 Intrinsic and acquired resistance to conventional and targeted therapeutics is a fundamental reason for treatment failure in many cancer patients.
Intrinsic and acquired resistance to conventional and targeted therapeutics is a fundamental reason for treatment failure in many cancer patients.
Abstract
Intrinsic and acquired resistance to conventional and targeted therapeutics is a fundamental reason for treatment failure in many cancer patients. Targeted approaches to overcome chemoresistance as well as resistance to targeted approaches require in depth understanding of the underlying molecular mechanisms. The anti-cancer activity of a drug can be limited by a broad variety of molecular events at different levels of drug action in a cell-autonomous and non-cell-autonomous manner. This review summarizes recent insights into the adaptive mechanisms used by tumours to resist therapy including cellular phenotypic plasticity, dynamic alterations of the tumour microenvironment, activation of redundant signal transduction pathways, modulation of drug target expression levels, and exploitation of pro-survival responses.
1. Introduction
Our understanding of the molecular drivers of cancer has increased remarkably during the past two decades. The Cancer Genome Atlas program (http://cancergenome.nih.gov) has identified a remarkable range of recurrent gene mutations and structural rearrangements underpinning tumourigenesis. In concert with this, the pharmaceutical industry has developed scores of molecularly targeted therapeutics with ever increasing precision that potently block these mechanisms. In spite of this progress, acquired resistance to chemotherapeutic and targeted agents is responsible for the lack of durable clinical responses for many cancer patients. Cancer remains a leading cause of death worldwide, and according to the WHO estimates more than half of current adults under the age of 65 years are expected to be diagnosed with cancer at some point during their lifetime.1
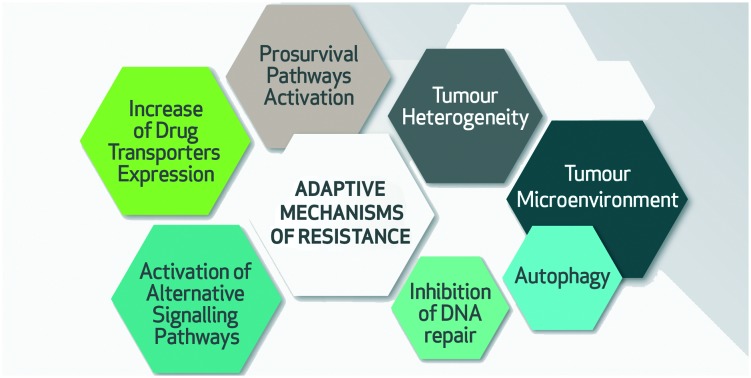
The confounding reality for anti-cancer drug development is the bewildering adaptive aptitude of tumour cells.2,3 In response to therapeutic challenge, tumour cells exploit both genetic (mutational) and epigenetic (phenotypic) evasive mechanisms. This is manifested in tumour heterogeneity; comprising oncogenic mutations that initially engendered uncontrolled clonal growth and acquired mutations during tumour clonal evolution as well as epigenetic reprogramming affecting the expression of hundreds of genes required for cellular phenotypic programs. Delineating these adaptive mechanisms of treatment resistance is crucial in order to develop durable therapies for cancer patients. In this review, we will discuss recent insights into adaptive anti-cancer drug resistance mechanisms that might enable us to refine current treatment strategies. These resistance mechanisms include altered tumour phenotypic heterogeneity, dynamic tumour microenvironmental changes, increased expression of drug transporters, activation of redundant and alternative signal transduction pathways, alterations in DNA damage repair, enhanced drug target expression levels and adaptive pro-survival cellular responses.
2. Tumour phenotypic heterogeneity
Cancer is caused by genetic mutations. However, in recent years it has been recognized that non-genetic (epigenetic) changes are critical contributors to malignant development. The heterogeneity of tumours is a key driver of drug resistance and therapeutic failure.2,4,5 This intra-tumour heterogeneity comprises both genetic and epigenetic components distinct from the founding immortalized cell. New agents that target proteins involved in chromatin regulation are important supplements to the approved small molecule drugs that inhibit specific mutant oncogenes (e.g. erlotinib for mutant EGFR). These new agents affect epigenetic regulators such as “chromatin readers” that comprise specialized binding domains and recognize distinct nucleosome modifications.6 For example, inhibitors targeting the bromodomain prevent interactions between BET proteins and acetylated histones that affect gene regulation.
Tumour heterogeneity supports a view of cancer as an abnormal tissue comprising a complex interplay between tumour cells and the normal cellular counterparts in the organ in which they reside.7 The deterioration of normal tissue structure during cancer progression presents tumour cells with unprecedented biophysical and nutritional challenges. In response, tumour cells coopt cellular plasticity programs that govern normal embryonic development, wound healing and adult organ homeostasis that allow access to adaptive cellular functions.8,9 Hence, tumours display remarkable phenotypic heterogeneity comprising cells of various levels of phenotypic plasticity. Importantly, the ability to adapt and transition between different cellular phenotypes will in turn increase the likelihood of tumour cell survival.10 For example, carcinomas, the epithelial-derived tumours that represent nearly 80% of human malignancies, have been shown to display a cellular phenotypic diversity reflective of the epithelial cell hierarchies present in normal tissues. Acquisition of such phenotypic diversity provides the carcinoma cells with an expanded repertoire of cellular functions, mirroring the hierarchical and diverse cell-type composition required to form and maintain homeostasis in adult organs.11–13
An important example of cellular phenotypic plasticity is the epithelial-to-mesenchymal transition (EMT). EMT is an evolutionary conserved process by which epithelial cells reversibly abandon their characteristic cell polarity and cell–cell adhesions, in favour of migratory and invasive properties typical of mesenchymal (fibroblast-like) cells during embryonic development and wound healing.9,14 The epithelial and mesenchymal cell states represent opposite ends of a spectrum differing in morphological characteristics and functional abilities. In a physiological context, epithelial cells are highly organized and closely connected to provide critical polarized barrier functions in tissues, while the mesenchymal cells are only loosely connected and display a spindle-shaped morphology reflective of their migratory behaviour. Cells undergoing the highly regulated molecular transition EMT, simultaneously alter the expression of hundreds of genes, including cytoskeletal, adhesion and signal transduction proteins.4,15
In cancer, EMT is associated with resistance to chemotherapeutics, immune evasion, metastasis and poor clinical outcome.14,16 Thomson et al. conducted a systems-level proteomic analysis of the changes that occur during EMT in lung cancer cells.15 They categorized the cells into three different EMT states; epithelial, “metastable” mesenchymal, and “epigenetically-fixed” mesenchymal, and further identified a number of transcriptional and protein regulatory changes during EMT including cell–cell junctional proteins, increase in pro-invasive and pro-migratory properties, changes in metabolic pathways and changes in secreted cytokines with the ability to modify the tumour microenvironment.15 During EMT they observed changes in pathways that promote cell survival and resistance to cancer therapies. These included a shift away from EGFR, IGF1R and Met/Ron signalling, and activation of autocrine survival networks including IL11/IL6/gp130/JAK2/STAT, fibronectin–integrin, Gas6-Axl/Tyro3, PDGFR/FGFR/RET and TGFβR.15 Their findings are of particular clinical relevance since the identified markers of the metastable mesenchymal state are also druggable targets.
The concept of epithelial-to-mesenchymal plasticity (EMP) includes EMT as well as the reverse transition; the mesenchymal-to-epithelial transition (MET), and indicates that tumour cells may exist in different stages along an epithelial to mesenchymal axis. Their ability to transit between the different states, i.e., to exist in intermediate states harbouring characteristics of both epithelial and mesenchymal cells is a hallmark of the most aggressive and therapy resistant cells.5,16,17 Multiple studies have broadly linked mesenchymal traits to drug resistance in different cancer types and against various cancer therapies including both chemotherapy and targeted therapies, implicating EMP as a key mediator of drug resistance.4,8,10–13,18–28 These studies highlight the fact that tumour plasticity enables phenotypic flexibility that underlie immune evasion, metastasis and acquired drug resistance, and impede durable treatment responses in patients.
Two recent studies have challenged the necessity of EMT for metastasis, and highlighted the requirement of EMP in chemoresistance.23,24 For example, Zheng et al. reported that genetic deletion of the EMT transcription factors Snail or Twist in KRAS-driven mouse models of spontaneous invasive pancreatic ductal adenocarcinoma (PDAC) did not affect metastasis, but increased sensitivity and overall survival in response to gemcitabine treatment.24 EMT was associated with reduced PDAC cell proliferation and drug transporter/concentrating protein expression (equilibrative nucleoside transporter 1 (ENT1), concentrating nucleoside transporter protein (Cnt3)) thus protecting tumours from the anti-proliferative effects of nucleoside analogs such as gemcitabine. Fischer et al. established an in vivo EMT lineage-tracing mouse model to monitor epithelial plasticity in mice.23 In this system they observed a small proportion of cells within an epithelial primary tumour that underwent EMT, and these cells preferentially survived cyclophosphamide treatment due to reduced proliferation, apoptotic tolerance and increased expression of genes related to chemoresistance.23 Interestingly, in untreated mice all metastases observed by Fisher et al. were derived from epithelial cells, as shown by lineage tracing. However, in the group of cyclophosphamide treated mice, they observed a significant contribution of cells that had undergone EMT in the metastatic lesions, indicating that EMT might be specifically involved in metastasis in the context of chemotherapy. It is unclear whether the cells in these studies represent the metastable state or the epigenetically fixed mesenchymal state. Taken together, these studies highlight the role of EMT in therapy resistance and emphasize the rationale for targeting EMP in order to overcome multiple drug resistance mechanisms.19,23,24,28 Indeed, tumour plasticity is a key advantage in a dynamic tumour microenvironment and represents an important target for new therapeutics. EMT is an embodiment of cellular plasticity where the transition to a mesenchymal/basal phenotype is strongly correlated with drug resistance.29 In other tumours derived from other cell types such as melanoma or glioblastoma, related stemness programs are similarly correlated with drug resistance.
Several studies report successful targeting of EMT in vitro and in vivo. Strategies include inhibiting the initiation of EMT, promoting MET, and selective depletion or functional inhibition of cells in the mesenchymal cell state.26 The multifunctional cytokine TGFβ is a major EMT inducer that is upregulated in several carcinomas and therefore considered a good target for therapeutic intervention.26 However, some TGFβ inhibitors have shown cardiovascular toxicity in preclinical studies.27 In hepatocellular carcinoma cells, TGFβ receptor inhibition was shown to block mesenchymal invasiveness and increase the expression of E-cadherin in vitro.28 In a subsequent phase I clinical study in glioblastoma the small molecule TGFβ receptor inhibitor LY2157299 demonstrated clinical benefit with no significant cardiac adverse events,30 and this inhibitor is currently in phase II clinical trials for patients with advanced hepatocellular carcinoma (NCT02178358, NCT01246986). A study by Tanaka et al. attempted to disrupt mesenchymal prostate cancer tumour cells by blocking the mesenchymal adhesion receptor N-cadherin with a monoclonal antibody. Results from this experiment showed that this strategy slowed the growth of xenografts, blocked local invasion, metastasis and castration resistance in vivo, and at high doses the compound led to complete regression of the xenografts.31
The Axl receptor tyrosine kinase, which is correlated with poor outcome and drug resistance in many cancer types, was shown to be necessary for maintaining tumour EMP.32–37 Multiple recent studies report that inhibition of Axl re-sensitizes cancer cells to cytotoxic and targeted cancer therapies in vitro and in vivo.36,38–44 Several compounds targeting Axl receptor are currently in clinical development. While most of these drugs are multi-kinase inhibitors, the small molecule inhibitor BGB324 was specifically developed to target Axl and is currently in clinical trials (NCT02488408, NCT02872259, NCT02424617).45
3. Dynamic tumour microenvironment in drug resistance
As briefly mentioned above, a tumour is much more than just bulk cancer cells. The tumour microenvironment comprises numerous normal cell types, extracellular matrix (ECM) components, and soluble growth factors. In healthy tissues, the microenvironment is a barrier to tumourigenesis by maintaining differentiated cell states and defined spatial boundaries.46 However, during cancer development, the breakdown of normal tissue architecture subverts this tumour suppressive function and the microenvironment is coopted by malignant cells and transformed into a supportive niche.47–49 It is increasingly appreciated that a dynamic tumour microenvironment facilitates many steps in the tumour development including cancer initiation, growth and metastasis.47,50,51 Indeed, the crosstalk between tumour and surrounding cells as well as the role of the ECM compartment, are important factors that contribute to both tumour development and therapy resistance. Normal cellular function is directed by cell–cell interactions, cell–ECM interactions, physiochemical properties of the microenvironment, and presentation of soluble factors. Novel combinations of biochemical and biophysical signals from the cell–cell and cell–ECM interactions in the tumour microenvironment regulate responsiveness to growth factors that determine cell behaviours and affect cancer therapy responses.52 The tumour microenvironment also determines therapeutic efficacy by influencing tumour blood flow, lymphatic drainage and intra-tumour pressure gradients.53 ECM proteins constitute significant barriers to interstitial drug transport by being a source of physical resistance to diffusional transport.53,54 An animal study in pancreatic ductal adenocarcinoma showed that degradation of stroma by enzymatic destruction of hyaluronan led to improved drug delivery and increased efficacy of the traditional chemotherapeutic agent gemcitabine.55
Stromal cells as drivers of drug resistance
Cancer cells secrete cytokines and chemokines that recruit stromal cells to the tumour. The tumour microenvironment comprises various stromal cell types including fibroblasts, pericytes, vascular cells and cells of the immune system. The tumour supportive cancer-associated fibroblasts (CAFs) and pericytes secrete growth factors that support cancer cell proliferation, evasion of apoptosis, tissue invasion and metastasis,50 and also pro-inflammatory and pro-angiogenic CAF-derived factors promote tumour growth.47,50,56,57 Stromal cell gene expression signatures are correlated with clinical responses to chemotherapy, various regimens of anti-angiogenic therapy and tyrosine kinase inhibitors,50,58–60 emphasizing a central role for stromal cells in regulating therapeutic outcome. For example, Wang et al. demonstrated that lung cancer cells became resistant to the EGFR inhibitor gefitinib when co-cultured together with HGF-producing fibroblasts, and combined treatment with anti-HGF antibody or the HGF antagonist NK4 successfully overcame the fibroblast induced resistance in vitro and in vivo.61
In order for a tumour to grow beyond the critical size of a few millimetres, the tumour cells must stimulate the sprouting of blood- and lymphatic vessels in order to ensure continued access to nutrients and oxygen and efficient waste exchange required for further growth. In general, tumours are characterized by a less organized and leakier vasculature than normal tissues that is reflected in lower blood flow and decreased drug delivery.53 Larger tumours usually display lower density of blood vessels in the centre compared to periphery making a heterogeneous drug distribution within larger tumours. Anti-angiogenic therapy has been suggested as a strategy to target the tumour microenvironment. Tumours induce angiogenesis by secreting various growth factors such as vascular endothelial growth factor (VEGF) or angiopoietin (Ang-2) in response to hypoxia. The necrotic centres of solid tumours and the onset of HIF1alfa has further been suggested to aid in the onset of tumour plasticity programs including EMP, which affects therapy response in multiple ways as discussed above. Targeting these critical factors has therefore been suggested as a strategy to inhibit cancer-induced angiogenesis and normalize the blood-flow of tumours. For example, bevacizumab (Avastin) is a monoclonal antibody that inhibits vascular angiogenesis by binding to VEGF. Excessive production of VEGF contributes to leaky vasculature and anti-angiogenic therapies aiming to normalize the blood-flow can thereby also increase the delivery of chemotherapeutic drugs. However, the clinical efficacy of anti-angiogenic therapies targeting VEGF has not met its expectations in terms of increased overall survival, and anti-VEGF therapy has on the contrary been associated with increased drug resistance and metastasis.62,63 However, the anti-angiogenic inhibitor sunitinib that targets VEGFR and other tyrosine kinases involved in angiogenesis pathway was effective in the treatment of renal cancers.64 A phase III randomized trial comparing sunitinib verses interferon-α demonstrated increased overall survival and progression-free survival with sunitinib as first-line treatment in patients with metastatic renal cell carcinoma (NCT00083889).65
Immune cells are critical components of the dynamic tumour microenvironment. Recent evidence demonstrates the notion that the immune system eliminates precancerous cells, a process called immunosurveillance.66 In fact, tumour-promoting inflammation and avoiding immune destruction represent two of the more recently established hallmarks of cancer.67 This initially protective anti-cancer mechanism of immunosurveillance needs to be evaded by tumours in establishment, in order for the tumour to develop and progress. Immune evasive cancer cells alter the tumour microenvironment and manipulate the immune cells of the innate and acquired immune system. Cancer cells and cells in the microenvironment can contribute to immune suppression of the host immune system in many ways, for example mediated by the immune suppressive properties of myeloid-derived suppressor cells and regulatory T-cells.47 Tumour associated macrophages (TAMs) can promote invasion by secretion of EGF,47 and chronic overexpression of inflammation mediators such as TNFα and TGFβ produced by TAMs are prevalent in cancer. Interestingly, it has been shown that certain types of traditional anti-cancer therapy including radiotherapy and several commonly used chemotherapies activate immunogenic cell death and require involvement of the immune system for efficacy.68 Targeting the communication between cancer cells and immune cells in the tumour microenvironment to unleash an efficient elimination of tumour cells by immune effector cells is currently a main focus in oncology.69 For example, androgen blockade therapy was shown to induce expression of macrophage colony-stimulating factor 1 (CSF1) and other cytokines recruiting and modulating TAMs resulting in increased TAM infiltration in a prostate cancer animal model.70 Targeting the receptor CSF1 in this model reverses macrophage-mediated resistance to androgen blockade therapy.71 Importantly, a recent study highlighted an overlap between melanoma resistance to immunotherapy and small molecule BRAF inhibitors suggesting a broader role for immune editing in acquired drug resistance.72
Many soluble factors, including growth factors, hormones and cytokines (including chemotactic cytokines; chemokines) are secreted by tumour cells and other cells in the tumour microenvironment such as CAFs or TAMs, mediate drug resistance. For example, stromal cell-derived factor 1 (SDF1) enhances tumour cell pro-survival and anti-apoptotic signalling pathways. In AML cells, SDF-1 mediated CXCR4 activation was shown to induce resistance to cytarabine.73 Soluble factors including EGF, PDGF and TGFβ also induce EMT, as discussed above, which is associated with drug resistance. In addition, TGFβ also stimulates CAFs to produce collagen I which indirectly is coupled with resistance because higher collagen content gives higher stiffness and decreased drug delivery.49,74 In summary, reciprocal interactions between tumour cells and reactive stromal cells create an unstable tumour microenvironment that triggers carcinoma cell plasticity gene programs, as discussed above which are related to normal regenerative epithelial homeostasis. A more comprehensive understanding of the crosstalk initiating these plasticity gene programs might substantially improve our ability to prevent development of aggressive malignancies, as well as to treat therapy resistant cancers with an EMT associated phenotype. Although still in its infancy, based on the recent clinical success in therapy resistant patients with advanced cancers, the emerging field of immuno-oncology holds great promise for the future treatment of cancer patients.
ECM as a driver of drug resistance
The ECM surrounding the cells consists mainly of polysaccharides and proteins. Common ECM components include collagen, laminin, fibronectin, and proteoglycans. Differentiation and maintenance of the different cell types require different signals that are restricted to a tissue specific microenvironment. Thus, the structure and function of ECM differ between different tissues, and the ECM components strongly affect the surrounding cells mediated by signalling through various cell adhesion molecules (CAMs).51 Integrins are the major types of CAMs responsible for bi-directional cell–ECM interactions, and they have a remarkable ability to respond to chemical, biological and physical signals from the ECM.51,75 Binding of integrin receptor pairs to ECM induces a signal transduction critical for cellular functions including proliferation, survival, differentiation and migration mediated by multi-molecular complexes formed between the cytoplasmic domains of the integrin and proteins involved in cell signalling and adapter proteins providing connections to the cytoskeleton.51 The ability to survive in the absence of normal ECM signalling input (anchorage independence) is a hallmark of malignancy, particularly with respect to the metastatic potential.
During metastasis, cancer cells pass multiple structural barriers including the extracellular matrix and basement membranes within the tissue, vasculature and at the metastatic site.76 During tumourigenesis normal tissue architecture breaks down, and changes in the ECM favour survival, angiogenesis and tumour spread.47,49 Cancer cells elaborate ECM remodeling factors such as metalloproteases and matricellular factors that degrade and remodel the ECM surrounding the growing tumour.76 Various types of ECM remodeling enzymes are often deregulated in human cancer.49,76,77 For example, increased MMP expression is responsible for angiogenesis and tumour progression, has been associated with poor prognosis in cancer patients and has been linked to metastasis.77–81 Altered ECM also affects the tumour progression indirectly by influencing the behavior of stromal cells including endothelial cells, immune cells and fibroblasts.49,82 ECM proteins such as collagen control the presentation and release of growth factors.81 Fibrosis, which involves excessive accumulation of ECM components (particularly type I collagen) in and around damaged tissues, is strongly correlated with cancer development.81 For example, chronic fibrosis in the liver constitutes a predisposition to carcinoma development.83–86
Multiple studies support a role for ECM in mediating cancer responses to cytotoxic agents and targeted therapies. In acute myeloid leukemia (AML), interactions between VLA-4 on AML cells and stromal fibronectin blocked drug-induced apoptosis by daunorubicin and cytarabine, and this resistance mechanism was overcome by VLA-4 specific antibodies.87 Matricellular proteins, ECM components whose primary role are not as structural components but as modulators of cell–matrix interactions, are correlated with drug resistance.88 Osteopontin (OPN) has been shown to promote tumour progression and therapy resistance. In prostate cancer cells, OPN caused increased expression of P-glycoprotein (P-gp) and knockdown of OPN increased cell sensitivity to various therapies including daunomycin, paclitaxel, doxorubicin, actinomycin-D, and rapamycin.89,90 Secreted protein acidic and rich in cysteins (SPARC) facilitates cell–matrix interactions and is a crucial mediator of cellular crosstalk during tissue remodeling and wound healing, and SPARC expression has been linked to various types of cancer.88 SPARC is associated with a highly aggressive phenotype in cancers such as melanomas and gliomas, while in others cancer types including ovarian, neuroblastomas and colorectal cancer it may function as a tumour suppressor.91 It has been suggested that SPARC increase chemosensitivity by enhancing drug delivery to the tumour making SPARC a possible therapeutic target to overcome chemoresistance in some cancer types.91–94
In addition to the cellular response to chemical and biological properties of the microenvironment, cells sense changes in the stiffness, force and spatial geometry of the surrounding matrix through mechanotransduction.49,95,96 The transcriptional regulators YAP/TAZ are key mediators of cellular mechanotransduction induced by ECM stiffness, cell geometry, and cytoskeletal tension, and are often upregulated in human malignancies.97,98 In general, the tumour microenvironment is considerably stiffer than normal tissue in which the tumours arise and higher ECM stiffness is hypothesized to be a driver of tumourigenesis.99–102 For example, Paszek et al. demonstrated that mechanical properties of the ECM can drive malignant behavior mediated by Rho-dependent integrin modulation.103 Oku et al. demonstrated that the small YAP/TAZ inhibiting molecules dasatinib, fluvastatin and pazopanib sensitizes breast cancer cells against the chemotherapeutic agents doxorubicin and paclitaxel.104 The increased stiffness in the tumour microenvironment can partly be described by increased activity of lysyl oxidase (LOX) which crosslinks collagens and other ECM proteins.99 LOX is upregulated in various cancers and has been associated with metastasis and poor prognosis.49,105,106 Levental at al. demonstrated that overexpression of LOX increases ECM stiffness and promotes tumour cell invasion and progression in a breast cancer mouse model.107 Inhibition of LOX reduced tissue fibrosis and tumour incident indicating that increased stiffness is not only a secondary effect but plays a causative role in cancer development.49,108 Physical properties of the ECM can also modulate drug resistance. For example, in breast cancer cells the stiffness of the microenvironment was shown to regulate the drug response to the Her-2 inhibitor lapatinib via the Hippo pathway.109
4. Increasing the expression of drug transporters
One of the obstacles behind successful chemotherapeutical treatment is the development of multidrug resistance (MDR). A well-established mechanism of MDR is the increase in the expression of a family of drug transporters, namely the ATP binding cassette (ABC) family of drug transporters. ABC transporters are ATP-dependent transporters which normal function is detoxifying and protecting cells from xenobiotics.110,111 Chemotherapy resistant tumours hijacking this mechanism, actively expel the cytotoxic drugs from cells, maintaining the drug concentration within the cells below the toxic level.112,113 Therefore, prevention of clinical multi-drug resistance should significantly improve therapeutic response in a large number of cancer patients. The three most extensively characterized ABC transporters include ABCB1 (also known as MDR1 or Pgp-P-glycoprotein), ABCC1 (also known as MRP1) and ABCG2 (also known as BCRP or MXR).112,113 Furthermore, multiple studies have linked the increased expression of these ABC transporters to poor outcome in a wide range of cancer types.114–119 Despite this fact, development of pharmacological modulators of MDR transporters yielded three generations of compounds showing poor clinical response. First-generation compounds included drugs that were already in clinical use like verapamil, quinine and cyclosporine-A. These modulators were shown to inhibit ABCB1 but revealed to be ineffective and extremely toxic. The second-generation of inhibitors were developed to circumvent toxicity and increase potency. Examples are valspodar or cyclosporine D, but similarly to first-generation compounds, they all showed little success in the clinic. Third-generation inhibitors (e.g. zosuquidar or tariquidar) were designed for high-transporter affinity and low pharmacokinetic interaction.120,121 Tariquidar, for example, has been shown to inhibit ABCB1 with low toxicity alone or in combination therapy. However, most clinical trials showed disappointing results and we are still far from obtaining a compound that efficaciously modulates ABC transporters. One of the reasons that might be behind the failure of MDR inhibitors is their functional redundancy, as different transporter family-members are often overexpressed in the same tumour. Later-generation inhibitors were developed to target multiple ABC transporters, like biricodar that targets ABCB1 and ABCC1. Although, this approach might present itself as beneficial for ABCB1-negative tumours, it can also increase the number of side effects.120,122 In an effort to minimize toxicity, alternative strategies are being explored to inhibit the expression of this class of transporters. These include the use of targeted antibodies/peptides, the use of nanoparticles capable of selectively deliver compounds to tumour cells or RNA interference (RNAi). The latter, appears to be an emerging application in cancer therapy. Unlike small-molecule drugs, which inhibit the mutant proteins that drive tumour growth, RNAi treatment inhibits protein production with great specificity. Several studies have successfully used this system to transcriptionally regulate ABCB1 expression with high specificity, although safe and efficient delivery of these constructs remains a limitation.123,124
5. Activating redundant and alternative signalling pathways
Another major mechanism of resistance to targeted chemotherapeutic agents is the activation of redundant or alternative signalling pathways that effectively control cancer cell growth and survival. This is often achieved as part of tumour phenotypic transitions (see above). Currently, the most druggable oncogenic drivers are kinases, including cell surface receptor tyrosine kinases125 and cytoplasmic kinases such as BRAF. Receptor tyrosine kinases (RTKs) are fundamental regulators of signalling pathways that control diverse cellular functions including growth, survival, differentiation and motility in normal cells and in numerous types of cancers. The EGFR kinase inhibitors gefitinib and erlotinib are two main drugs used to treat non-small cell lung cancers (NSCLCs) that carry EGFR activating mutations. In spite of impressive initial responses to EGFR inhibitors, most patients eventually relapse due to acquired drug resistance. Engelman et al. showed that c-Met (also known as MET or hepatocyte growth factor receptor; HGFR) is commonly amplified in NSCLC126 cells in agreement to what has been described earlier for gastric and esophageal cancers.127,128 Furthermore, increased c-Met signalling trough ERBB3 activation and consequent PI3K-AKT pathway reactivation underlies the acquired resistance to gefitinib. This provides an example of a resistance mechanism characterized by gene amplification of a kinase that is not a direct or downstream target of gefitinib or erlotinib.126 Lapatinib is a dual EGFR/HER2 tyrosine kinase inhibitor (TKI) used in combination therapy for advanced or metastatic ErbB2/HER2 positive breast cancer patients. Inhibition of HER2 TK with lapatinib results in compensatory upregulation of HER3 mRNA and protein, mediated by PI3K/AKT signalling pathway.129,130 To circumvent this problem, an alternative would be to include HER3 therapeutic inhibitors in combination with HER2 targeted drugs and PI3K inhibitors. However, the development of HER3 specific drugs has been challenging since HER3 lacks a functional kinase domain. As an alternative, HER2 specific antibodies, trastuzumab and pertuzumab, can be used to target the HER2-HER3 dimer. The formation of this dimer is essential to drive breast tumour formation and maintenance.131,132 Another good example refers to vemurafenib, an ATP-competitive RAF inhibitor that is commonly used in melanoma patients. The majority of these patients exhibit constitutive activation of the ERK pathway and a frequent number (50–70%) is due to the presence of a missense mutation of codon 600 in BRAF leading to the substitution of a valine for a glutamate (BRAFV600E).133 Patients harbouring this mutation and treated with RAF inhibitors very often relapse after initial response to treatment. PTEN loss appears to be concomitant with V600E mutations in 10–30% of patients. The tumour suppressor PTEN negatively regulates PI3K signaling pathway, a main regulator of cell growth, metabolism and survival. Intriguingly, RAF inhibitors have a paradoxical activating effect on the RAF/MEK/ERK pathway when wild-type BRAF is expressed instead of the mutant form.134–136 One way to overcome this problem would be the administration of a MEK or ERK inhibitor in conjunction with the RAF inhibitor.137 However, Miller et al.,138 recently reported that MEK inhibition in melanoma decreases the activity of surface metalloproteases responsible for the proteolytic shedding of RTKs, including HER4, c-Met and AXL. The resultant increased RTK expression drives bypass signaling and acquired resistance to BRAF/MEK inhibitors. Selective targeting of AXL or disrupting surface protease inhibition reversed this acquired drug resistance established at a post-translational level. A different study by Montero-Conde et al. described that the majority of BRAF-mutant thyroid cancer cell lines show a transient RAF inhibition when treated with vemurafenib, and that this is largely due to HER3 activation.139
Rapamycin also known as sirolimus, is an inhibitor of the mammalian target of rapamycin (mTOR) and has been shown to have anti-cancer properties. Recognition of these properties and the fact that mTOR pathway is often deregulated, led to the development of drug analogues that could act as chemotherapeutic agents against different types of tumours. Despite promising clinical benefits, resistance to rapamycin or other rapalogs is one of the reasons for the limited anti-cancer efficacy. The majority of the resistance mechanisms associated with this drug refer to context-dependent feedback loops. mTORC1 inhibition leads to upregulation of receptor tyrosine kinases (RTKs or substrates) such as platelet-derived growth factor receptors (PDGFRs) and insulin receptor substrate 1 (IRS-1), resulting in increased PI3K-dependent AKT phosphorylation (Ser473).140,141 A study from Carracedo et al. showed that mTORC1 inhibition by RAD001 (rapamycin derivative) leads to PI3K-Ras activation and consequent mitogen-activated protein kinase (MAPK) signaling activation.142 These findings limit the potential of combined therapy using mTORC1 and MAPK inhibitors in the clinic.
6. Inhibition of DNA damaging repair
The capacity of tumour cells to recover from DNA damage after cancer treatments, such as radiation and chemotherapy, may play a crucial role as a drug resistance mechanism.143 In order to survive, cells must protect and repair their genome from insults to their DNA, especially from double-strand breaks (DSB), which are very cytotoxic. The DNA damage response (DDR) is a well-orchestrated network of proteins constituting a signaling pathway that aims to detect DNA defects, repair them or induce cell cycle arrest to allow time to repair, or apoptosis in order to prevent propagation of damaged cells.143,144 The DDR consists of sensor proteins that detect the DNA damage and recruit signal transducing proteins increasing the signal to the effectors which will then be responsible for the adequate response to the type of damage.144 Under physiological circumstances, DNA repair is characterized by redundant pathways that aid in the maintenance of genomic stability. But as tumour cells progressively accumulate defects in DNA and DNA repair mechanisms, some DNA repair pathways become dysfunctional whilst others are maintained and even over-activated, very often contributing to intrinsic and acquired resistance to DNA disrupting cancer treatments.145 Thus, inhibition of key components of these pathways has the potential to prevent the emergence of therapy resistance.
Most tumours rely on a single functional pathway that can be targeted therapeutically (chemo- and radio-sensitization).146,147 Defects in a particular DDR-pathway may lead to a dependence on a complimentary pathway. Thus, inhibition of this complimentary pathway may induce tumour cell specific killing. A strategy commonly used is to target a main DNA repair pathway, knowing that those tumour cells display a dysfunctional alternate pathway, therefore causing tumour cell death (synthetic lethality). Synthetic lethality can be defined as cell death caused by inactivation of two genes simultaneously, whereas inactivation of either gene does not alter the cell viability, and this can be applied to target specifically tumour cells.148 One successful example refers to the poly(ADP) ribose polymerase (PARP) inhibitors. These inhibitors prevent normal function of the base excision repair (BER) pathway, leading to an increase of single-strand breaks that consequently will stall replication forks and cause the appearance of double-strand breaks. These latter, cannot be repaired by homologous recombination (HR) pathway in the context of BRCA-mutated cancers.145,149 The most successful PARP inhibitors are olaparib, veliparib and niraparib. Olaparib is approved for monotherapy in relapsed BRCA-mutated ovarian cancer patients150 and is currently in clinical trials for combined therapy.143 Veliparib is presently in phase 3 clinical trials and shows promising biological activity and tolerability.151 Niraparib is currently being tested in BRCA-mutated ovarian cancer patients following conventional chemotherapy. This study is currently in phase 3 clinical trials and the outcome of this study will definitely shed some light on the efficacy of this drug.152 Additionally, these inhibitors are being tested as combined therapy in BRCA-mutated ovarian tumours, and more recently, also being tested in non-mutated BRCA tumours, of which a subset of the tumours have responded well to PARP inhibitors.143,152
To target the DDR signaling pathway, several other types of inhibitors against DDR main players are currently being investigated. ATM (ataxia-telangiectasia mutated) is activated by DSBs and will affect cell cycle and cytotoxicity by phosphorylating downstream targets such as Chk2 (checkpoint kinase 2) and p53. KU-559403, currently at pre-clinical stage, is the first potent and specific ATM inhibitor with adequate solubility and bioavailability to allow for in vivo studies, confers radiosensitivity in vitro, and improved anti-tumour activity of etoposide and irinotecan in xenograft models.153 ATR (ataxia-telangiectasia and Rad3-related protein) is activated by single-strand breaks (SSB) and inhibition of ATR with its downstream effector Chk1 (checkpoint kinase 1) radiosensitizes some cancers, induces cell cycle arrest and increases DSBs.154 AZD6738, is an ATR inhibitor that is currently in phase 1 clinical trials, displays good solubility, pharmacodynamics and bioavailability, significantly increases anti-tumour activity of radiation or carboplatin in vivo and shows monotherapy-agent activity in ATM-deficient xenograft models.153 RAD51 is involved in HR, together with BRCA, to repair DNA.155 To prevent DNA repair in cancer cells, some inhibitors of RAD51 have been developed, such as RI-1 that binds to RAD51 protein, preventing it from assembling onto the DNA.156 RI-1 is currently on pre-clinical trials,156 but has already shown that it has an anti-cancer effect on gliobastoma cells combined with alkylating drugs, by disrupting HR.157 Many tumours rely on non-homologous end joining (NHEJ) repair pathway, and novel NHEJ inhibitors are being developed and tested, such as dual DNA-dependent protein kinase (DNA-PK) and mammalian target of rapamycin (mTOR) inhibitor, Cc-115, that promotes caspase-dependent cell death in chronic lymphocytic leukemia and blocks proliferation of tumour cells (phase 1 clinical trial).158
DDR sensor proteins, after detection of foreign DNA damage in the cytoplasm, can activate the immune system response159 and persistent DNA damage signaling can trigger a chronic inflammatory response and even tumourigenesis.160 For example, Ku70 and DNA-PK after detecting foreign DNA, will lead to an increase of IFN-λ1 production by Ku70, and IFN-β, cytokine, and chemokine genes by DNA-PK. Furthermore, PARP-1 affects TNFα, IL-6, and iNOS, leading to NF-κB transference to the nucleus.159 PARP inhibitors have recently started to be tested with immunotherapy for cancer treatment. For example, Higuchi et al. used an immune-competent BRCA1-deficient murine ovarian cancer model to evaluate the therapeutic outcome of the treatment with CTLA-4 antibody (immune checkpoint blockade) alone or combined with a PARP inhibitor. Results show induced long-term survival and increase of tumour cell cytotoxicity, mediated by anti-tumour immunity and increased levels of IFN-γ. These data provides support for the development of future clinical trials.161
DDR inhibitors have still a long way to go, as many are currently in pre-clinical trials and there is still a need to investigate their efficacy for combined therapy. However, PARP inhibitors, especially olaparib have shown promising results, and NHEJ inhibitors seem to be the next focus.
7. Activating prosurvival signalling
Cells displaying increased resistance to stress are characterized by the requirement for higher levels of apoptotic stimuli for cell death to be induced. A wide range of alterations may account for the ability to evade apoptosis including protein tyrosine kinases (PTKs) and their downstream pro-survival mediators (i.e. Erk, AKT) significantly contribute to drug resistance,162 as does inactivation or downregulation of pro-survival molecules (i.e. Fas receptor, Bax). EGFR signal transduction involves both PI3k/AKT and STAT pathways that enhance pro-survival traits. PI3K activates AKT that subsequently affects regulators of apoptosis and downregulates p53. Overexpression of EGFR or mutated PTEN (one of the most commonly lost tumour suppressors) increases AKT activity that modulates chemotherapy-induced apoptosis. Ovarian cancer cells with upregulated AKT were highly resistant to paclitaxel, contrasting with cancer cells expressing low AKT levels.163 Therapies targeting EGFR have resulted in limited clinical success, in some cases less than 10% of the patients showed a positive response when treated with cetuximab or panitumumab as single agents. This can be due to resistance mechanisms such as mutations in the KRAS gene and other components that interact with the EGFR pathway.164 However, combined therapies are being investigated namely with the ADAM17 inhibitor INCB3619, a sheddase enzyme that regulates RTKs.165 Other studies are investigating low molecular weight compounds and monoclonal antibodies that target ADAM17, such as INCB7839 and MED13622 respectively. The first blocks enzymatic activity and the latter blocks the release of ADAM17 substrates. Indeed, a phase I/II clinical trial is currently evaluating the efficacy of INCB7839 in combination with rituximab when administered to patients with diffuse large B cell non-Hodgkin lymphoma (NCT02141451).166 STAT proteins transcriptionally activate their target genes according to signalling originated from cytokine and growth factor receptors.167 STATS are often activated in many cancers167 and it has been demonstrated that STAT3 can induce Bcl2 overexpression, preventing apoptosis that would otherwise be induced by chemotherapy in breast cancer cells.168 Inhibiting STAT3 in head and neck cancer cells increases sensitivity to the effects of 5-FU.169 Although promising, no targeted STAT3 inhibitors have yet been approved by the FDA for use in the clinic. Pre-clinical small molecule STAT3 inhibitors are currently under investigation, such as S31-201 (NSC74859) that specifically inhibits STAT3 by preventing its phosphorylation and dimerization.170 Another prosurvival signalling mechanism involves NF-kB, a transcription factor that inhibits apoptosis, promotes cell proliferation and is activated by AKT.171 NF-kB is activated in various cancer cells targeted by several drugs, namely 5-FU, cisplatin, doxorubicin and paclitaxel, and it seems to be a critical player in preventing cell death induced by chemotherapy. Patients suffering from oesophageal cancer exhibited decreased clinical response to combined therapy (chemotherapy and radiation therapy) when they displayed high levels of activated NF-kB.172 Clinical trials have been investigating bortezomib, a proteasome inhibitor that inhibits NF-kB by preventing NF-kB translocation to the nucleus173 and established results regarding bortezomib administration in B-cell malignancies against NF-kB also confirmed its efficacy.174 Inhibiting NF-kB pathway may then be a good strategy to tackle drug resistance by increasing tumour sensitivity to apoptosis-inducing therapy. Overall, improving our understanding of the molecular mechanisms of anti-apoptotic and prosurvival signalling will hopefully lead us to improve clinical outcome for cancer patients by enabling clinicians to stratify the patients with predictive biomarkers followed by combined therapy that also target the activated prosurvival signalling pathways.
8. Autophagy
The role of autophagy in chemotherapy resistance has emerged in several cancers, particularly later tumour development.175 The autophagy process, denominated autophagic flux, is a conserved adaptive cellular survival mechanism that begins with a membrane vesicle (phagophore) that envelops cellular components and organelles to form a mature autophagosome. This structure fuses with the lysosome, allowing degradation and eventual release of constituents into the cytoplasm for anabolic recycling.175–178 Autophagy is essential for cell survival in response to cellular stress elicited by hypoxia, genomic instability, endoplasmic reticulum stress, and nutrient depletion.177 During malignant progression, tumour cells experience multiple forms of metabolic stresses that are exacerbated by anti-cancer treatment, resulting in upregulated autophagy processes that stimulate cell prosurvival and resistance mechanisms towards anticancer treatments.177,178 Indeed, autophagy is strongly correlated with tumour plasticity and drug resistance. A good example refers to autophagy induction by Aurora kinase A inhibition that ultimately triggers drug resistance in breast cancer.160,179 Autophagy is regulated by metabolic processes including ER stress and glucose deprivation, as well as by molecular processes such as PI3K/mTOR, AMPK and p53 signalling pathways.180,181 The process of autophagy is mediated by several autophagy gene (ATG) effector proteins, and many interact with mTORC1 and AMPK proteins. PI3K/mTOR pathway is often upregulated in cancer, which inhibits autophagy, however AMPK will inhibit mTOR under starvation stress hence promoting autophagy during tumour growth.175 Among the several strategies to improve anti-cancer therapy, combining chemotherapy or targeted therapy with inhibitors of autophagy increases tumour sensitivity to treatment and may improve clinical outcome.175,178 Inhibitors of autophagy include substances that inhibit endosomal acidification, or prevent fusion of autophagosomes with acidic endosomes by inhibiting the proton pump, and are currently being studied.182 Particularly, some clinical trials are evaluating the effects of autophagy inhibition combined with chemotherapy against solid tumours, using hydroxychloroquine (HCQ) that impairs lysosomal function and autophagic flux. However the HCQ therapeutic dose is currently a limiting factor due to side effects such as nausea and abdominal pain.175 As such, more potent autophagy inhibitors are needed and some compounds are being studied, such as Lys05, a lysosomal autophagy inhibitor that is a chloroquine derivative that is more potent and effective,183 and liensinine which is a isoquinoline alkaloid that inhibits late-stage autophagy by blocking autophagosome–lysosome fusion.184 Targeting autophagy to improve clinical outcome on anticancer therapies may thus become an encouraging therapeutic approach.
9. Conclusions
Resistance to conventional and targeted anti-cancer drugs is a major obstacle in successful therapy of cancer. The identification of molecular mechanisms involved in drug resistance or sensitization to targeted therapy is of enormous clinical importance. Today's era of targeted therapeutics and personalized medicine, and now effective immune therapies, achieving durable clinical responses will require mechanistic understanding of underlying causes of acquired resistance. New therapeutic modalities are needed to address these mechanisms. In recent years, significant progress in understanding the molecular basis of resistance to targeted therapies has been made and many overlapping mechanisms underlying the resistance to both standard chemotherapeutic drugs and to targeted agents have been identified. As a result an increasing number of therapeutic strategies to overcome resistance have been proposed or are being tested. Means to inhibit EMT induction, promoting MET or depleting cells in the mesenchymal cell state have been reported. As the tumour microenvironment has been increasingly recognized as a key contributor for drug resistance, agents specifically directed against targets in benign cells in pathologically active niches have become an attractive therapeutic option. On the other hand, inhibitors of activated redundant pathways or alternative components of survival signaling, as well as agents capable of interfering with autophagy might increase sensitivity to treatment and improve clinical outcome. Many of these therapeutic options are being evaluated as part of combination therapies in the clinic and can open new avenues of effective cancer treatment.
Acknowledgments
This work was supported by Fundação para a Ciência e Tecnologia (FCT), Research Center Grant UID/BIM/04773/2013 Centre for Biomedical Research 1334. Norwegian Research Council, Norwegian Cancer Society and Helse Vest Health Authority. B. I. Ferreira is the recipient of a FCT 2014 research grant SFRH/BPD/100434/2014. S. Machado is the recipient of a ProRegem grant PD/BD/114258/2016.
Footnotes
†The authors declare no competing interests.
References
- Ahmad A. S., Ormiston-Smith N., Sasieni P. D. Br. J. Cancer. 2015;112:943–947. doi: 10.1038/bjc.2014.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila M. R., de Sauvage F. J. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- Meacham C. E., Morrison S. J. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Weinberg R. A. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunert S., Jechlinger M., Beug H. Nat. Rev. Mol. Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- Dawson M. A., Kouzarides T., Huntly B. J. N. Engl. J. Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- Egeblad M., Nakasone E. S., Werb Z. Dev. Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C. J., Li X., Landis M., Dixon J. M., Neumeister V. M., Sjolund A., Rimm D. L., Wong H., Rodriguez A., Herschkowitz J. I., Fan C., Zhang X., He X., Pavlick A., Gutierrez M. C., Renshaw L., Larionov A. A., Faratian D., Hilsenbeck S. G., Perou C. M., Lewis M. T., Rosen J. M., Chang J. C. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay E. D. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- Cheng Q., Chang J. T., Gwin W. R., Zhu J., Ambs S., Geradts J., Lyerly H. K. Breast Cancer Res. 2014;16:407. doi: 10.1186/s13058-014-0407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveras-Ferraros C., Corominas-Faja B., Cufi S., Vazquez-Martin A., Martin-Castillo B., Iglesias J. M., Lopez-Bonet E., Martin A. G., Menendez J. A. Cell Cycle. 2012;11:4020–4032. doi: 10.4161/cc.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam T., Ramachandran V., Fournier K. F., Wang H., Marquis L., Abbruzzese J. L., Gallick G. E., Logsdon C. D., McConkey D. J., Choi W. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto C. A., Widodo E., Waltham M., Thompson E. W. Cancer Lett. 2013;341:56–62. doi: 10.1016/j.canlet.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Nieto M. A. Annu. Rev. Cell Dev. Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Thomson S., Petti F., Sujka-Kwok I., Mercado P., Bean J., Monaghan M., Seymour S. L., Argast G. M., Epstein D. M., Haley J. D. Clin. Exp. Metastasis. 2011;28:137–155. doi: 10.1007/s10585-010-9367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Weinberg R. A. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Bardia A., Wittner B. S., Stott S. L., Smas M. E., Ting D. T., Isakoff S. J., Ciciliano J. C., Wells M. N., Shah A. M., Concannon K. F., Donaldson M. C., Sequist L. V., Brachtel E., Sgroi D., Baselga J., Ramaswamy S., Toner M., Haber D. A., Maheswaran S. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani Y., Okimura A., Sato K., Nakagiri T., Kadota Y., Inoue M., Sawabata N., Minami M., Ikeda N., Kawahara K., Matsumoto T., Matsuura N., Ohta M., Okumura M., Ann. Thorac. Surg., 2011, 92 , 1794 –1804 , – discussion 1804 . [DOI] [PubMed] [Google Scholar]
- Singh A., Settleman J. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kono E., Tran C. P., Miyazaki H., Yamashiro J., Shimomura T., Fazli L., Wada R., Huang J., Vessella R. L., An J., Horvath S., Gleave M., Rettig M. B., Wainberg Z. A., Reiter R. E. Nat. Med. 2010;16:1414–1420. doi: 10.1038/nm.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li Y., Ahmad A., Azmi A. S., Kong D., Banerjee S., Sarkar F. H. Drug Resist. Updates. 2010;13:109–118. doi: 10.1016/j.drup.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Feng M., Zheng G., Chen Y., Wang X., Pen B., Yin J., Yu Y., He Z. Biochem. Biophys. Res. Commun. 2012;417:679–685. doi: 10.1016/j.bbrc.2011.11.142. [DOI] [PubMed] [Google Scholar]
- Fischer K. R., Durrans A., Lee S., Sheng J., Li F., Wong S. T., Choi H., El Rayes T., Ryu S., Troeger J., Schwabe R. F., Vahdat L. T., Altorki N. K., Mittal V., Gao D. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Carstens J. L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C. C., LeBleu V. S., Kalluri R. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F. M., Stewart T. A., Thompson E. W., Monteith G. R. Trends Pharmacol. Sci. 2014;35:479–488. doi: 10.1016/j.tips.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Marcucci F., Stassi G., De Maria R. Nat. Rev. Drug Discovery. 2016;15:311–325. doi: 10.1038/nrd.2015.13. [DOI] [PubMed] [Google Scholar]
- Anderton M. J., Mellor H. R., Bell A., Sadler C., Pass M., Powell S., Steele S. J., Roberts R. R., Heier A. Toxicol. Pathol. 2011;39:916–924. doi: 10.1177/0192623311416259. [DOI] [PubMed] [Google Scholar]
- Fransvea E., Angelotti U., Antonaci S., Giannelli G. Hepatology. 2008;47:1557–1566. doi: 10.1002/hep.22201. [DOI] [PubMed] [Google Scholar]
- Nieto M. A., Huang R. Y., Jackson R. A., Thiery J. P. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Rodon J., Carducci M. A., Sepulveda-Sanchez J. M., Azaro A., Calvo E., Seoane J., Brana I., Sicart E., Gueorguieva I., Cleverly A. L., Pillay N. S., Desaiah D., Estrem S. T., Paz-Ares L., Holdhoff M., Blakeley J., Lahn M. M., Baselga J. Clin. Cancer Res. 2015;21:553–560. doi: 10.1158/1078-0432.CCR-14-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kono E., Tran C. P., Miyazaki H., Yamashiro J., Shimomura T., Fazli L., Wada R., Huang J., Vessella R. L., An J., Horvath S., Gleave M., Rettig M. B., Wainberg Z. A., Reiter R. E. Nat. Med. 2010;16:1414–1420. doi: 10.1038/nm.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerdrum C., Tiron C., Hoiby T., Stefansson I., Haugen H., Sandal T., Collett K., Li S., McCormack E., Gjertsen B. T., Micklem D. R., Akslen L. A., Glackin C., Lorens J. B. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1124–1129. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Batalla I., Schultze A., Wroblewski M., Erdmann R., Heuser M., Waizenegger J. S., Riecken K., Binder M., Schewe D., Sawall S., Witzke V., Cubas-Cordova M., Janning M., Wellbrock J., Fehse B., Hagel C., Krauter J., Ganser A., Lorens J. B., Fiedler W., Carmeliet P., Pantel K., Bokemeyer C., Loges S. Blood. 2013;122:2443–2452. doi: 10.1182/blood-2013-03-491431. [DOI] [PubMed] [Google Scholar]
- Shieh Y. S., Lai C. Y., Kao Y. R., Shiah S. G., Chu Y. W., Lee H. S., Wu C. W. Neoplasia. 2005;7:1058–1064. doi: 10.1593/neo.05640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J., Peng D., Chen Z., Sehdev V., Belkhiri A. Cancer Res. 2013;73:331–340. doi: 10.1158/0008-5472.CAN-12-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lee J. C., Lin L., Olivas V., Au V., LaFramboise T., Abdel-Rahman M., Wang X., Levine A. D., Rho J. K., Choi Y. J., Choi C. M., Kim S. W., Jang S. J., Park Y. S., Kim W. S., Lee D. H., Lee J. S., Miller V. A., Arcila M., Ladanyi M., Moonsamy P., Sawyers C., Boggon T. J., Ma P. C., Costa C., Taron M., Rosell R., Halmos B., Bivona T. G. Nat. Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang X., Bi S., Zhao K., Yu C. Biochem. Biophys. Res. Commun. 2015;457:461–466. doi: 10.1016/j.bbrc.2015.01.017. [DOI] [PubMed] [Google Scholar]
- Liu L., Greger J., Shi H., Liu Y., Greshock J., Annan R., Halsey W., Sathe G. M., Martin A. M., Gilmer T. M. Cancer Res. 2009;69:6871–6878. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]
- Fleuren E. D., Hillebrandt-Roeffen M. H., Flucke U. E., Te Loo D. M., Boerman O. C., van der Graaf W. T., Versleijen-Jonkers Y. M. Oncotarget. 2014;5:12753–12768. doi: 10.18632/oncotarget.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C., Ye X., Pham T., Lin E., Chan S., McNamara E., Neve R. M., Belmont L., Koeppen H., Yauch R. L., Ashkenazi A., Settleman J. Cancer Res. 2014;74:5878–5890. doi: 10.1158/0008-5472.CAN-14-1009. [DOI] [PubMed] [Google Scholar]
- Brand T. M., Iida M., Stein A. P., Corrigan K. L., Braverman C. M., Coan J. P., Pearson H. E., Bahrar H., Fowler T. L., Bednarz B. P., Saha S., Yang D., Gill P. S., Lingen M. W., Saloura V., Villaflor V. M., Salgia R., Kimple R. J., Wheeler D. L. Clin. Cancer Res. 2015;21:2601–2612. doi: 10.1158/1078-0432.CCR-14-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabets M., Pazarentzos E., Juric D., Sheng Q., Pelossof R. A., Brook S., Benzaken A. O., Rodon J., Morse N., Yan J. J., Liu M., Das R., Chen Y., Tam A., Wang H., Liang J., Gurski J. M., Kerr D. A., Rosell R., Teixido C., Huang A., Ghossein R. A., Rosen N., Bivona T. G., Scaltriti M., Baselga J. Cancer Cell. 2015;27:533–546. doi: 10.1016/j.ccell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufies M., Jacquel A., Belhacene N., Robert G., Cluzeau T., Luciano F., Cassuto J. P., Raynaud S., Auberger P. Oncotarget. 2011;2:874–885. doi: 10.18632/oncotarget.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan D., Theiss N., Morales C., Stejskal A. E., Cooke L. S., Zhu M., Kurtzman D., Swart R., Ong E., Qi W. Oncotarget. 2015;6:1954–1966. doi: 10.18632/oncotarget.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. Nat. Biotechnol. 2013;31:775–776. doi: 10.1038/nbt0913-775a. [DOI] [PubMed] [Google Scholar]
- Bissell M. J., Hines W. C. Nat. Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Wei H., Lu T. Oncotarget. 2016;7:n32. doi: 10.18632/oncotarget.9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick N. A., Moses H. L. Curr. Opin. Genet. Dev. 2005;15:97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Weaver V. M., Werb Z. J. Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras K., Ostman A. Exp. Cell Res. 2010;316:1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- Berrier A. L., Yamada K. M. J. Cell. Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- Labarge M. A., Parvin B., Lorens J. B. Adv. Drug Delivery Rev. 2014;69-70:123–131. doi: 10.1016/j.addr.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au J. L., Yeung B. Z., Wientjes M. G., Lu Z., Wientjes M. G. Adv. Drug Delivery Rev. 2016;97:280–301. doi: 10.1016/j.addr.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netti P. A., Berk D. A., Swartz M. A., Grodzinsky A. J., Jain R. K. Cancer Res. 2000;60:2497–2503. [PubMed] [Google Scholar]
- Provenzano P. P., Cuevas C., Chang A. E., Goel V. K., Von Hoff D. D., Hingorani S. R. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez N., Truitt M., Olson P., Arron S. T., Hanahan D. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- Boersma B. J., Reimers M., Yi M., Ludwig J. A., Luke B. T., Stephens R. M., Yfantis H. G., Lee D. H., Weinstein J. N., Ambs S. Int. J. Cancer. 2008;122:1324–1332. doi: 10.1002/ijc.23237. [DOI] [PubMed] [Google Scholar]
- Finak G., Bertos N., Pepin F., Sadekova S., Souleimanova M., Zhao H., Chen H., Omeroglu G., Meterissian S., Omeroglu A., Hallett M., Park M. Nat. Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Farmer P., Bonnefoi H., Anderle P., Cameron D., Wirapati P., Becette V., Andre S., Piccart M., Campone M., Brain E., Macgrogan G., Petit T., Jassem J., Bibeau F., Blot E., Bogaerts J., Aguet M., Bergh J., Iggo R., Delorenzi M. Nat. Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- Crawford Y., Kasman I., Yu L., Zhong C., Wu X., Modrusan Z., Kaminker J., Ferrara N. Cancer Cell. 2009;15:21–34. doi: 10.1016/j.ccr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Wang W., Li Q., Yamada T., Matsumoto K., Matsumoto I., Oda M., Watanabe G., Kayano Y., Nishioka Y., Sone S., Yano S. Clin. Cancer Res. 2009;15:6630–6638. doi: 10.1158/1078-0432.CCR-09-1001. [DOI] [PubMed] [Google Scholar]
- Ribatti D. Oncotarget. 2016;7:n29. doi: 10.18632/oncotarget.8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers E. J., van Beijnum J. R., Thijssen V. L., Sabrkhany S., Nowak-Sliwinska P., Griffioen A. W. Drug Resist. Updates. 2016;25:26–37. doi: 10.1016/j.drup.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Coppin C. Biologics. 2008;2:97–105. doi: 10.2147/btt.s1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer R. J., Hutson T. E., Tomczak P., Michaelson M. D., Bukowski R. M., Oudard S., Negrier S., Szczylik C., Pili R., Bjarnason G. A., Garcia-del-Muro X., Sosman J. A., Solska E., Wilding G., Thompson J. A., Kim S. T., Chen I., Huang X., Figlin R. A. J. Clin. Oncol. 2009;27:3584–3590. doi: 10.1200/JCO.2008.20.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn G. P., Bruce A. T., Ikeda H., Old L. J., Schreiber R. D. Nat. Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Inoue H., Tani K. Cell Death Differ. 2014;21:39–49. doi: 10.1038/cdd.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol J., Vacchelli E., Aranda F., Castoldi F., Eggermont A., Cremer I., Sautes-Fridman C., Fucikova J., Galon J., Spisek R., Tartour E., Zitvogel L., Kroemer G., Galluzzi L. OncoImmunology. 2015;4:e1008866. doi: 10.1080/2162402X.2015.1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla J., Schokrpur S., Liu C., Priceman S. J., Moughon D., Jiang Z., Pouliot F., Magyar C., Sung J. L., Xu J., Deng G., West B. L., Bollag G., Fradet Y., Lacombe L., Jung M. E., Huang J., Wu L. Cancer Res. 2015;75:950–962. doi: 10.1158/0008-5472.CAN-14-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla J., Schokrpur S., Liu C., Priceman S. J., Moughon D., Jiang Z., Pouliot F., Magyar C., Sung J. L., Xu J., Deng G., West B. L., Bollag G., Fradet Y., Lacombe L., Jung M. E., Huang J., Wu L. Cancer Res. 2015;75:950–962. doi: 10.1158/0008-5472.CAN-14-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W., Zaretsky J. M., Sun L., Song C., Moreno B. H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., Seja E., Lomeli S., Kong X., Kelley M. C., Sosman J. A., Johnson D. B., Ribas A., Lo R. S. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Hui H., Yang H., Zhao K., Qin Y., Gu C., Wang X., Lu N., Guo Q. Blood. 2013;121:3682–3691. doi: 10.1182/blood-2012-11-466219. [DOI] [PubMed] [Google Scholar]
- Baronzio G., Parmar G., Baronzio M. Front. Oncol. 2015;5:165. doi: 10.3389/fonc.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber D. E. Annu. Rev. Physiol. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- Chambers A. F., Matrisian L. M. J. Natl. Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- Tsunezuka Y., Kinoh H., Takino T., Watanabe Y., Okada Y., Shinagawa A., Sato H., Seiki M. Cancer Res. 1996;56:5678–5683. [PubMed] [Google Scholar]
- Kessenbrock K., Plaks V., Werb Z. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieszalo K., Lawicki S., Szmitkowski M. Pol. Merkuriusz Lek. 2016;40:193–197. [PubMed] [Google Scholar]
- Sounni N. E., Noel A. Clin. Chem. 2013;59:85–93. doi: 10.1373/clinchem.2012.185363. [DOI] [PubMed] [Google Scholar]
- Wahyudi H., Reynolds A. A., Li Y., Owen S. C., Yu S. M. J. Controlled Release. 2016;240:323–331. doi: 10.1016/j.jconrel.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick N. A., Neilson E. G., Moses H. L. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybinski B., Franco-Barraza J., Cukierman E. Physiol. Genomics. 2014;46:223–244. doi: 10.1152/physiolgenomics.00158.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldacre M. J., Wotton C. J., Yeates D., Seagroatt V., Collier J. Eur. J. Gastroenterol. Hepatol. 2008;20:384–392. doi: 10.1097/MEG.0b013e3282f4489f. [DOI] [PubMed] [Google Scholar]
- Farazi P. A., DePinho R. A. Nat. Rev. Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- Campbell J. S., Hughes S. D., Gilbertson D. G., Palmer T. E., Holdren M. S., Haran A. C., Odell M. M., Bauer R. L., Ren H. P., Haugen H. S., Yeh M. M., Fausto N. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3389–3394. doi: 10.1073/pnas.0409722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T., Takemoto N., Sato T., Takimoto R., Tanaka I., Fujimi A., Akiyama T., Kuroda H., Kawano Y., Kobune M., Kato J., Hirayama Y., Sakamaki S., Kohda K., Miyake K., Niitsu Y. Nat. Med. 2003;9:1158–1165. doi: 10.1038/nm909. [DOI] [PubMed] [Google Scholar]
- Sawyer A. J., Kyriakides T. R. Adv. Drug Delivery Rev. 2016;97:56–68. doi: 10.1016/j.addr.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevde L. A., Samant R. S. Matrix Biol. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh I. S., Huang W. H., Liou H. C., Chuang W. J., Yang R. S., Fu W. M. Mol. Pharmacol. 2013;83:968–977. doi: 10.1124/mol.112.082339. [DOI] [PubMed] [Google Scholar]
- Tai I. T., Tang M. J. Drug Resist. Updates. 2008;11:231–246. doi: 10.1016/j.drup.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Cheetham S., Tang M. J., Mesak F., Kennecke H., Owen D., Tai I. T. Br. J. Cancer. 2008;98:1810–1819. doi: 10.1038/sj.bjc.6604377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai I. T., Dai M., Owen D. A., Chen L. B. J. Clin. Invest. 2005;115:1492–1502. doi: 10.1172/JCI23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz J., Ansari D., Sasor A., Andersson R. Pancreas. 2015;44:1024–1035. doi: 10.1097/MPA.0000000000000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V., Sheetz M. Nat. Rev. Mol. Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Guo W. H., Frey M. T., Burnham N. A., Wang Y. L. Biophys. J. 2006;90:2213–2220. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Low B. C., Pan C. Q., Shivashankar G. V., Bershadsky A., Sudol M., Sheetz M. FEBS Lett. 2014;588:2663–2670. doi: 10.1016/j.febslet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Lu P., Weaver V. M., Werb Z. J. Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tredan O., Galmarini C. M., Patel K., Tannock I. F. J. Natl. Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- Ng M. R., Brugge J. S. Cancer Cell. 2009;16:455–457. doi: 10.1016/j.ccr.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D., Hammer D. A., Weaver V. M. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D., Hammer D. A., Weaver V. M. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Oku Y., Nishiya N., Shito T., Yamamoto R., Yamamoto Y., Oyama C., Uehara Y. FEBS Open Bio. 2015;5:542–549. doi: 10.1016/j.fob.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Q. T., Harris J., Magliocco A. M., Kong C. S., Diaz R., Shin B., Cao H., Trotti A., Erler J. T., Chung C. H., Dicker A., Pajak T. F., Giaccia A. J., Ang K. K. J. Clin. Oncol. 2009;27:4281–4286. doi: 10.1200/JCO.2008.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler J. T., Bennewith K. L., Nicolau M., Dornhofer N., Kong C., Le Q. T., Chi J. T., Jeffrey S. S., Giaccia A. J. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- Levental K. R., Yu H., Kass L., Lakins J. N., Egeblad M., Erler J. T., Fong S. F., Csiszar K., Giaccia A., Weninger W., Yamauchi M., Gasser D. L., Weaver V. M. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K. R., Yu H., Kass L., Lakins J. N., Egeblad M., Erler J. T., Fong S. F., Csiszar K., Giaccia A., Weninger W., Yamauchi M., Gasser D. L., Weaver V. M. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H., Pelissier F. A., Zhang H., Lakins J., Weaver V. M., Park C., LaBarge M. A. Mol. Biol. Cell. 2015;26:3946–3953. doi: 10.1091/mbc.E15-07-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki A., Ierano C., Szakacs G., Robey R. W., Bates S. E. Essays Biochem. 2011;50:209–232. doi: 10.1042/bse0500209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland I. B. Essays Biochem. 2011;50:1–17. doi: 10.1042/bse0500001. [DOI] [PubMed] [Google Scholar]
- Fletcher J. I., Haber M., Henderson M. J., Norris M. D. Nat. Rev. Cancer. 2010;10:147–156. doi: 10.1038/nrc2789. [DOI] [PubMed] [Google Scholar]
- Gottesman M. M., Fojo T., Bates S. E. Nat. Rev. Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- Ho M. M., Ng A. V., Lam S., Hung J. Y. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- Raaijmakers M. H. G. P., de Grouw E. P. L. M., van der Reijden B. A., de Witte T. J. M., Jansen J. H., Raymakers R. A. P. Clin. Cancer Res. 2006;12:3452–3458. doi: 10.1158/1078-0432.CCR-05-1945. [DOI] [PubMed] [Google Scholar]
- Evers R., Cnubben N. H., Wijnholds J., van Deemter L., van Bladeren P. J., Borst P. FEBS Lett. 1997;419:112–116. doi: 10.1016/s0014-5793(97)01442-7. [DOI] [PubMed] [Google Scholar]
- Mitra P., Oskeritzian C. A., Payne S. G., Beaven M. A., Milstien S., Spiegel S. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16394–16399. doi: 10.1073/pnas.0603734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschmann-Jax C., Foster A. E., Wulf G. G., Nuchtern J. G., Jax T. W., Gobel U., Goodell M. A., Brenner M. K. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14228–14233. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Balch C., Chan M. W., Lai H.-C., Matei D., Schilder J. M., Yan P. S., Huang T. H.-M., Nephew K. P. Cancer Res. 2008;68:4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakacs G., Paterson J. K., Ludwig J. A., Booth-Genthe C., Gottesman M. M. Nat. Rev. Drug Discovery. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- Guns E. S., Denyssevych T., Dixon R., Bally M. B., Mayer L. Eur. J. Drug Metab. Pharmacokinet. 2002;27:119–126. doi: 10.1007/BF03190426. [DOI] [PubMed] [Google Scholar]
- Minderman H., O'Loughlin K. L., Pendyala L., Baer M. R. Clin. Cancer Res. 2004;10:1826–1834. doi: 10.1158/1078-0432.ccr-0914-3. [DOI] [PubMed] [Google Scholar]
- Susa M., Iyer A. K., Ryu K., Choy E., Hornicek F. J., Mankin H., Milane L., Amiji M. M., Duan Z. PLoS One. 2010;5:e10764. doi: 10.1371/journal.pone.0010764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A., Zelcer N., Prior J. L., Kuil A. J., Piwnica-Worms D. Clin. Cancer Res. 2005;11:4487–4494. doi: 10.1158/1078-0432.CCR-05-0038. [DOI] [PubMed] [Google Scholar]
- Lemmon M. A., Schlessinger J. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C.-M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., Lee C., Johnson B. E., Cantley L. C., Jänne P. A. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Smolen G. A., Sordella R., Muir B., Mohapatra G., Barmettler A., Archibald H., Kim W. J., Okimoto R. A., Bell D. W., Sgroi D. C., Christensen J. G., Settleman J., Haber D. A. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. T., Lin L., Casper A. M., Lim J., Thomas D. G., Orringer M. B., Chang A. C., Chambers A. F., Giordano T. J., Glover T. W., Beer D. G. Oncogene. 2006;25:409–418. doi: 10.1038/sj.onc.1209057. [DOI] [PubMed] [Google Scholar]
- Garrett J. T., Olivares M. G., Rinehart C., Granja-Ingram N. D., Sánchez V., Chakrabarty A., Dave B., Cook R. S., Pao W., McKinely E., Manning H. C., Chang J., Arteaga C. L. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergina N. V., Rausch M., Wang D., Blair J., Hann B., Shokat K. M., Moasser M. M. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro T., Beerli R. R., Maurer F., Koziczak M., Barbas C. F., Hynes N. E. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gala K., Chandarlapaty S. Clin. Cancer Res. 2014;20:1410–1416. doi: 10.1158/1078-0432.CCR-13-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B. A., Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G. J., Bigner D. D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J. W. C., Leung S. Y., Yuen S. T., Weber B. L., Seigler H. F., Darrow T. L., Paterson H., Marais R., Marshall C. J., Wooster R., Stratton M. R., Futreal P. A. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Heidorn S. J., Milagre C., Whittaker S., Nourry A., Niculescu-Duvas I., Dhomen N., Hussain J., Reis-Filho J. S., Springer C. J., Pritchard C., Marais R. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou G., Song K., Yen I., Brandhuber B. J., Anderson D. J., Alvarado R., Ludlam M. J. C., Stokoe D., Gloor S. L., Vigers G., Morales T., Aliagas I., Liu B., Sideris S., Hoeflich K. P., Jaiswal B. S., Seshagiri S., Koeppen H., Belvin M., Friedman L. S., Malek S. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- Poulikakos P. I., Zhang C., Bollag G., Shokat K. M., Rosen N. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K. T., Infante J. R., Daud A., Gonzalez R., Kefford R. F., Sosman J., Hamid O., Schuchter L., Cebon J., Ibrahim N., Kudchadkar R., Burris H. A., Falchook G., Algazi A., Lewis K., Long G. V., Puzanov I., Lebowitz P., Singh A., Little S., Sun P., Allred A., Ouellet D., Kim K. B., Patel K., Weber J. N. Engl. J. Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Oudin M. J., Sullivan R. J., Wang S. J., Meyer A. S., Im H., Frederick D. T., Tadros J., Griffith L. G., Lee H., Weissleder R., Flaherty K. T., Gertler F. B., Lauffenburger D. A. Cancer Discovery. 2016;6:382–399. doi: 10.1158/2159-8290.CD-15-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Conde C., Ruiz-Llorente S., Dominguez J. M., Knauf J. A., Viale A., Sherman E. J., Ryder M., Ghossein R. A., Rosen N., Fagin J. A. Cancer Discovery. 2013;3:520–533. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A., Baselga J., Pandolfi P. P. Cell Cycle. 2008;7:3805–3809. doi: 10.4161/cc.7.24.7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly K. E., Rojo F., She Q.-B., Solit D., Mills G. B., Smith D., Lane H., Hofmann F., Hicklin D. J., Ludwig D. L., Baselga J., Rosen N. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A., Ma L., Teruya-Feldstein J., Rojo F., Salmena L., Alimonti A., Egia A., Sasaki A. T., Thomas G., Kozma S. C., Papa A., Nardella C., Cantley L. C., Baselga J., Pandolfi P. P. J. Clin. Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. A., Lawrence T. S. Clin. Cancer Res. 2015;21:2898–2904. doi: 10.1158/1078-0432.CCR-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Elledge S. J. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley M. R., Logsdon D., Fishel M. L. Future Oncol. 2014;10:1215–1237. doi: 10.2217/fon.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin N. J. Br. J. Pharmacol. 2013;169:1745–1765. doi: 10.1111/bph.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velic D., Couturier A. M., Ferreira M. T., Rodrigue A., Poirier G. G., Fleury F., Masson J.-Y. Biomolecules. 2015;5:3204–3259. doi: 10.3390/biom5043204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W. G. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Livraghi L., Garber J. E. BMC Med. 2015;13:188. doi: 10.1186/s12916-015-0425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe M., Kolesar J. Am. J. Health-Syst. Pharm. 2016;73:1037–1041. doi: 10.2146/ajhp150550. [DOI] [PubMed] [Google Scholar]
- Wagner L. M. OncoTargets Ther. 2015;8:1931–1939. doi: 10.2147/OTT.S69935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. F., Matulonis U. A. Curr. Oncol. Rep. 2016;18:29. doi: 10.1007/s11912-016-0515-z. [DOI] [PubMed] [Google Scholar]
- Weber A. M., Ryan A. J. Pharmacol. Ther. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Gavande N. S., VanderVere-Carozza P. S., Hinshaw H. D., Jalal S. I., Sears C. R., Pawelczak K. S., Turchi J. J. Pharmacol. Ther. 2016;160:65–83. doi: 10.1016/j.pharmthera.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Nicolai C., Ehlen A., Martin C., Zhang X., Carreira A. Nat. Commun. 2016;7:12813. doi: 10.1038/ncomms12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B., Logan H. L., Kalin J. H., Zelivianskaia A. S., Cameron McGuire W., Miller L. L., Stark J. M., Kozikowski A. P., Bishop D. K., Connell P. P. Nucleic Acids Res. 2012;40:7347–7357. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berte N., Piee-Staffa A., Piecha N., Wang M., Borgmann K., Kaina B., Nikolova T. Mol. Cancer Ther. 2016 doi: 10.1158/1535-7163.MCT-16-0176. [DOI] [PubMed] [Google Scholar]
- Thijssen R., Ter Burg J., Garrick B., van Bochove G. G., Brown J. R., Fernandes S. M., Rodriguez M. S., Michot J. M., Hallek M., Eichhorst B., Reinhardt H. C., Bendell J., Derks I. A., van Kampen R. J., Hege K., Kersten M. J., Trowe T., Filvaroff E. H., Eldering E., Kater A. P. Blood. 2016;128:574–583. doi: 10.1182/blood-2016-02-700328. [DOI] [PubMed] [Google Scholar]
- Nakad R., Schumacher B. Front. Genet. 2016;7:147. doi: 10.3389/fgene.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Dai H., Zhou M., Li M., Singh P., Qiu J., Tsark W., Huang Q., Kernstine K., Zhang X., Lin D., Shen B. Nat. Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- Higuchi T., Flies D. B., Marjon N. A., Mantia-Smaldone G., Ronner L., Gimotty P. A., Adams S. F. Cancer Immunol. Res. 2015;3:1257–1268. doi: 10.1158/2326-6066.CIR-15-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P., Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Page C., Lin H. J., Jin Y., Castle V. P., Nunez G., Huang M., Lin J. Anticancer Res. 2000;20:407–416. [PubMed] [Google Scholar]
- Van Emburgh B. O., Sartore-Bianchi A., Di Nicolantonio F., Siena S., Bardelli A. Mol. Oncol. 2014;8:1084–1094. doi: 10.1016/j.molonc.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan C., Van Schaeybroeck S., Longley D. B., Johnston P. G. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- Mullooly M., McGowan P., Crown J., Duffy M. J. Cancer Biol. Ther. 2016:0. doi: 10.1080/15384047.2016.1177684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Jove R. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Real P. J., Sierra A., De Juan A., Segovia J. C., Lopez-Vega J. M., Fernandez-Luna J. L. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- Masuda M., Toh S., Koike K., Kuratomi Y., Suzui M., Deguchi A., Komiyama S., Weinstein I. B. Jpn. J. Cancer Res. 2002;93:329–339. doi: 10.1111/j.1349-7006.2002.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Li H., Lin H.-J., Yang S., Lin J., Liang G. Trends Pharmacol. Sci. 2016;37:47–61. doi: 10.1016/j.tips.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Longley D. B., Johnston P. G. J. Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- Abdel-Latif M. M., O'Riordan J., Windle H. J., Carton E., Ravi N., Kelleher D., Reynolds J. V. Ann. Surg. 2004;239:491–500. doi: 10.1097/01.sla.0000118751.95179.c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P. Cancer Treat. Rev. 2003;29(Suppl 1):33–39. doi: 10.1016/s0305-7372(03)00080-x. [DOI] [PubMed] [Google Scholar]
- Holkova B., Grant S. Expert Rev. Hematol. 2011;4:483–486. doi: 10.1586/ehm.11.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A. R., Konig H., Johnson D. E., Tang D., Amaravadi R. K., Boyiadzis M., Lotze M. T. Leukemia. 2015;29:517–525. doi: 10.1038/leu.2014.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A. M., Ryter S. W., Levine B. N. Engl. J. Med. 2013;368:1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- Sui X., Chen R., Wang Z., Huang Z., Kong N., Zhang M., Han W., Lou F., Yang J., Zhang Q., Wang X., He C., Pan H. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Saggar J. K., Yu M., Wang M., Tannock I. F. Cancer J. 2015;21:254–262. doi: 10.1097/PPO.0000000000000131. [DOI] [PubMed] [Google Scholar]
- Chouaib S., Janji B., Tittarelli A., Eggermont A., Thiery J. P. Crit. Rev. Immunol. 2014;34:91–102. doi: 10.1615/critrevimmunol.2014010183. [DOI] [PubMed] [Google Scholar]
- Yu L., McPhee C. K., Zheng L., Mardones G. A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., Hailey D. W., Oorschot V., Klumperman J., Baehrecke E. H., Lenardo M. J. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Klionsky D. J. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Joshua A. M., Saggar J. K., Yu M., Wang M., Kanga N., Zhang J. Y., Chen X., Wouters B. G., Tannock I. F. Br. J. Cancer. 2015;112:832–840. doi: 10.1038/bjc.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Amaravadi R. K., Winkler J. D. Autophagy. 2012;8:1383–1384. doi: 10.4161/auto.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li G., Zheng Y., Shen H.-M., Hu X., Ming Q.-L., Huang C., Li P., Gao N. Autophagy. 2015;11:1259–1279. doi: 10.1080/15548627.2015.1056970. [DOI] [PMC free article] [PubMed] [Google Scholar]


