 This review discusses potent molecular targets for cancer stem cells and recently developed small molecules against MELK, TOPK, and TTK.
This review discusses potent molecular targets for cancer stem cells and recently developed small molecules against MELK, TOPK, and TTK.
Abstract
Cancer stem cells (CSCs) are indicated to play critical roles in drug resistance, recurrence, and metastasis of cancer. Although molecular targeted therapies have contributed to the improvement of cancer treatments by targeting vulnerable pathways indispensable to the proliferation and survival of cancer cells, no relevant therapeutic modalities targeting CSCs have been developed yet. This review focuses on MELK (maternal embryonic leucine zipper kinase), TOPK (T-lymphokine-activated killer cell-originated protein kinase), and TTK (tyrosine threonine kinase), which are over-expressed frequently in human cancers and play indispensable roles in the development and maintenance of cancer stem cells. In addition, we will discuss recently developed small molecules for these protein targets, which have shown remarkable anti-tumor efficacies in several preclinical studies.
Introduction
Cancer stem cells (CSCs) can be defined as a small subpopulation of tumor cells having self-renewal capability and multi-lineage differentiation potential. CSCs are also called tumor initiating cells because of their potential to initiate and sustain tumor progression, and play critical roles in cancer metastasis and relapse.1 CSCs are often resistant to chemo/radiotherapies as well as currently-developed molecular-targeted therapies because they have unique properties in a slow cell cycle, the ability to detoxify or increase the efflux of cytotoxic agents, and a rapid response to DNA damage.2 As a result, CSCs have been enriched in the minimal residual disease of several malignancies after the initial response to the conventional cancer treatments.3 Therefore, new treatment strategies targeting CSCs will be required to achieve complete eradication of tumors and to reduce the risk of tumor relapse.4 For the development of anti-CSC drugs, certain molecules that are only present or specifically overexpressed in CSCs and functionally involved in the replication and/or survival of CSCs can be ideal targets.5,6 In this review, we will discuss promising molecular targets in CSCs and recently developed small molecular compounds that have shown remarkable anti-tumor efficacies in preclinical studies.
Promising molecular targets in CSCs
We defined ideal targets for the development of novel anti-cancer drugs by the following criteria: (i) molecules are frequently and highly up-regulated in cancer cells, (ii) they are not or barely expressed in normal organs, particularly in the vital organs such as heart, lung, liver, and kidney, and (iii) they must play indispensable roles in the proliferation and/or survival of cancer cells. Based on these criteria, our laboratory has identified 84 novel molecular targets matched to the above criteria through microarray expression profiles of more than 1300 clinical specimens, multiple tissue northern blot analysis, and knockdown experiments to confirm the significant roles of each gene in cancer cell growth.7–20 Considering their critical roles in cancer cell proliferation/survival and restrictive expression pattern in normal tissues except testis, fetal organs and placenta, it is not surprising that 14 of our molecular candidates (Table 1) are highly ranked in the “consensus stemness ranking (CSR) signature” genes which were derived in silico from expression data sets of mouse and human embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and CD44+CD24–/low CSC-like breast cancer cells.21 The CSR signature genes are up-regulated in cancer stem cell-enriched samples at advanced tumor stages and associated with poor prognosis in multiple cancer types, and thus regarded to play key roles in cancer stemness.21 Among them, since three molecules, MELK, TOPK, and TTK, are considered to have kinase activity, we decided to screen small molecular compounds that may effectively suppress the growth of cancer stem cells. From here on, we will focus on these kinase targets for CSCs and their small molecular compound inhibitors that we developed.
Table 1. Promising molecular targets for CSCs.
MELK inhibitor
MELK (maternal embryonic leucine zipper kinase) is a serine/threonine protein kinase and highly up-regulated in multiple types of cancer, including leukemia, as well as breast, lung, and kidney cancers,12,22–26 but its expression in normal organs is restricted to the testis although very low expression was observed in the thymus and small intestine.12 Knockdown of MELK expression by short hairpin RNA (shRNA) or small interfering RNA (siRNA) significantly inhibited growth of human breast cancer cells,12 acute myeloid leukemia (AML) cells,24 small cell lung cancer (SCLC) cells,25 and kidney cancer cells.26 As one of the possible substrates of MELK, Bcl-GL was identified by an immunocomplex kinase assay, which revealed that MELK physically interacted with Bcl-GL through its amino-terminal region and directly phosphorylated Bcl-GL in vitro.12 Bcl-GL is a pro-apoptotic member of the Bcl-2 family, and it was suggested that MELK-mediated phosphorylation could suppress the pro-apoptotic function of Bcl-GL. Indeed, overexpression of wild-type MELK suppressed Bcl-GL-induced cell apoptosis, while that of inactive MELK mutant (D150A) did not.12 Similarly, knockdown of MELK significantly induced apoptosis in AML, SCLC, and kidney cancer cells,24–26 indicating that the kinase activity of MELK is indispensable for the survival of cancer cells. In addition, we and others have reported that MELK plays important roles in CSCs with evidence that exogenous induction of MELK increased expression of a well-known stem cell marker Oct3/4.22 MELK was up-regulated in the tumor initiating cells isolated from a genetically engineered mouse model of breast cancer, and its function was required for mammary tumorigenesis in vivo.27
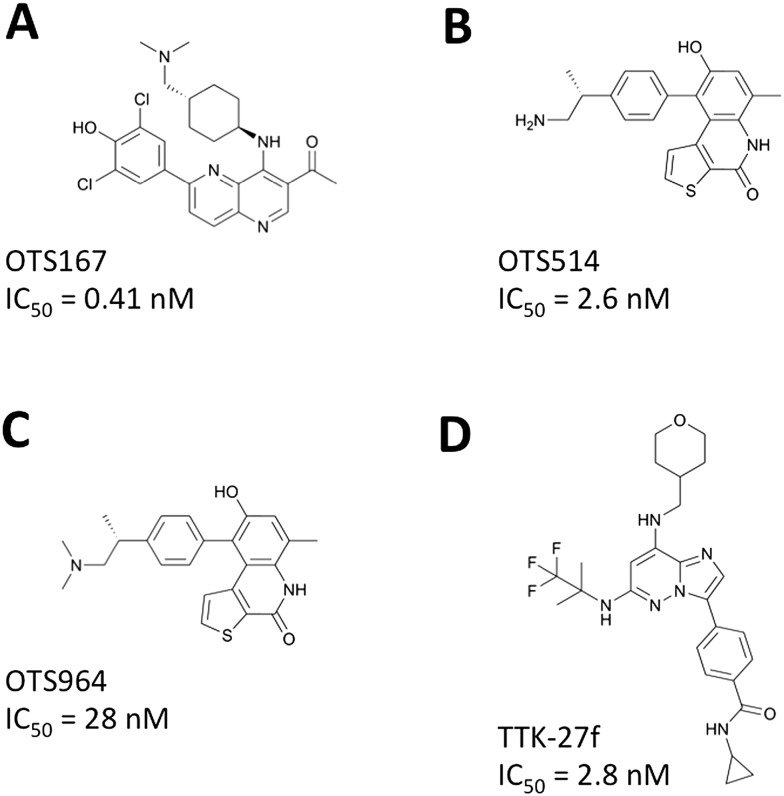
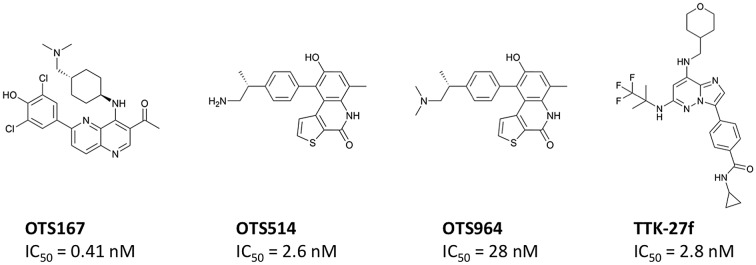
High-throughput screening of a library consisting of over 100 000 small molecular compounds and subsequent intensive structure–activity relationship (SAR) studies developed a potent MELK inhibitor, OTS167 (Fig. 1A), which has a high potential to inhibit MELK enzyme activity with a very low half maximal inhibitory concentration (IC50 = 0.41 nM).22 Treatment with OTS167 effectively suppressed growth of multiple types of MELK-positive human cancer cell lines with very low IC50 values ranging from 0.8 nM to 69.8 nM (Table 2). In mouse xenograft models of human cancers, such as breast cancer (MDA-MB-231), lung cancer (A549), pancreatic cancer (MIAPaCa-2), and prostate cancer (DU-145), intravenous injection of OTS167 showed significant tumor growth inhibitory (TGI) effects without any detectable toxicities at effective doses.22 No growth suppressive effect was observed on the MELK-negative bladder cancer xenograft model, indicating the MELK-dependent growth-suppressive effect of OTS167. Moreover, the high bioavailability of OTS167 allowed oral administration of this compound, which showed significant TGI effects in the mouse xenograft models of human cancers.22 The in vivo effects of OTS167 in mouse models are summarized in Table 2. Gastric cancer is another target disease for OTS67 because up-regulation of MELK is significantly associated with poor prognosis of gastric cancer patients.28 A recent preclinical study in gastric cancer patient-derived xenograft (PDX) mouse models showed significant suppression of tumor growth along with drastic elimination of MELK-positive gastric cancer cells.28 At present, the safety and tolerability of OTS167 are examined by two phase I clinical trials of solid tumors (Clinical Trial No. NCT01910545) and hematologic malignancies (Clinical Trial No. NCT02795520).
Fig. 1. Small molecular compounds targeting CSCs. Chemical structures and enzymatic IC50 values are shown for the MELK inhibitor (A), TOPK inhibitors (B and C), and TTK inhibitor (D).
Table 2. Tumor growth inhibitory effects of small molecular compounds.
| Target | Compound | Cancer type | Cellular effects (IC50, nM) | In vivo effects | Ref. |
| MELK | OTS167 | Multiple | DU4475 (2.3), T47D (4.3), 22Rv1 (6.0), A549 (6.7) | In A549 xenograft, 10 mg kg–1 IV injection once a day for 14 days → 108% TGI on day 15; 10 mg kg–1 PO administration once a day for 14 days → 124% TGI on day 15 | 22 |
| AML | MV4-11 (7.9), THP-1 (15.6), KG1 (16.9), KOCL-48 (16.9), MOLM13 (17.5), KG1a (26.4), K562 (33.0), ML-2 (33.8), U937 (69.8) | 24 | |||
| Gastric cancer | BGC823 (25), SGC7901 (25) | In two MELK-positive PDX models, 15 mg kg–1 IV injection once every two days for 14 days → 106% and 112% TGI on day 15 | 28 | ||
| SCLC | H69 (0.8), H82 (1.7), H524 (1.9), DMS114 (2.1), SBC5 (2.8), SBC3 (4.3), H69AR (4.4), H2171 (4.7), H446 (6.2), H146 (7.9), DMS273 (8.4) | 25 | |||
| Kidney cancer | Caki-1 (12.4), 769-P (20.1), 786-O (23.9), VMRC-RCW (25.5), Caki-2 (38.7) | 26 | |||
| TOPK | OTS514 | Multiple | LU-99 (1.5), Daudi (3.7), MKN1 (4.6), HCT-116 (4.8), MIAPaca-2 (5.2), UM-UC-3 (5.7), MKN45 (6.1), DU4475 (6.4), A549 (6.5), 22Rv1 (8.0), T47D (8.4), HepG2 (8.5), MDA-MB-231 (14) | In A549 xenograft, 5 mg kg–1 IV injection once a day for 14 days → 65% TGI on day 15; in LU-99 xenograft, 5 mg kg–1 IV injection once a day for 14 days → 104% TGI on day 15, but caused reduction of WBCs | 34 |
| AML | KOCL-48 (7.1), MOLM13 (9.0), MV4-11 (10.7), ML-2 (17.9), U937 (19.2), THP-1 (23.5), KG1a (26.2), K562 (36.8), KG1 (48.7) | In MV4-11 engrafted xenograft, 7.5 mg kg–1 IV injection once a day for 21 days → significantly elongated overall survival of mice (P < 0.001) | 29 | ||
| Kidney cancer | VMRC-RCW (19.9), Caki-2 (20.1), 769-P (20.7), Caki-1 (27.8), 786-O (44.1) | 26 | |||
| Ovarian cancer | CaOV3 (5.1), PA-1 (5.0), ES-2 (5.6), SKOV3 (6.3), OV90 (8.8), SW626 (9.4), OVCAR3 (12), A2780 (10), OVSAHO (13), RMG-I (15), OVTOKO (46) | In ES-2 abdominal dissemination xenograft, 25 mg kg–1 PO administration once a day for 14 days → significantly elongated overall survival of mice (P < 0.001) | 30 | ||
| OTS964 | Multiple | LU-99 (7.6), HepG2 (19), Daudi (25), MIAPaca-2 (30), A549 (31), UM-UC-3 (32), HCT-116 (33), MKN1 (38), MKN45 (39), 22Rv1 (50), DU4475 (53), T47D (72), MDA-MB-231 (73) | In LU-99 xenograft, 40 mg kg–1 IV injection of liposomal OTS964 five times for 14 days → 110% TGI on day 15, and complete regression of tumors in 5 of 6 mice by day 29; 100 mg kg–1 PO administration once a day for 14 days → 113% TGI on day 15, and complete regression of tumors in all 6 mice by day 29 | 34 | |
| TTK | TTK-27f | Multiple | MKN45 (3.3), LU-99 (4.7), GSS (5.3), A549 (6.0), LU-116 (6.1), HCT15 (6.3), MIAPaca-2 (8.1), HL60 (9.1), HCT116 (9.4), PC-14 (9.8), NCI-H460 (10), HT29 (16), LC2/ad (17) | In NCI-H460 xenograft, 10 mg kg–1 PO administration once a day for 14 days → 81% TGI on day 15, but caused body weight loss and 3 of 6 mice died on day 11 | 46 |
In vitro tumor sphere formation assay by selecting and culturing CSC population is a simple and useful method to determine the anti-proliferative effects of small molecular compounds in the CSCs. A previous report showed that shRNA-mediated knockdown of MELK abrogated mammosphere formation in mouse primary mammary tumor cells.27 In concordance with this finding, our cell-based assays showed that OTS167 could suppress mammosphere formation in breast cancer cells as well as lung sphere formation in SCLC cells,22,25 indicating that OTS167 preferentially suppresses the growth of CSCs. Using a potent CSC marker (CD133) and a neuronal differentiation marker (CD56), our results further showed that OTS167 suppressed growth of SCLC CSCs and also facilitated differentiation of SCLC CSCs to neuron-like cells.25 Moreover, treatment with OTS167 also induced differentiation of patient-derived primary AML blasts as assessed by CD11b staining.24 Collectively, these results suggest that OTS167 has preferential growth suppressive effects on CSCs and might be applied as a differentiation therapy, which aims to resume the process of cell differentiation from the progenitor cell phenotype.
TOPK inhibitor
TOPK (T-lymphokine-activated killer cell-originated protein kinase) is a serine/threonine protein kinase and highly up-regulated in multiple types of cancer, including leukemia as well as breast, kidney and ovarian cancers,9,23,26,29,30 but its expression was hardly detectable in normal organs except the testis and a very low level in the thymus.9 Knockdown of TOPK expression by shRNA or siRNA significantly inhibited growth of human breast cancer cells,9 AML cells,29 kidney cancer cells,26 and ovarian cancer cells.30 TOPK is highly activated during cell mitosis by autophosphorylation and suppression of its function abrogated the cytokinesis of cancer cells, leading to formation of intercellular bridges between two dividing cells followed by apoptosis.31 As one of the possible substrates of TOPK, the p47/p97 protein complex was identified by a pull-down assay with GST (glutathione S-transferase)-tagged TOPK. A subsequent in vitro kinase assay revealed that p97 could be directly phosphorylated by recombinant TOPK protein.31 The p97 protein is an ATPase enzyme and known to regulate spindle disassembly at the end of mitosis and then coordinate cellular morphologies from M to the next G1 phase.32 Indeed, our previous results showed that knockdown of p97 also induced cytokinesis defects in breast cancer cells, which were in concordance with knockdown effects of TOPK.31 These findings strongly suggested that TOPK is a mitotic kinase required to terminate cancer cell mitosis through phosphorylation of essential substrates involved in the cytokinesis, like p97, histone H3, protein regulator of cytokinesis 1 (PRC1), and G-protein signaling modulator 2 (GPSM2).18,33
By high-throughput screening of a small molecular compound library and extensive SAR studies, OTS514 (Fig. 1B) and its dimethylated derivative OTS964 (Fig. 1C) were developed as potent TOPK inhibitors. These two compounds have high potential to inhibit TOPK enzyme activity with very low IC50 values of 2.6 nM and 28 nM for OTS514 and OTS964, respectively.34 The selectivities of OTS514 and OTS964 were confirmed by examination of a panel of 60 diverse human protein kinases, which revealed the highest inhibition against TOPK itself whereas the mean and the standard deviation (SD) of the inhibitory effects against other kinases were 11.5% and 18.5% for OTS514 or 18.5% and 24.6% for OTS964.34 Treatment with OTS514 effectively suppressed the growth of multiple types of TOPK-positive human cancer cell lines with very low IC50 values ranging from 1.5 nM to 48.7 nM (Table 2). In solid cancer cell lines, OTS514 treatment caused unique cell morphological changes, intercellular bridges between dividing daughter cells, which represented cytokinesis defects caused by strong inhibition of TOPK activity.26,34 Our previous in vivo study showed that intravenous injection of OTS514 not only generated significant TGI effects in lung cancer xenograft models (Table 2), but also caused unusual hematopoietic toxicities (leukocytopenia associated with thrombocytosis) probably due to skewed lineage differentiation of hematopoietic stem cells.34 To improve both the safety and efficacy of TOPK inhibition, we developed a liposomal formulation OTS964, which is a dimethylated derivative of OTS514 with enhanced bioavailability. Moreover, the liposomal formulation was expected to increase drug concentration in tumors by the “enhanced permeability and retention (EPR)” effect that can enhance preferential extravasation of drugs from tumor vessels and accumulation in the tumor area.35 Expectedly, our LU-99 lung cancer xenograft studies demonstrated complete regression in five of six tumors after only five shots of liposomal OTS964 treatment twice per week, which caused tumor shrinkage even after discontinuation of treatment and resulted in complete tumor regression without observing any hematopoietic toxicity as described above.34 Similarly, oral administration of free OTS964 compound led to complete regression of tumors in all 6 treated mice.34 Although this treatment caused some hematopoietic toxicity, all mice spontaneously recovered after termination of the treatment. Other preclinical studies also showed anti-cancer effects of OTS514 in mouse xenograft models of AML and ovarian cancer.29,30 The in vivo effects of OTS514 and OTS964 in mouse xenograft models are summarized in Table 2.
Another important downstream molecule of TOPK is forkhead box protein M1 (FOXM1), an important transcriptional factor and a master regulator of mitosis in cancer stem cells.36 For example, either siRNA-mediated knockdown of TOPK or treatment with OTS514 in kidney cancer cells resulted in down-regulation of FOXM1 at the transcriptional level, which in turn reduced the transcriptional level of MELK.26 This kind of FOXM1-mediated effect on MELK expression might be the reason why expression levels of MELK and TOPK genes are strongly correlated with each other in various types of human cancer.25 It also indicated that targeting TOPK would be an effective approach to inhibit both FOXM1 and MELK activities in CSCs. Actually, critical roles of TOPK in CSCs and TOPK-targeting strategies were proposed in glioblastoma stem cells. For example, integrated genomic and proteomic analyses identified TOPK as one of nine representative molecules in glioblastoma stem cells.37 In another study, either knockdown of TOPK or treatment with a TOPK inhibitor (HI-TOPK-32) decreased the viability and sphere formation ability in human brain tumor-derived glioblastoma stem cells.38 Therefore, these findings strongly support that TOPK is a potential therapeutic target in CSCs and the patients who have CSC-enriched tumors would benefit from treatment with TOPK inhibitors.
TTK inhibitor
TTK (tyrosine threonine kinase), also known as Mps1 (monopolar spindle 1), is a dual specificity protein kinase that phosphorylates tyrosine and serine/threonine residues. TTK is also regarded as a mitotic kinase required for proper chromosome alignment, thereby inhibition of its kinase activity forced cancer cells to exit mitosis prematurely with missegregated chromosomes, eventually leading to cell death.39 The TTK gene is highly expressed in various types of cancer, including esophageal cancer, while its transcript is hardly detectable in normal organs except the testis.40 Due to this cancer-testis expression pattern of TTK, TTK was applied for the development of cancer-peptide immunotherapy. Treatment with a combination of multiple peptide vaccines, including HLA-A24-restricted epitope peptides derived from TTK, lymphocyte antigen 6 complex locus K (LY6K), and insulin-like growth factor-II mRNA binding protein 3 (IMP-3), could be applied to advanced esophageal cancer patients and be confirmed to effectively induce peptide specific T-cell responses and significantly improve the overall survival of patients.41,42
The TTK gene was ranked in the top CSR signature genes,21 and its biological functions in CSCs have been suggested by many studies. For example, the TTK gene was overexpressed in the CSC-like cell population isolated from human esophageal carcinoma cell lines as well as in the human multiple myeloma stem cells sorted by aldehyde dehydrogenase 1 (ALDH1).43,44 In addition, human kinase siRNA library (targeting 691 kinases) screening in triple-negative breast cancer cells (TNBCs) revealed that knockdown of TTK reduced the number of CD44-positive TNBC CSCs.45 However, the biological functions of TTK are not yet fully understood in CSCs. Identification of TTK's protein substrates and their signaling pathways will provide better understanding of the critical roles of TTK in CSCs.
High throughput screening of kinase-focused small molecular compound libraries and subsequent SAR studies identified potent TTK inhibitors including compound 27f (called “TTK-27f” in this article; Fig. 1D), as previously reported.46 TTK-27f has high potential to inhibit TTK enzyme activity with a very low IC50 value of 2.8 nM.46 Compound TTK-27f displayed excellent selectivity against TTK over 192 protein kinases. Although only two kinases (CAMK1 and CLK2) were inhibited more than 50% by this compound at 1 μM, no significant inhibition (>50%) against 192 kinases was observed at 0.2 μM.46 Treatment with TTK-27f effectively suppressed the growth of multiple types of TTK-positive human cancer cell lines with very low IC50 values ranging from 3.3 nM to 17 nM (Table 2). In the NCI-H460 xenograft, oral administration of 10 mg kg–1 of TTK-27f daily for 14 days could induce 81% of TGI effects on day 15, but caused significant body weight loss of mice and 3 of 6 mice died on day 11.46 This toxicity might be due to off-target interactions of TTK-27f with unknown proteins, either kinase or non-kinase protein, therefore a detailed mechanism of action for this compound should be fully elucidated.
In parallel, another group developed CFI-401870 that is a potent TTK inhibitor with enzymatic IC50 at 3.1 nM and cellular IC50 at 17 nM in the HCT116 colon cancer cell line.47 In the HCT116 xenograft model, oral administration of 35 mg kg–1 of CFI-401870 daily for 21 days could induce 77% of TGI effects on day 22, but 1 of 8 mice died on day 14 with body-weight loss.47 The cause of TTK inhibitor-driven toxicity was not fully elucidated in these two preclinical studies, but a lower dose treatment of TTK inhibitor in combination with other cancer therapies or applications in drug delivery systems may provide an opportunity to reduce the toxicity with improved anti-tumor efficacy of TTK inhibitors.46
Challenges and future perspectives
CSCs represent a distinct cancer cell population that has clonal long-term repopulation, self-renewal, and tumor initiating capacity, which generally define the characteristics of CSCs. However, it has not been possible to completely distinguish CSCs from non-CSCs because some cancer cells have similar features to CSCs in tumors of very aggressive type or advanced stage. Moreover, recent evidence suggested that certain cancer cells might exhibit plasticity by reversibly transitioning between a stem and non-stem-cell state.1 The most commonly used CSC markers are CD34, CD38, CD44, CD90, CD133, and ALDH. However, their expressions are dependent on cancer types and some of them are also expressed in normal stem cells, such as ESCs, iPSCs, hematopoietic stem cells, neural stem cells, and mesenchymal stem cells.48 Therefore, CSCs should be clearly distinguished from both non-CSC cancer cells and normal stem cells, by well-defined CSC surface markers as well as transcriptional gene signatures specific to CSCs. In addition, the properties of CSCs should be confirmed by experimental approaches, such as in vitro tumor sphere formation and clonogenic assays, and in vivo transplantation and lineage-tracing experiments coupled with CSC-specific markers.1 Using well-defined CSC markers and experimental models, we should be able to intensively validate promising small molecular compounds targeting CSCs.
Considering the emergence of personalized medicine, development of novel cancer therapies will be able, along with identification of biomarkers, to predict human tumor response to each therapy, which thus will enable us to choose the most appropriate treatment for individual cancer patients. For example, our recent efforts in the analysis of the MELK pathway revealed that treatment with OTS167 induced reduction of Snail, a marker for CSC and EMT (epithelial-mesenchymal transition), as well as two representative oncogenic proteins, DEPDC1 (DEP domain containing 1) and FOXM1, accompanied by drastic induction of tumor-suppressive proteins, p21 and p53, in xenograft tumor tissues.49 These downstream proteins of MELK can be utilized as possible biomarkers in the assessment of the clinical efficacy of OTS167 treatment.
In conclusion, we have identified three promising molecular targets, MELK, TOPK, and TTK, which are overexpressed almost exclusively in cancer cells, particularly those with CSC properties. Their functions as oncogenic protein kinases can be effectively inhibited by the small molecular compounds, OTS167, OTS514/OTS964, and TTK-27f, which are expected to have potent drug-like properties (Table 3) and preferentially suppress CSCs, and thus may offer novel strategies for CSC-targeted therapy.
Table 3. Drug-likeness of compounds evaluated by Lipinski's rule of five.
| Target | Compound | MW | HBD | HBA | log P | N |
| MELK | OTS167 | 487.43 | 2 | 6 | 5.20 | 1 |
| TOPK | OTS514 | 364.46 | 4 | 4 | 4.49 | 0 |
| OTS964 | 392.52 | 2 | 4 | 5.04 | 1 | |
| TTK | TTK-27f | 516.57 | 3 | 8 | 3.16 | 1 |
Acknowledgments
We thank Dr. Takashi Miyamoto at OncoTherapy Science, Inc. for providing chemical information on small molecular compounds.
Biographies

Jae-Hyun Park
Jae-Hyun Park received a PhD in Molecular Pathology at The University of Tokyo, Japan, in 2008, and finished postdoctoral training at RIKEN Yokohama Institute and Cold Spring Harbor Laboratory, before joining The University of Chicago in 2013. He is now a Research Associate/Assistant Professor at the Department of Medicine, The University of Chicago, and his major research interests are identification/characterization of molecular targets and development of novel therapeutics for cancer treatment.

Suyoun Chung
Suyoun Chung received her PhD in Cancer Biology from The University of Tokyo, Japan, in 2011 and undertook postdoctoral research at The University of Chicago, USA, under the supervision of professor Yusuke Nakamura. After a period as a researcher at the National Cancer Center in Japan, she joined OncoTherapy Science, Inc., a biopharmaceutical company in Japan. Her research interests are the evaluation of signal transduction, biomarker identification, and clinical development of novel cancer therapy.

Yo Matsuo
Yo Matsuo received a PhD in Biophysics from Kyoto University, Japan, in 1996. After undertaking postdoctoral research on bioinformatics and protein structure data analysis at the National Center for Biotechnology Information, NIH, USA, he worked at RIKEN Yokohama Institute, Japan, from 2000 to 2007, and then joined OncoTherapy Science, Inc., in 2007. His current major research field is computational chemistry and computer-aided design of small molecule drugs.

Yusuke Nakamura
Yusuke Nakamura is a Professor at the Department of Medicine and Surgery, and has been a Deputy Director at the Center for Personalized Therapeutics, The University of Chicago since 2012. Before joining The University of Chicago, he had been a full-time professor at The University of Tokyo since 1994, and a Secretary General in the Japanese Government's Office of Medical Innovation in 2011. He has published over 1300 research articles and his recent works focus on the development of novel molecular targeted therapies and immunopharmacogenomics to understand tumor-immune cell networks by employing next-generation DNA sequencing.
Footnotes
†J. P. is a scientific advisor of OncoTherapy Science, Inc. S. C. and Y. M. are employees of OncoTherapy Science, Inc. Y. N. is a stockholder and a scientific advisor of OncoTherapy Science, Inc.
References
- Kreso A., Dick J. E. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Yoshida G. J., Saya H. Cancer Sci. 2016;107:5–11. doi: 10.1111/cas.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiaur G., Gerber J., Jones R. J. Stem Cells. 2012;30:89–93. doi: 10.1002/stem.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank N. Y., Schatton T., Frank M. H. J. Clin. Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Fojo T., Bates S. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Takebe N., Miele L., Harris P. J., Jeong W., Bando H., Kahn M., Yang S. X., Ivy S. P. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K., Iinuma H., Akutsu Y., Tanaka H., Hayashi N., Uchikado Y., Noguchi T., Fujii H., Okinaka K., Fukushima R., Matsubara H., Ohira M., Baba H., Natsugoe S., Kitano S., Takeda K., Yoshida K., Tsunoda T., Nakamura Y. J. Transl. Med. 2012;10:141. doi: 10.1186/1479-5876-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro T., Tanikawa C., Furukawa Y., Katagiri T., Nakamura Y., Matsuda K. Oncogene. 2008;27:1562–1571. doi: 10.1038/sj.onc.1210799. [DOI] [PubMed] [Google Scholar]
- Park J. H., Lin M. L., Nishidate T., Nakamura Y., Katagiri T. Cancer Res. 2006;66:9186–9195. doi: 10.1158/0008-5472.CAN-06-1601. [DOI] [PubMed] [Google Scholar]
- Hayama S., Daigo Y., Kato T., Ishikawa N., Yamabuki T., Miyamoto M., Ito T., Tsuchiya E., Kondo S., Nakamura Y. Cancer Res. 2006;66:10339–10348. doi: 10.1158/0008-5472.CAN-06-2137. [DOI] [PubMed] [Google Scholar]
- Taniwaki M., Takano A., Ishikawa N., Yasui W., Inai K., Nishimura H., Tsuchiya E., Kohno N., Nakamura Y., Daigo Y. Clin. Cancer Res. 2007;13:6624–6631. doi: 10.1158/1078-0432.CCR-07-1328. [DOI] [PubMed] [Google Scholar]
- Lin M. L., Park J. H., Nishidate T., Nakamura Y., Katagiri T. Breast Cancer Res. 2007;9:R17. doi: 10.1186/bcr1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimo A., Nishidate T., Ohta T., Fukuda M., Nakamura Y., Katagiri T. Cancer Sci. 2007;98:174–181. doi: 10.1111/j.1349-7006.2006.00381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi K., Nakagawa H., Nakamura T., Eguchi H., Ohigashi H., Ishikawa O., Katagiri T., Nakamura Y. Cancer Res. 2005;65:105–112. [PubMed] [Google Scholar]
- Hirata D., Yamabuki T., Miki D., Ito T., Tsuchiya E., Fujita M., Hosokawa M., Chayama K., Nakamura Y., Daigo Y. Clin. Cancer Res. 2009;15:256–266. doi: 10.1158/1078-0432.CCR-08-1672. [DOI] [PubMed] [Google Scholar]
- Ueki T., Nishidate T., Park J. H., Lin M. L., Shimo A., Hirata K., Nakamura Y., Katagiri T. Oncogene. 2008;27:5672–5683. doi: 10.1038/onc.2008.186. [DOI] [PubMed] [Google Scholar]
- Shimo A., Tanikawa C., Nishidate T., Lin M. L., Matsuda K., Park J. H., Ueki T., Ohta T., Hirata K., Fukuda M., Nakamura Y., Katagiri T. Cancer Sci. 2008;99:62–70. doi: 10.1111/j.1349-7006.2007.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukukawa C., Ueda K., Nishidate T., Katagiri T., Nakamura Y. Genes, Chromosomes Cancer. 2010;49:861–872. doi: 10.1002/gcc.20795. [DOI] [PubMed] [Google Scholar]
- Koinuma J., Akiyama H., Fujita M., Hosokawa M., Tsuchiya E., Kondo S., Nakamura Y., Daigo Y. Cancer Sci. 2012;103:577–586. doi: 10.1111/j.1349-7006.2011.02167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa M., Takehara A., Matsuda K., Eguchi H., Ohigashi H., Ishikawa O., Shinomura Y., Imai K., Nakamura Y., Nakagawa H. Cancer Res. 2007;67:2568–2576. doi: 10.1158/0008-5472.CAN-06-4356. [DOI] [PubMed] [Google Scholar]
- Shats I., Gatza M. L., Chang J. T., Mori S., Wang J., Rich J., Nevins J. R. Cancer Res. 2011;71:1772–1780. doi: 10.1158/0008-5472.CAN-10-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Suzuki H., Miyamoto T., Takamatsu N., Tatsuguchi A., Ueda K., Kijima K., Nakamura Y., Matsuo Y. Oncotarget. 2012;3:1629–1640. doi: 10.18632/oncotarget.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Yoshimaru T., Matsuo T., Kiyotani K., Miyoshi Y., Tanahashi T., Rokutan K., Yamaguchi R., Saito A., Imoto S., Miyano S., Nakamura Y., Sasa M., Shimada M., Katagiri T. Int. J. Oncol. 2013;42:478–506. doi: 10.3892/ijo.2012.1744. [DOI] [PubMed] [Google Scholar]
- Alachkar H., Mutonga M. B., Metzeler K. H., Fulton N., Malnassy G., Herold T., Spiekermann K., Bohlander S. K., Hiddemann W., Matsuo Y., Stock W., Nakamura Y. Oncotarget. 2014;5:12371–12382. doi: 10.18632/oncotarget.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Kato T., Olugbile S., Tamura K., Chung S., Miyamoto T., Matsuo Y., Salgia R., Nakamura Y., Park J. H. Oncotarget. 2016;7:13621–13633. doi: 10.18632/oncotarget.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Inoue H., Imoto S., Tamada Y., Miyamoto T., Matsuo Y., Nakamura Y., Park J. H. Oncotarget. 2016;7:17652–17664. doi: 10.18632/oncotarget.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbard L. W., Maurer J., Miller A., Lesperance J., Hassell J., Oshima R. G., Terskikh A. V. Cancer Res. 2010;70:8863–8873. doi: 10.1158/0008-5472.CAN-10-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li Z., Guo T., Xing X. F., Cheng X., Du H., Wen X. Z., Ji J. F. Oncotarget. 2016;7:6266–6280. doi: 10.18632/oncotarget.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alachkar H., Mutonga M., Malnassy G., Park J. H., Fulton N., Woods A., Meng L., Kline J., Raca G., Odenike O., Takamatsu N., Miyamoto T., Matsuo Y., Stock W., Nakamura Y. Oncotarget. 2015;6:33410–33425. doi: 10.18632/oncotarget.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Park J. H., Miyamoto T., Takamatsu N., Kato T., Iwasa A., Okabe S., Imai Y., Fujiwara K., Nakamura Y., Hasegawa K. Clin. Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-0207. [DOI] [PubMed] [Google Scholar]
- Park J. H., Nishidate T., Nakamura Y., Katagiri T. Cancer Sci. 2010;101:403–411. doi: 10.1111/j.1349-7006.2009.01400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Nakajima R., Meyer H. H., Zheng Y. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- Abe Y., Takeuchi T., Kagawa-Miki L., Ueda N., Shigemoto K., Yasukawa M., Kito K. J. Mol. Biol. 2007;370:231–245. doi: 10.1016/j.jmb.2007.04.067. [DOI] [PubMed] [Google Scholar]
- Matsuo Y., Park J. H., Miyamoto T., Yamamoto S., Hisada S., Alachkar H., Nakamura Y. Sci. Transl. Med. 2014;6:259ra145. doi: 10.1126/scitranslmed.3010277. [DOI] [PubMed] [Google Scholar]
- Matsumura Y. Adv. Drug Delivery Rev. 2012;64:710–719. doi: 10.1016/j.addr.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Joshi K., Banasavadi-Siddegowda Y., Mo X., Kim S. H., Mao P., Kig C., Nardini D., Sobol R. W., Chow L. M., Kornblum H. I., Waclaw R., Beullens M., Nakano I. Stem Cells. 2013;31:1051–1063. doi: 10.1002/stem.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B., Mughal A. A., Grieg Z., Sandberg C. J., Joel M., Nygård S., Meling T., Murrell W., Vik Mo E. O., Langmoen I. A. Oncotarget. 2015;6:26192–26215. doi: 10.18632/oncotarget.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel M., Mughal A. A., Grieg Z., Murrell W., Palmero S., Mikkelsen B., Fjerdingstad H. B., Sandberg C. J., Behnan J., Glover J. C., Langmoen I. A., Stangeland B. Mol. Cancer. 2015;14:121. doi: 10.1186/s12943-015-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Brauer C., Thu K. L., Mason J. M., Blaser H., Bray M. R., Mak T. W. Mol. Cell. 2015;60:524–536. doi: 10.1016/j.molcel.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Kono K., Daigo Y., Takano A., Tsunoda T., Kawaguchi Y., Nakamura Y., Fujii H. Cancer Sci. 2008;99:1448–1454. doi: 10.1111/j.1349-7006.2008.00844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K., Mizukami Y., Daigo Y., Takano A., Masuda K., Yoshida K., Tsunoda T., Kawaguchi Y., Nakamura Y., Fujii H. Cancer Sci. 2009;100:1502–1509. doi: 10.1111/j.1349-7006.2009.01200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K., Iinuma H., Akutsu Y., Tanaka H., Hayashi N., Uchikado Y., Noguchi T., Fujii H., Okinaka K., Fukushima R., Matsubara H., Ohira M., Baba H., Natsugoe S., Kitano S., Takeda K., Yoshida K., Tsunoda T., Nakamura Y. J. Transl. Med. 2012;10:141. doi: 10.1186/1479-5876-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Gao Q., Guo L., Zhang C., Jiang W., Li H., Wang J., Han X., Shi Y., Lu S. H. Stem Cells Dev. 2009;18:465–473. doi: 10.1089/scd.2008.0033. [DOI] [PubMed] [Google Scholar]
- Zhou W., Yang Y., Gu Z., Wang H., Xia J., Wu X., Zhan X., Levasseur D., Zhou Y., Janz S., Tricot G., Shi J., Zhan F. Leukemia. 2014;28:1155–1158. doi: 10.1038/leu.2013.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Law J. H., Fotovati A., Dunn S. E. Breast Cancer Res. 2012;14:R22. doi: 10.1186/bcr3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe K., Ide N., Daigo Y., Itoh T., Yamamoto T., Hashizume H., Nozu K., Yoshida H., Tadano G., Tagashira S., Higashino K., Okano Y., Sato Y., Inoue M., Iguchi M., Kanazawa T., Ishioka Y., Dohi K., Kido Y., Sakamoto S., Ando S., Maeda M., Higaki M., Baba Y., Nakamura Y. J. Med. Chem. 2015;58:1760–1775. doi: 10.1021/jm501599u. [DOI] [PubMed] [Google Scholar]
- Liu Y., Lang Y., Patel N. K., Ng G., Laufer R., Li S. W., Edwards L., Forrest B., Sampson P. B., Feher M., Ban F., Awrey D. E., Beletskaya I., Mao G., Hodgson R., Plotnikova O., Qiu W., Chirgadze N. Y., Mason J. M., Wei X., Lin D. C., Che Y., Kiarash R., Madeira B., Fletcher G. C., Mak T. W., Bray M. R., Pauls H. W. J. Med. Chem. 2015;58:3366–3392. doi: 10.1021/jm501740a. [DOI] [PubMed] [Google Scholar]
- Pattabiraman D. R., Weinberg R. A. Nat. Rev. Drug Discovery. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Kijima K., Kudo A., Fujisawa Y., Harada Y., Taira A., Takamatsu N., Miyamoto T., Matsuo Y., Nakamura Y. Oncotarget. 2016;7:18171–18182. doi: 10.18632/oncotarget.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. Adv. Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]