Abstract
Understanding the direction of information flow is essential for characterizing how genetic networks affect phenotypes. However, methods to find genetic interactions largely fail to reveal directional dependencies. We combine two orthogonal Cas9 proteins from Streptococcus pyogenes and Staphylococcus aureus to carry out a dual screen in which one gene is activated while a second gene is deleted in the same cell. We analyse the quantitative effects of activation and knockout to calculate genetic interaction and directionality scores for each gene pair. Based on the results from over 100,000 perturbed gene pairs, we reconstruct a directional dependency network for human K562 leukemia cells and demonstrate how our approach allows the determination of directionality in activating genetic interactions. Our interaction network connects previously uncharacterised genes to well-studied pathways and identifies targets relevant for therapeutic intervention.
Genetic interaction mapping approaches compare single gene loss-of-function phenotypes against combinations of loss-of-function to identify aggravating or alleviating interactions1. However, studies to address the directionality of genetic interactions have been largely limited to lower eukaryotes2–6, despite the fact that elucidating human pathway directionality is key to properly interpreting functional genetic dependencies. Such basic information offers rational approaches for therapeutic intervention, precision medicine, and evading drug resistance in human cancers. At the most fundamental level, knowing the directional flow of genetic information is critical to properly reconstruct networks and assemble a cohesive picture of gene function.
The recently discovered bacterial CRISPR phage-defence system has remarkably advanced RNA interference and related gene perturbation technologies7. A growing CRISPR toolbox offers a diversity of approaches to perform a highly parallel functional interrogation of every single gene in the human genome8,9. However, whereas single perturbation (e.g. knockout or overexpression) approaches have proven highly successful to systematically attribute function to individual mammalian genes, they typically do not provide a deeper understanding of how these genes function together in complex genetic signalling networks.
To reconstruct directional regulatory networks in human cells, we developed an orthogonal CRISPR system comprising two Cas9 enzymes derived from different species. This system allows the simultaneous and asymmetric activation of one gene and deletion of a second gene in the same cell. When compared to conventional symmetrical loss-of-function experiments in which the function of both interaction partners is lost, our orthogonal asymmetric platform allowed us to determine whether the activated gene functionally depends on, or can compensate for the loss of a deleted gene. Using this platform, we identified directional genetic interactions between genes whose activation or ablation altered the fitness of human chronic myeloid leukaemia (CML) cells. We demonstrate that the orthogonal screening approach can quantify loss- and gain-of-function phenotypes from the same cell, and that it is suitable to systematically identify genetic interactions between cancer relevant genes. We reconstruct a substantial number of directional dependencies, connecting previously uncharacterised genes to well-studied pathways.
Results
CRISPRa screen identifies cancer pathway genes
CML is a leukaemia characterised by a reciprocal translocation between chromosome 9 and 22. This translocation creates the BCR-ABL fusion oncogene, a constitutively active tyrosine kinase oncogene that causes myeloid precursor cells to divide in an uncontrolled fashion10. Application of BCR-ABL tyrosine kinase inhibitors (e.g. imatinib) have revolutionised treatment for this cancer, and decades of study have yielded fundamental information on the genes critical for BCR-ABL dependent signalling. We thus chose CML to benchmark our method to identify directional genetic interactions, using the K562 CML cell line to systematically quantify genes that function as negative or positive regulators of cancer cell fitness.
To ascertain CRISPR screening conditions capable of identifying the full gamut of imatinib dependent phenotypes, we characterised K562 cell response to a broad range of imatinib drug concentrations. We found that K562 cells respond to a wide range of imatinib concentrations (10 – 1,000 nM), and that CRISPR mediated activation (CRISPRa) of the imatinib efflux transporter ABCB1 using the SunTag system11 can result in an approximately 2-fold increase in the IC50 after 3 days of treatment (Supplemental Fig. 1a). However 3 days of drug treatment did not provide the dynamic range needed to identify genes displaying weaker phenotypes in a screen. To optimise conditions, we analysed the influence of repeated imatinib treatment cycles at IC50 on cell viability. We observed increased cell viability for ABCB1 overexpressing cells, 31.5-fold (sgABCB1-1) and 23.5-fold (sgABCB1-2) over negative controls (sgNTC), after three cycles of 100 nM imatinib (day 9) (Supplemental Fig. 1b). These results show that repeated exposure to low imatinib doses allows for much greater enrichment of cells with activated resistance genes than a single treatment.
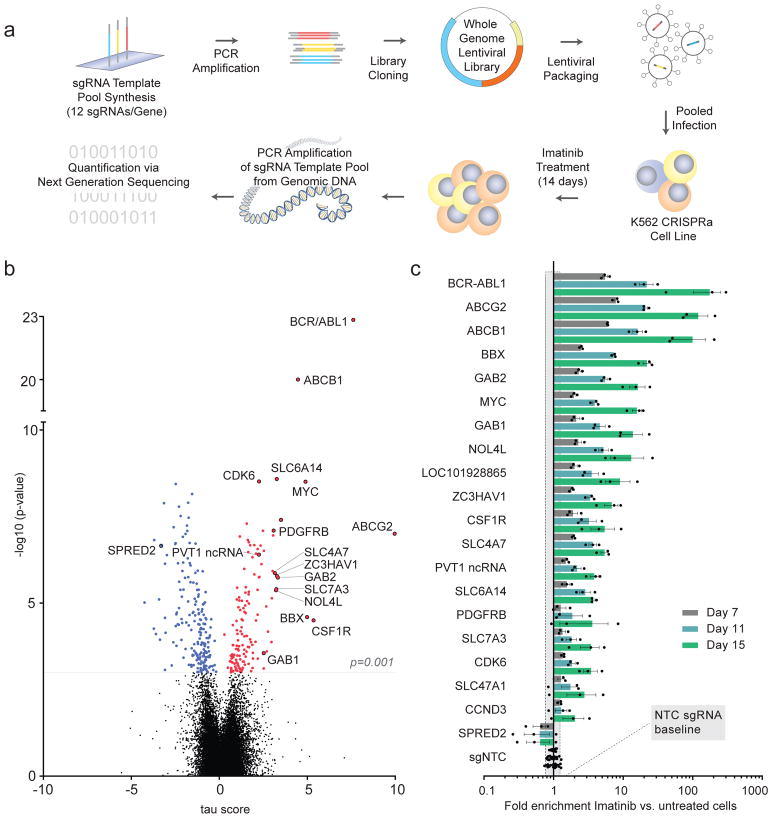
To systematically identify genes whose activation can alter imatinib drug response, we created an ultra-complex, genome-scale sgRNA library consisting of over 260,000 total sgRNAs targeting every coding and over 4,000 non-coding Refseq annotated (hg19) transcripts in the human genome. Quality-controlled sgRNA libraries (Supplemental Fig. 2) were introduced into K562 CRISPRa target cells11 followed by 14 days of imatinib treatment with escalating doses of imatinib ranging from 100 nM (IC50) to 300 nM (IC80) (Fig. 1a). Abundance of sgRNA encoding sequences was determined via next generation sequencing (NGS), comparing the beginning (baseline) and endpoint (day 14) of the screen (Supplemental Table 1). NGS read count ratios of the top 25% most enriched/disenriched sgRNAs were normalized to define an enrichment score (τ) for each gene (Supplemental Table 2). Activation phenotypes were found to be highly reproducible (r>0.98) between technical screen replicates (Supplemental Fig. 3). From a total of 26,700 targeted transcripts, we observed that the activation of 332 genes significantly (FDR<0.05, p<0.001) altered the fitness of imatinib treated K562 cells, with 57% (188 genes) causing significant depletion (blue) and 43% (144 genes) driving cell enrichment (Fig. 1b).
Figure 1. Ultra-complex CRISPRa screen identifies hundreds of genes involved in cancer signalling pathways.
a, Schematic of genome-scale CRISPRa screening approach (see text for details). b, Overview of CRISPRa screen results. Negative τ values indicate depletion and positive values enrichment of cells following imatinib selection. Significant candidate genes (FDR<0.05, p<0.001) are in colour (blue = depleted, red = enriched). Validated candidate genes are labelled in black. Mann-Whitney U test was used to calculate p-values as described previously44. To correct for multiple hypothesis testing, we first performed random sampling with replacement among the set of τ values for non-targeting control sgRNAs and calculated p-values for each sampling. Then, we calculated the false discover rate (FDR) based on the distribution of p-values for all genes in the library and for non-targeting controls generated above. c, Candidate gene validation. Enrichment of candidate sgRNA expressing cells was measured over time. Values represent the mean of three different sgRNAs targeting each gene with s.e.m. Grey shading = two standard deviations of sgNTCs at day 15. All values from separate sgRNAs on days 7, 11 and 15 normalised to baseline or untreated cells are shown in Supplemental Table 3.
A key advantage of the gain-of-function approach used here, as opposed to more commonly employed loss-of-function approaches, is that genes exhibiting no- to very low-expression can also be investigated. We found that out of the 332 candidate genes, 21% were not expressed in K562 cells (FPKM<100) indicating that imatinib responsive genes could be identified from the full spectrum of endogenous gene expression levels (Supplemental Table 2 and Supplemental Fig. 4). This approach allows for the study of genes which may have functional relevance in other cell types, i.e. different types of cancers including non-CML leukemias.
To assess the quality of the screening data on a global level, we executed a gene set enrichment analysis (GSEA)12,13 using the above 332 target genes identified in the imatinib CRISPRa screen. GSEA identified the strongest gene enrichment in leukaemia and other cancer-related KEGG signalling pathways (Supplemental Fig. 5a), illustrating the ability to identify positive and negative regulators of cancer cell survival pathways. A graphic summary of the CRISPRa screen results, assembled into their relevant oncogenic pathways is shown in Supplemental Fig. 5b. The three strongest hits, namely ABCB1, ABCG2 and BCR-ABL are well known to be overexpressed in CML patients with high tolerance to imatinib14. Additionally, we identified BCR-ABL binding partners CBL and CRKL 15, and downstream effectors SOS1, SOS2, GAB2, RAF1, MYC, PIM1, PIM2 and STAT5B 16, the c-Abl phosphatase PTPN12 17, the Ras-GAPs NF1, RASA1 and RASA3 18, the cell cycle regulators CDK6 19 and CCND3 20 and receptor tyrosine kinases having well documented roles in imatinib resistance, specifically PDGFRB 21, FGF1R 22, CSF1R 23 and AXL 24.
To evaluate the reproducibility of the CRISPRa screen data, we tested three sgRNAs per gene, against 20 of the 332 significant candidate genes individually in an arrayed 96 well plate validation assay. Genes selected for validation included the five most significantly enriched candidate genes BCR-ABL, ABCB1, SLC6A14, CDK6 and MYC as well as 15 genes whose activation produced less significant phenotypes (Supplemental Table 3). The sgRNAs targeting these 20 genes were selected based on CRISPRa screen enrichment (Supplemental Table 1). The results showed a high degree of quantitative reproducibility when compared to screen enrichment data (r=0.78), displaying a wide dynamic enrichment range over several orders of magnitude (Supplemental Fig. 6). In addition to the large number of aforementioned genes with well-established roles in leukaemia and imatinib resistance, we identified and validated a set of candidate genes with uncharacterised roles in cancer therapy resistance (Fig. 1c), including numerous solute carriers, the non-coding RNAs PVT1 and LOC101928865, as well as BBX, NOL4L and ZC3HAV1 for which upregulation following sgRNA expression was further confirmed via qRT-PCR (Supplemental Fig. 7). In total, these experiments yielded a highly reproducible list of target genes, some having well-established functions in cancer pathways, while others are completely uncharacterised. These results gave us the opportunity to study the functional relationships between the genes; hence, we sought to develop an orthogonal CRISPR platform that could illuminate genetic interactions and directional dependencies for drug resistance.
The orthogonal CRISPR system
To enable the scalable investigation of directional dependencies, we conceptualized an orthogonal CRISPR system that would allow the simultaneous activation and deletion of two genes in the same cell. To test this concept, we developed a K562 cell line harbouring the Streptococcus pyogenes based SunTag CRISPRa system and Cas9 nuclease from Staphylococcus aureus (SaCas9). These two Cas9 proteins have different PAM requirements and structural studies have shown that each enzyme recognises different constant regions of the cognate sgRNA25,26. These observations suggest that each Cas9 enzyme is not likely to cross-react with the cognate sgRNA engineered for the other Cas9 enzyme.
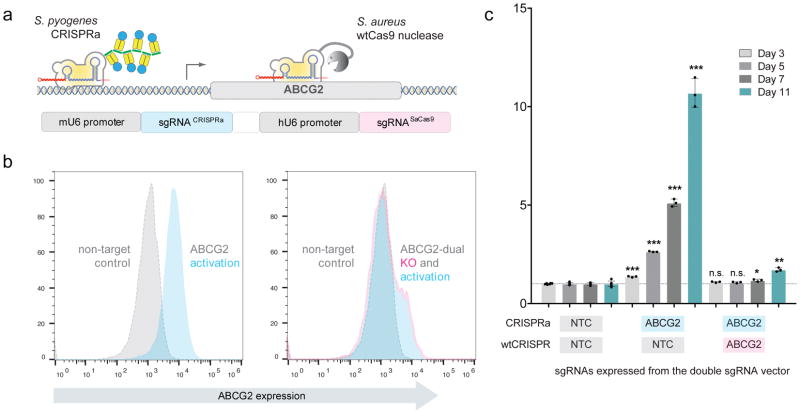
To test whether both CRISPR systems can work in parallel to activate and delete genes in the same cell without cross-interference, we designed orthogonal sgRNA expression constructs to activate the imatinib efflux transporter ABCG2 in combination with a non-target control sgRNA, or alternatively, in combination with an sgRNA that deletes ABCG2 (Fig. 2a). As expected, ABCG2 protein levels increased following the expression of a CRISPRa sgRNA against ABCG2 (Fig. 2b, left panel). In contrast, ABCG2 expression is almost completely abolished when the same CRISPRa sgRNA is used in combination with an sgRNA that deletes ABCG2 via SaCas9 nuclease (Fig. 2b, right panel). A small residual population of ABCG2 expressing cells can be observed, which we suspect likely represents cells harbouring non-edited or in-frame indels of the ABCG2 gene (Fig. 2b, right panel). To compare drug resistance profiles of these sgRNA constructs, we analysed cell enrichment following 11 days of imatinib treatment (Fig. 2c). As shown in Figure 1c, ABCG2 activation confers imatinib resistance; however, resistance is almost completely reversed in cells that concomitantly express an sgRNA that deletes ABCG2 (Fig. 2c). Taken together, these data demonstrate the ability of the orthogonal CRISPR platform to simultaneously functionally activate and delete genes in the same cell.
Figure 2. The orthogonal CRISPR system.
a, Schematic of the orthogonal system on the example of imatinib efflux transporter ABCG2. Combination of CRISPR systems from S.pyogenes (CRISPRa) and S.aureus (Cas9 nuclease) allows the simultaneous activation and knockout of genes in the same cell simply by expressing two appropriate sgRNAs. b, Orthogonal system is able to modulate ABCG2 protein levels. Flow cytometry analysis of ABCG2 levels following CRISPRa mediated activation of ABCG2 without (left) or with (right) SaCas9 nuclease mediated knockout of ABCG2 (grey histogram = sgNTC for both CRISPR systems). A representative result from n>10 independent experiments with similar results is shown. c, Orthogonal system can control imatinib response. Enrichment of imatinib treated cells with activated ABCG2 with/out SaCas9 nuclease mediated knockout of ABCG2. Values represent the mean of independent experiments (n=3) with s.e.m. and statistical significance was determined via two-tailed, homoscedastic t-test with * = p<0.05, ** =p<0.01 and *** = p<0.001.
Since our orthogonal system is based on two completely independent CRISPR systems, it opens the door to combinations of any two CRISPR-based technologies, such as transcriptional silencing27 or targeted DNA methylation28, which represents a substantial advance compared to the only other orthogonal CRISPR-based method published to date by Dahlman et al.29, which achieves gene activation and knockout using ‘catalytically dead’ sgRNAs engineered to bind the MS2:P65:HSF1 (MPH) activation complex in combination with a catalytically active wt CRISPR Cas9 nuclease from S.pyogenes. In contrast, our approach is based on Cas enzymes from two different bacterial species – S.pyogenes and S.aureus – both of which recognize distinct sgRNAs and PAMs. Consequently, the co-expressed sgRNAs for either CRISPR system do not compete for common protein factors or target sites within the same cell.
Systematic quantification of genetic interactions
To establish a high-throughput screen using the above described orthogonal CRISPR platform, we created an orthogonal dual sgRNA library composed of selected combinations of CRISPRa and SaCas9 nuclease sgRNAs. This library combined activating sgRNAs targeting 87 enriched or depleted candidate genes from the primary screen (2 sgRNAs/gene for a total of 174 sgRNAs) and knockout sgRNAs targeting 1,327 genes (8 sgRNAs/gene for a total of 11,594 sgRNAs). The knockout sgRNA population targeted all KEGG annotated cancer-relevant signalling pathway genes. The final dual orthogonal sgRNA library contained a total of over 2 million sgRNA or 100,000 gene combinations (annotated library sequences are provided in Supplemental Tables 4 and 5) targeting well-established and dark matter genes.
To promote rigour and reproducibility, the complex sgRNA combination expression library was transduced into two independently derived clonal lines of orthogonal K562 cells. The clonal lines were screened in parallel, in two separate bioreactors in the presence of escalating doses of imatinib. After 19 days, cells from both bioreactors were harvested and sgRNA representation was compared between baseline cells (day 0) and imatinib-treated cells (day 19, see Methods for details). Before calculating genetic interactions, it was imperative that we first calculated the effects of single gene activation-only and knockout-only phenotypes. To facilitate this calculation, we included a large number of non-target control sequences in the combination library; 18 non-target controls in the CRISPRa position and nearly 900 in the SaCas9 nuclease position (see Supplemental Tables 4 and 5). Dissecting these two populations of control vectors allowed the clean evaluation of replicate performance for both single gene activation (r=0.96, Fig. 3b) and single gene knockout phenotypes (r=0.98, Fig. 3c). Both single gene activation and single gene knockout phenotypes included negative and positive regulators of cell fitness in the presence of imatinib. These results were highly reproducible between replicates (Fig. 3b and c), and CRISPRa values correlated (r=0.9267) with values from the initial CRISPRa screen (Supplemental Fig. 8).
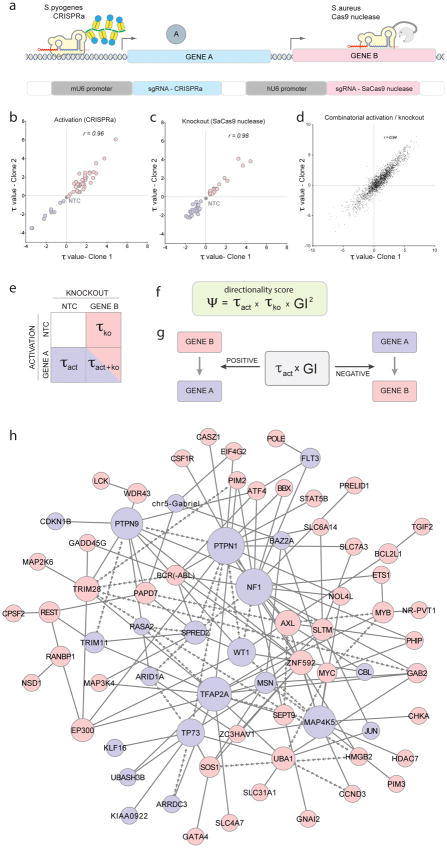
Figure 3. Orthogonal CRISPR screens can quantify directional genetic interactions.
a, Concept of the application of the orthogonal system for directional gene interaction studies. In the same cell, one gene is activated (CRISPRa) while another gene in knocked out (SaCas9 nuclease). b–d, Correlation of τ values from two clonal cell line replicates is shown for b, gene activation, c, gene knockout and d, all possible combinations thereof. Correlation values (r) are Pearson product-moment correlation coefficients. e, Schematic of perturbation data set from each gene pair (blue = depleted, red = enriched, NTC = non-target control sgRNA) f, Formula for calculating Ψ scores. Negative Ψ scores define interactions in which directionality could be inferred. g, To determine which of both interaction partners acts up- or downstream, τactivation values were multiplied with genetic interaction scores. Positive values indicate a downstream function, negative values an upstream function of the activated gene. h, Based on GI and Ψ scores determined by the full orthogonal interaction screen, a genetic interaction model was constructed. For positive regulators of cell fitness, nodes are shown in red and negative regulators in blue. Arrow-shaped edges indicate inferred directional interactions between nodes. Line-shaped edges symbolise genetic interactions where directionality could not be inferred. Node sizes are proportional to the degree of connectivity. In total, 2258 gene:gene combinations that passed the filter criteria were considered for the construction of the directional genetic interaction network.
Notably, phenotypic measurements derived from all possible combinations of sgRNAs in the activation and knockout position with one another were also found to be highly reproducible between clonal screen replicates (r=0.94), allowing a quantitative comparison of all possible combinations of sgRNAs in their full operationally functional orthogonal context (Fig. 3d). Taken together these data confirm: 1) the ability for both Cas9 systems to work in parallel to produce activation and knockout phenotypes in the same cell, and 2) the suitability of our NGS analysis pipeline to accurately quantify phenotypes from combinatorial gene perturbations.
Deducing directional dependencies
In genetic interactions where a gene activates its partner, gene activation and knockout produce opposing phenotypes (τact and τko) and the double perturbation phenotype (τact+ko) can lie in the full spectrum between both individual perturbation phenotypes (Fig. 3e). To systematically identify and quantitate directional genetic dependencies from the screen, we determined genetic interaction (GI) scores from individual and combinatorial τ values and based on those, defined a single directionality score Ψ (Fig. 3f). In essence, Ψ displays a negative value when enrichment scores τ from gene activation and knockout have opposing signs, as would be expected in activating interactions. For interactions with negative Ψ scores, the τact and GI scores were multiplied to determine whether the activated gene functions downstream (positive value) or upstream (negative value) of the deleted gene (Fig. 3g).
To maintain rigor, we assigned directionality only in reproducible genetic interactions (GI scores that exceeded a 1x standard deviation in both clonal cell line replicates) that exhibited a negative Ψ score. A summary of all calculated τ, GI and Ψ values is shown in Supplemental Table 6. Based on the determined GI and Ψ scores, we derived a directional genetic interaction network de novo. We assembled the network from the most significant and reproducible directional and non-directional interactions determined by the orthogonal screen (Fig. 3h; Supplemental Table 7 and Supplemental Figure 9). As explained above, directionality among genetic interactions can be inferred only when the activated gene displayed the opposite phenotype of the knocked-out gene; but not if activation and knockout both resulted in the same phenotype (Supplemental Fig.10). The resulting directional-edge model connects a total of 70 cancer-centric nodes, via 137 gene:gene interactions determined from both clonal replicates, for 26 of which directionality could be directly inferred. For clarity, interactions for all 70 individual nodes are depicted in Supplemental Figure 11.
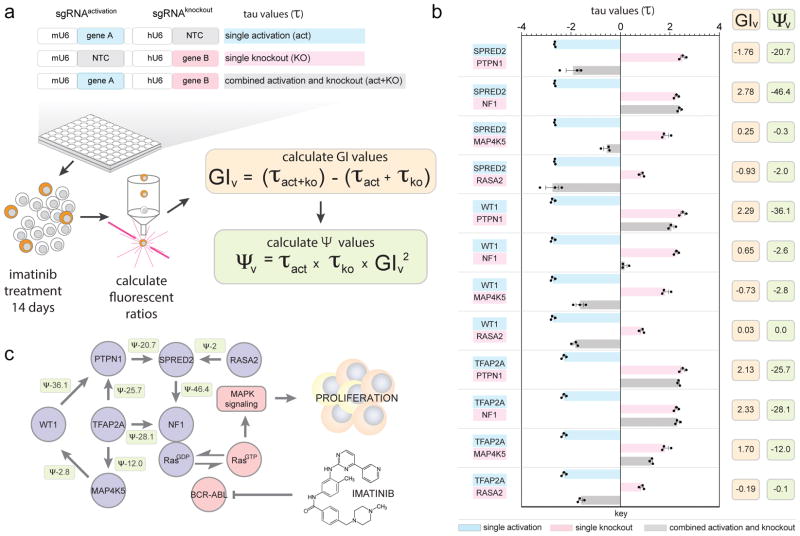
To quantitatively evaluate the orthogonal screen data, we validated the performance and calculated pathway directionalities in several independent assays. First, a sample set of predicted directional interactions between selected genes with negative Ψ scores which passed the cut-off in clonal replicate 2 (Supplemental Table 6) were re-tested in an arrayed validation assay using the same orthogonal clonal line. Single as well as combinatorial activation and knockout τ values from the arrayed validation experiments were determined and used to derive GIv and Ψv scores (Fig. 4a). Control single gene activation and knockout phenotypes validated for all re-tested genes. Activation of SPRED2, WT1 and TFAP2A had a sensitizing effect to imatinib treatment, while deletion of PTPN1, NF1, MAP4K5 and RASA2 caused cells to steadily enrich in the culture over time (Fig. 4b and Supplemental Table 8). Figure 4b shows single and double perturbation τ values of twelve gene:gene combinations, determined on day 14 of the arrayed validation along with calculated GIv and Ψv scores.
Figure 4. Validation of a directional Ras-centric genetic sub-network.
a, Relative fitness (τ) was measured over 14 days following gene activation, knockout or the combination of both. From those values, genetic interaction (GIv) as well as directionality (Ψv) scores were calculated. NTC = non-target control sgRNA. b, Twelve activation/knockout combinations were re-tested in an arrayed format from which ten were predicted by the orthogonal screen to show a directional genetic interaction. Eight combinations displayed the same trend of directional interactions predicted by the orthogonal screen data while two interactions did not reproduce (see also Supplemental Table 9). Single perturbation, and combinatorial τ values are shown following 14 days of imatinib selection (mean with s.e.m. from technical replicates (n=3)) along with calculated GIv and Ψv scores for each gene:gene combination. c, A directional genetic interaction model was assembled based on validated interactions from b. Arrows indicate the direction of the functional dependencies as explained in the text but do not suggest direct physical interactions. Values represent Ψv scores calculated from τ values in b. Each directional interaction was reproduced three times independently.
Overall, GIv scores were found to be in good agreement (r=0.72) with GI scores determined by the orthogonal screen (Supplemental Table 9). In five cases the activated gene was unable to execute its sensitizing function following the deletion of its interaction partner (τact x GI = negative: SPRED2-NF1, WT1-PTPN1 and TFAP2A-PTPN1/-NF1/-MAP4K5), supporting a model for an upstream function of the activated gene. In contrast, we observed three instances where the activated gene could compensate for the loss of its interaction partner (τact x GI = positive: SPRED2-PTPN1/-RASA2 and WT1-MAP4K5), supporting a model for a downstream function of the activated gene. Out of the total of twelve tested combinations, ten were predicted by the orthogonal screen to show a directional interaction, of which the aforementioned eight were confirmed by our arrayed validation while two interactions (WT1-NF1 and TFAP2A-RASA2) did not reproduce (Fig. 4b and Supplemental Table 9). The inability to validate those two interactions might be explained by the markedly different experimental conditions between the orthogonal screen in a 14 L agitated bioreactor with precisely controlled culture conditions versus validation in a 96-well plate. Moreover, GI and Ψ scores from the screen were calculated based on multiple sgRNAs for gene activation and knockout whereas validation was performed with one selected sgRNA in either position.
Based on the validated interactions, we reconstructed a Ras-centric high-confidence directional genetic interaction model with Ψv scores calculated from the validation data (Fig. 4c). This model is further supported by our findings that SPRED2 cannot sensitise NF1-deleted cells to imatinib treatment, despite showing a similar increase in mRNA levels following its activation (Supplemental Fig. 12a and b). At this point it is important to mention that the relative changes in gene expression, as detected by qRT-PCR, do not necessarily translate into equivalent phenotypes. In other words, although the significant increase in SPRED2 mRNA levels following its CRISPRa mediated activation might seem modest (approx. 2 fold), the detected phenotype at 8 days after imatinib treatment is a remarkably significant 3-fold decrease in cell numbers (Supplemental Data Figure 12). Moreover, SPRED2 overexpression in HEK293T cells lowered Ras-GTP levels only in the presence of NF1, confirming that the ability of SPRED2 to suppress Ras activity depends on NF1 (Supplemental Figure 12c). These observations build upon previous observations that double knockdown of SPRED1/2 leads to increased Ras-GTP levels due to disruption of an NF1/SPRED2 complex30.
Exploiting genetic vulnerabilities for cancer therapy
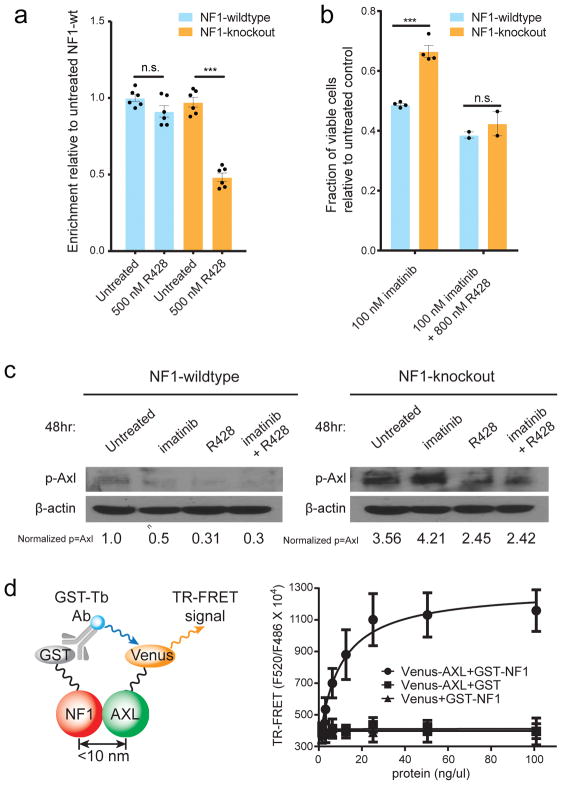
Given the potential to discover genetic dependencies of therapeutic relevance, we investigated the observed interaction between NF1 and the TAM receptor tyrosine kinase AXL31 (Fig. 3h and Supplemental Table 7). Targeting AXL-mediated signaling pathways can lead to regained drug sensitivity and improved therapeutic efficacy, defining AXL as a promising target for cancer therapeutics32,33. However, a key issue for therapeutic intervention is the selection of appropriate biomarkers and potential synergistic drug targets for combination-based regimes. To evaluate therapeutic applicability and potential synergies, we applied R428 (a specific AXL inhibitor which is currently being evaluated in clinical trials34) to a population of Cas9 NF1-knockout sgRNA treated cells as well as control cells. We found that NF1-knockout cells were highly sensitive to R428, whereas NF1-wildtype control cells did not show a significant response to 8 days of treatment with 500 nM R428 (Fig. 5a). Given that the NF1-knockout sgRNA treated cells contain sub-populations of non- and in-frame edited cells (Supplemental Fig.13) we anticipate that these observations likely underestimate R428 drug sensitivity. Moreover, these observations were also extended to lung epithelial cells using RNAi mediated knockdown of NF1 in BEAS-2B cells, which displayed significantly increased drug sensitivity (p=3x10−3 at 500 nM R428, p=6x10−5 at 1000 nM R428) when compared to matched control cells (Supplemental Fig. 14). Finally, we confirmed that NF1-knockout K562 cells are more resistant to treatment with imatinib, but that these cells can be re-sensitised to imatinib by R428 treatment (Fig. 5b).
Figure 5. Exploiting genetic dependencies for cancer therapy.
a, NF1-knockout K562 cells are significantly more sensitive to the AXL kinase inhibitor R428 than NF1-wildtype cells. Cells were treated for 8 days with 500 nM R428 on day 0 and day 4. (mean with s.e.m. from technical replicates (n=6)). b, NF1-knockout K562 cells are significantly more resistant to imatinib but can be re-sensitised by R428 treatment (mean with s.e.m. from technical replicates (n=4 for imatinib and n=2 for imatinib + R428 treated cells). In panel a and b, statistical significance was determined via two-tailed, homoscedastic t-test with * = p<0.05, **=p<0.01 and *** = p<0.001. c, NF1-knockout cells accumulate elevated levels of phosphorylated AXL kinase (p-AXL) which can be reduced by treatment with the AXL kinase inhibitor R428. Quantification of the ratio of band intensity from p-AXL/β-actin, normalised to p-AXL levels in NF1-wt untreated cells is shown. The experiment was performed once. d, TR-FRET assay shows direct interaction between NF1 and AXL in HEK293T cells. Shown is the mean with s.d. from three independent experiments.
To explore the nature of the selective AXL dependency of NF1-deficient cells, we quantitated phosphorylated AXL kinase (p-AXL) levels in control untreated wildtype cells and NF1-knockout sgRNA treated cells. NF1-knockout sgRNA cells displayed markedly higher levels of p-AXL than NF1-wildtype cells, indicating that these cells had accumulated higher levels of AXL activity and p-AXL levels were reduced upon R428 treatment in both, NF1-wildtype and knockout cells (Fig. 5c). To further investigate the interaction between NF1 and AXL, we performed a homogeneous time-resolved Förster resonance energy transfer (TR-FRET) assay where the stringent proximity (<10 nm) based idiosyncrasy allows the detection of direct physical interactions35. These experiments provided additional support that NF1 and AXL physically interact with each other in a cell based assay (Fig. 5d). Additionally, we show that both proteins bind to all three Ras isoforms N-Ras, H-Ras and K-Ras (Supplemental Figure 15), supporting a model where NF1 deficient cells become increasingly dependent on AXL signalling, and that these cells can be selectively targeted by the AXL inhibitor R428. Given the high recurrence of NF1 mutations and AXL activation in a variety of human cancers, our data provide an informed basis for therapeutic intervention.
Discussion
Inferring the direction of genetic interactions has been a long-standing challenge. While previously described genetic interaction studies are based on simple dual loss-of-function36–39,40–43, the orthogonal approach combines the power of CRISPR mediated activation of one interaction partner with the functional loss of a second gene in the same cell. Here we establish the full methodology and reagents necessary to conduct highly parallel directional CRISPR screens in human cancer cells, including stable CRISPRa-SaCas9 nuclease cell lines, dual sgRNA libraries, and a barcode-free next generation sequencing strategy to quantify sgRNA combinations in orthogonal screens. As a general concept, our described inference of directionality strategy is readily applicable to numerous other dual activation/inhibition expression embodiments, notwithstanding other transcriptional, post-transcriptional, and post-translational regulatory modules.
Approaches to construct quantifiable directional models for genetic interactions have been limited and there have been no established technologies to efficiently specify directionality within pathways. This is particularly a problem in fields such as cancer biology where a major ongoing focus is to identify synergistic genetic vulnerabilities that provide a sound basis for the design of rational polytherapies to help prevent drug resistance. Here, we provide a comprehensive dataset consisting of single and combinatorial gain- and loss-of-function phenotypes in CML cells, and a high-confidence network of genetic interactions that will help researchers to build hypotheses to further understand why some patients respond well to tyrosine kinase inhibitors like imatinib, whereas others acquire resistance. In many cases, directional dependencies need to be considered when designing a treatment plan for patients harbouring multiple genetic lesions, and the described orthogonal platform offers a fresh new approach to uncovering key dependencies in pathways critical for human gene function and disease.
Methods
Vector maps
For the single sgRNA (sgLenti), dual sgRNA (sgLenti-orthogonal) and SaCas9 nuclease vector, vector maps are provided in Genbank format (Supplemental 1–3) and have been deposited along with the plasmids at Addgene
CRISPRa and orthogonal K562 cell lines
K562 CRISPRa cells11,46 were kindly provided by Luke Gilbert and cultured in RPMI 1640 medium, supplemented with 10% fetal bovine serum and 1x Anti-Anti (Gibco). Via lentiviral transduction, S. aureus Cas9 under the control of an EF1α promoter, was introduced into K562 CRISPRa cells (see Supplemental 3 for vector map). Successfully transduced cells were selected with hygromycin (200 ug/mL) and single clones were expanded for 14 days. To test functionality of the expanded clonal orthogonal lines, cells were transduced with sgRNAs to activate the imatinib efflux transporter ABCG2 via CRISPRa (5′-GCCACTGCGTTCAGCTCTGG-3′) or to knock it out in combination with SaCas9 (5′-CATCTGCTATCGAGTAAAACTG -3′). Four weeks post introduction of the SaCas9 expression cassette, clonal lines were screened for functionality of both CRISPR systems (CRISPRa and SaCas9 nuclease) via flow cytometry analysis of >10,000 cells stained with CD338 (ABCG2) antibodies (Miltenyi, 130-104-960). Out of a total of 28 screened orthogonal lines, all 28 retained the functional CRISPRa system but only four lines displayed stable function of SaCas9 nuclease. Two of those lines were used for the orthogonal CRISPR screen.
CRISPRa and orthogonal sgRNA library design
For the initial CRISPRa screen, a genome-scale sgRNA library consisting of over 260,000 total sgRNAs targeting every coding, and over 4,000 non-coding, Refseq annotated (hg19) transcripts in the human genome, as well as every unique protein coding isoform with up to 12 sgRNAs, plus 7,700 non-target control sequences (NTC).
The promoter regions for coding transcripts targeted windows 25 to 500bp upstream of the Refseq-annotated transcription start sites. SgRNAs were designed against targets in the promoters that are of the format (N)20NGG, and selected sgRNAs must pass the following off-targeting criteria: 1) the 11bp-seed must not have an exact match in any other promoter region, and 2) if there is an exact off-target seed match, then the rest of the sgRNA must have at least 7 mismatches with the potential off-target site. Regions outside a window of 25 to 500 bp upstream of the TSS were not considered for off-targeting since the employed CRISPRa system was shown to work only in proximity to the TSS of genes46 and to not further limit the number of designable sgRNAs for the narrow on-target space. After all sgRNAs that pass off-targeting criteria were generated, up to 12 sgRNAs/transcript were selected that were nearest to the transcription start sites. All sgRNA sequences are shown in Supplemental Table 1. In addition to the sgRNA sequence, every plasmid contained a unique 20 nt barcode sequence (see Supplemental 1 for vector map). This sequence allowed the distinction between sgRNAs expressed from different plasmids and hence in different sub-populations of cells and was used to bin cells into mutually exclusive barcode bins to create technical screen replicates after sequencing.
For the orthogonal genetic interaction screen, a focused nuclease-active S. aureus Cas9 library was generated targeting 1327 genes. For the selected genes, sgRNAs targeting coding exons were generated using Cas-Designer 47, generating sgRNAs that were adjacent to the PAM sequence ‘NHGRRT’ (H = A, C, or T), which allows for targeting with S. aureus Cas9 but not with S. pyogenes Cas9. Potential off-targets against the human genome were identified using Cas-OFFinder48. To score sgRNA sequences by Cas-OFFinder, sgRNAs that have perfect-seed off-targets and 5 mismatches or less in potential off-target regions were penalised. The 20% of sgRNAs with the highest off-target penalties and bottom 20% of sgRNAs with the lowest out-of-frame scores from Cas-Designer were eliminated. From the resulting list of sgRNAs, up to 8 sgRNAs/gene were selected, targeting the most 5′ constitutive exons for each gene.
CRISPRa and orthogonal sgRNA library cloning
For the CRISPRa library, the designed 20 nt target specific sgRNA sequences were synthesised as a pool, on microarray surfaces (CustomArray, Inc.), flanked by overhangs compatible with Gibson Assembly49 into the pSico based sgLenti sgRNA library vector (see Supplemental 1 for vector map). The synthesised sgRNA template sequences were of the format: 5′-GGAGAACCACCTTGTTGG-(N)20-GTTTAAGAGCTATGCTGGAAAC-3′. Template pools were PCR amplified using Phusion Flash High-Fidelity PCR Master Mix (ThermoFisher Scientific) according to the manufacturers protocol with 1 ng/uL sgRNA template DNA, 1 uM forward primer (5′-GGAGAACCACCTTGTTGG-3′), 1 uM reverse primer (5′-GTTTCCAGCATAGCTCTTAAAC-3′) and the following cycle numbers: 1x (98C for 3 min), 15x (98C for 1 sec, 55C for 15 sec, 72C for 20 sec) and 1x (72C for 5 min). PCR products were purified using Minelute columns (Qiagen). The library vector sgLenti was preapred by restriction digest with AarI (Thermo-Fischer) at 37C overnight, followed by 1% agarose gel excision of the digested band and purification via NucleoSpin columns (Macherey-Nagel). Using Gibson Assmbly Master Mix (NEB), 1000 ng digested sgLenti and 100 ng amplified sgRNA library insert were assembled in a total 200 uL reaction volume. The reaction was purified using P-30 buffer exchange columns (Biorad) that were equilibrated 5x with H2O and the total eluted volume was transformed into three vials of Electromax DH5α (ThermoFisher). E.coli were recovered, cultured overnight in 500 mL LB (100 ug/mL ampicillin) and used for Maxiprep (Qiagen). In parallel, a fraction of the transformation reaction was plated and used to determine the total number of transformed clones. The coverage was determined to be 70x clones per sgRNA ensuring even representation of all library sgRNA sequences and their narrow distribution (Supplemental Fig. 2). Fidelity of sgRNA sequences was confirmed with a more than 90% perfect Bowtie alignment rate and narrow distribution of sgRNA sequences, with read counts for 87% of sgRNA sequences falling within a single order of magnitude.
For orthogonal CRISPR libraries, CRISPRa sgRNA pools of 174 sgRNA against 87 selected target genes (2 sgRNAs/gene) plus 18 non-target control sgRNAs were cloned into position 1 of the AarI-digested plasmid sgLenti-orthogonal exactly as described for the CRISPRa library. Off target analysis using Cas-OFF finder48 showed that out of the total 192 sgRNAs, only two had an additional perfectly matched genomic target site which was outside of the defined relevant CRISPRa off-target space (25 to 500 nt upstream of the TSS) while the rest had exactly one target site. In addition, 6 sgRNAs had off-target sites with 1 mis-match and 65 sgRNAs had off-target sites with 2 mis-matches outside the defined off-target space. A full summary of CRISPRa sgRNA sequences with the number and nature of determined off-target sites is shown in Supplemental Table 4.
Following amplification in E.coli, library plasmids with the first position cloned were digested with BfuAI (NEB) to allow cloning of SaCas9 sgRNAs into the second position. To remove undigested orthogonal sgRNA library plasmid from the pool, the purified (Nucleospin, Macherey-Nagel) BfuAI digested plasmid was subsequently digested with AscI for which restriction sites exist in the stuffer sequences in both sgRNA positions 1 and 2. BfuAI/AscI digested plasmid was extracted from 1% Agarose gel (Nucleospin, Macherey-Nagel).
Synthesised SaCas9 sgRNA template sequences (12,500 total, 8 sgRNAs/gene) were of the format: 5′-GAAAGGACGAAACACCGTG-(N)22-GTTTTAGTACTCTGGAAACAGAATCT-3′. PCR amplification of the SaCas9 template pool was performed as described above using primer sequences: 5′-GAAAGGACGAAACACCGTG-3′ and 5′-AGATTCTGTTTCCAGAGTACTAAAAC-3′ and the purified PCR product was cloned into BfuAI digested sgLenti-orthogonal (see Supplemental 2 for vector map) via Gibson Assembly as described above. The resulting orthogonal sgRNA library was transformed into Electromax cells at 30x coverage as described above and the plasmid sgRNA library pool was purified (Qiagen Plasmid Maxi kit). From the resulting plasmid pool, sgRNA sequences were recovered via PCR as described below and sequenced for quality control. At a read depth of 94x, 2.389 million out of the total possible 2.394 million combinations (>99%) were read at least once, with less than 5% of the library elements read 20 or less times.
Lentivirus production
HEK293T cells were seeded at 65,000 cells per ccm in 15 cm dishes in 20 mL media (DMEM, 10% fetal bovine serum) and incubated overnight at 37C, 5% CO2. The next morning, 8 ug sgRNA library plasmid, 4 ug psPAX2 (Addgene #12260), 4 ug pMD2.G (Addgene #12259) and 40 uL jetPRIME (Polyplus) were mixed into 1 mL serum free OptiMEM (Gibco) with 1x jetPRIME buffer, vortexed and incubated for 10 min at RT and added to the cells. 24 h later, 40U DNAseI (NEB) were added to each plate in order to remove untransfected plasmid and at 72h post-transfection, supernatant was harvested, passed through 0.45 um filters (Millipore, Stericup) and aliquots were stored at −80C.
Genome-wide and orthogonal CRISPR screens
Imatinib selection conditions for all screens were optimized by activating the imatinib efflux transporter ABCB1 using sgABCB1-1 (5′-CAGGAACAGCGCCGGGGCGT-3′) and sgABCB1-2 (5′-AGCATTCAGTCAATCCGGGC-3′) (Supplemental Figure 1). K562 CRISPRa/orthogonal cells were transduced with lentivirally packaged sgRNA libraries at MOI=0.3 and 500x coverage. The low MOI was used to reduce the frequency of multiple-infected cells; thus, only one gene was activated in each cell. Cells were then cultured in RPMI with 10% FBS and 1x Anti-Anti (Gibco) in a 37C incubator with 5%CO2. 48h post transduction, cells were selected with puromycin (2 ug/mL) for 96h. Following selection, aliquots of 300 million cells each, were frozen down in FBS with 10% DMSO for later analysis via NGS (see below). Fully selected cells (300 million) were transferred into a 14 liter CelligenBlu bioreactor (Eppendorf) and sub-cultured at 37C, pH=7.4 and 2% oxygen. Coverage at cell level was kept above 1000x throughout the entire screen and the culture was diluted with fresh medium when cell density reached 1 mio/mL.
For the genome-wide CRISPRa screen: 14 days post transduction, aliquots of 300 mio cells from the beginning of the screen were frozen down (baseline sample) as described above and an IC50 concentration of 100 nM imatinib (Sigma) was added to the bioreactor vessel. Imatinib was refreshed on day 17 (IC60 = 150 nM) and day 19 (IC80 = 300 nM) after initial sgRNA library transduction and cells for the analysis of the final time point were harvested on day 28. For the orthogonal genetic interaction screen: Puromycin selected cells (2 ug/mL) at 8 days post transduction (2.5 billion per sample) were frozen down as described above and 100 nM imatinib (IC50) were added to the bioreactor vessel. Imatinib concentrations were increased throughout the screen to the IC60 concentration of 150 nM (day 10), the IC80 of 300 nM (day 13 and 15) and finally the IC90 of 500 nM (day 17). On day 19 2.5 billion cells per sample were harvested for downstream analysis via NGS as described below.
Genomic DNA (gDNA) extraction
Cell pellets from baseline and imatinib treated samples were resuspended in 20 mL P1 buffer (Qiagen) with 100 ug/mL RNase A and 0.5% SDS followed by incubation at 37C for 30 min. After that, Proteinase K was added (100 ug/mL final) followed by incubation at 55C for 30 min. After digest, samples were homogenised by passing them three times through a 18G needle followed by three times through a 22G needle. Homogenised samples were mixed with 20 mL Phenol:Chlorophorm:Isoamyl Alcohol (Invitrogen #15593-031), transferred into 50 mL MaXtract tubes (Qiagen) and thoroughly mixed. Samples were then centrifuged at 1,500g for 5 min at room temperature (RT). The aqueous phase was transferred into ultracentrifuge tubes and thoroughly mixed with 2 mL 3M sodium acetate plus 16 mL isopropanol at RT before centrifugation at 15,000g for 15 min. The gDNA pellets were carefully washed with 10 mL 70% ethanol and dried at 37C. Dry pellets were resuspended in H2O and gDNA concentration was adjusted to 1 ug/uL. The degree of gDNA shearing was assessed on a 1% agarose gel and gDNA was sheared further by boiling at 95C until average size was between 10–20 kb.
PCR recovery of sgRNA sequences from gDNA
Multiple PCR reactions were prepared to allow amplification of the total harvested gDNA from a 1000x cell coverage for each sample. For the first round of two nested PCRs, the total volume was 100 uL containing 50 ug sheared gDNA, 0.3 uM forward (5′-ggcttggatttctataacttcgtatagca-3) and reverse (5′-cggggactgtgggcgatgtg-3′) primer, 200 uM each dNTP, 1x Titanium Taq buffer and 1 uL Titanium Taq (Clontech). PCR cycles were: 1x (94C - 3 min), 16x (94C - 30 sec, 65C – 10 sec, 72C – 20 sec), 1x (68C – 2 min). All first round PCRs were pooled and a fraction was used as template for the second round PCR. The total volume of the second round PCR was 100 uL containing 2 uL pooled first round PCR, 0.5 uM forward (5′-AATGATACGGCGACCACCGAGATCCACAAAAGGAAACTCACCCTAAC-3′) and reverse (5′-CAAGCAGAAGACGGCATACGAGAT-(N)6-GTGACTGGAGTTCAGACGTG-3′) primer where (N)6 is a 6 nt index for sequencing on the Illumina HiSeq platform, 200 uM each dNTP, 1x Titanium Taq buffer and 1 uL Titanium Taq (Clontech). PCR cycles were: 1x (94C - 3 min), 16x (94C - 30 sec, 55C – 10 sec, 72C – 20 sec), 1x (68C – 2 min). The resulting PCR product (344 bp) was extracted from a 1% agarose gel. For the orthogonal genetic interaction screen, conditions for the first round PCR were slightly modified to: total reaction volume 80 uL containing 20 ug sheared gDNA and the second round PCR product was 887 bp.
Gel extracted bands from the primary CRISPRa screen were submitted for sequencing on an Illumina HiSeq 2500 platform using paired end 50 kits with the custom sequencing primer 5′-GAGACTATAAGTATCCCTTGGAGAACCACCTTGTTGG-3′ for reading the sgRNA sequence and the Truseq Illumina reverse primer to read out 20 nt random barcode sequences used for generation of technical screen replicates (separation of sgRNA reads into three groups with mutually exclusive barcode sequence bins). For orthogonal dual sgRNA library analysis, single end 50 kits were used and read cycles were split, 25 cycles for Read1 with the sequencing primer above (reading the S.pyogenes sgRNA) and 25 read cycles for the ‘Illumina indexing read’ with the custom indexing primer 5′-TTGGCTTTATATATCTTGTGGAAAGGACGAAACACCGTG-3′ (reading the S.aureus sgRNA).
Data analysis
Total read counts of sgRNA sequences from each NGS sample were collapsed and quantified via alignment to the sgRNA library reference sequences using Bowtie 2.014. Data analysis was conducted similarly as described previously38,44. Briefly, for the primary CRISPRa screen, the frequency of sgRNAs was determined by deep sequencing and the average read count of three technical replicates was used. The phenotype τ was calculated to quantify the effect of an sgRNA on cell growth in the presence of imatinib. Specifically, τ values were calculated as:
where Nx denotes the frequency of sgRNA x and NNTC denotes the frequency of non-targeting control sgRNAs at baseline (t0) or after imatinib treatment (t). Gene-level phenotypes were calculated by averaging the phenotypes of the top 25% most extreme sgRNAs targeting this gene. The statistical significance for each gene is determined by comparing the set of τ values for sgRNAs targeting it with the set of τ values for non-targeting control sgRNAs using the Mann-Whitney U test, as described previously44. To correct for multiple hypothesis testing, we first performed random sampling with replacement among the set of τ values for non-targeting control sgRNAs and calculated p values for each sampling. Then, we calculated the false discover rate (FDR) based on the distribution of P values for all genes in the library and for non-targeting controls generated above. The P-value cutoff was chosen based on an FDR < 0.05.
For the orthogonal double-sgRNA screen, combinations of non-targeting control sgRNAs served as negative control, combinations of one non-targeting control sgRNA and one targeted sgRNA were used to determine single-sgRNA phenotypes and combinations of two targeted sgRNAs were used to calculate double phenotypes. Raw read counts used for analysis are shown in Supplemental Table 10. We then implemented a series of filtering steps on the sgRNA level. First of all, on the SaCas9 nuclease side, p values were calculated for each gene as described above. Only the sgRNAs targeting genes that have significant editing phenotypes (P value < cutoff) were retained. Subsequently, GI scores were calculated using the ‘force-fit’ definition for genetic interactions on the sgRNA level and sgRNAs were further filtered by GI correlation as described previously 44. On the CRISPRa side, if two sgRNAs targeting the same gene have low correlation, the gene was excluded for further analysis. After the filtering process, gene-level phenotypes and GI scores were calculated by averaging all double-sgRNAs targeting the same gene-gene combinations.
Directional genetic interaction network model
Genetic interactions whose GI scores exceeded a 1x standard deviation consistently in both clonal screen replicates were used to construct a GI network (Supplemental Table 7). To quantify directionality in these reproducible genetic interactions, a directionality score (Ψ) was calculated as
resulting in a negative Ψ when gene activation and knockout had opposing phenotypes. Negative Ψ values below a negative 1x standard deviation of all calculated Ψ values were used to infer the direction of genetic interactions. The network analysis software platform Cytoscape50 was used to visualise the genetic interaction model. Where applicable, directionality in GIs was indicated by arrow shaped edges and line shaped edges indicate significant GIs for which directionality could not be inferred. Nodes were coloured according to gene function with blue symbolizing genes that act to decrease and red to increase cell fitness of imatinib treated cells.
Arrayed competitive growth validation experiments
Individual CRISPRa or orthogonal dual sgRNA sequences for validation experiments were sub-cloned into the same vector as the respective libraries. For that purpose oligonucleotides encoding the sgRNA sequence as well as the reverse complementary sgRNA sequence were synthesised with compatible 4 nucleotide 5′-overhangs for cloning into the SpCas9 (top strand: TTGG, bottom strand: AAAC) or SaCas9 (top strand: GCTG, bottom strand: AAAC) position of the target vector respectively (for vector preparation see above). Oligonucleotides were adjusted to 100 uM and reverse complementary strands were mixed, heated to 99°C and cooled down to 4°C at a ramp rate of −0.1°C/sec in a thermocycler. Annealed oligonucleotide double strands were diluted 1:200 and 1 uL was mixed with 50 ng digested vector, 1 uL 10x T4 ligase buffer (NEB) and 0.5 uL T4 ligase (2000U/uL, NEB) in a total volume of 10 uL. Following incubation for 30 min at room temperature, 1 uL ligation reaction was transformed into 20 uL chemically competent DH5α E.coli, plated on LB-amp (100 ug/mL) agar plates and incubated at 37°C overnight. Individual clones were picked and sgRNA sequences were validated via Sanger sequencing.
All library vectors co-expressed mCherry which was used to track the abundance of sgRNA expressing cell populations in growth competition assays. For this purpose, sgRNA expressing cells were mixed with parental - mCherry-negative – cells at ratios between 1:1 and 1:3 in 96-well plates before repeated treatment with imatinib, R428 or no drug for indicated time periods. Enrichment or depletion of the mCherry positive (sgRNA expressing) cell population, indicating an increase or decrease of fitness over time following sgRNA expression and could conveniently be followed via FACS quantification of the mCherry-positive (sgRNA expressing) versus mCherry negative (parental, no sgRNA expressing) population. For screen validation experiments, τ values for each sgRNA were calculated equivalent to screen τ values:
where N represents the fraction of sgRNA expressing (mCherry-positive/mCherry-negative) baseline cells (t0) or indicated time points (t) and X represents a given sgRNA while NTC represents a scrambled non-target control sgRNA. Each value was quantified from three technical replicates. Where indicated, fold-enrichment or fold-change values were calculated as 2 to the power of τ (2τ). The sgRNA sequences used for validation of candidate genes from the primary CRISPRa screen are shown in Supplemental Table 3.
For validation of genetic interactions, dual sgRNA expression constructs were cloned into sgLenti-orthogonal using CRISPRa sgRNAs for SPRED2 (5′-GATTCGGAGCCAGACGGTCG-3′), WT1 (5′-GGACTCACTGCTTACCTGAA-3′), TFAP2A (5′-AGGGGAATGTGGCGGAATTG-3′) and non-target control (5′-CCCTGCCGTCCTCTACGAAT-3′) and SaCas9 nuclease sgRNAs for NF1 (5′-TTGTCTTTGGGTGTATTAGCAA-3′), MAP4K5 (5′-AGCAGGACTACGAACTCGTCCA-3′), PTPN1 (5′-ACTTTCTTGATATCAACGGAAG-3′), RASA2 (5′-CCCACTAGAGAAACTGTTGCAT-3′) and non-target control (5′-ACGCGTGCGTAATGAGAGGATC-3′). Combinatorial sgRNA expression vectors were transduced into the orthogonal clonal line 2 used in the orthogonal screen. For arrayed validation experiments, genetic interaction scores (GIv) were calculated as:
Ψ scores from arrayed validation data (Ψv) were calculated as:
Cell viability assays
Cells were seeded at 10,000 cells per 96-well in 200 uL RPMI-1640 (10% FBS, 1% Anti-Anti) with indicated imatinib and/or R428 concentrations. Viability was determined at indicated time points by mixing 100 uL cell suspension with 50 uL resazurine medium (50 ug/mL, Acros Organics). After 2h incubation, fluorescence was quantified on a plate reader (BMG Labtech) at excitation: 530 nm and emmision: 590 nm.
RNAseq
RNA from K562 CRISPRa cells was extracted using RNeasy Mini kit (Qiagen). Sequencing libraries were prepared using the TruSeq mRNA stranded kit (Illumina) and sequenced via SE 50bp RNAseq on a HiSeq2000 platform. Reads were aligned to Homo sapiens Ensembl GRCm38v.78 using STAR_2.4.2a.
Quantitative RT-PCR
Total RNA from sgRNA expressing cells was purified using Rneasy Mini columns (Qiagen). Taqman probe assays (Applied Biosystems) were used with FAM labelled probes for target genes and VIC labelled probes for the housekeeping gene HPRT1. Reactions were carried out using the one step qRT-PCR master mix TaqMan RNA-to-CT (Applied Biosystems) according to the manufacturer’s instructions on the 2900 HT Fast RT-PCR machine (Applied Biosystems).
Western blot analyses
NF1-Null HEK293T cells were generated using SpCas9 and sgRNA targeting exon 2 with the sequence 5′-AGTCAGTACTGAGCACAACA-3′ (Shalem, O., et al., 2013). Following single cell cloning, target sequence amplification by PCR, TOPO cloning, and Sanger sequencing, both NF-1 alleles were confirmed deleted by a 1bp insertion resulting in NF1(N39fs) and a 11bp deletion resulting in NF1(S35fs). HEK293T cells were transfected with pcDNA3.1 Flag-eGFP (CTRL) and Flag-SPRED2 (SPRED2) using Lipofectamine 2000 (ThermoFisher Scientific, 11668019), serum starved for 24 hours, and stimulated with 20ng/ml recombinant human EGF (Invitrogen, PHG0311). Cells were washed with PBS and lysed in TNM buffer (0.2 M Tris pH 7.5, 1% Triton X-100, 1.5M NaCl, 50 mM MgCl2, 1mM DTT, protease and phosphatase inhibitor cocktails. Lysate was cleared and 1,000ug protein was subject GST-Raf1 RBD agarose beads (McCormick lab, in house) for 1.5 hours. Samples were analysed by Western blot using the following antibodies: NF1 (SCBT, sc-67 [D]), Flag (Sigma, F1804), pan-Ras (Cytoskeleton, Inc, AESA02), β-Actin (Sigma, A5441).
K562 orthogonal cells were transduced with lentivirus expressing S.aureus sgRNAs: sgNTC 5′-ACGCGTGCGTAATGAGAGGATC-3′ (NF1-wildtype) or sgNF1 5′-TTGTCTTTGGGTGTATTAGCAA-3′ (NF1-knockout). At 2 days post transduction, NF1-wildtype and -knockout cells were selected with puromycin (2 ug/mL) for 5 days and recovered for 2 additional days before treatment with vehicle (DMSO), 300nM imatinib, 1μM R428, or imatinib + R428 for 48hrs. Whole cell lysates were collected by lysis in RIPA buffer (150 mM NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0) and lysate concentration determined by BCA assay (ThermoFisher). 20 μg of lysate corresponding to each sample was separated by SDS-PAGE and transferred to a PVDF membrane by wet transfer (250mA, 2hrs). Western blot analysis was carried out following standard conditions using p-AXL antibody (R&D Technologies; Y779), followed by re-probing with β-actin antibody (Sigma; AC-74) to confirm equal loading.
Time-resolved fluorescence resonance energy transfer (TR-FRET) assay
TR-FRET assay utilizing the terbium/Venus as energy donor/acceptor was performed as described previously35. Briefly, HEK293T cells were transfected with GST- and Venus-tagged genes. Cells were lysed in FRET buffer (20 mM Tris, pH 7.0, 0.01% Nonidet-P40, and 50 mM NaCl with proteinase and phosphatase inhibitors) followed by three freeze-and-thaw cycles. Terbium conjugated Anti-GST antibody (Cisbio Bioassays, Codolet, France) was 1:1000 diluted in FRET buffer and dispensed into each well with MultidropTM Combi Reagent Dispenser (ThermoScientific). The lysate-antibody mixtures were incubated at 4°C before the TR-FRET signal was recorded (EnVision reader setting: Ex 337 nm, Em1: 520 nm, Em2: 486 nm; mirror: D400/D505 dual; time delay: 50 μs). The TR-FRET signal is expressed as the FRET ratio (F520/F486 × 104).
Data availability
Sequencing data from the CRISPRa screen and RNAseq are available at Sequence Read Archive accession number SRP127017 under BioProject ID PRJNA422995. All relevant additional data has been published with the manuscript, either as part of the main text or in the supplement. Plasmids and their sequences are deposited at Addgene.
Supplementary Material
Map of sgLenti vector for CRISPRa screen sgRNA expression.
Map sgLenti orthogonal vector for orthogonal screen sgRNA expression.
Map of S.aureus Cas9 nuclease vector.
MAGeCK normalized read count values for each separate sgRNAs from primary CRISPRa screen together with sgRNA nucleotide sequences.
Orthogonal screen raw read counts from baseline and Day19 (imatinib treated) of clonal replicates 1 and 2. Dual sgRNA construct names are in the format: CRISPRa target gene symbol (two sgRNA per gene labelled gene-A and gene-B) followed by SaCas9 nuclease target gene symbol, RefSeq accession number and sgRNA sequence. All CRISPRa sgRNA sequences are shown in Supplemental Table 4. All SaCas9 nuclease sequences are shown in Supplemental Table 5.
Transcript level analysis of primary CRISPRa screen with τ and p-values as well as expression levels (FPKM) for each transcript are shown. Mann-Whitney U test was used to calculate p-values as described previously44. To correct for multiple hypothesis testing, we first performed random sampling with replacement among the set of τ values for non-targeting control sgRNAs and calculated p-values for each sampling. A total of 300 million cells and 26,718 transcripts were analysed.
Arrayed validation of 20 candidate genes from primary CRISPRa screen are shown. Each gene was targeted by three separate sgRNAs and six non-target control sgRNAs (sgNTC) were included. Enrichment values (2τ) are shown for each sgRNA separately in the presence and absence of imatinib on days 7, 11 and 15. Values are means from three replicates with s.e.m.
Nucleotide sequences of sgRNAs used in the CRISPRa position of the orthogonal library are shown together with numbers of potential off-target sites as determined by Cas-OFFinder.
Nucleotide sequences for sgRNAs used in the SaCas9 nuclease (knockout) position of the orthogonal library are shown.
For gene:gene combinations from the orthogonal screen single activation and knockout τ values, expected and measured double perturbation τ values as well as calculated GI and Ψ scores are shown from each clonal replicate separately.
Reproducible gene:gene combinations that passed the filter criteria and that were used to construct the genetic interaction network in Figure 3h are shown.
Individual relative fitness values (τ) from arrayed validation of selected gene:gene interactions. Shown are enrichment values of cells expressing the indicated combination of sgRNAs in the absence (no drug) or presence of imatinib (IM) on days 3, 6, 9, 12 and 14. Values are means from three replicates with s.e.m.
GIv scores for each gene:gene combination and time point were calculated based on τ values in Supplemental Table 8. Correlation between GIv scores from each arrayed validation time point and the orthogonal screen in clonal replicate 2 are shown. For day 14 of the arrayed validation in the presence of imatinib, Ψ scores are shown which are also indicated in the genetic interaction network model in Figure 4c.
Extended Data Figure 1 | a, Dose response curve on K562-CRISPRa cells with non-target control sgRNA (sgNTC) or sgRNAs targeting the imatinib efflux transporter ABCB1 for activation (sgABCB1-1 and sgABCB1-2). IC50 values are shown in brackets. Values are mean with s.e.m. from independent experiments (n=3) b, K562-CRISPRa cells expressing sgRNAs from a or no sgRNA treated repeatedly with 100 nM imatinib on days 0, 3 and 6. Fractions of viable cells are shown as mean with s.e.m. and resistance factors from three independent experiments (RF = fraction of viable ABCB1 overexpressing cells / fraction of viable control cells) are shown from days 3, 6 and 9.
Extended Data Figure 2 | CRISPRa sgRNA library distribution. Shown is the cumulative fraction of sgRNAs in the CRISPRa library ranked by their normalised read counts. Shaded area highlights one order of magnitude around median read count.
Extended Data Figure 3 | CRISPRa screen reproducibility. Correlation between sgRNA phenotypes from three technical screen replicates (R1–3). Shown are fold-changes from sgRNAs (cutoff >100 reads per sgRNA in baseline sample) targeting all 332 significantly depleted/enriched CRISPRa screen candidate genes. Correlation values (r) are Pearson product-moment correlation coefficients.
Extended Data Figure 4 | CRISPRa works independent of endogenous gene expression levels. Comparison of absolute gene expression levels (FPKM) to results from the CRISPRa screen (−log10 p-value) showed a span of approximately five orders of magnitude in FPKM levels for the top 332 hit genes. These include undetectable genes (21%, FPKM<100), for example, ABCB1, ABCG2, PDGFRB and CSF1R which are frequently found activated in drug resistant cancer cells from leukaemia patients14,21,23. Other candidate genes also include expressed genes (79% with FPKM>100) with well described roles in leukaemia, like MYC, BCR-ABL or CDK6 45. Underlying data is summarised in Supplemental Table 2. Mann-Whitney U test was used to calculate p-values as described previously44. One sample of a total of 300 million cells was analysed.
Extended Data Figure 5 | a, GSEA using all 332 significant candidate genes identified by CRISPRa screen performed via DAVID analysis tool12. b, Subset of identified candidate genes mapped onto known signalling pathways (blue = negative, red = positive regulator of fitness).
Extended Data Figure 6 | CRISPRa screen results reproduce in arrayed format. sgRNA sequences against 20 significant candidate genes from pooled CRISPRa screen (n=3 sgRNAs/gene) were individually sub-cloned and re-assayed in an arrayed 96-well format. Validation results show high levels of correlation between even subtle phenotypes (e.g. CDK6, PVT1) from the pooled screen. Non-target controls (NTC) show no enrichment. Mean fold-enrichment values with s.e.m. are shown from independent replicates (n=3). Correlation values (r) are Pearson product-moment correlation coefficients.
Extended Data Figure 7 | CRISPRa efficiently activates target genes. Significantly increased target gene mRNA levels determined via quantitative RT-PCR from indicated CRISPRa sgRNAs with Taqman probes against a, BBX. b, NOL4L and c, ZC3HAV1. Mean fold-enrichment values with s.e.m. are shown from independent technical replicates (n=2). Statistical significance was determined via two-tailed, homoscedastic t-test with * = p<0.05, **=p<0.01 and *** = p<0.001.
Extended Data Figure 8 | CRISPRa τ values from the primary screen and CRISPRa τ values from the orthogonal screen (non-target control in SaCas9 nuclease position) show a high level of correlation (r = 0.9267). Correlation values (r) are Pearson product-moment correlation coefficients. τ values were calculated from two sgRNAs/gene at n=3 independent replicates (primary CRISPRa screen) and n=2 independent replicates (orthogonal screen).
Extended Data Figure 9 | Total raw GI scores from both clonal replicates (grey) with GI scores used to build de-novo genetic interaction network (Figure 3f, Extended Data Figure 11) highlighted in red.
Extended Data Figure 10 | Schematic showing both scenarios in which directionality of genetic interactions was inferred. When the activated gene displayed the opposite phenotype of the knocked out gene, the gene whose perturbation phenotype dominated the combinatorial phenotype was inferred to act downstream. Blue = decreased cell fitness, red = increased cell fitness.
Extended Data Figure 11 | All genetic interactions identified from the orthogonal CRISPR screen separated by central nodes (highlighted in yellow). Shown are results from independent clonal cell line replicates (n=2).
Extended Data Figure 12 | a, NF1 and SPRED2 activation significantly sensitises NF1 wildtype but not NF1 knockout cells to imatinib treatment (mean with s.e.m. is shown, n=3, x-axis indicates CRISPRa sgRNAs). b, Significantly increased SPRED2 mRNA levels were detected via qRT-PCR in NF1 wildtype and knockout cells that expressed SPRED2 activating CRISPRa sgRNAs (mean with s.e.m. is shown, n=3, x-axis indicates expressed CRISPRa sgRNAs). Statistical significance in panel a and b was determined via two-tailed, homoscedastic t-test with * = p<0.05, **=p<0.01 and *** = p<0.001, c, Overexpression of SPRED2 suppresses Ras activation only in NF1-wildtype cells, as determined by Ras-GTP levels. Quantified Ras-GTP level fold-changes of EGF-induced versus non-induced cells are shown. Negative control (CTRL) and SPRED2 overexpression constructs were transiently transfected into HEK293T cells 24h before EGF stimulation. The experiment was performed once.
Extended Data Figure 13 | a, NF1 protein levels from K562 cells expressing a non-target control (NF1-wildtype) or NF1 targeting sgRNA (NF1-knockout) are shown. The experiment was performed once. b, Quantification of phosphorylated AXL kinase levels (p-AXL) of vehicle (untreated), imatinib, R428 or imatinib + R428 treated cells from western blots in Fig. 5c are shown.
Extended Data Figure 14 | a, BEAS-2B lung epithelial cells with reduced levels of NF1 are more sensitive to R428. Mean fold-enrichment values with s.e.m. are shown from independent replicates (n=3). Statistical significance was determined via two-tailed, homoscedastic t-test with * = p<0.05, **=p<0.01 and *** = p<0.001. b, Western blot for Nf1 protein levels in BEAS-2B cells transduced with scrambled Mission control shRNA or shRNA against NF1 mRNA (TRCN0000039713, Sigma). The experiment was performed once.
Extended Data Figure 15 | TR-FRET assay shows that AXL (upper panels) as well as NF1 (lower panels) physically interact with NRAS, HRAS and KRAS. (Shown is the mean±s.d. from three independent experiments).
Acknowledgments
Special thanks go to members of the McManus Lab who provided critical feedback during the course of this project and Elizabeth Cahill for excellent technical support. We also thank Luke A. Gilbert and Marvin E. Tanenbaum for sharing the CRISPRa cell line ahead of its publication. M.T.M. was supported by NIH/CTD2 (U01CA168370) and IDG (1U01MH105028). M.K. was supported by NIH/NIGMS New Innovator Award DP2 GM119139, NIH/NCI K99/R00 CA181494, a Stand Up to Cancer Innovative Research Grant and the Chan Zuckerberg Biohub. J.B. was supported by NIH Training grant T32 GM00715 and an AFPE Predoctoral Fellowship. H.F. was supported by NIH/CTD2 (U01CA168449).
Footnotes
Author Contributions
The project was conceived and directed by M.B. and M.T.M. Screen optimisation was performed by M.B. and D.W. Libraries were designed by J.A.B. with guidance from M.B. and cloned by M.B. Orthogonal vectors and cell lines were created by M.B. All screens were performed by M.B., with A.B. assisting in CRISPRa screen analysis. R.T. developed the computational pipelines and the statistical framework for data analysis for screens with guidance from M.K. R.T. also selected the best-performing sgRNAs for arrayed validation. M.B. and R.T.W. conducted and analysed arrayed validation experiments. E.M. and R.T.W. performed western blot analyses. X.M. and H.F. conducted and analysed TR-FRET experiments. M.B. and M.T.M. wrote the manuscript with critical input from R.T., M.K., N.Z. and F.M. All authors read and approved the final manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischer B, et al. A map of directional genetic interactions in a metazoan cell. eLife. 2015;4 doi: 10.7554/eLife.05464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drees BL, et al. Derivation of genetic interaction networks from quantitative phenotype data. Genome biology. 2005;6:R38. doi: 10.1186/gb-2005-6-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.St Onge RP, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nature genetics. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends in genetics : TIG. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016:353. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nature reviews. Genetics. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boettcher M, McManus MT. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Molecular cell. 2015;58:575–585. doi: 10.1016/j.molcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Hare T, Deininger MW, Eide CA, Clackson T, Druker BJ. Targeting the BCR-ABL signaling pathway in therapy-resistant Philadelphia chromosome-positive leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:212–221. doi: 10.1158/1078-0432.CCR-09-3314. [DOI] [PubMed] [Google Scholar]
- 11.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014 doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 13.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 14.Milojkovic D, Apperley J. Mechanisms of Resistance to Imatinib and Second-Generation Tyrosine Inhibitors in Chronic Myeloid Leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:7519–7527. doi: 10.1158/1078-0432.CCR-09-1068. [DOI] [PubMed] [Google Scholar]
- 15.de Jong R, ten Hoeve J, Heisterkamp N, Groffen J. Crkl is complexed with tyrosine-phosphorylated Cbl in Ph-positive leukemia. The Journal of biological chemistry. 1995;270:21468–21471. doi: 10.1074/jbc.270.37.21468. [DOI] [PubMed] [Google Scholar]
- 16.Cilloni D, Saglio G. Molecular pathways: BCR-ABL. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:930–937. doi: 10.1158/1078-0432.CCR-10-1613. [DOI] [PubMed] [Google Scholar]
- 17.Cong F, et al. Cytoskeletal protein PSTPIP1 directs the PEST-type protein tyrosine phosphatase to the c-Abl kinase to mediate Abl dephosphorylation. Molecular cell. 2000;6:1413–1423. doi: 10.1016/s1097-2765(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 18.Calvisi DF, et al. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. Journal of hepatology. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo TC, Chavarria-Smith JE, Huang D, Schlissel MS. Forced expression of cyclin-dependent kinase 6 confers resistance of pro-B acute lymphocytic leukemia to Gleevec treatment. Molecular and cellular biology. 2011;31:2566–2576. doi: 10.1128/MCB.01349-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer discovery. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheah CY, et al. Patients with myeloid malignancies bearing PDGFRB fusion genes achieve durable long-term remissions with imatinib. Blood. 2014;123:3574–3577. doi: 10.1182/blood-2014-02-555607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, et al. FGFR-Mediated Reactivation of MAPK Signaling Attenuates Antitumor Effects of Imatinib in Gastrointestinal Stromal Tumors. Cancer discovery. 2015;5:438–451. doi: 10.1158/2159-8290.CD-14-0763. [DOI] [PubMed] [Google Scholar]
- 23.Chase A, et al. Imatinib sensitivity as a consequence of a CSF1R-Y571D mutation and CSF1/CSF1R signaling abnormalities in the cell line GDM1. Leukemia. 2009;23:358–364. doi: 10.1038/leu.2008.295. [DOI] [PubMed] [Google Scholar]
- 24.Dufies M, et al. Mechanisms of AXL overexpression and function in Imatinib-resistant chronic myeloid leukemia cells. Oncotarget. 2011;2:874–885. doi: 10.18632/oncotarget.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi LS, et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vojta A, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic acids research. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlman JE, et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nature biotechnology. 2015;33:1159–1161. doi: 10.1038/nbt.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stowe IB, et al. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes & development. 2012;26:1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham DK, DeRyckere D, Davies KD, Earp HS. The TAM family: phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nature reviews. Cancer. 2014;14:769–785. doi: 10.1038/nrc3847. [DOI] [PubMed] [Google Scholar]
- 32.Gay CM, Balaji K, Byers LA. Giving AXL the axe: targeting AXL in human malignancy. British journal of cancer. 2017;116:415–423. doi: 10.1038/bjc.2016.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Postel-Vinay S, Ashworth A. AXL and acquired resistance to EGFR inhibitors. Nat Genet. 2012;44:835–836. doi: 10.1038/ng.2362. [DOI] [PubMed] [Google Scholar]
- 34.Sheridan C. First Axl inhibitor enters clinical trials. Nature biotechnology. 2013;31:775–776. doi: 10.1038/nbt0913-775a. [DOI] [PubMed] [Google Scholar]
- 35.Li Z, et al. The OncoPPi network of cancer-focused protein-protein interactions to inform biological insights and therapeutic strategies. Nature communications. 2017;8:14356. doi: 10.1038/ncomms14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laufer C, Fischer B, Billmann M, Huber W, Boutros M. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat Methods. 2013;10:427–431. doi: 10.1038/nmeth.2436. [DOI] [PubMed] [Google Scholar]
- 37.Roguev A, et al. Quantitative genetic-interaction mapping in mammalian cells. Nat Methods. 2013;10:432–437. doi: 10.1038/nmeth.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizeacoumar FJ, et al. A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities. Molecular systems biology. 2013;9:696. doi: 10.1038/msb.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen JP, et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nature methods. 2017 doi: 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han K, et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nature biotechnology. 2017 doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 43.Du D, et al. Genetic interaction mapping in mammalian cells using CRISPR interference. Nature methods. 2017 doi: 10.1038/nmeth.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kampmann M, Bassik MC, Weissman JS. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2317–2326. doi: 10.1073/pnas.1307002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quintas-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113:1619–1630. doi: 10.1182/blood-2008-03-144790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014 doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaMontagne KR, Jr, Flint AJ, Franza BR, Jr, Pandergast AM, Tonks NK. Protein tyrosine phosphatase 1B antagonizes signalling by oncoprotein tyrosine kinase p210 bcr-abl in vivo. Molecular and cellular biology. 1998;18:2965–2975. doi: 10.1128/mcb.18.5.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burk O, Klempnauer KH. Myb and Ets transcription factors cooperate at the myb-inducible promoter of the tom-1 gene. Biochimica et biophysica acta. 1999;1446:243–252. doi: 10.1016/s0167-4781(99)00097-4. [DOI] [PubMed] [Google Scholar]
- 50.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Map of sgLenti vector for CRISPRa screen sgRNA expression.
Map sgLenti orthogonal vector for orthogonal screen sgRNA expression.
Map of S.aureus Cas9 nuclease vector.
MAGeCK normalized read count values for each separate sgRNAs from primary CRISPRa screen together with sgRNA nucleotide sequences.
Orthogonal screen raw read counts from baseline and Day19 (imatinib treated) of clonal replicates 1 and 2. Dual sgRNA construct names are in the format: CRISPRa target gene symbol (two sgRNA per gene labelled gene-A and gene-B) followed by SaCas9 nuclease target gene symbol, RefSeq accession number and sgRNA sequence. All CRISPRa sgRNA sequences are shown in Supplemental Table 4. All SaCas9 nuclease sequences are shown in Supplemental Table 5.
Transcript level analysis of primary CRISPRa screen with τ and p-values as well as expression levels (FPKM) for each transcript are shown. Mann-Whitney U test was used to calculate p-values as described previously44. To correct for multiple hypothesis testing, we first performed random sampling with replacement among the set of τ values for non-targeting control sgRNAs and calculated p-values for each sampling. A total of 300 million cells and 26,718 transcripts were analysed.
Arrayed validation of 20 candidate genes from primary CRISPRa screen are shown. Each gene was targeted by three separate sgRNAs and six non-target control sgRNAs (sgNTC) were included. Enrichment values (2τ) are shown for each sgRNA separately in the presence and absence of imatinib on days 7, 11 and 15. Values are means from three replicates with s.e.m.
Nucleotide sequences of sgRNAs used in the CRISPRa position of the orthogonal library are shown together with numbers of potential off-target sites as determined by Cas-OFFinder.
Nucleotide sequences for sgRNAs used in the SaCas9 nuclease (knockout) position of the orthogonal library are shown.
For gene:gene combinations from the orthogonal screen single activation and knockout τ values, expected and measured double perturbation τ values as well as calculated GI and Ψ scores are shown from each clonal replicate separately.
Reproducible gene:gene combinations that passed the filter criteria and that were used to construct the genetic interaction network in Figure 3h are shown.
Individual relative fitness values (τ) from arrayed validation of selected gene:gene interactions. Shown are enrichment values of cells expressing the indicated combination of sgRNAs in the absence (no drug) or presence of imatinib (IM) on days 3, 6, 9, 12 and 14. Values are means from three replicates with s.e.m.
GIv scores for each gene:gene combination and time point were calculated based on τ values in Supplemental Table 8. Correlation between GIv scores from each arrayed validation time point and the orthogonal screen in clonal replicate 2 are shown. For day 14 of the arrayed validation in the presence of imatinib, Ψ scores are shown which are also indicated in the genetic interaction network model in Figure 4c.
Extended Data Figure 1 | a, Dose response curve on K562-CRISPRa cells with non-target control sgRNA (sgNTC) or sgRNAs targeting the imatinib efflux transporter ABCB1 for activation (sgABCB1-1 and sgABCB1-2). IC50 values are shown in brackets. Values are mean with s.e.m. from independent experiments (n=3) b, K562-CRISPRa cells expressing sgRNAs from a or no sgRNA treated repeatedly with 100 nM imatinib on days 0, 3 and 6. Fractions of viable cells are shown as mean with s.e.m. and resistance factors from three independent experiments (RF = fraction of viable ABCB1 overexpressing cells / fraction of viable control cells) are shown from days 3, 6 and 9.
Extended Data Figure 2 | CRISPRa sgRNA library distribution. Shown is the cumulative fraction of sgRNAs in the CRISPRa library ranked by their normalised read counts. Shaded area highlights one order of magnitude around median read count.
Extended Data Figure 3 | CRISPRa screen reproducibility. Correlation between sgRNA phenotypes from three technical screen replicates (R1–3). Shown are fold-changes from sgRNAs (cutoff >100 reads per sgRNA in baseline sample) targeting all 332 significantly depleted/enriched CRISPRa screen candidate genes. Correlation values (r) are Pearson product-moment correlation coefficients.
Extended Data Figure 4 | CRISPRa works independent of endogenous gene expression levels. Comparison of absolute gene expression levels (FPKM) to results from the CRISPRa screen (−log10 p-value) showed a span of approximately five orders of magnitude in FPKM levels for the top 332 hit genes. These include undetectable genes (21%, FPKM<100), for example, ABCB1, ABCG2, PDGFRB and CSF1R which are frequently found activated in drug resistant cancer cells from leukaemia patients14,21,23. Other candidate genes also include expressed genes (79% with FPKM>100) with well described roles in leukaemia, like MYC, BCR-ABL or CDK6 45. Underlying data is summarised in Supplemental Table 2. Mann-Whitney U test was used to calculate p-values as described previously44. One sample of a total of 300 million cells was analysed.
Extended Data Figure 5 | a, GSEA using all 332 significant candidate genes identified by CRISPRa screen performed via DAVID analysis tool12. b, Subset of identified candidate genes mapped onto known signalling pathways (blue = negative, red = positive regulator of fitness).
Extended Data Figure 6 | CRISPRa screen results reproduce in arrayed format. sgRNA sequences against 20 significant candidate genes from pooled CRISPRa screen (n=3 sgRNAs/gene) were individually sub-cloned and re-assayed in an arrayed 96-well format. Validation results show high levels of correlation between even subtle phenotypes (e.g. CDK6, PVT1) from the pooled screen. Non-target controls (NTC) show no enrichment. Mean fold-enrichment values with s.e.m. are shown from independent replicates (n=3). Correlation values (r) are Pearson product-moment correlation coefficients.
Extended Data Figure 7 | CRISPRa efficiently activates target genes. Significantly increased target gene mRNA levels determined via quantitative RT-PCR from indicated CRISPRa sgRNAs with Taqman probes against a, BBX. b, NOL4L and c, ZC3HAV1. Mean fold-enrichment values with s.e.m. are shown from independent technical replicates (n=2). Statistical significance was determined via two-tailed, homoscedastic t-test with * = p<0.05, **=p<0.01 and *** = p<0.001.
Extended Data Figure 8 | CRISPRa τ values from the primary screen and CRISPRa τ values from the orthogonal screen (non-target control in SaCas9 nuclease position) show a high level of correlation (r = 0.9267). Correlation values (r) are Pearson product-moment correlation coefficients. τ values were calculated from two sgRNAs/gene at n=3 independent replicates (primary CRISPRa screen) and n=2 independent replicates (orthogonal screen).
Extended Data Figure 9 | Total raw GI scores from both clonal replicates (grey) with GI scores used to build de-novo genetic interaction network (Figure 3f, Extended Data Figure 11) highlighted in red.
Extended Data Figure 10 | Schematic showing both scenarios in which directionality of genetic interactions was inferred. When the activated gene displayed the opposite phenotype of the knocked out gene, the gene whose perturbation phenotype dominated the combinatorial phenotype was inferred to act downstream. Blue = decreased cell fitness, red = increased cell fitness.
Extended Data Figure 11 | All genetic interactions identified from the orthogonal CRISPR screen separated by central nodes (highlighted in yellow). Shown are results from independent clonal cell line replicates (n=2).
Extended Data Figure 12 | a, NF1 and SPRED2 activation significantly sensitises NF1 wildtype but not NF1 knockout cells to imatinib treatment (mean with s.e.m. is shown, n=3, x-axis indicates CRISPRa sgRNAs). b, Significantly increased SPRED2 mRNA levels were detected via qRT-PCR in NF1 wildtype and knockout cells that expressed SPRED2 activating CRISPRa sgRNAs (mean with s.e.m. is shown, n=3, x-axis indicates expressed CRISPRa sgRNAs). Statistical significance in panel a and b was determined via two-tailed, homoscedastic t-test with * = p<0.05, **=p<0.01 and *** = p<0.001, c, Overexpression of SPRED2 suppresses Ras activation only in NF1-wildtype cells, as determined by Ras-GTP levels. Quantified Ras-GTP level fold-changes of EGF-induced versus non-induced cells are shown. Negative control (CTRL) and SPRED2 overexpression constructs were transiently transfected into HEK293T cells 24h before EGF stimulation. The experiment was performed once.
Extended Data Figure 13 | a, NF1 protein levels from K562 cells expressing a non-target control (NF1-wildtype) or NF1 targeting sgRNA (NF1-knockout) are shown. The experiment was performed once. b, Quantification of phosphorylated AXL kinase levels (p-AXL) of vehicle (untreated), imatinib, R428 or imatinib + R428 treated cells from western blots in Fig. 5c are shown.
Extended Data Figure 14 | a, BEAS-2B lung epithelial cells with reduced levels of NF1 are more sensitive to R428. Mean fold-enrichment values with s.e.m. are shown from independent replicates (n=3). Statistical significance was determined via two-tailed, homoscedastic t-test with * = p<0.05, **=p<0.01 and *** = p<0.001. b, Western blot for Nf1 protein levels in BEAS-2B cells transduced with scrambled Mission control shRNA or shRNA against NF1 mRNA (TRCN0000039713, Sigma). The experiment was performed once.
Extended Data Figure 15 | TR-FRET assay shows that AXL (upper panels) as well as NF1 (lower panels) physically interact with NRAS, HRAS and KRAS. (Shown is the mean±s.d. from three independent experiments).
Data Availability Statement
Sequencing data from the CRISPRa screen and RNAseq are available at Sequence Read Archive accession number SRP127017 under BioProject ID PRJNA422995. All relevant additional data has been published with the manuscript, either as part of the main text or in the supplement. Plasmids and their sequences are deposited at Addgene.