion active site (MIDAS domain), both synergistically cooperating to the activation of LFA-1 integrin responsible for the lymphocyte function. The strength behind this work was the capability to demonstrate a synergistic interplay between the MIDAS domain region and the downward α7 helix motion necessary for the biological activity.
MD, in the past years, has been widely applied to the identification of new scaffolds and new chemical entities, integrating the classical virtual screening techniques to prioritise hit molecules and elucidate their binding mode within the protein cavity. Some mitotic kinesin Eg5 allosteric modulators have been identified, thanks to MD application by Makala and Ulaganathan in their work published in 2017.65 Eg5 is a well-known target for cancer therapy, but all the discovered compounds so far were designed for the canonical orthosteric binding site because of its ease of access for small molecules and the availability of structural data. In this work, the authors firstly docked some free available molecular libraries on the allosteric site (site 2) of the target and ranked molecules prioritising them based on the docking score. The 5 best poses retrieved from the docking were then subjected to MD to evaluate the conformational rearrangement and stability of the ligand–protein complex. The results obtained from this study suggested the pyridazine scaffold as an optimal starting point for further development of Eg5 allosteric modulators.
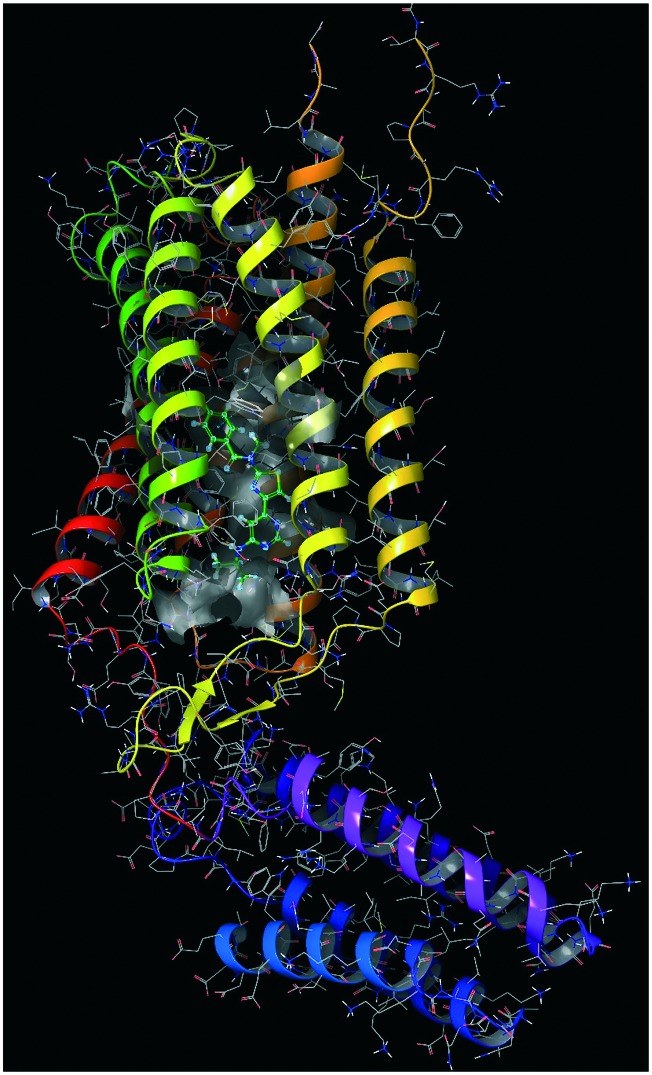
MD as a strategy for unveiling undruggable sites in membrane ion channels was employed by Martin et al. and Guan et al.66,67 to study pentameric ligand-gated ion-channels and calcium channels, respectively. In the first work, the authors presented an extensive point of view on a glutamate-gated chloride channel (GluCl) responsible for the intercellular synaptic communication. A biochemical mechanism of activation/deactivation for this target is still not well-recognised and this work was a first attempt to elucidate the shift from the open state to the closed state of the channel. The experiment was conducted using the protein in its active state bound to l-glutamate and the positive allosteric modulator (PAM) ivermectin. On this system, a μs-long simulation was run in order to explore the structural relaxation upon PAM modulator ejection. By analysing the MD trajectory, two different transition states were retrieved, and most importantly, it was clarified that the structural global twisting observed is the unique responsible feature for the channel pore closing, acting on the M2–M3 loop at the interface between the extracellular and transmembrane domains. Further simulations under equally restrained conditions showed the same structural rearrangement opening up to a pharmacological mechanism clarification of PAMs in this neurotransmitter receptor family. The rearranged structure observed during the dynamics simulation was comparable to the X-ray crystal structure of GluCl, thus enforcing the reliability of the molecular dynamics-based method. Glutamate receptors (PDB ID: ; 4OR2, Fig. 3) represent one of the most important targets involved in neurological diseases and, in certain cases, they show low subtype selectivity for orthosteric modulators. For this reason, it is necessary to address new investigation strategies such as the allosteric modulation. Starting from this hypothesis, Jiang et al. led a study on the mGluR1 receptor in order to discover new putative negative allosteric modulators (NAM) derived from Chinese herbs components.68 In this research work, the authors started from the crystal structure of the seven-transmembrane domain of mGluR1 to detect the putative allosteric binding sites and run some pharmacophore screening. The authors combined the structure-based interaction pharmacophore with a ligand-based approach in order to increase the specificity of virtual screening.
Fig. 3. Metabotropic glutamate receptor 1 in complex with a negative allosteric modulator (green molecule).
The ligand-based approach exploited different scaffolds of known NAMs, thus allowing pharmacophore models differentiated into molecular classes. Starting from these first results, the models were validated using a test set and the most reliable model was then used for the prospective screening campaigns. The best ranked compounds based on pharmacophore feature matching were then docked into the allosteric binding site of the protein in order to evaluate their binding pose. Later, the best poses were subjected to MD to evaluate the effective stability of the ligand–protein complex and the stability of interactions of the selected molecules.
One of the most important outcomes of this study was the identification of some key protein residues (Leu757, Asn760, Trp798, Phe801, Tyr805 and Thr815) that were revealed to be crucial for NAM selectivity and binding stabilisation.
Another comprehensive view of the possible use of MD to discover otherwise unrevealed undruggable sites has been proposed by Pabon and Camacho in a very exhaustive work published in 2017.69 In this study, the attention was focused on the apo form of the anticancer target PD1, a particularly hard-to-drug protein. The use of MD revealed a new hidden hydrophilic cavity around Asn66 and Ile126 residues participating in the ligand recognition pattern.
The use of two PD1 ligands, L1 and L2, permitted the discovery that, while the unbound PD1 presented a hard-to-target hydrophilic interface to host the ligand, the recognition of both L1 and L2 induced a complex conformational shift with the consequent opening up of a hydrophobic cavity otherwise unreachable by any small molecule. These outcomes opened up new strategies to rationally design selective compounds and suggest a possible efficient biophysical approach to the evaluation of the binding pathways as a means for targeting undruggable proteins.
As was recently shown in a paper published by Marko Novinec,70 computational investigations also helped in the discovery of allosteric effectors active on cathepsin K and other related endopeptidases. In his work, Novinec presented for the first time an interesting scenario about allosteric targeting as a progressively winning strategy in drug discovery, even though not yet well explored mainly because of lack of structural information. Papain-like cysteine peptidase cathepsin K allosteric modulation was investigated through MD, together with other peptidases, to catch any putative conformational shift primarily important for the protein activation process. In this approach, the MD-derived conformational space was plotted for different cathepsin endopeptidases (L, K, S and V), using principal component analysis in order to explore possible conformation “clusters”. Proteins of the same family adopted similar conformations during the MD trajectories. At a later stage, the author proceeded with a deeper analysis of cathepsin K to show how some known allosteric modulators, NSC13345 and NSC94914, affect conformational changes. During the simulations, these effectors were able to influence the active site conformational shift, affecting the region nearby pockets of sites S1 and S2. This portion is the tightest part of the active site cleft and it is responsible for specificity in the ligand binding process. Using molecular docking on structures retrieved from MD, it was supposed that allosteric modulation starts stabilizing the pre-existing conformational state prior to influencing the binding of the substrate to the orthosteric site. Comparison of these results to those of related enzymes confirmed that this as a possible mechanism for allosteric modulators, thus demonstrating the usefulness of MD in unveiling binding sites otherwise difficult to be recognised and targeted.
Another interesting application of MD to unveiling the allostery phenomenon was published in 2017 by Latallo et al. In this work, the authors used simulations to predict allosteric mutations responsible for increasing antibiotic resistance mediated by beta-lactamase.71 This research topic starts from the evidence that allosteric mutations are really difficult to predict prospectively. In this work, allosteric mutants of the CTX-M9 enzyme have been used for MD simulations examining a wide range of antibiotics. Experimentally, mutated isoforms of CTX-M9 showed an increase in their catalytic activity and efficiency. When the same mutants were studied “statically”, starting from crystal structures, no differences were noted in comparison with the wild-type form of the enzyme. Based on this outcome, researchers concluded that the activity increase could be related to conformational changes within the structure, which are not observable with canonical static in silico screening, allowing the enzyme to enhance its activity. The use of machine learning techniques applied to MD trajectories allowed the discovery of the most important allosteric mutations influencing the conformational rearrangement of the catalytic site. This study highlighted how the conformation shift was important for the increase in the catalytic activity of the enzyme in developing drug resistance, not affecting the minimum free energy. There are different theories about the conformational change propagation from the allosteric sites to the core catalytic pocket.72 In their work, Latallo et al. showed that such mutations substantially did not affect the catalytic site conformation in the apo form of the protein, leaving the general structure conformation unaltered. Thanks to the application of machine learning techniques to trajectory derived conformations, the authors found out that the catalytic activity variation could be connected to a particular set of protein residues involved in coordinating catalytic water within the substrate binding site or directly involved in the substrate positioning. These outcomes open up some interesting hints in the rational design of new antibiotics.
Molecular dynamics simulations were also applied to address the problem of allosteric modulation in the A2A adenosine receptor. Recently, an interesting work has been produced by Caliman et al.73 as a follow-up to a previous one where the same authors had already explored the conformational analysis of the apo A2A adenosine receptor through MD.74 In the previous work, molecular dynamics analysis allowed the identification of different non-orthosteric sites on the active conformations, on both two intermediate conformations observed and on the inactive conformations of the protein. In this more recent work, the authors started from different structures retrieved from the previous dynamics simulations and 20 different X-ray structures of the same protein family, in order to map the previously retrieved allosteric sites using the fragment-based approach accessible via FTMap software.73 This software, which is usually used to identify available non-orthosteric sites, was revealed to be especially helpful for transmembrane proteins and especially for compounds that cannot target the extracellular part of the protein but go to the lipid bilayer of the membrane. The use of MD combined with FTMap allowed them to mainly identify five allosteric binding sites that are present in both active, intermediate and inactive protein conformations. We here report a table of the five sites identified, as was reported in the original paper (Table 1). Such kind of study represents invaluable help for designing new allosteric modulators for difficult targets where selectivity is difficult to reach. As in the previous case, the Bartuzi research group extensively adopted MD on a GPCR receptor to simulate the activation and the interplay between allosteric sites in the human μ-opioid receptor (MOR).75,76
Table 1. Data taken from original paper73.
| Site | Location | Regions | Residues |
| 1 | Intracellular crevice | TM3/TM4/TM5 | I3.40, F3.41, L3.44, A3.45, D3.49 (TM3); I4.45, I4.48, C4.49 (TM4); Y112 (ICL1); C5.46, P5.50 (TM5) |
| 2 | G protein coupling site | TM2/TM3/TM6/TM7 | N39 (ICL1); T2.39, N2.40 (TM2); D3.49, R3.50 (TM3); H6.32, S6.36, F6.44 (TM6); Y7.53 (TM7); I292 (C-term) |
| 3 | The lipid interface | TM5/TM6 | P5.50, M5.54 (TM5); V6.41, F6.44, W6.48 (TM6) |
| 4 | C-Terminus cleft | TM1/TM7 | L1.45, G1.49, L1.52 (TM1); V7.47, P7.50, F7.51 (TM7) |
| 5 | Extracellular cleft | TM3/TM4 | C3.30, F3.31, V3.34 (TM3); L4.58, G4.57, F4.54 (TM4) |
In their first work, the authors started from evidence on negative modulators of the μ-opioid receptor, a GPCR receptor, for the identification of allosteric sites and modulation mechanisms. In detail, starting from homology modeling, known compounds were docked into the allosteric sites and then conformations and complex stability was evaluated using MD. In particular, salvinorin A, a negative allosteric MOR modulator, was used to evaluate key residues in the binding process using site-directed mutagenesis. According to what was reported in the literature, as experimentally proved, the residues Ile316 (7.39), Tyr320 (7.43), Gln115 (2.60), Tyr312 (7.35), Tyr313 (7.36), and Tyr119 (2.64) were demonstrated to be essential for the recognition and binding of allosteric modulators in the κ-opioid receptor. From the results obtained, the authors suspected possible overlapping regions between the salvinorin A interacting portion (TM I-TM II-TM VI interface) and orthosteric binding site. The main mechanism of action underlined for salvinorin A consists in interfering with orthosteric ligands. The same authors went deeply in order to better understand the possible interplay between two allosteric sites and the agonist binding process.
This time, 200 ns replica molecular simulations were run using a positive modulator, BMS986122 (BMS), to evaluate its influence on the binding process of the agonist (R)-methadone (RME) and Na+. The simulations were able to differentiate between BMS and Na+ orientations within TM VII. On the different trajectories, PCA was applied to investigate on possible clusters of common conformations and TM rearrangements. From the simulation analysis, it was deduced that the agonist binding process was negatively influenced by the presence of the sodium ion interacting with conserved Asp2.50, as was already proposed both experimentally77 and computationally.78,79 Focusing on the BMS, it was noted that its role was the stabilisation of the target–agonist interaction, whereas the allosteric binding of the Na+ was instead disrupted. These data were in complete agreement with experimental findings, that is, a BMS-induced increase in the affinity of full agonists towards MOR.80 Moreover, these results highlighted a possible binding mode of RME involving Asp3.32 with a consequent rearrangement of the TM VII position. This conformational change seems to be driven by an influence on Trp7.35 in the binding pocket, causing the rotation of TM VII. This hypothesis was then confirmed by adding BMS, which stabilised the RME binding through direct interaction with Trp7.35.
Furthermore, during the revision process, we found a “just published” work by Meng et al.,81 where a great approach based on the use of the MD/MSM approach combined with a multi-source seeding strategy was used to explore different possible conformations and transition states of Abl tyrosine kinase. The use of a multi-source seeding strategy consists in using different source protein conformations (X-ray, homologs, “piecewise-mixing” conformations from different crystal structures, previous MSM retrieved conformations). The use of such an approach allowed researchers to widely explore the conformational space of Abl tyrosine kinase, including the myristoyl-binding pocket situated at the C-terminus of the protein. This portion was identified as an allosteric site responsible for modulating conformational transitions between active and inactive states of the protein. From MSM outcomes, researchers found a specific conformational state of the allosteric site, promoting the DFG-out conformation and maintaining the protein in the inactive state. These findings represent a great value in the design of possible Abl allosteric inhibitors.
Protein–protein interactions as undruggable targets
In the past decades, PPI drew the attention of the scientific research community across academia and industry, because of the increasing knowledge about their relevant role in cells for signalling and regulation of cellular life cycle and vital functions (e.g. cellular growth, differentiation and apoptosis). The PPI target space is consistently larger than that of classical protein targets, with putative relevant PPIs comprising between 130 000 and 650 000.82–85 Such a huge amount of potential targets is associated with biological implications in several diseases, for example cancer and neurodegenerative disorders. Unfortunately, to date, less than 0.01% of PPIs belonging to the human interactome present approved modulators.86 Protein–protein interactions are particularly complex targets, usually labelled as undruggable, and thus there are few discovery programs as these are considered high-risk failure therapies.82 Actually, cells are crowded environments where proteins behave as promiscuous macromolecules, i.e. able to take part in interaction networks, binding more than one partner, and in this way making it difficult to achieve specificity during a drug design process. Hence, targeting bi-molecular complexes requires an interdisciplinary approach to identify binding determinants at PPI interfaces and overcome issues tightly linked to the intrinsic nature and structural features of protein–protein interactions.87,88 Indeed, PPI interfaces are often shallow and lack deep grooves able to accommodate a ligand and recognise its shape in a complementary manner. While in a classical receptor–ligand interaction the first one presents a well-defined pocket with clear complementary binding recognition motifs, a protein–protein interaction occurs when both protein partners establish few high-affinity contacts through the so-called “hot spots” residues, exploiting complementary regions as well as several weaker interactions. However, hot spots are residues widely deployed within protein surfaces, and are thus sequentially not connected among them within the same protein, creating a discontinuous epitope.87,89 Another consideration is related to the size of a PPI interface, which is wider (on average 1500 to 3000 Å2) than receptor–ligand contact areas of classical targets (about 300 to 1000 Å2) (Table 2).87 Furthermore, if a protein target takes part in the interaction with another protein, mainly providing protrusions and not just pockets for accommodation, managing the design of a small molecule able to cover a protrusion of the protein target appears rather unlikely.82
Table 2. Data from original paper88.
| Host–guest interaction pocket attributes | ||
| Protein–ligand | Protein–protein | |
| Shape | Deep | Shallow, flat |
| Size | ∼300 to 1000Å2 | 1150–1200 Å2 small interfaces97,98 |
| 1200–2000 Å2 medium interfaces | ||
| 2000–4660 Å2 large interfaces97,99,100 | ||
| Types of interaction | Electrostatic interactions, hydrogen bonds, hydrophobic contacts, π-stacking | Hydrophobic contacts (for protein–protein complexes formation) and electrostatic interactions (for PPI stabilization) |
Despite the above difficulties and the shortage of structural information about protein–protein complexes, PPIs are becoming more accepted and popular targets,90 thanks to promising computational techniques, such as MD. Here, we provide an overview of some case studies where molecular dynamics techniques proved their usefulness to the drug design and discovery process for thoroughly studying protein–protein interactions but above all facing the intrinsic difficulties of this type of target.
Molecular dynamics simulations for the identification of potential pockets and binding hot spots
To date, many theoretical and computational tools have been developed to map a potential protein–protein binding site; some examples are AnchorQuery™91 and FTMap.92 The limitation of these techniques relies on the static structures to be analysed. In this context, MD can represent a valuable tool. Actually, it has been used by numerous research groups to identify hot spots responsible for the interaction between two proteins.93
The hot spots within a PPI interface represent less than 50% of the contact area between proteins and they are defined as those amino acid residues replaced with alanine by alanine scanning mutagenesis which provoke a decrease in binding free energy of at least 2 kcal mol–1.94,95 The hot spots more frequently found in PPI interfaces are the amino acids Tyr, Trp and Arg.95,96 All these three residues take part in hydrophobic interactions, since the main driving force of protein–protein complex formation is precisely the hydrophobicity.88
In a work published in 2014, Sing Tan et al. showed how MD simulations present a remarkable potential for detecting hydrophobic hot spots and for ligand-mapping. Indeed, these computational techniques allowed the unveiling of cryptic binding sites on specific protein (RAD-51 and MDM2) surfaces by MD simulations of 5 ns and 20 ns. The authors suggest the use of shorter simulations (5 ns) for mapping protein X-ray crystal structures and simulations of 20 ns for NMR-resolved structures to ensure careful exploration of protein cavity.
While shorter simulations allowed the identification of pockets buried by amino acid side chains and the protein backbone in MDM2 and RAD-51, respectively, those of 20 ns were able to unveil the hot spot Leu26, which is the most buried of the three MDM2 residues responsible for p53-binding. Furthermore, in the same paper, the authors focused on the capacity of MD techniques to unveil hydrophobic regions able to bind hydrocarbon-stapled peptides at interfaces of proteins, such as MCL-1 and BH3 α-helices of Bcl-2 family proteins.93 The hydrocarbon-stapled peptides are peptides folded as an α-helix, which present all-hydrocarbon “braces” (staples) that make them suitable pharmacological candidates for disrupting protein–protein interactions.101 For this purpose, small molecule probes (benzene, propane and isopropanol) were used to mimic most of the protein residue interactions between hydrocarbon-stapled peptides. In particular, by exploiting the ligand-mapping MD simulations, it was possible to detect a new binding site previously unexplored and design a peptide inhibitor (SAHB8–12) of the hydrophobic interactions between two protein partners.93
Another example of MD application to PPI modulator design is reported in the work of Saez and co-workers published in 2015. They carried out MD studies concerning the investigation of hot spots involved in the interaction of the acid-sensing ion channel 1a (ASIC1a) and its selective inhibitor psalmotoxin-1 (PcTx1), a peptide extracted from spider venom. ASIC1a is a member of the degenerin/epithelial sodium channel family,102,103 which is involved in several diseases including chronic pain104 and ischaemic stroke.105 To date, the most potent and selective inhibitor discovered for this ion channel is precisely the peptide psalmotoxin-1, which binds and fixes the desensitized state of ASIC1a.106,107 Saez et al. analysed the interaction of these two protein partners to elucidate the nature of the binding contacts and to identify the crucial hot spots.108 Starting from a crystal structure resolved by Dawson et al. in 2012,109 the authors observed that the two protein partners established 57 intermolecular contacts, which the authors defined as “pairwise interactions <5 Å”. The bulk of this network was reduced by MD simulations of 30 ns carried out using GROMACS 3.3.3 (GROMOS54a7 force field). The interaction network rate was analysed out of 600 frames for each simulation, setting a cut-off of 5 Å for non-bonded interactions and 2.5 Å for hydrogen bonds. The MD trajectory analysis halved the pairwise interactions to almost 31 intermolecular contacts. These outcomes were further examined in depth by alanine scanning mutagenesis, which led to the identification of a smaller number of hot spots, thus paving the way to the design of novel selective compounds for ASIC1a.108
Furthermore, in a recent article, Biswas et al. presented a study on TRAF6/Basigin interaction implicated in melanoma metastasis. Basigin (BSG) protein is able to stimulate the overexpression of matrix metalloproteinases (MMPs), which contribute to cancer development, and to interact with tumour necrosis factor receptor-associated factor 6 (TRAF6), promoting the invasiveness of melanoma cells.110–113 Biswas and collaborators performed MD simulations of individual proteins and complexes using the GROMACS 5.0.5 (ref. 114) package with the CHARMM force field. The simulation times were 70 ns (BSG), 50 ns (TRAF-6), and 120 ns (BSG-TRAF6 complex). MD results allowed them to observe a conformational change in the BSG transmembrane region participating in the PPI, which as a consequence of TRAF6 binding, acquired a helical conformation. Besides, these simulations provided information about the interacting hot spots of TRAF6; recognizing residues contributing more to binding free energies, MD proved to be a useful instrument for recognising residue contacts previously not identified between protein partners, as a compelling aid for drug design and development.
Another interesting work published in 2017 by Xue and collaborators concerns the use of a recent MD technique, steered molecular dynamics (SMD). This work aimed at obtaining information about an interaction between two chaperones, Hsp70 (also called Hsc70) and Hsp40 (or auxilin).115 These two proteins take part in a cellular ATP-consuming network, which ensures the correct folding of proteins, membrane translocation and protein degradation.115–117 In order to extensively analyse the Hsp70 nucleotide-binding domain (NBD) and Hsp40 J domain interaction, SMD turned out to be useful. SMD is a methodology which involves an unequilibrated system and consists in applying external forces to the system under consideration forcing the protein complex dissociation.115,118–121 During the simulations, binding energy changes were registered and values were plotted into a curve against the simulation time. In an early phase, starting from a PDB file (; 1Q2G) containing the Hsc70–auxilin complex, Xue et al. equilibrated the system performing a classical MD simulation of 20 ns at pH 7.0 and 300 K (using the GROMACS 4.5 program,115,122 under constant NPT and periodic boundary conditions). The RMSD (root-mean-square deviation) was calculated and the system allowed the extrapolation of the stabilised structure at 7 ns of simulation as a starting point for SMD studies. Later, the equilibrated system was subjected to a SMD simulation of 2 ns, applying a spring constant of 300 kJ mol–1 nm–2. This value was chosen as it was more suitable for the complex under consideration, given that it produced reliable and detectable rupture forces responsible for Hsc70–auxilin complex dissociation. The binding energies (kJ mol–1 nm–2) against simulation time (ps) were reported in two curves according to the different types of interaction involved (electrostatic interactions and van der Waals interactions) and the sums of all the points of these two curves were plotted into another curve. This plot suggested that, in the early phases of SMD simulation, electrostatic interactions out-numbered van der Waals (VdW) interactions; in contrast, in the second part of the simulation, VdW interactions dominated. At the end of SMD simulation, the binding energies of the complex achieved the value of zero, revealing a complete protein complex dissociation. Furthermore, in order to deeply study which residues and how much these affect electrostatic and VdW interactions within the complex, Xue and co-workers also reported the residues involved in binding domains of proteins and the related binding energies. In this way, it was possible to identify key residues in the J domain of auxilin and in NBD of Hsc70. In view of the above, a host–guest complex dissociation process using SMD is not intended to be the opposite of the binding process between two interacting partners. Instead, this technique represents an aid to deeply study the nature of established interactions and the related effect within a given complex.115 A very recent and modern approach based on MD, together with MSMs has been presented by Martinez-Rosell et al.123 In their work, the authors adopted a MD-driven approach for fragment screening on CXCL12, a “hard-to-drug” chemokine because of its partial shallowness. The interesting and innovative point of view presented by the authors consists in using combined methods for targeting CXCL12 and not its receptor CXCR4, as was already done by others.124 In their work, Martinez-Rossell et al. used a series of short MD simulations on the target to explore possible conformations. On the obtained conformations, docking was run using fragment libraries. Then, different cycles of short MD–MSM-adaptive respawning, based on the use of the target and solvated ligands, were run to produce putative binding poses to be analysed. One of the strengths of the method used is based on the short simulations as starting points for an adaptive sampling scheme, consisting of their concatenation using the MSM approach in order to enhance the sampling capability. The use of a stochastic method such as MSM allows the reduction of single timescale simulations and at the same time opens up the possibility of deeply studying a wide landscape, allowing the creation of aggregate simulations of 29 to 45 μs. Furthermore, the use of MSM in this work permitted the observation of unbound and different bound states for the ligands screened, together with probabilities of complex creation and associated free energies. Besides, this approach allowed researchers to evaluate the kinetics parameters (kon and koff) for the unbound/bound state of the ligands screened.
Conformational shift analysis as starting point for the design of PPI modulators
The structures obtained by X-ray crystallography do not permit deep exploration of each protein's cavity, especially those transient, which could be responsible for protein–protein interactions. Furthermore, X-ray crystal structures do not account for flexibility and adaptation induced by both proteins that often result in complementarity between two protein partners.125 In this case, starting from PDB files or homology models, the MD methodology provides a dynamic trajectory over time of the positions and the velocities of all atoms in a system, allowing the investigation of the protein surface, reproducing conformational changes occurring in the cellular environment, and detecting potential shallow pockets able to accommodate ligands.88
In 2015, Cau et al. reported a study on the g14-3-3 protein of the protozoan parasite Giardia duodenalis, which colonises the upper regions of small intestines in mammals, causing severe consequences to the host's health.126,127 At present, there are no vaccines available and the number of useful drugs is rather limited, presenting refractory cases as well.127,128 As a consequence, targeting the interaction between g14-3-3 protein, which is responsible for triggering the invasive activity of the parasite, and host protein partners (phosphorylated Ser and Thr proteins – pSer/pThr proteins) represents a priority solution to address this unmet medical need. G14-3-3 protein is able to explicate its activity only upon phosphorylation on the Thr214 residue, producing conversion from the “open” conformation of the apo form to the “closed” one of the phosphorylated form.127,129,130 In early phases, Cau and co-workers conducted SGLD (self-guided Langevin dynamics) simulations using SANDER on nine tripeptides belonging to the phosphorylation region. At a later time, classical MD studies were performed using the PMEMD (particle mesh Ewald molecular dynamics) module of AMBER12 with the AMBER force field ff12SB in an explicit water solvent on wild-type g14-3-3 protein, pThr214-g14-3-3 protein (g14-3-3 protein with phosphorylated Thr214) and T214E-g14-3-3 protein (with phosphomimetic T214E mutation). The authors concluded that in the closed protein conformation (i.e. the phosphorylated form) the structural rearrangement at the expense of the α8–α9 flexible loop containing Thr214 is stabilised not so much by interaction between the loop and neighbouring residues but rather by a steric hindrance of side chains, which provokes a dihedral angle restraint, allowing the protein to interact with its partners. These results were fundamental to permit the authors to investigate the chemical and physico-chemical properties of the interaction region through X-ray crystallographic studies, and finally identify the key hot spot residues of g14-3-3 protein.127
In an article released in December 2017, Vin Chan et al. reported a MD study on murine double minute 4 protein (MDMX) and tumour suppressor protein p53 interaction. MDMX is a regulation factor of protein p53, which under cellular stress conditions undergoes phosphorylation on the tyrosine 99 (Tyr99 or Y99) residue and releases protein p53, resulting in cell cycle arrest and apoptosis.131–133 In their work, MD is applied to integrate a study previously carried out by Zuckerman et al.134 This study suggested that the release of protein p53 from MDMX was caused by a steric bump produced by the phosphate group of pTyr99 in the p53-binding site. This assumption didn't take into account the negative feedback mechanism, which occurs upon Tyr99 phosphorylation, whereby another phosphorylation on Tyr55 brings MDMX back to a conformation able to rebind protein p53 and inhibit its activity. Through MD simulations on MDMX-pTyr99 and MDMX-pTyr99–pTyr55, Vin Chan and co-workers suggested that, in addition to a steric clash, there is a MDMX region, the N-terminal lid, which takes part in protein p53 release through a salt bridge formation between pTyr99 (phosphorylated Tyr99) and Arg18 (or R18) of the lid. Indeed, this salt bridge stabilises the lid in a “closed state”, preventing MDMX–p53 interaction.133 Such results were validated by mutagenesis studies using a glutamate residue, as a phosphomimetic.133,135,136 At the same time, MD simulations on MDMX–pTyr99–pTyr55 unveiled the formation of electrostatic interactions between the phosphate group of pTyr55 and three residues of the N-terminus of the lid (Met1, Thr2, and Ser3), resulting in a shift of the lid towards an “open state” (away from the p53-binding site). The starting structure for MD simulations was built with homology modelling using a PDB file of MDMX (missing N-terminal lid) as the template. 200 ns MD simulations were performed as the NPT ensemble, using the PMEMD module of AMBER14 with the ff99SB force field.137,138 These studies turned out to be a valuable support, because they highlighted likely interactions not yet experimentally discovered, paving the way to further studies for digging out MD information and efficiently designing potential drugs.133
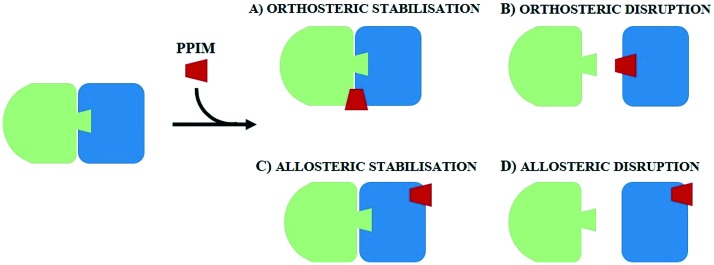
Unlike classical targets, such as membrane receptors or enzymes, the complex nature of PPIs impacts on designing modulators and their chemical and physicochemical features. In most cases, protein–protein interactions do not have natural ligands or known active compounds to be exploited as templates for drug design, rendering the hit identification phase of PPI modulators (PPIMs) complementary to receptor binding pocket difficult in terms of shape and chemical attributes.87,88 Besides, classical virtual screening of drug-like compound libraries against a protein–protein binding site is not always able to provide reliable results both for structure- and fragment-based approaches. Successful PPI modulators, in fact, usually have molecular weights two or three times larger than those of traditional drugs, and hence they have wider sizes.86,139 In addition, due to shallowness and quite broad solvent exposure of PPI binding clefts, generally hits show low affinities for protein–protein interactions, with a Kd value of 0.1–5 mM.86,140,141 Generally, PPIMs are classified according to their mechanism of action in disruptor or stabiliser modulators. PPI disruptors are able to compete in binding one of the two protein partners (orthosteric disruptors) or destabilise a protein–protein interaction through an interaction with a distal or proximal site on the protein surface (allosteric disruptors), eliciting a decrease in PP affinity. In contrast, PPI stabilisers increase the protein complex binding affinity and stability either by acting directly at the interaction interface (orthosteric stabilisation) or by binding to a remote site of the protein and causing an increase in PP affinity142 (Fig. 4). MD has become a valuable tool for validating the stability of a protein–protein-modulator complex and for unveiling the binding modes of PPIMs. An example of complex stability validation can be found in a recent paper. In this article, Gupta and collaborators143 showed how MD came to support other techniques, confirming the virtual screening results. In particular, the target under consideration is an NH(3)-dependent nicotinamide adenine dinucleotide synthetase protein (GEM_3202 or NadE144) involved in a protein–protein interaction network responsible for the infective activity of the opportunistic Burkholderia cepacia pathogen complex.143,145–148 Gupta et al. exploited MD simulations to check the stability of the interaction between the drug target and two potential hits (ZINC83103551 and ZINC38008121) identified through virtual screening. MD simulations were performed in triplicate on a validated homology model of the protein, using the GROMACS 4.5.5 package.
Fig. 4. Mechanism of action of PPI modulators.
The radius of gyration (Rg) fluctuations were calculated for both NadE-ZINC83103551 and NadE-ZINC38008121 interactions and revealed that both complexes showed high structural solidity, obtaining on average 1.87–1.95 nm. Moreover, the measured RMSD values of protein with ligands were low (∼0.41–0.45 nm for NadE-ZINC83103551 and ∼0.15 nm for NadE-ZINC38008121), confirming the high stability of the complexes.143 In December 2017, another interesting work on PPI modulator design was published by Nath Jha et al.149
It concerns α-synuclein (α-syn) protein, which is responsible for the death of dopaminergic neurons in Parkinson's disease. Recent studies demonstrated that the decrease of oligomeric α-synuclein species and the acceleration of amyloid fibril formation represent a potential entry point target strategy for the design of novel drugs.149–152 Based on this hypothesis, Nath Jha et al. conducted MD studies on three hexapeptides in the presence of full-length α-synuclein. The three peptides were designed on the basis of the hydrophobic region of α-synuclein responsible for self-association and aggregation, the non-amyloid-β component (NAC) region (α-syn-71-82).153
The rationally designed peptide sequences were VAQKTV (or peptide C, corresponding to amino acids 77–82 of NAC), VRQKTV (called R78, due to the mutation of A78R), and VPQKTV (named P78, due to the mutation of A78P). MD simulations for all three hexapeptides were performed in an explicit solvent (TIP3P), keeping the ratio at 1 : 5 (8 full-length α-synuclein molecules and 40 hexapeptides) and a random orientation without interactions between proteins. After an early step of minimization and equilibration of 20 ns, MD simulations were performed for 100 ns under NPT conditions. The results suggested that the hexapeptide-containing arginine (VRQKTV) showed a major capacity to establish hydrogen bonds with α-synuclein, with a larger number of H-bond interactions and a higher occupancy of contacts than the other two peptides. Furthermore, MD trajectory analysis brought about the identification of contact areas between full-length α-synuclein and peptide R78, i.e. the negatively charged C-terminus of α-syn and the positively charged Arg78 of peptide R78. These results paved the way to the further study and design of novel specific peptide ligands able to stimulate α-syn oligomer aggregation and prevent their cytotoxic activity.149 Besides the PPI disruptors and stabilisers, Fischer et al.142 provided a further category of PPI modulators, which are the modulators of protein dynamics (or interfacial dynamic modulators). These, in fact, do not necessarily affect the binding affinity of a protein–protein complex, but they bind to clefts produced through homo- or hetero-oligomerisation, modifying the dynamic properties of the individual protein partners.
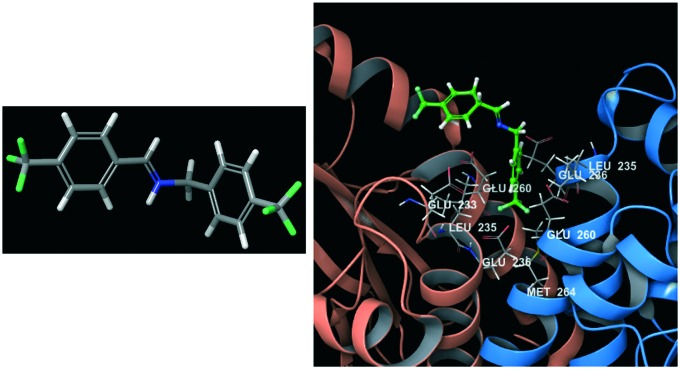
An example of an MD application to support the identification of these types of modulators was provided by Hammoudeh et al.154 in an article released in 2014. The target considered was the dimeric form of the bacterial enzyme dihydropteroate synthase (DHPS), which is implicated in a key step of the folate biosynthetic pathway. The most commonly used drugs for this target are now beginning to show an antimicrobial resistance phenomenon and for this reason, targeting DHPS became a health emergence. Hammoudeh et al. were able to discover an allosteric PPI inhibitor (compound 11, Fig. 5), which at low micromolar concentrations reduces the enzyme Km and Vmax. In this study, MD simulations were used to observe the fluctuations of loop1 and loop2 involved in the active site in four different situations: without any binder, in the presence of a natural substrate, with compound 11, and in the presence of both a natural ligand and an inhibitor. MD simulations highlighted that compound 11 was able to bind the dimeric interface of DHPS and loop7 through its distal half.
Fig. 5. Compound 11 (on the left); binding mode of compound 11 (green molecule) at the dimeric DHPS interface (on the right).
This interaction causes a conformational change which is transmitted to the enzyme active site, thus considerably reducing loop1 and loop2 fluctuations responsible for natural substrate binding.
An interesting work concerning analysis of the thermodynamics and kinetics of protein–protein association was published by Plattner et al. in 2017.155 The authors carried out a cutting-edge study using all-atom MD simulations and MSMs to explore the states and properties, unlikely to be experimentally captured, of two proteins known to form a tightly bound complex, namely, barnase and barstar. Plattner et al. performed a large number of aggregate MD simulations of 2 milliseconds overall, consisting of 1.7 milliseconds of individual trajectories and 0.30 milliseconds of multiple parallel adaptive MD runs. One of the main advantages of adaptive MD runs is that they allow the speeding up of biological processes presenting high energy barriers, decomposing them into smaller paths with relative lower energy barriers. Starting from the unbound state of the two proteins, the hidden Markov model (HMM) was used to explore the states of proteins (early intermediates, late intermediates, pre-bound and bound states), the related interacting contacts, the binding free energies and the transition rates, to obtain the bound complex of the two protein partners. The latter was a complex formed under equilibrium conditions between “loosely bound” (5% population) and “tightly bound” states (95% population), with the last one being consistent with the crystal structure in the PDB database (PDB ID: ; 1BRS). The average heavy atom RMSD values between the PDB crystal structure and the “loosely bound” form were 0.3 nm and 0.21 nm in comparison with the “tightly bound” form. These results are remarkably important for medicinal chemists, demonstrating the good reliability of the Markov modelling method in reproducing information potentially consistent with experimental data and in exploring the conformational space of a finite number of biological molecule states and the related transition rates.
Conclusions and future perspectives
The main goal of medicinal chemistry is identifying chemical entities with optimal affinity, selectivity and safety for patients. Nowadays, computational techniques applied to drug discovery represent an invaluable tool for rationally designing novel chemical entities. Indeed, at present these methodologies have allowed researchers to discover small molecules or small peptides with good drug-like properties,2 speeding up the drug discovery process and decreasing research-associated costs. However, research activities in the field of medicinal chemistry have not yet been able to drug a wide range of biological targets responsible for the aetiology of several diseases, labelling them as undruggable. Nevertheless, in the past years, there has been a huge rise in interest in these pharmacological targets together with the increased use of molecular dynamics simulations to explore related binding sites. Medicinal chemists were thus able to deeply investigate shallow and/or buried grooves, particularly those transient, which are impossible to detect through only the observation of NMR and X-ray crystal data.12 In this review, we have provided an overview of successful MD techniques mainly adopted in the last decade, to deeply study undruggable target-related drug discovery issues, such as protein–ligand/protein–protein interaction stability,21–24 binding kinetics25–29 and interaction mode.30–32 In particular, we have focused on the application of MD to two specific types of considerably important undruggable targets, allosteric sites and protein–protein interactions.
In this work, we have reported several case studies, whereby MD techniques resulted in the identification of binding cavities, contact areas, or ligand binding modes. In conclusion, we would like to underline an important point of view as a future perspective, which was recently presented by Jiménez et al. in their latter current opinion paper.156 In this work, a scenario about the use of MD in synergism with machine learning (ML) approaches is presented. The combination of these two methods seems to be crucial in order to improve the accuracy of predictions and speed up the MD analysis process. The ML/MD combined approach, together with a continuously increasing simulation timescale, as was observed by Martinez-Rosell et al.157 in their recent overview, depicts without any doubt an encouraging landscape for researchers, paving the way for “drugging the undruggable targets”.
Conflicts of interest
The authors declare no competing interests.
Biographies

Ugo Perricone
Ugo Perricone graduated with a degree in Pharmaceutical Chemistry and Technology at the University of Palermo. He completed his studies first with a specialisation in mass spectrometry and then with a Ph.D. in Molecular and Biomolecular Sciences. During his doctorate, he attended the University of Vienna, under the guidance of Prof. Thierry Langer, where he carried out most of the studies aimed at the development of dynamic pharmacophores to be used in the processes of molecular virtual screening. In 2016 he was selected as Scientist by the Fondazione Ri.MED and included in the Drug Design team. Since 2018, he has been the reference resource of the newly formed Computer Aided Drug Design group.

Maria Rita Gulotta
Maria Rita Gulotta graduated with a degree in Pharmaceutical Chemistry and Technology at the University of Palermo and completed her Master's degree with an experimental thesis in synthetic medicinal chemistry. From 2016 to 2017, she worked as a junior scientist in Computer Aided Drug Design (CADD) at Fondazione Ri.MED in Palermo. Then she got a PhD fellowship in Molecular and Biomolecular Sciences funded by Fondazione Ri.MED. She is actually working on a protein–protein interaction study through CADD techniques.

Jessica Lombino
Jessica Lombino graduated with a degree in Pharmacy and completed a Master's thesis in synthetic medicinal chemistry. She is actually a PhD student in Molecular and Biomolecular Science at the University of Palermo with an Italian Ministry funded industrial PhD grant. Her research includes rational drug design of small molecules targeting the epigenetics pathway using computational approaches, completed by the synthesis of active molecules.

Barbara Parrino
Barbara Parrino graduated with a degree in Medicinal Chemistry and Technology at the University of Palermo with full marks and honors in 2007 and obtained her “Doctor Europaeus” Ph.D. degree in Pharmaceutical Sciences in March 2012 for which she was awarded the P. Ehrlich MedChem Euro-PhD Network Certificate in September 2012. From November 2012 to July 2017, she worked as a postdoc research fellow at the University of Palermo, and in March 2017, she received the National Academic Qualification as Associate Professor. Since November 2017, she has been working as Researcher at the University of Palermo, and her research interest is focused on the synthesis and reactivity studies of polycondensed nitrogen heterocycles with antitumor activity. She is the author of 37 papers and one patent.

Stella Cascioferro
Stella Cascioferro graduated with a degree in Pharmacy with full marks and honors in 1999 and obtained her PhD in Medicinal Chemistry in 2004 at the University of Palermo. In 2004 she joined the group of Professor Graham Richards at the Physical and Theoretical Chemistry Laboratory, University of Oxford. Her research interests include the design, synthesis, and biological evaluation of heterocyclic compounds as antitumoral, anti-infective, and anti-inflammatory agents. Currently she is working at the Department of Biological, Chemical and Pharmaceutical Science and Technology (University of Palermo). She is the author of 41 scientific papers published in peer reviewed international journals and a reviewer for several journals in the field of medicinal chemistry.
References
- Mandal S., Moudgil M., Mandal S. K. Eur. J. Pharmacol. 2009;625:90–100. doi: 10.1016/j.ejphar.2009.06.065. [DOI] [PubMed] [Google Scholar]
- Shirai H., Prades C., Vita R., Marcatili P., Popovic B., Xu J., Overington J. P., Hirayama K., Soga S., Tsunoyama K., Clark D., Lefranc M. P., Ikeda K. Biochim. Biophys. Acta, Proteins Proteomics. 2014;1844:2002–2015. doi: 10.1016/j.bbapap.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Pei J., Lai L. Curr. Pharm. Des. 2013;19:2326–2333. doi: 10.2174/1381612811319120019. [DOI] [PubMed] [Google Scholar]
- Sheridan R. P., Maiorov V. N., Holloway M. K., Cornell W. D., Gao Y. D. J. Chem. Inf. Model. 2010;50:2029–2040. doi: 10.1021/ci100312t. [DOI] [PubMed] [Google Scholar]
- Edfeldt F. N. B., Folmer R. H. A., Breeze A. L. Drug Discovery Today. 2011;16:284–287. doi: 10.1016/j.drudis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Dang C. V., Reddy E. P., Shokat K. M., Soucek L. Nat. Rev. Cancer. 2017;17:502–508. doi: 10.1038/nrc.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. Clin. Cancer Res. 2015;21:1797–1801. doi: 10.1158/1078-0432.CCR-14-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield J. R., Beaulieu M.-E., Soucek L. Front. Cell Dev. Biol. 2017;5:10. doi: 10.3389/fcell.2017.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayal M., Honig B. Proteins: Struct., Funct., Bioinf. 2006;63:892–906. doi: 10.1002/prot.20897. [DOI] [PubMed] [Google Scholar]
- Cheng A. C., Coleman R. G., Smyth K. T., Cao Q., Soulard P., Caffrey D. R., Salzberg A. C., Huang E. S. Nat. Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- Hajduk P. J., Huth J. R., Fesik S. W. J. Med. Chem. 2005;48(7):2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- Li Y., Kang C. Molecules. 2017;22(9):E1399. doi: 10.3390/molecules22091399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelananda S. P., Lindert S. Beilstein J. Org. Chem. 2016;12:2694–2718. doi: 10.3762/bjoc.12.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanovic I. M. Chem.-Biol. Interact. 2008;171:165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou-Yang S.-S., Lu J.-Y., Kong X.-Q., Liang Z.-J., Luo C., Jiang H. Acta Pharmacol. Sin. 2012;33:1131–1140. doi: 10.1038/aps.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. A., Ai R., Gutierrez M., Marsella M. J. Methods. 2012;819:3–12. doi: 10.1007/978-1-61779-465-0_35. [DOI] [PubMed] [Google Scholar]
- Schneider G., Fechner U. Nat. Rev. Drug Discovery. 2005;4:649–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- Jhoti H. and Leach A. R., Structure-based drug discovery, 2007. [Google Scholar]
- Schneider M., Fu X., Keating A. E. Proteins. 2009;77:97–110. doi: 10.1002/prot.22421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vivo M., Masetti M., Bottegoni G., Cavalli A. J. Med. Chem. 2016;59:4035–4061. doi: 10.1021/acs.jmedchem.5b01684. [DOI] [PubMed] [Google Scholar]
- Wieder M., Perricone U., Seidel T., Boresch S., Langer T. Monatsh. Chem. 2016;147:553–563. doi: 10.1007/s00706-016-1674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieder M., Perricone U., Boresch S., Seidel T., Langer T. Biochem. Biophys. Res. Commun. 2016;470:685–689. doi: 10.1016/j.bbrc.2016.01.081. [DOI] [PubMed] [Google Scholar]
- Perricone U., Wieder M., Seidel T., Langer T., Padova A., Almerico A. M., Tutone M. ChemMedChem. 2017;12(16):1399–1407. doi: 10.1002/cmdc.201600526. [DOI] [PubMed] [Google Scholar]
- Gorska-Ponikowska M., Perricone U., Kuban-Jankowska A., Lo Bosco G., Barone G. J. Cell. Physiol. 2017;232(11):3030–3049. doi: 10.1002/jcp.25888. [DOI] [PubMed] [Google Scholar]
- Decherchi S., Berteotti A., Bottegoni G., Rocchia W., Cavalli A. Nat. Commun. 2015;6:6155. doi: 10.1038/ncomms7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollica L., Decherchi S., Zia S. R., Gaspari R., Cavalli A., Rocchia W. Sci. Rep. 2015;5:11539. doi: 10.1038/srep11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistrato A., Sgrignani J., Krause R., Cavalli A. J. Phys. Chem. Lett. 2017;8:2036–2042. doi: 10.1021/acs.jpclett.7b00697. [DOI] [PubMed] [Google Scholar]
- Bernetti M., Masetti M., Pietrucci F., Blackledge M., Jensen M. R., Recanatini M., Mollica L., Cavalli A. J. Phys. Chem. B. 2017;121:9572–9582. doi: 10.1021/acs.jpcb.7b08925. [DOI] [PubMed] [Google Scholar]
- Gaspari R., Rechlin C., Heine A., Bottegoni G., Rocchia W., Schwarz D., Bomke J., Gerber H.-D., Klebe G., Cavalli A. J. Med. Chem. 2016;59:4245–4256. doi: 10.1021/acs.jmedchem.5b01643. [DOI] [PubMed] [Google Scholar]
- Cuzzolin A., Sturlese M., Deganutti G., Salmaso V., Sabbadin D., Ciancetta A., Moro S. J. Chem. Inf. Model. 2016;56:687–705. doi: 10.1021/acs.jcim.5b00702. [DOI] [PubMed] [Google Scholar]
- Deganutti G., Welihinda A., Moro S. ChemMedChem. 2017;12:1319–1326. doi: 10.1002/cmdc.201700200. [DOI] [PubMed] [Google Scholar]
- Deganutti G., Moro S. Molecules. 2017;22:818. doi: 10.3390/molecules22050818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E. Ber. Dtsch. Chem. Ges. 1894;27:2985–2993. [Google Scholar]
- Ma B., Shatsky M., Wolfson H. J., Nussinov R. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague S. J. Nat. Rev. Drug Discovery. 2003;2:527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- Seco J., Luque F. J., Barril X. J. Med. Chem. 2009;52:2363–2371. doi: 10.1021/jm801385d. [DOI] [PubMed] [Google Scholar]
- Lexa K. W., Carlson H. A. J. Am. Chem. Soc. 2011;133(2):200–202. doi: 10.1021/ja1079332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan A., Nevins N., Lakdawala A. S., Bahar I. J. Chem. Theory Comput. 2012;8:2435–2447. doi: 10.1021/ct300117j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchillo R., Pinto-Gil K., Michel J. J. Chem. Theory Comput. 2015;11:1292–1307. doi: 10.1021/ct501072t. [DOI] [PubMed] [Google Scholar]
- Ferruz N., Doerr S., Vanase-Frawley M. A., Zou Y., Chen X., Marr E. S., Nelson R. T., Kormos B. L., Wager T. T., Hou X., Villalobos A., Sciabola S., De Fabritiis G. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-19345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Némethy G., Filmer D. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Monod J., Wyman J., Changeux J.-P. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Motlagh H. N., Wrabl J. O., Li J., Hilser V. J. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler-Wildman K. A., Thai V., Lei M., Ott M., Wolf-Watz M., Fenn T., Pozharski E., Wilson M. A., Petsko G. A., Karplus M., Hübner C. G., Kern D. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Xiao X., Zeng X., Yuan Y., Gao N., Guo Y., Pu X., Li M. Phys. Chem. Chem. Phys. 2015;17:2512–2522. doi: 10.1039/c4cp04528a. [DOI] [PubMed] [Google Scholar]
- Wolf R. M. J. Comput.-Aided Mol. Des. 2015;29:1025–1034. doi: 10.1007/s10822-015-9863-2. [DOI] [PubMed] [Google Scholar]
- Perdih A., Kotnik M., Hodoscek M., Solmajer T. Proteins: Struct., Funct., Bioinf. 2007;68:243–254. doi: 10.1002/prot.21374. [DOI] [PubMed] [Google Scholar]
- Deganutti G., Cuzzolin A., Ciancetta A., Moro S. Bioorg. Med. Chem. 2015;23:4065–4071. doi: 10.1016/j.bmc.2015.03.039. [DOI] [PubMed] [Google Scholar]
- Klepeis J. L., Lindorff-Larsen K., Dror R. O., Shaw D. E. Curr. Opin. Struct. Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- Anderson J. S., Mustafi S. M., Hernández G., LeMaster D. M. Biophys. Chem. 2014;192:41–48. doi: 10.1016/j.bpc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczek E. E., Reefschläger L. G., Dehling M., Struller T. J., Häusler E., Seidl A., Kaila V. R. I., Buchner J. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E3189–E3198. doi: 10.1073/pnas.1424342112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da L.-T., Sheong F. K., Silva D.-A. and Huang X., in Advances in experimental medicine and biology, 2014, vol. 805, pp. 29–66. [DOI] [PubMed] [Google Scholar]
- Chodera J. D., Noé F. Curr. Opin. Struct. Biol. 2014;25:135–144. doi: 10.1016/j.sbi.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman G. R., Huang X., Pande V. S. Methods. 2009;49:197–201. doi: 10.1016/j.ymeth.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y. S., Reeks J., Brown C. J., Thean D., Ferrer Gago F. J., Yuen T. Y., Goh E. T. L., Lee X. E. C., Jennings C. E., Joseph T. L., Lakshminarayanan R., Lane D. P., Noble M. E. M., Verma C. S. J. Phys. Chem. Lett. 2016;7:3452–3457. doi: 10.1021/acs.jpclett.6b01525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernychova L., Man P., Verma C., Nicholson J., Sharma C.-A., Ruckova E., Teo J. Y., Ball K., Vojtesek B., Hupp T. R. Proteomics. 2013;13:2512–2525. doi: 10.1002/pmic.201300029. [DOI] [PubMed] [Google Scholar]
- Luo Q., Chen L., Cheng X., Ma Y., Li X., Zhang B., Li L., Zhang S., Guo F., Li Y., Yang H. Nat. Commun. 2017;8:378. doi: 10.1038/s41467-017-00499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbadin D., Moro S. J. Chem. Inf. Model. 2014;54:372–376. doi: 10.1021/ci400766b. [DOI] [PubMed] [Google Scholar]
- Laio A., Parrinello M. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazioso G., Sgrignani J., Capelli R., Matera C., Dallanoce C., De Amici M., Cavalli A. J. Chem. Inf. Model. 2015;55:2528–2539. doi: 10.1021/acs.jcim.5b00459. [DOI] [PubMed] [Google Scholar]
- Gomez-Gutierrez P., Rubio-Martinez J., Perez J. J. J. Chem. Inf. Model. 2017;57:2566–2574. doi: 10.1021/acs.jcim.7b00439. [DOI] [PubMed] [Google Scholar]
- Hamelberg D., Mongan J., McCammon J. A. J. Chem. Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- Abdullahi M., Olotu F. A., Soliman M. E. Biotechnol. Lett. 2017;39(12):1843–1851. doi: 10.1007/s10529-017-2432-0. [DOI] [PubMed] [Google Scholar]
- Makala H., Ulaganathan V. J. Recept. Signal Transduction. 2017;0:1–8. doi: 10.1080/10799893.2017.1387922. [DOI] [PubMed] [Google Scholar]
- Martin N. E., Malik S., Calimet N., Changeux J. P., Cecchini M. PLoS Comput. Biol. 2017;13:1–25. doi: 10.1371/journal.pcbi.1005784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R., Zhou H., Li J., Xiao S., Pang C., Chen Y., Du X., Ke S., Tang Q., Su J., Zhan Y., An H. PLoS One. 2017;12(9):e0182067. doi: 10.1371/journal.pone.0182067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhang X., Chen X., He Y., Qiao L., Zhang Y., Li G., Xiang Y. Molecules. 2015;20:12769–12786. doi: 10.3390/molecules200712769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabon N. A., Camacho C. J. eLife. 2017;6:1–24. doi: 10.7554/eLife.22889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novinec M. PLoS One. 2017;12:1–22. doi: 10.1371/journal.pone.0182387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latallo M. J., Cortina G. A., Faham S., Nakamoto R. K., Kasson P. M. Chem. Sci. 2017;8:6484–6492. doi: 10.1039/c7sc02676e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh H. N., Wrabl J. O., Li J., Hilser V. J. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliman A. D., Miao Y., McCammon J. A. Chem. Biol. Drug Des. 2017:5–16. doi: 10.1111/cbdd.13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliman A. D., Swift S. E., Wang Y., Miao Y., McCammon J. A. Protein Sci. 2015;24:1004–1012. doi: 10.1002/pro.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi D., Kaczor A. A., Matosiuk D. J. Chem. Inf. Model. 2015;55:2421–2434. doi: 10.1021/acs.jcim.5b00280. [DOI] [PubMed] [Google Scholar]
- Bartuzi D., Kaczor A. A., Matosiuk D. J. Chem. Inf. Model. 2016;56:563–570. doi: 10.1021/acs.jcim.5b00705. [DOI] [PubMed] [Google Scholar]
- Liu W., Chun E., Thompson A. A., Chubukov P., Xu F., Katritch V., Han G. W., Roth C. B., Heitman L. H., IJzerman A. P., Cherezov V., Stevens R. C. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., LeRouzic V., Schneider S., Bisignano P., Pasternak G. W., Filizola M. Biochemistry. 2014;53:5140–5149. doi: 10.1021/bi5006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Caliman A. D., McCammon J. A. Biophys. J. 2015;108:1796–1806. doi: 10.1016/j.bpj.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston K. E., Traynor J. R. Proc. Natl. Acad. Sci. U. S. A. 2014;111:18369–18374. doi: 10.1073/pnas.1415013111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Gao C., Clawson D. K., Atwell S., Russell M., Vieth M. and Roux B., Predicting the Conformational Variability of Abl tyrosine kinase using Molecular Dynamics Simulations and Markov State Models, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry D. C. Biopolymers. 2006;84:535–552. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- Venkatesan K., Rual J., Vazquez A., Lemmens I., Hirozane-kishikawa T., Hao T., Xin X., Goh K., Yildirim M. A., Heinzmann K., Gebreab F., Sahalie J. M., Simon C., De Smet A., Dann E., Vinayagam A., Yu H., Szeto D., Dricot A., Klitgord N., Murray R. R., Lin C., Lalowski M., Timm J., Rau K., Boone C., Braun P., Michael E., Roth F. P., Hill D. E., Tavernier J., Wanker E. E., Barabási L., Vidal M. Syst. Biol. 2010;6:83–90. [Google Scholar]
- Zhang Q. C., Petrey D., Deng L., Qiang L., Shi Y., Thu C. A., Bisikirska B., Lefebvre C., Accili D., Hunter T., Maniatis T., Califano A., Honig B. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M. P. H., Thorne T., de Silva E., Stewart R., An H. J., Lappe M., Wiuf C. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell A. E., Blosser S. L., Arora P. S. Trends Pharmacol. Sci. 2016;37:702–713. doi: 10.1016/j.tips.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz A., Ciglia E., Gohlke H. Curr. Pharm. Des. 2012;18:4630–4647. doi: 10.2174/138161212802651553. [DOI] [PubMed] [Google Scholar]
- Rakers C., Bermudez M., Keller B. G., Mortier J., Wolber G. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2015;5:345–359. [Google Scholar]
- Cukuroglu E., Engin H. B., Gursoy A., Keskin O. Prog. Biophys. Mol. Biol. 2014;116:165–173. doi: 10.1016/j.pbiomolbio.2014.06.003. [DOI] [PubMed] [Google Scholar]
- Zhang X., Betzi S., Morelli X., Roche P. Future Med. Chem. 2014;6:1291–1307. doi: 10.4155/fmc.14.57. [DOI] [PubMed] [Google Scholar]
- Koes D. R., Dömling A., Camacho C. J. Protein Sci. 2018;27:229–232. doi: 10.1002/pro.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D., Grove L. E., Hall D. R., Bohnuud T., Mottarella S. E., Luo L., Xia B., Beglov D., Vajda S. Nat. Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y. S., Spring D. R., Abell C., Verma C. S. J. Chem. Theory Comput. 2015;11:3199–3210. doi: 10.1021/ct5010577. [DOI] [PubMed] [Google Scholar]
- Clackson T., Wells J. A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Sable R., Jois S. Molecules. 2015;20:11569–11603. doi: 10.3390/molecules200611569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan A. A., Thorn K. S. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Moreira I. S., Fernandes P. A., Ramos M. J. Proteins: Struct., Funct., Bioinf. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- Lo Conte L., Chothia C., Janin J. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Janin J., Chothia C. J. Biol. Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- Horton N., Lewis M. Protein Sci. 1992;1:169–181. doi: 10.1002/pro.5560010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky L. D., Bird G. H. J. Med. Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S., Schild L. Physiol. Rev. 2002;82:735–767. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- Izumi M., Ikeuchi M., Ji Q., Tani T. J. Biomed. Sci. 2012;19:77. doi: 10.1186/1423-0127-19-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval E., Gasull X., Noël J., Salinas M., Baron A., Diochot S., Lingueglia E. Pharmacol. Ther. 2010;128:549–558. doi: 10.1016/j.pharmthera.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Xiong Z.-G., Zhu X.-M., Chu X.-P., Minami M., Hey J., Wei W.-L., MacDonald J. F., Wemmie J. A., Price M. P., Welsh M. J., Simon R. P. Cell. 2004;118:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Escoubas P., De Weille J. R., Lecoq A., Diochot S., Waldmann R., Champigny G., Moinier D., Ménez A., Lazdunski M. J. Biol. Chem. 2000;275:25116–25121. doi: 10.1074/jbc.M003643200. [DOI] [PubMed] [Google Scholar]
- Sherwood T. W., Lee K. G., Gormley M. G., Askwith C. C. J. Neurosci. 2011;31:9723–9734. doi: 10.1523/JNEUROSCI.1665-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez N. J., Deplazes E., Cristofori-Armstrong B., Chassagnon I. R., Lin X., Mobli M., Mark A. E., Rash L. D., King G. F. Br. J. Pharmacol. 2015;172:4985–4995. doi: 10.1111/bph.13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson R. J. P., Benz J., Stohler P., Tetaz T., Joseph C., Huber S., Schmid G., Hügin D., Pflimlin P., Trube G., Rudolph M. G., Hennig M., Ruf A. Nat. Commun. 2012;3:936. doi: 10.1038/ncomms1917. [DOI] [PubMed] [Google Scholar]
- Biswas R., Ghosh S., Bagchi A. J. Mol. Recognit. 2017;30:e2643. doi: 10.1002/jmr.2643. [DOI] [PubMed] [Google Scholar]
- Li R., Huang L., Guo H., Toole B. P. J. Cell. Physiol. 2001;186:371–379. doi: 10.1002/1097-4652(2000)9999:999<000::AID-JCP1042>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Liao C.-G., Yao L., Xie W., Liu L., Wu S.-D., Lu N., Huang J.-G., Kong L.-M., Zhang H.-L. Cancer Cell Int. 2016;16:28. doi: 10.1186/s12935-016-0302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Zhang X., Zeng W., Su J., Yang K., Lu L., Lim C. B., Tang W., Wu L., Zhao S., Jia X., Peng C., Chen X. Oncotarget. 2016;7:7179–7192. doi: 10.18632/oncotarget.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Xue Y. L., Zhou L., Sun Y., Li H., Jones G. W., Song Y. J. Mol. Model. 2017;23(11):320. doi: 10.1007/s00894-017-3453-2. [DOI] [PubMed] [Google Scholar]
- Fink A. L. Physiol. Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- Walsh P., Bursać D., Law Y. C., Cyr D., Lithgow T. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalikka J., Akola J. Eur. Biophys. J. 2011;40:181–194. doi: 10.1007/s00249-010-0638-3. [DOI] [PubMed] [Google Scholar]
- González A., Perez-Acle T., Pardo L., Deupi X. PLoS One. 2011;6:e23815. doi: 10.1371/journal.pone.0023815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Hasin N., Shen M., He J., Xue Y., Zhou X., Perrett S., Song Y., Jones G. W. PLoS Comput. Biol. 2013;9:e1002896. doi: 10.1371/journal.pcbi.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Guan J., Xu L., Yu Y., He J., Jones G. W., Song Y. J. Biomol. Struct. Dyn. 2012;30:652–661. doi: 10.1080/07391102.2012.689698. [DOI] [PubMed] [Google Scholar]
- Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M. R., Smith J. C., Kasson P. M., van der Spoel D., Hess B., Lindahl E. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rosell G., Harvey M. J., De Fabritiis G. J. Chem. Inf. Model. 2018;58(3):683–691. doi: 10.1021/acs.jcim.7b00625. [DOI] [PubMed] [Google Scholar]
- Choi W.-T., Yang Y., Xu Y., An J. Curr. Top. Med. Chem. 2014;14:1574–1589. doi: 10.2174/1568026614666140827143541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin M. M. R., Wells J. A. Nat. Rev. Drug Discovery. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- Halliez M. C., Buret A. G. World J. Gastroenterol. 2013;19:8974. doi: 10.3748/wjg.v19.i47.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau Y., Fiorillo A., Mori M., Ilari A., Botta M., Lalle M. J. Chem. Inf. Model. 2015;55:2611–2622. doi: 10.1021/acs.jcim.5b00452. [DOI] [PubMed] [Google Scholar]
- Lalle M. Infect. Disord.: Drug Targets. 2010;10:283–294. doi: 10.2174/187152610791591610. [DOI] [PubMed] [Google Scholar]
- Lalle M., Bavassano C., Fratini F., Cecchetti S., Boisguerin P., Crescenzi M., Pozio E. Int. J. Parasitol. 2010;40:201–213. doi: 10.1016/j.ijpara.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Fiorillo A., di Marino D., Bertuccini L., Via A., Pozio E., Camerini S., Ilari A., Lalle M. PLoS One. 2014;9:e92902. doi: 10.1371/journal.pone.0092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D., Levine A. Cold Spring Harbor Perspect. Biol. 2010;2:1–10. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts A., Steegenga W. T., Riteco N., van Laar T., Dekker P., Bazuine M., van Ham R. C., van der Houven van Oordt W., Hateboer G., van der Eb A. J., Jochemsen A. G. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Chan J. V., Koh D. X. P., Liu Y., Joseph T. L., Lane D. P., Verma C. S., Tan Y. S. Oncotarget. 2017;8:112825–112840. doi: 10.18632/oncotarget.22829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman V., Lenos K., Popowicz G. M., Silberman I., Grossman T., Marine J. C., Holak T. A., Jochemsen A. G., Haupt Y. J. Biol. Chem. 2009;284:4031–4039. doi: 10.1074/jbc.M809211200. [DOI] [PubMed] [Google Scholar]
- Dissmeyer N., Schnittger A. Methods Mol. Biol. 2011;779:93–138. doi: 10.1007/978-1-61779-264-9_6. [DOI] [PubMed] [Google Scholar]
- Stateva S. R., Salas V., Benaim G., Menéndez M., Solís D., Villalobo A. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0120798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D., Babin V., Berryman J., Betz R., Cai Q., Cerutti D., Cheatham T., Darden T., Duke R., Gohlke H., Götz A., Gusarov S., Homeyer N., Janowski P., Kaus J., Kolossváry I., Kovalenko A., Lee T.-S. and Kollman P. A., 10.13140/RG.2.2.17892.37766. [DOI]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Proteins: Struct., Funct., Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio O., Reynès C. H., Camproux A.-C., Villoutreix B. O. Drug Discovery Today. 2010;15:220–229. doi: 10.1016/j.drudis.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Valkov E., Sharpe T., Marsh M., Greive S. and Hyvönen M., in Topics in current chemistry, 2011, vol. 317, pp. 145–179. [DOI] [PubMed] [Google Scholar]
- Magee T. V. Bioorg. Med. Chem. Lett. 2015;25:2461–2468. doi: 10.1016/j.bmcl.2015.04.089. [DOI] [PubMed] [Google Scholar]
- Fischer G., Rossmann M., Hyvönen M. Curr. Opin. Biotechnol. 2015;35:78–85. doi: 10.1016/j.copbio.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Chauhan R., Prasad Y., Wadhwa G., Jain C. K. Comput. Biol. Chem. 2016;65:80–90. doi: 10.1016/j.compbiolchem.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Pankiewicz K. W., Petrelli R., Singh R., Felczak K. Curr. Med. Chem. 2015;22(34):3991–4028. doi: 10.2174/0929867322666150821100720. [DOI] [PubMed] [Google Scholar]
- Jordan I. K., Rogozin I. B., Wolf Y. I., Koonin E. V. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Dandekar T., Diaz-Lazcoz Y., Eisenhaber F., Huynen M., Yuan Y. J. Mol. Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- Remm M., Storm C. E. V., Sonnhammer E. L. L. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Goymer P. Nat. Rev. Genet. 2008;9:650. doi: 10.1038/nrg2450. [DOI] [PubMed] [Google Scholar]
- Jha N. N., Ranganathan S., Kumar R., Mehra S., Panigrahi R., Navalkar A., Ghosh D., Kumar A., Padinhateeri R., Maji S. K. Biochemistry. 2018;57(5):791–804. doi: 10.1021/acs.biochem.7b01090. [DOI] [PubMed] [Google Scholar]
- Conway K. A., Lee S. J., Rochet J. C., Ding T. T., Williamson R. E., Lansbury P. T. Proc. Natl. Acad. Sci. U. S. A. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Uversky V. N., Fink A. L. Biochemistry. 2001;40:11604–11613. doi: 10.1021/bi010616g. [DOI] [PubMed] [Google Scholar]
- Fares M.-B., Ait-Bouziad N., Dikiy I., Mbefo M. K., Jovi i A., Kiely A., Holton J. L., Lee S.-J., Gitler A. D., Eliezer D., Lashuel H. A. Hum. Mol. Genet. 2014;23:4491–4509. doi: 10.1093/hmg/ddu165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson B. I., Murray I. V. J., Trojanowski J. Q., Lee V. M.-Y. J. Biol. Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- Hammoudeh D. I., Date M., Yun M.-K., Zhang W., Boyd V. A., Follis V., Griffith E., Lee R. E., Bashford D., White S. W. ACS Chem. Biol. 2014;9(6):1294–1302. doi: 10.1021/cb500038g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner N., Doerr S., De Fabritiis G., Noé F. Nat. Chem. 2017;9:1005–1011. doi: 10.1038/nchem.2785. [DOI] [PubMed] [Google Scholar]
- Jiménez J., Škalič M., Martínez-Rosell G., De Fabritiis G. J. Chem. Inf. Model. 2018;58:287–296. doi: 10.1021/acs.jcim.7b00650. [DOI] [PubMed] [Google Scholar]
- Martinez-Rosell G., Giorgino T., Harvey M. J., de Fabritiis G. Curr. Top. Med. Chem. 2017;17:2617–2625. doi: 10.2174/1568026617666170414142549. [DOI] [PubMed] [Google Scholar]