 A library of forty 5,6-benzoflavone derivatives was synthesized and evaluated for their inhibitory potential against cholesterol esterase (CEase) enzyme.
A library of forty 5,6-benzoflavone derivatives was synthesized and evaluated for their inhibitory potential against cholesterol esterase (CEase) enzyme.
Abstract
In a continued effort to develop potent cholesterol esterase (CEase) inhibitors, a series of 5,6-benzoflavone derivatives was rationally designed and synthesized by changing the position of the benzene ring attached to the flavone skeleton in previously reported 7,8-benzoflavones. All the synthesized compounds were checked for their inhibitory potential against cholesterol esterase (CEase) using a spectrophotometric assay. Among the series of forty compounds, seven derivatives (B-10 to B-16) exhibited above 90 percent inhibition against CEase in an in vitro enzymatic assay. Compound B-16 showed the most promising activity with an IC50 value of 0.73 nM against cholesterol esterase. To determine the type of inhibition, enzyme kinetic studies were carried out for B-16, which revealed its mixed-type inhibition approach. Moreover, to figure out the key binding interactions of B-16 with the amino acid residues of the enzyme's active site, molecular protein–ligand docking studies were also performed. B-16 completely blocks the catalytic assembly of CEase and prevents it from participating in the ester hydrolysis mechanism. The favorable binding conformation of B-16 suggests its prevailing role as a CEase inhibitor. Overall, the study showed that the cis-orientation of ring A with respect to the carbonyl group of ring C is responsible for the potent CEase inhibitory activity of the newly synthesized compounds.
Introduction
Cholesterol is a vital component of the cell membrane and possesses many physiological functions. Plasma cholesterol levels are linked to many diseases such as coronary artery disease, cancer, obesity, and diabetes, which are major health concerns worldwide.1 In this respect, the matter of cholesterol level control has gained much attention.
Pancreatic cholesterol esterase (CEase) is an important serine hydrolase that plays an imperative role in the absorption of dietary cholesterol. The transport of cholesterol micelles to enterocytes is also performed by this enzyme.2 Due to its dual role in absorption and transportation, the inhibition of CEase is very important and thus provides a potential approach to treat hypercholesterolemia and atherosclerosis.3 In the recent past, several classes of potent CEase inhibitor have been developed including aryl phosphates and phosphonates,4 carbamates,5 chloroisocoumarins,6 6-chloro-2-pyrones,7 2-(1H-indol-3-yl)-4-phenylquinolines,8 3-phenyl substituted 1,3,4-oxadiazol-2(3H)-ones,9 phosphaisocoumarins,10 phosphorylated flavonoids,11 thiazolidinediones,2 and thieno[1,3]-oxazin-4-ones12 (Fig. 1). However, most of the reported inhibitors are not highly selective for CEase and could also inhibit other serine hydrolases, such as acetylcholinesterase (AChE), butylcholinesterase (BuChE), Pseudomonas species lipase, chymotrypsin and trypsin.3,13–16 One of the major reasons behind their lack of selectivity is that all serine enzymes share a similar catalytic triad of Ser–His–Asp (Glu) and mechanism of acylation–deacylation.17
Fig. 1. Reported CEase inhibitors.18.
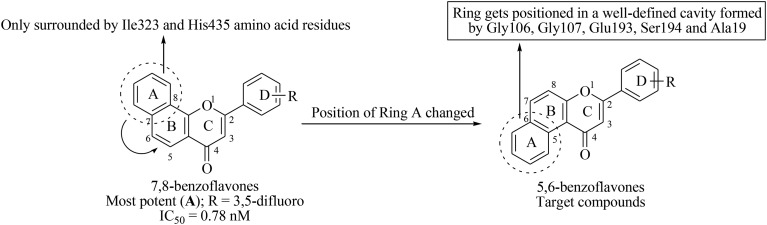
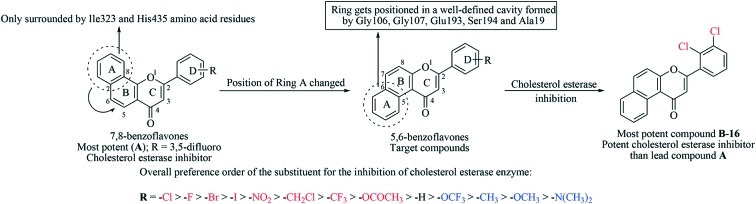
Inspired by various biological attributes of flavones, we have recently reported a series of 7,8-benzoflavone derivatives as potential CEase inhibitors.18 In the whole series of compounds, twenty seven molecules were found to exhibit more than 60% inhibition against CEase enzyme with IC50 values ranging from 0.78 to 47.80 nM. Compound A was the most promising molecule among the series of reported molecules (Fig. 2). Prompted by these significant in vitro results, a new series of compounds has been designed by simply changing the orientation of ring A, from trans- to cis-configuration with respect to the carbonyl group of ring C (Fig. 2). The docking study demonstrated that ring A is positioned in a well-defined cavity formed by Gly106, Gly107, Glu193, Ser194 and Ala19 within the active site of CEase, while ring A in the 7,8-benzoflavone derivatives is only surrounded by the Ile323 and His435 amino acid residues. We concluded from these particular findings that the cis-orientation of ring A with respect to the carbonyl group of ring C might be responsible for the good CEase inhibitory activity of the newly designed compounds. In the present study, the designed compounds were synthesized in order to evaluate the inhibitory potential against CEase enzyme using an in vitro spectrophotometric assay. The type of inhibition and the various types of interaction of the most potent inhibitor with CEase have also been investigated.
Fig. 2. Design strategy for target compounds.
Results and discussion
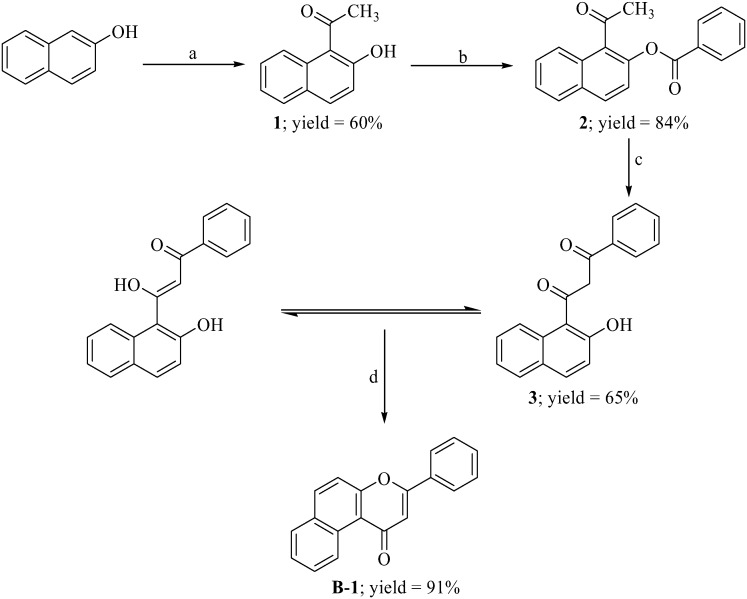
5,6-Benzoflavone derivatives were synthesized as shown in Scheme 1. β-Naphthol was subjected to Fries rearrangement and the product (1) was benzoylated using benzoylchloride to obtain 2. Product 2 was then subjected to a Baker–Venkataraman rearrangement. The Baker–Venkataraman rearranged product (3) existed in enol form (confirmed by the appearance of a singlet for two D2O exchangeable protons at 11.35 ppm along with the vinylic protones which appeared as a merged signal in a multiplet at 7.26–7.36 ppm). Compound 3 was then cyclized by treatment with sulphuric acid to yield the desired 5,6-benzoflavone (B-1). Some of the synthesized molecules have been previously reported by various research groups (as mentioned below) but most of the molecules reported herein are novel to the best of our knowledge. All the reactions proceeded smoothly with diverse benzoylchlorides (Table 1) and products were obtained in good yields. No retro-Diels–Alder fragmentation was observed for derivatives in the mass spectrum. The structures of the synthesized compounds were elucidated by 1H NMR, 13C NMR and mass spectrometry. All spectral data were in accordance with assumed structures.
Scheme 1. Synthesis of 5,6-benzoflavone. Reagents and conditions: (a) MW, ZnCl2, CH3COOH, 20 min; (b) benzoyl chloride, pyridine, stirring, RT, 1 h; (c) KOH, pyridine, warm, 15 min; (d) a drop of conc. H2SO4, CH3COOH, reflux, 30 min.
Table 1. Various substituted 5,6-benzoflavone derivatives and their CEase inhibitory activity.
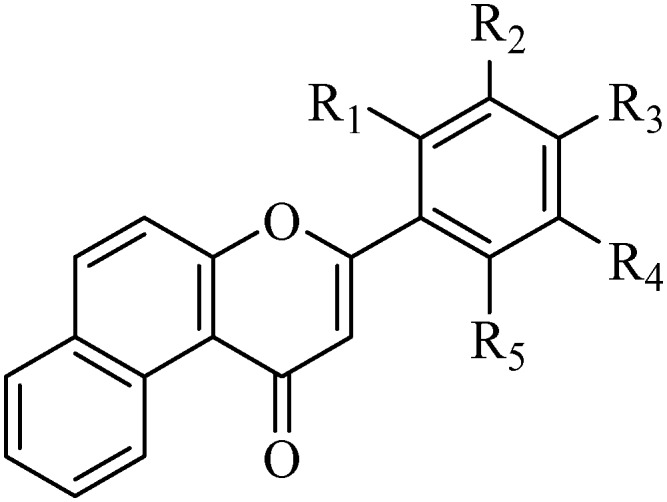
| ||||||
| Code | R1 | R2 | R3 | R4 | R5 | IC50 (nM ± SD) |
| B-1 | H | H | H | H | H | 51.06 ± 2.33 |
| B-2 | H | H | OCH3 | H | H | ND |
| B-3 | H | OCH3 | OCH3 | H | H | ND |
| B-4 | OCH3 | H | OCH3 | H | H | ND |
| B-5 | H | OCF3 | H | H | H | ND |
| B-6 | H | H | OCF3 | H | H | ND |
| B-7 | F | H | H | H | H | 2.59 ± 0.33 |
| B-8 | H | F | H | H | H | 3.90 ± 0.43 |
| B-9 | H | H | F | H | H | 4.50 ± 0.54 |
| B-10 | F | H | H | H | F | 1.19 ± 0.24 |
| B-11 | F | H | F | H | H | 1.46 ± 0.34 |
| B-12 | H | F | H | F | H | 0.99 ± 0.17 |
| B-13 | F | H | H | F | H | 1.08 ± 0.19 |
| B-14 | H | F | F | H | H | 1.15 ± 0.23 |
| B-15 | H | Cl | H | H | H | 0.83 ± 0.14 |
| B-16 | Cl | Cl | H | H | H | 0.73 ± 0.09 |
| B-17 | Br | H | H | H | H | 4.69 ± 0.76 |
| B-18 | H | Br | H | H | H | 6.01 ± 0.54 |
| B-19 | H | H | Br | H | H | 8.15 ± 0.56 |
| B-20 | I | H | H | H | H | 9.55 ± 0.67 |
| B-21 | H | H | I | H | H | 13.11 ± 0.56 |
| B-22 | H | NO2 | H | H | H | 16.44 ± 0.76 |
| B-23 | H | H | NO2 | H | H | 18.01 ± 0.65 |
| B-24 | H | NO2 | H | NO2 | H | 9.80 ± 0.53 |
| B-25 | H | H | CH3 | H | H | ND |
| B-26 | H | H | CF3 | H | H | ND |
| B-27 | H | CF3 | H | H | H | ND |
| B-28 | CF3 | H | H | H | H | 31.35 ± 0.99 |
| B-29 | H | CF3 | H | CF3 | H | ND |
| B-30 | CF3 | H | F | H | H | ND |
| B-31 | F | H | CF3 | H | H | 23.11 ± 0.75 |
| B-32 | F | H | H | H | CF3 | 29.17 ± 0.66 |
| B-33 | CF3 | H | H | F | H | 27.79 ± 0.88 |
| B-34 | H | F | H | CF3 | H | 29.11 ± 0.34 |
| B-35 | H | CF3 | F | H | H | ND |
| B-36 | H | H | CH2Cl | H | H | 22.23 ± 0.85 |
| B-37 | H | CH2Cl | H | H | H | 20.05 ± 0.94 |
| B-38 | H | H | N(CH3)2 | H | H | ND |
| B-39 | H | OCOCH3 | H | H | H | ND |
| B-40 | H | H | OCOCH3 | H | H | ND |
| PF | 0.72 ± 0.06 | |||||
In vitro screening
A CEase inhibition assay of all the synthetics was performed using a spectrophotometric assay as described in the literature8 and the results were compared with the potent cholesterol esterase inhibitor (PF) reported by Wei Y et al.11
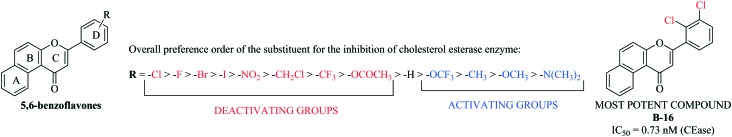
In vitro results showed that among the series of forty compounds, nine compounds exhibited a significant percentage inhibition against the CEase enzyme (more than 90% inhibition). Compound B-16 was found to be endowed with the most potent percentage inhibition against CEase with 100% inhibition. Careful examination of Table 1 revealed an interesting structure activity relationship similar to that of the reported benzoflavone derivatives (7,8-benzoflavones) used as CEase inhibitors. Any substitution on ring D (phenyl at 2nd position of 5,6-benzoflavone) significantly influences the cholesterol esterase inhibitory activity. Placement of halogen atoms on this phenyl ring considerably increases the potency against cholesterol esterase enzyme. It is also clear that as the size of halogen atom increases, the inhibitory potency significantly decreases. Ring D with deactivating groups (nitro, cholomethyl, trifloromethyl and acetoxy) favors the inhibitory activity whereas a ring D with activating groups (dimethylamino, methoxy, methyl and trifloromethoxy) disfavors the CEase inhibitory activity. Thus, the overall order of preference of the substituent on the phenyl ring at the 2nd position of the 5,6-benzoflavone moiety for the inhibition of cholesterol esterase enzyme is as follows: –Cl > –F > –Br > –I > –NO2 > –CH2Cl > –CF3 > –OCOCH3 > –H > –OCF3 > –CH3 > –OCH3 > –N(CH3)2 (Fig. 3). Compounds with a CEase enzyme inhibition of more than 60% at 50 nM were further evaluated at four different concentrations (1, 5, 10 and 25 nM) in order to calculate their IC50 values. Exceptionally, the IC50 value of unsubstituted compound B-1 was also calculated to better describe the structure activity relationship. The IC50 value of the most potent compound, B-16 (0.73 nM), was found to be comparable to that of the literature value for a phosphorylated flavonoid (PF, IC50 = 0.72 nM)11 (Table 1). Most excitingly, the whole series was found to be more active compared to the previous series of compounds (7,8-benzoflavones) with IC50 values ranging from 0.72–31.35 nM. Moreover, experiments for the evaluation of the specificity of the most potent compound towards the cholesterol esterase enzyme are also in progress.
Fig. 3. Structure activity relationship.
Enzyme kinetics study
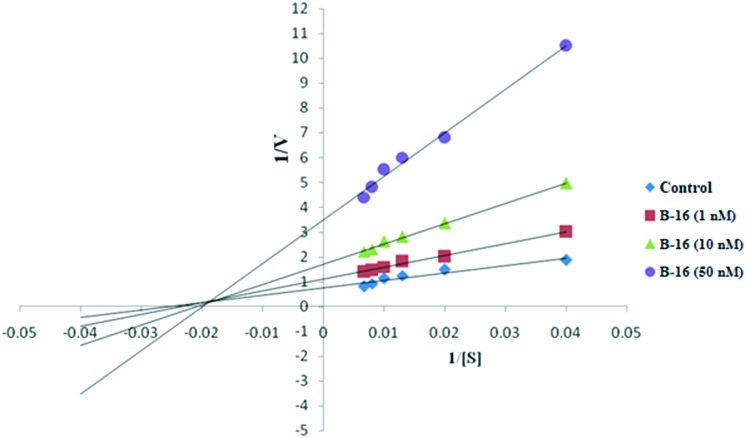
The most potent compound among the series (B-16) was further investigated to determine the type of inhibition by performing enzyme kinetic studies.19 The Lineweaver–Burk plot (Fig. 4) reveals that compound B-16 is a mixed-type CEase inhibitor. The pattern of the graph shows that it is a form of mixed inhibition scenario. The Km, Vmax and slope are all affected by the inhibitor. The inhibitor has increased Km and the slope (Km/Vmax) while decreasing Vmax. Moreover, by carefully observing Fig. 4 it was found that the intersecting lines on the graph converge to the left of the y-axis and above the x-axis which indicates that the value of α (a constant that defines the degree to which inhibitor binding affects the affinity of the enzyme for the substrate) is greater than 1. This confirms that the inhibitor preferentially binds to the free enzyme and not to the enzyme substrate complex.
Fig. 4. Lineweaver–Burk plot of B-16.
Docking studies
Various types of the binding interaction of the most potent compound, B-16, within the active site of human cholesterolesterase enzyme (hCEase), were also streamlined using molecular modeling studies. The active site apparatus of hCEase consists of a catalytic triad and an oxyanion hole.20 The catalytic triad is made up of Ser194, Asp320, and His435 residues and serves as a general acid–base and nucleophilic catalytic entity along with an oxyanion hole consisting of Gly107, Ala108, and Ala195 residues.20,21 The hydroxyl group of Ser194 acts as a nucleophile and is necessary for the hydrolytic reaction. The serine lipases and serine proteases also possess the Ser–Asp–His catalytic triad and share the catalytic mechanism of hCEase. In the present docking study, the hCEase residues within a radius of 10 Å around the hydroxyl function of Ser194 were defined as forming the active site of the enzyme.21
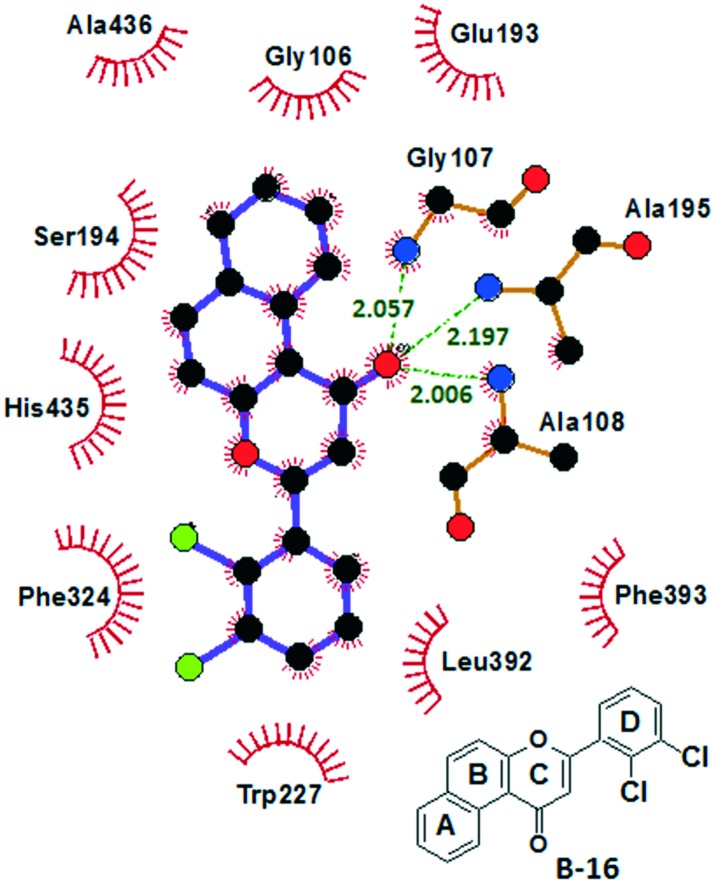
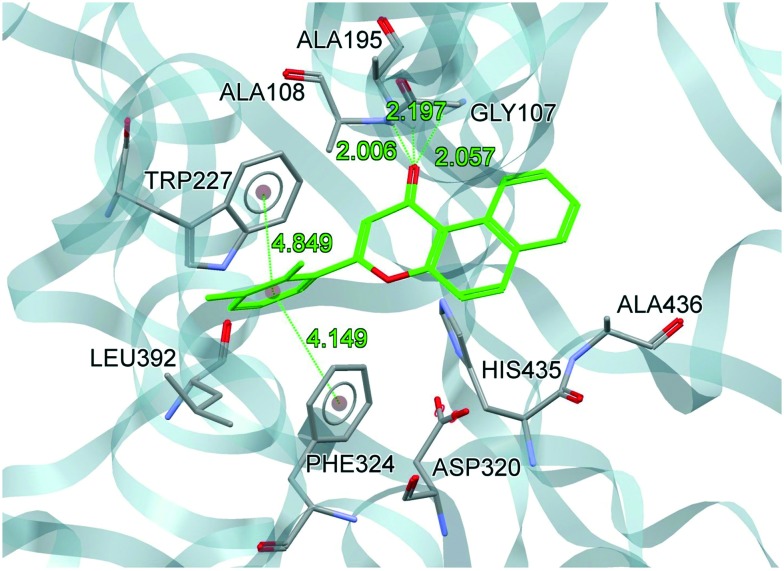
B-16 fits well at the catalytic site and is stabilized by H-bonds and polar and van der Waals interactions (Fig. 5 and 6). Interestingly, the Gly107, Ala108, and Ala195 residues of the oxyanion hole were involved in H-bond interactions with the carbonyl oxygen of ring C (H-bond acceptor; d = 2.01 to 2.19 Å). The three H-bonds showed their significance in the tight binding of B-16 with hCEase. Rings A, B and C were stabilized by van der Waals interactions with Ser194, His435 and Ala436. In addition to this, the ring C of B-16 is placed in a well-defined cavity formed by Gly106, Gly107, Glu193, Ser194 and Ala195 and is suggested to be stabilized by dispersion interactions. Ring D (dichlorophenyl) is positioned in a hydrophobic cavity created by Trp227, Phe324, Leu392 and Phe393 residues and is involved in face-to-face pi–pi stacking interactions with Trp227 and Phe324. The study showed that B-16 completely blocks the catalytic assembly of hCEase. Its binding with the oxyanion hole prevents it from participating in the ester hydrolysis mechanism. The favorable binding conformation of B-16 suggests its prevailing role as a hCEase inhibitor.
Fig. 5. Schematic 2D representations of the hCEase-B-16 complex showing H-bond and van der Waals interactions (figure generated by LIGPLOT27).
Fig. 6. Docked conformation of B-16 at the catalytic site of hCEase (B-16: carbon atoms are shown in green; only hydrogens which are involved in H-bond interactions are shown in white).
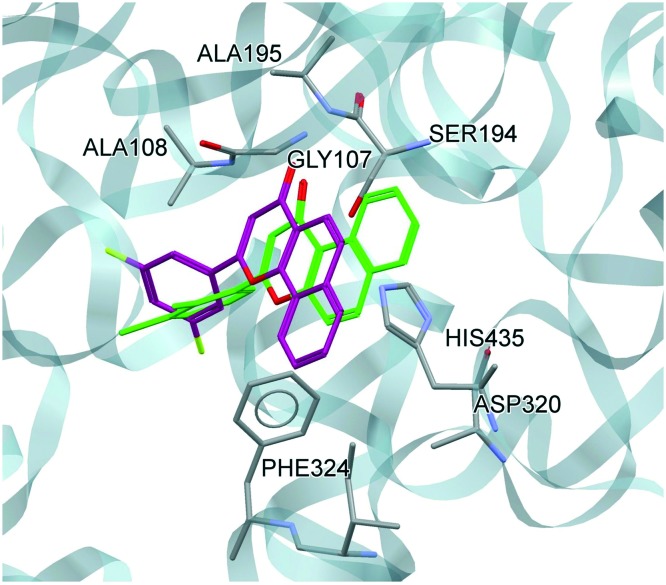
While comparing the docking conformation of B-16 and A (lead compound), B-16 showed stronger binding with hCEase as shown by a higher GoldScore than that of A (GoldScore = 54.35 and 46.06 for B-16 and A, respectively). Both compounds shared common pharmacophoric features except for the position of ring A (Fig. 2). In contrast to B-16, ring A in “compound A” is in a trans conformation with respect to the carbonyl function at ring C and is surrounded by Ile323 and His435 only. Therefore, this suggests that the cis conformation of ring A and the carbonyl function at ring C is more favorable for activity than the trans conformation. This pattern of docking is in full agreement with the in vitro results. (Fig. 7).
Fig. 7. Docked conformation of B-16 and A (lead compound) at the catalytic site of hCEase (B-16: shown in green; A: shown in purple).
In silico studies
Furthermore, physico-chemical properties like the absorption, distribution, metabolism and excretion (ADME) of the synthesized compounds were determined in silico using the web based applications MarvinSketch (; http://www.chemaxon.com/) and PreADMET (; http://preadmet.bmdrc.org/). The Caco-2 cell, MDCK cell, blood brain barrier (BBB) & skin permeabilities, human intestinal absorption values and plasma protein binding affinities are predicted and are summarized in Table 2. Results indicated that the compounds are predicted to have lower blood brain barrier permeation, which is less likely to cause neurotoxicity. In other words we can say that the synthesized compounds cannot alter the normal activity of the neuronal cells. The basicity and lipophilicity of the synthesized compounds were determined with the ChemAxon software MarvinSketch and the results are shown in Table 3, which describes the compliance of all the synthesized compounds with the Lipinski rule of five. Tabular values indicated that a) all the molecules have molecular weights within the limits of 286–408 which lies in the range of 180–500, b) the compounds have no H-bond donating properties, c) the compounds followed the H-bond acceptor criteria (<10), d) the molar refractivity was found to be very consistent in the range of 83.64–98.07 cm3 mol–1, which lies well in the accepted value range of 40–130, and e) the log P of all the compounds was found to be lower than 5.6 indicating that the compounds are not very lipophilic. These results suggest that all the compounds follow the Lipinski rule of five and have ADME properties which make them pharmacologically efficient for clinical use in the future.
Table 2. In silico ADME properties of 5,6-benzoflavone derivatives.
| Absorption | Distribution | |||||
| Compound | Human intestinal absorption (HIA)% | In vitro Caco-2 cell permeability (nm s–1) | In vitro MDCK cell permeability (nm s–1) | In vitro skin permeability (log Kp) cm h–1 | In vitro plasma protein binding (%) | In vivo blood brain barrier penetration (Cbrain/Cblood) |
| B-1 | 100.00 | 56.73 | 50.02 | –2.65 | 95.97 | 2.75 |
| B-2 | 98.80 | 57.28 | 2.31 | –2.80 | 95.48 | 0.08 |
| B-3 | 97.64 | 56.64 | 4.46 | –2.93 | 93.63 | 0.04 |
| B-4 | 97.64 | 56.64 | 4.46 | –2.93 | 93.03 | 0.04 |
| B-5 | 98.80 | 27.98 | 7.46 | –1.73 | 96.49 | 1.70 |
| B-6 | 98.80 | 27.90 | 0.13 | –1.73 | 99.05 | 1.36 |
| B-7 | 100.00 | 54.71 | 20.11 | –2.91 | 98.40 | 2.17 |
| B-8 | 100.00 | 54.75 | 27.06 | –2.94 | 100.00 | 0.28 |
| B-9 | 100.00 | 55.39 | 3.48 | –2.94 | 100.00 | 0.24 |
| B-10 | 100.00 | 53.72 | 6.62 | –3.06 | 99.69 | 1.82 |
| B-11 | 100.00 | 54.73 | 0.87 | –3.09 | 100.00 | 0.30 |
| B-12 | 100.00 | 73.77 | 10.53 | –3.12 | 100.00 | 0.30 |
| B-13 | 100.00 | 53.76 | 8.28 | –3.09 | 100.00 | 0.39 |
| B-14 | 100.00 | 54.70 | 23.36 | –3.09 | 100.00 | 0.38 |
| B-15 | 100.00 | 47.91 | 26.61 | –2.70 | 96.47 | 0.42 |
| B-16 | 100.00 | 50.14 | 20.56 | –2.62 | 100.00 | 0.80 |
| B-17 | 100.00 | 46.44 | 0.16 | –2.57 | 100.00 | 1.27 |
| B-18 | 100.00 | 46.57 | 0.12 | –2.58 | 100.00 | 0.45 |
| B-19 | 100.00 | 46.46 | 0.01 | –2.58 | 100.00 | 0.46 |
| B-20 | 100.00 | 46.45 | 0.23 | –2.62 | 100.00 | 1.07 |
| B-21 | 100.00 | 46.43 | 0.18 | –2.64 | 100.00 | 0.36 |
| B-22 | 98.49 | 21.43 | 0.42 | –2.86 | 95.25 | 0.01 |
| B-23 | 98.49 | 15.17 | 0.33 | –2.86 | 96.00 | 0.01 |
| B-24 | 96.03 | 20.38 | 0.06 | –2.83 | 94.55 | 0.04 |
| B-25 | 100.00 | 56.22 | 5.38 | –2.56 | 95.75 | 0.36 |
| B-26 | 100.00 | 41.23 | 0.93 | –1.89 | 99.68 | 0.95 |
| B-27 | 100.00 | 41.36 | 0.07 | –1.89 | 98.73 | 1.11 |
| B-28 | 100.00 | 41.61 | 3.42 | –1.89 | 97.32 | 0.58 |
| B-29 | 100.00 | 42.14 | 0.04 | –1.47 | 95.99 | 3.89 |
| B-30 | 100.00 | 48.21 | 0.15 | –1.96 | 99.97 | 0.85 |
| B-31 | 100.00 | 47.41 | 0.07 | –1.96 | 100.00 | 1.20 |
| B-32 | 100.00 | 47.73 | 1.05 | –1.95 | 100.00 | 0.72 |
| B-33 | 100.00 | 47.72 | 1.22 | –1.96 | 98.13 | 0.77 |
| B-34 | 100.00 | 48.21 | 0.06 | –1.96 | 100.00 | 1.49 |
| B-35 | 100.00 | 48.63 | 0.04 | –1.96 | 100.00 | 1.61 |
| B-36 | 100.00 | 25.60 | 2.79 | –2.60 | 100.00 | 0.39 |
| B-37 | 100.00 | 25.58 | 18.06 | –2.60 | 100.00 | 0.49 |
| B-38 | 100.00 | 57.87 | 0.37 | –2.81 | 92.67 | 0.16 |
| B-39 | 97.37 | 45.13 | 2.63 | –2.79 | 94.14 | 0.03 |
| B-40 | 97.37 | 48.99 | 0.22 | –2.79 | 93.78 | 0.02 |
Table 3. Physicochemical parameters of 5,6-benzoflavone derivatives.
| Compound | Molecular weight | No. of H-bond donors | No. of H-bond acceptors | Molar refractivity (cm3 mol–1) | Log P | No. of Lipinski violation |
| B-1 | 272 | 0 | 2 | 83.42 | 3.90 | 0 |
| B-2 | 302 | 0 | 3 | 89.88 | 3.80 | 0 |
| B-3 | 332 | 0 | 4 | 96.35 | 3.64 | 0 |
| B-4 | 332 | 0 | 4 | 96.35 | 3.64 | 0 |
| B-5 | 356 | 0 | 2 | 89.40 | 4.83 | 0 |
| B-6 | 356 | 0 | 2 | 89.40 | 4.83 | 0 |
| B-7 | 290 | 0 | 2 | 83.64 | 4.10 | 0 |
| B-8 | 290 | 0 | 2 | 83.64 | 4.10 | 0 |
| B-9 | 290 | 0 | 2 | 83.64 | 4.10 | 0 |
| B-10 | 308 | 0 | 2 | 83.85 | 4.24 | 0 |
| B-11 | 308 | 0 | 2 | 83.85 | 4.24 | 0 |
| B-12 | 308 | 0 | 2 | 83.85 | 4.24 | 0 |
| B-13 | 308 | 0 | 2 | 83.85 | 4.24 | 0 |
| B-14 | 308 | 0 | 2 | 83.85 | 4.24 | 0 |
| B-15 | 306 | 0 | 2 | 88.23 | 4.56 | 0 |
| B-16 | 341 | 0 | 2 | 93.03 | 5.16 | 0 |
| B-17 | 351 | 0 | 2 | 91.04 | 4.73 | 0 |
| B-18 | 351 | 0 | 2 | 91.04 | 4.73 | 0 |
| B-19 | 351 | 0 | 2 | 91.04 | 4.73 | 0 |
| B-20 | 398 | 0 | 2 | 96.78 | 4.89 | 0 |
| B-21 | 398 | 0 | 2 | 96.78 | 4.89 | 0 |
| B-22 | 317 | 0 | 4 | 90.75 | 3.90 | 0 |
| B-23 | 317 | 0 | 4 | 90.75 | 3.90 | 0 |
| B-24 | 362 | 0 | 6 | 98.07 | 3.84 | 0 |
| B-25 | 286 | 0 | 2 | 88.46 | 4.47 | 0 |
| B-26 | 340 | 0 | 2 | 89.40 | 4.83 | 0 |
| B-27 | 340 | 0 | 2 | 89.40 | 4.83 | 0 |
| B-28 | 340 | 0 | 2 | 89.40 | 4.83 | 0 |
| B-29 | 408 | 0 | 2 | 95.37 | 5.71 | 0 |
| B-30 | 358 | 0 | 2 | 89.61 | 4.98 | 0 |
| B-31 | 358 | 0 | 2 | 89.61 | 4.98 | 0 |
| B-32 | 358 | 0 | 2 | 89.61 | 4.98 | 0 |
| B-33 | 358 | 0 | 2 | 89.61 | 4.98 | 0 |
| B-34 | 358 | 0 | 2 | 89.61 | 4.98 | 0 |
| B-35 | 358 | 0 | 2 | 89.61 | 4.98 | 0 |
| B-36 | 320 | 0 | 2 | 93.29 | 4.54 | 0 |
| B-37 | 320 | 0 | 2 | 93.29 | 4.54 | 0 |
| B-38 | 315 | 0 | 3 | 97.85 | 4.06 | 0 |
| B-39 | 330 | 0 | 3 | 94.55 | 3.56 | 0 |
| B-40 | 330 | 0 | 3 | 94.55 | 3.56 | 0 |
Conclusion
A series of 5,6-benzoflavone derivatives was rationally designed, synthesized and characterized using 1H NMR, 13C NMR, mass spectrometry and elemental analysis. All the synthetics were evaluated for in vitro cholesterol esterase inhibitory activity. Among all the derivatives, B-16 was found to be endowed with the most potent enzyme inhibitory activity with an IC50 value of 0.73 nM. Enzyme kinetic studies confirmed that the inhibitor B-16 preferentially binds to the free enzyme and not to the enzyme substrate complex (mixed type inhibition). Docking studies suggested that compound B-16 fits well in the active site of cholesterol esterase enzyme and completely blocks its catalytic assembly. The study also concluded that the cis conformation of ring A and carbonyl function at ring C is favorable for CEase inhibition. In silico parameters revealed that the compounds with improved CEase inhibitory potential could act as hit lead molecules for the further development of a pharmacologically active CEase inhibitory framework.
Experimental
Materials and measurements
The reagents were purchased from Sigma Aldrich, Loba and CDH, India and used without further purification. The porcine cholesterol esterase enzyme was also procured from Sigma Aldrich. All yields refer to isolated products after purification. Products were characterized by comparison with authentic samples and by spectroscopic data (1H, 13C NMR and mass spectrometry). 1H NMR and 13C NMR spectra were recorded on a JEOL AL 300 NMR spectrometer. The spectra were measured in CDCl3 relative to TMS (0.00 ppm). During 1H NMR, chemical shifts were reported in δ values using tetramethylsilane as an internal standard with the number of protons, multiplicities (s – singlet, d – doublet, t – triplet, q – quartet, m – multiplet, dd – double doublet) and coupling constants (J) in Hz (Hertz) in the solvent indicated. HRMS was recorded on a micrOTOF-QII Bruker Daltonik LC-MS/MS high resolution mass spectrometer. Melting points were determined in open capillaries and were uncorrected.
Procedure for synthesis of 1-(2-hydroxynaphthalen-1-yl)ethanone (1)
β-Naphthol (1 mmol) was treated with glacial acetic acid (1.2 mmol) in the presence of ZnCl2 (0.41 mmol) under microwave irradiation for 20 min at 200 °C. The crude mixture was dissolved in methanol and adsorbed on silica (60–120 #). The desired product was purified by column chromatography with an increasing percentage of ethyl acetate in hexane as the eluting solvent. The characterization data for 1-(2-hydroxynaphthalen-1-yl)ethanone is as follows:
Yield: 60%, mp: 62–68 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 13.48 (1H, s, OH), 8.09 (1H, d, J = 8.4 Hz, ArH), 7.89 (1H, d, J = 9.0 Hz, ArH), 7.78 (1H, d, J = 8.1 Hz, ArH), 7.55–7.60 (1H, m, ArH), 7.37–7.42 (1H, m, ArH), 7.14 (1H, d, J = 9.0 Hz, ArH), 2.87 (3H, s, COCH3). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 35.22, 117.55, 122.59, 126.49, 127.06, 130.85, 131.28, 132.31, 134.63, 140.25, 166.81, 207.44. Anal. calcd. For C12H10O2: C, 77.40; H, 5.41; O, 17.18; found: C, 77.32; H, 5.55.
Procedure for synthesis of 1-acetylnaphthalen-2-yl benzoate (2)
To a solution of 1-(2-hydroxynaphthalen-1-yl)ethanone (0.01 mol) in pyridine (5 ml), benzoylchloride (1 eq.) was added and stirred for 1 h at room temperature. The mixture was poured on ice and the precipitated solid was collected and dried.
Yield: 84%, mp: 49–53 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 8.19–8.21 (2H, m, ArH), 7.81–7.96 (3H, m, ArH), 7.64–7.67 (1H, m, ArH), 7.53–7.55 (4H, m, ArH), 7.36–7.40 (1H, m, ArH), 2.62 (3H, s, COCH3); 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 35.01, 124.08, 127.11, 128.84, 130.34, 131.05, 131.33, 131.44, 132.44, 132.95, 133.65, 134.25, 136.76, 147.57, 167.52, 205.70. Anal. calcd. for C19H14O3: C, 78.61; H, 4.86; found: C, 78.93; H, 4.61.
Procedure for synthesis of 1-(2-hydroxynaphthalen-1-yl)-3-phenylpropane-1,3-dione (3)
The mixture of 1-acetylnaphthalen-2-yl benzoate (0.01 mmol), KOH (1 mmol) and pyridine (2 ml) was warmed in a water bath for 15 min. Acetic acid solution (10%, 1.3 ml) was added to the cooled mixture. The crude mixture was dissolved in ethyl acetate and adsorbed on silica (60–120 #). The desired product was purified by column chromatography with an increasing percentage of ethyl acetate in hexane as the eluting solvent. The characterization data for 1-(2-hydroxynaphthalen-1-yl)-3-phenylpropane-1,3-dione is as follows:
Yield: 65%; 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 11.35 (1H, s, D2O exchangeable proton), 8.27–8.30 (1H, d, J = 8.4 Hz, ArH), 7.90 (1H, s, ArH), 7.67–7.70 (1H, m, ArH), 7.39–7.42 (1H, m, ArH), 7.15–7.26 (5H, m, ArH), 6.66 (1H, s, ArH), 1.29 (2H, s, –CH2–). Anal. calcd. for C19H14O3: C, 78.61; H, 4.86; O, 16.53; found: C, 78.72; H, 4.73.
Procedure for synthesis of 5,6-benzoflavones (B-1)
To a solution of 1-(2-hydroxynaphthalen-1-yl)-3-phenylpropane-1,3-dione (1 mmol) in acetic acid (5 ml), a drop of concentrated sulfuric acid was added and the mixture was refluxed for 1 hour. The cooled mixture was poured onto ice and the product was collected by simple filtration. The characterization data for the synthesized derivatives is given below:
3-Phenyl-1H-benzo[f]chromen-1-one (B-1):22–24 yield 91%, mp 157–162 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 9.78 (1H, d, J = 9.0 Hz, ArH), 7.77–7.79 (1H, m, ArH), 7.73–7.74 (1H, m, ArH), 7.56–7.61 (3H, m, ArH), 7.36–7.47 (5H, m, ArH), 6.93 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 108.64, 119.15, 120.43, 122.56, 124.45, 125.32, 126.45, 127.54, 128.32, 129.34, 129.21, 131,65, 131.34, 136.65, 153.34, 162.23, 178.34. MS: m/z: 273 (M+ + 1). Anal. calcd for C19H12O2: C, 83.81; H, 4.44; found: C, 83.59; H, 4.54.
All the 5,6-benzoflavone derivatives were synthesized with the above given procedure and their characterization data is given below:
3-(4-Methoxyphenyl)-1H-benzo[f]chromen-1-one (B-2):22,25 yield 88%, mp 72–78 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.11 (1H, m, ArH), 8.13–8.17 (3H, m, ArH), 8.07 (1H, m, ArH), 7.97 (1H, m, ArH), 7.41 (2H, d, J = 8.5 Hz, ArH), 7.01–7.03 (2H, m, ArH), 6.95 (1H, s, –CH–), 3.96 (3H, s, OCH3). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 56.93, 108.27, 114.16, 118.45, 119.54, 121.66, 123.45, 123.99, 126.75, 127.16, 128.34, 130.12, 130.67, 135.76, 153.34, 162.56, 177.76. MS: m/z: 303 (M+ + 1). Anal. calcd for C20H14O3: C, 79.46; H, 4.67; found: C, 79.57; H, 4.42.
3-(3,4-Dimethoxyphenyl)-1H-benzo[f]chromen-1-one (B-3):22 yield 84%, mp 99–105 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.07 (1H, d, J = 8.9 Hz, ArH), 8.12–8.19 (3H, m, ArH), 7.93–7.99 (2H, m, ArH), 7.72–7.78 (3H, m, ArH), 6.94 (1H, s, –CH–), 3.92 (3H, s, OCH3), 3.96 (3H, s, OCH3). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 55.83, 56.35, 101.43, 102.99, 1105.54, 107.34, 110.76, 119.45, 122.76, 123.34, 123.76, 123.45, 128.76, 128.34, 129.65, 130.56, 135.34, 148.76, 153.34, 162.54, 177.34. MS: m/z: 322 (M+ + 1). Anal. calcd for C21H16O4: C, 75.89; H, 4.85; found: C, 75.91; H, 4.71.
3-(2,4-Dimethoxyphenyl)-1H-benzo[f]chromen-1-one (B-4):25 yield 89%, mp 105–113 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.11 (1H, m, ArH), 8.12–8.14 (3H, m, ArH), 8.06–8.09 (2H, m, ArH), 7.76 (2H, m, ArH), 7.22–7.24 (1H, m, ArH), 7.18 (1H, s, –CH–), 3.94 (3H, s, OCH3), 3.98 (3H, s, OCH3). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 55.95, 56.40, 101.34, 102.35, 105.43, 107.22, 119.97, 122.60, 123.74, 123.91, 128.10, 128.44, 128.76, 129.45, 130.66, 135.88, 153.68, 158.75, 160.88, 162.20, 177.33. MS: m/z: 322 (M+ + 1). Anal. calcd for C21H16O4: C, 75.89; H, 4.85; found: C, 75.66; H, 4.91.
3-(3-Trifluoromethoxyphenyl)-1H-benzo[f]chromen-1-one (B-5): yield 83%, mp: 96–101 °C, 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.08 (1H, d, J = 8.5 Hz, ArH), 8.14–8.18 (3H, m, ArH), 8.02–8.04 (2H, m, ArH), 7.21–7.26 (2H, m, ArH), 7.09–7.16 (2H, m, ArH), 6.95 (1H, s, –CH–): 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.55, 111.42, 113.67, 118.34, 120.21, 120.65, 122.76, 123.45, 123.65, 128.67, 128.99, 129.54, 129.99, 130.43, 131.78, 135.45, 153.76, 159.34, 162.55, 177.23. MS: m/z: 357 (M+ + 1). Anal. calcd for C20H11F3O3: C, 67.42; H, 3.11; F, 16.00; Found C, 67.55; H, 2.99; F, 16.09.
3-(4-Trifluoromethoxyphenyl)-1H-benzo[f]chromen-1-one (B-6): yield 87%, mp: 115–120 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.09 (1H, d, J = 8.6 Hz, ArH), 8.14–8.19 (3H, m, ArH), 8.06–8.09 (2H, m, ArH), 7.88–7.91 (2H, m, ArH), 7.23 (2H, d, J = 8.5 Hz, ArH), 6.98 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.76, 115.34, 118.95, 120.76, 120.34, 122.23, 123.21, 123.56, 125.11, 128.17, 128.34, 129.76, 130.45, 135.34, 153.87, 162.45, 177.45. MS: m/z: 357 (M+ + 1). Anal. calcd for C20H11F3O3; C, 67.42; H, 3.11; F, 16.00; found: C, 67.36; H, 3.24; F, 16.01.
3-(2-Fluorophenyl)-1H-benzo[f]chromen-1-one (B-7):23,24 yield 76%, mp 105–109 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.06 (1H, d, J = 8.4 Hz, ArH), 8.13 (1H, d, J = 9.0 Hz, ArH), 7.91–7.99 (2H, m, ArH), 7.75–7.81 (1H, m, ArH), 7.35–7.66 (5H, m, ArH), 7.13 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 115.16, 115.26, 116.90, 117.08, 117.52, 123.78, 124.36, 124.66, 126.71, 127.20, 128.17, 128.90, 129.33, 129.56, 130.41, 130.62, 132.70, 132.77, 135.65, 137.47, 156.40, 157.53, 180.28. MS: m/z: 291 (M+ + 1). Anal. calcd. for C19H11FO2: C, 78.61; H, 3.82; F, 6.54; found: C, 78.55; H, 3.91; F, 6.44.
3-(3-Fluorophenyl)-1H-benzo[f]chromen-1-one (B-8): yield 78%, mp 123–128 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.03 (1H, d, J = 8.6 Hz, ArH), 8.12–8.14 (1H, m, ArH), 7.92–7.95 (2H, m, ArH), 7.75–7.79 (1H, m, ArH), 7.62–7.66 (3H, m, ArH), 7.34–7.36 (2H, m, ArH), 7.14 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 114.13, 115.45, 116.96, 117.56, 117.56, 123.74, 124.33, 124.68, 126.75, 127.27, 128.13, 128.96, 129.34, 129.87, 130.45, 130.34, 132.77, 132.45, 135.67, 137.44, 156.45, 157.56, 180.45. MS: m/z: 291 (M+ + 1). Anal. calcd. for C19H11FO2: C, 78.61; H, 3.82; F, 6.54; found: C, 78.74; H, 3.75; F, 6.55.
3-(4-Fluorophenyl)-1H-benzo[f]chromen-1-one (B-9): yield 77%, mp 130–136 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.04 (1H, d, J = 8.5 Hz, ArH), 8.13–8.15 (1H, m, ArH), 7.91–7.94 (2H, m, ArH), 7.75–7.79 (2H, m, ArH), 7.62–7.66 (2H, m, ArH), 7.22 (2H, d, J = 8.9 Hz, ArH), 7.13 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 114.16, 115.43, 116.94, 117.57, 117.54, 123.75, 124.38, 124.64, 126.76, 127.23, 128.16, 128.94, 129.33, 129.84, 130.47, 130.33, 132.72, 132.44, 135.63, 137.45, 156.42, 157.53, 180.42. MS: m/z: 291 (M+ + 1). Anal. calcd. for C19H11FO2: C, 78.61; H, 3.82; F, 6.54; found: C, 78.72; H, 3.64; F, 6.59.
3-(2,6-Difluorophenyl)-1H-benzo[f]chromen-1-one (B-10):23,24 yield 73%, mp 89–93 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.05 (1H, d, J = 8.6 Hz, ArH), 8.13–8.16 (3H, m, ArH), 8.05–8.09 (2H, m, ArH), 7.55–7.58 (1H, m, ArH), 7.23–7.29 (2H, m, ArH), 6.93 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 114.22, 115.46, 116.93, 117.57, 117.53, 123.74, 128.12, 128.64, 129.96, 130.46, 135.34, 153.05, 158.44, 162.26, 179.99. MS: m/z: 309 (M+ + 1). Anal. calcd. for C19H10F2O2: C, 74.03; H, 3.27; F, 12.33; found: C, 74.22; H, 3.17; F, 12.44.
3-(2,4-difluorophenyl)-1H-benzo[f]chromen-1-one (B-11): yield 76%, mp 103–108 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.07 (1H, d, J = 8.4 Hz, ArH), 8.10–8.15 (3H, m, ArH), 7.99–8.03 (2H, m, ArH), 7.64–7.69 (1H, m, ArH), 7.21–7.25 (2H, m, ArH), 6.91 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 115.21, 115.88, 116.97, 117.53, 117.56, 123.73, 128.17, 128.63, 129.93, 130.47, 135.39, 153.03, 158.47, 162.24, 180.34. MS: m/z: 309 (M+ + 1). Anal. calcd. for C19H10F2O2: C, 74.03; H, 3.27; F, 12.33; found: C, 74.22; H, 3.36; F, 12.23.
3-(3,5-Difluorophenyl)-1H-benzo[f]chromen-1-one (B-12): yield 78%, mp 124–139 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 9.80 (1H, d, J = 8.4 Hz, ArH), 7.97 (1H, d, J = 9.0 Hz, ArH), 7.75 (1H, d, J = 8.1 Hz, ArH), 7.42–7.58 (4H, m, ArH), 7.29–7.33 (2H, m, ArH), 6.76–6.85 (1H, m, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 114.56, 115.41, 116.93, 117.54, 117.56, 123.77, 128.18, 128.69, 129.94, 130.43, 135.32, 153.01, 158.42, 162.23, 179.94. MS: m/z: 309 (M+ + 1). Anal. calcd. for C19H10F2O2: C, 74.03; H, 3.27; F, 12.33; found: C, 73.99; H, 3.34; F, 12.15.
3-(2,5-Difluorophenyl)-1H-benzo[f]chromen-1-one (B-13): yield 81%, mp 131–136 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.09 (1H, m, ArH), 8.15–8.19 (3H, m, ArH), 8.02–8.05 (2H, m, ArH), 7.33–7.42 (3H, m, ArH), 6.85 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 106.64, 109.09, 109.31, 111.43, 117.36, 126.90, 127.11, 128.24, 129.49, 130.33, 130.71, 135.96, 137.01, 157.27, 162.43, 164.31, 179.84. MS: m/z: 309 (M+ + 1). Anal. calcd. for C19H10F2O2: C, 74.03; H, 3.27; F, 12.33; found: C, 74.21; H, 3.08; F, 12.66.
3-(3,4-Difluorophenyl)-1H-benzo[f]chromen-1-one (B-14): yield 79%, mp 104–107 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.06 (1H, m, ArH), 8.12–8.17 (3H, m, ArH), 8.02–8.05 (2H, m, ArH), 7.42–7.45 (1H, m, ArH), 7.38 (1H, s, ArH), 7.29–7.32 (1H, m, ArH), 6.94 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 115.23, 115.65, 116.96, 117.53, 117.55, 123.71, 128.16, 128.67, 129.93, 130.42, 135.37, 153.08, 158.41, 162.23, 179.95. MS: m/z: 309 (M+ + 1). Anal. calcd. for C19H10F2O2: C, 74.03; H, 3.27; F, 12.33; found: C, 74.32; H, 3.45; F, 12.10.
3-(3-Chlorophenyl)-1H-benzo[f]chromen-1-one (B-15): yield 78%, mp 160–166 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.07 (1H, m, ArH), 8.11–8.17 (3H, m, ArH), 8.00–8.04 (2H, m, ArH), 7.66 (1H, s, ArH), 7.23–7.30 (3H, m, ArH), 6.96 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 115.45, 117.45, 117.65, 126.26, 127.64, 128.36, 129.08, 130.33, 131.64, 132.37, 133.35, 134.34, 135.43, 136.55, 153.54, 161.18, 180.05. MS: m/z: 307 (M+ + 1). Anal. calcd. for C19H11ClO2: C, 74.40; H, 3.61; Cl, 11.56; found: C, 74.50; H, 3.59; Cl, 11.60.
3-(2,3-Dichlorophenyl)-1H-benzo[f]chromen-1-one (B-16): yield 84%, mp 160–165 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.08 (1H, m, ArH), 8.15–8.19 (3H, m, ArH), 7.98–8.03 (2H, m, ArH), 7.32–7.36 (3H, m, ArH), 6.81 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 115.97, 117.26, 117.55, 126.80, 127.16, 127.64, 128.21, 129.02, 129.40, 130.41, 130.69, 131.67, 132.51, 133.71, 134.61, 135.83, 157.79, 159.70, 179.79. MS: m/z: 341 (M+ + 1). Anal. calcd. for C19H10Cl2O2: C, 66.89; H, 2.95; Cl, 20.78; found: C, 66.99; H, 2.85; Cl, 20.91.
3-(2-Bromophenyl)-1H-benzo[f]chromen-1-one (B-17): yield 75%, mp 153–159 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.08 (1H, d, J = 8.7 Hz, ArH), 8.13 (1H, d, J = 9.0 Hz, ArH), 7.94 (1H, d, J = 7.5 Hz, ArH), 7.40–7.80 (7H, m, ArH), 6.78 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 115.12, 116.89, 117.07, 117.11, 117.51, 119.90, 124.64, 124.66, 126.71, 127.19, 128.18, 128.90, 129.33, 130.61, 132.71, 132.79, 135.68, 157.54, 180.32. MS: m/z: 350 (M+ + 1). Anal. calcd. for C19H11BrO2: C, 64.98; H, 3.16; Br, 22.75; found: C, 65.05; H, 3.03; Br, 22.88.
3-(3-Bromophenyl)-1H-benzo[f]chromen-1-one (B-18): yield 78%, mp 168–171 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.04 (1H, d), 8.15 (1H, d, J = 9.0 Hz, ArH), 7.95 (1H, m, ArH), 7.77–7.83 (3H, m, ArH), 7.63 (1H, s, ArH), 7.23–7.33 (3H, m ArH), 6.98 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.53, 117.23, 117.43, 126.08, 126.79, 127.18, 127.47, 128.16, 129.35, 130.34, 130.42, 130.67, 132.35, 135.66, 157.33, 159.72, 179.97. MS: m/z: 350 (M+ + 1). Anal. calcd. for C19H11BrO2: C, 64.98; H, 3.16; Br, 22.75; found: C, 65.02; H, 3.09; Br, 22.79.
3-(4-Bromophenyl)-1H-benzo[f]chromen-1-one (B-19): yield 80%, mp 220–225 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.06 (1H, d, J = 8.7 Hz, ArH), 8.14 (1H, d, J = 9.0 Hz, ArH), 7.93 (1H, d, J = 8.1 Hz, ArH), 7.78–7.85 (3H, m, ArH), 7.61–7.70 (4H, m, ArH), 6.97 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.57, 117.28, 117.46, 126.05, 126.74, 127.16, 127.49, 128.18, 129.34, 130.34, 130.43, 130.65, 132.37, 135.65, 157.32, 159.76, 180.07. MS: m/z: 350 (M+ + 1). Anal. calcd. for C19H11BrO2: C, 64.98; H, 3.16; Br, 22.75; found: C, 64.78; H, 3.00; Br, 22.65.
3-(2-Iodophenyl)-1H-benzo[f]chromen-1-one (B-20): yield 69%, mp 141–146 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.05 (1H, m, ArH), 8.19–8.25 (3H, m, ArH), 8.11–8.13 (2H, m, ArH), 7.82–7.84 (1H, m, ArH), 7.31–7.36 (3H, m, ArH), 6.96 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.32, 113.65, 119.25, 120.65, 122.82, 124.16, 125.62, 127.26, 128.12, 128.56, 129.47, 130.81, 131.94, 136.03, 138.07, 140.53, 154.02, 165.25, 180.12. MS: m/z: 398 (M+ + 1). Anal. calcd. for C19H11IO2: C, 57.31; H, 2.78; I, 31.87; found: C, 57.42; H, 2.88; I, 31.93.
3-(4-Iodophenyl)-1H-benzo[f]chromen-1-one (B-21): yield 72%, mp 133–139 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.08 (1H, m, ArH), 8.19–8.25 (3H, m, ArH), 8.11–8.13 (2H, m, ArH), 7.88 (2H, d, J = 8.9 Hz, ArH), 7.25–7.29 (2H, m, ArH), 6.95 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.55, 113.69, 119.28, 120.62, 122.87, 124.13, 125.67, 127.22, 128.16, 128.53, 129.42, 130.87, 131.92, 136.34, 138.09, 140.51, 154.07, 165.29, 180.18. MS: m/z: 398 (M+ + 1). Anal. calcd. for C19H11IO2: C, 57.31; H, 2.78; I, 31.87; found: C, 57.28; H, 2.79; I, 31.77.
3-(3-Nitrophenyl)-1H-benzo[f]chromen-1-one (B-22): yield 79%, mp 139–141 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.15 (1H, m, ArH), 8.42 (1H, m, ArH), 8.11–8.14 (3H, m, ArH), 7.78–7.85 (2H, m, ArH), 7.62–7.65 (2H, m, ArH), 7.07 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 112.48, 115.93, 117.35, 124.25, 124.30, 126.77, 126.99, 127.14, 128.20, 128.31, 129.31, 129.62, 136.14, 136.16, 137.45, 157.45, 158.11, 179.74. MS: m/z: 318 (M+ + 1). Anal. calcd. for C19H11NO4: C, 71.92; H, 3.49; N, 4.41; found: C, 72.00; H, 3.33; N, 4.34.
3-(4-Nitrophenyl)-1H-benzo[f]chromen-1-one (B-23): yield 76%, mp: 102–106 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.14 (1H, d, J = 8.4 Hz, ArH), 8.41 (1H, d, J = 8.7 Hz, ArH), 8.14–8.19 (3H, m, ArH), 7.95 (1H, d, J = 8.4 Hz, ArH), 7.78–7.80 (1H, m, ArH), 7.65–7.68 (2H, m, ArH), 7.08 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 112.48, 115.93, 117.35, 124.25, 124.30, 126.77, 126.99, 127.14, 128.20, 128.31, 129.31, 129.62, 136.14, 136.16, 137.45, 157.45, 158.11, 179.74. MS: m/z: 318 (M+ + 1). Anal. calcd. for C19H11NO4: C, 71.92; H, 3.49; N, 4.41; found: C, 71.85; H, 3.35; N, 4.65.
3-(3,5-Dinitrophenyl)-1H-benzo[f]chromen-1-one (B-24): yield 81%, mp 101–106 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.15 (1H, s, ArH), 9.08 (1H, s, ArH), 8.65–8.70 (2H, m, ArH), 8.11 (1H, m, ArH), 7.97–7.99 (1H, m, ArH), 7.77–7.84 (3H, m, ArH), 7.07 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 112.43, 115.97, 117.39, 124.22, 124.36, 126.73, 126.93, 127.17, 128.24, 128.36, 129.37, 129.67, 136.18, 136.13, 137.46, 157.47, 158.18, 180.75 MS: m/z: 363 (M+ + 1). Anal. calcd. for C19H10N2O6: C, 62.76; H, 2.93; N, 7.35; found: C, 62.66; H, 3.03; N, 7.30.
3-p-Tolyl-1H-benzo[f]chromen-1-one (B-25):24 yield 85%, mp: 96–102 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.04 (1H, m, ArH), 8.12–8.18 (3H, m, ArH), 8.01–8.06 (2H, m, ArH), 7.33 (2H, d, J = 8.4 Hz, ArH), 7.11–7.15 (2H, m, ArH), 7.01 (1H, s, –CH–), 2.45 (3H, s, –CH3). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 26.45, 110.26, 119.93, 122.65, 123.77, 123.95, 126.33, 127.46, 128.18, 128.63, 129.07, 129.93, 130.45, 135.32, 138.55, 153.04, 162.25, 180.24. MS: m/z: 287 (M+ + 1). Anal. calcd. for C20H14O2: C, 83.90; H, 4.93; found: C, 83.80; H, 4.98.
3-(4-(Trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-26): yield 85%, mp: 115–119 °C, 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.04 (1H, m, ArH), 8.14–8.19 (3H, m, ArH), 8.01–8.05 (2H, m, ArH), 7.88 (2H, d, J = 8.9 Hz, ArH), 7.41–7.45 (2H, m, ArH) 6.97 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.221, 119.91, 122.62, 123.73, 123.95, 126.52, 128.12, 128.64, 129.93, 130.25, 130.46, 133.57, 135.31, 153.03, 162.22, 177.25. MS: m/z: 341 (M+ + 1). Anal. calcd. for C20H11F3O2: C, 70.59; H, 3.26; F, 16.75; found: C, 70.68; H, 3.06; F, 16.82.
3-(3-(Trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-27): yield 74%, mp 106–110 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.07 (1H, m, ArH), 8.13–8.17 (3H, m ArH), 8.00–8.04 (2H, m, ArH), 7.88 (1H, s, ArH), 7.21–7.33 (3H, m, ArH), 6.97 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 111.45, 119.95, 122.66, 122.83, 123.76, 123.93, 124.05, 124.67, 128.17, 128.67, 129.74, 129.93, 130.45, 130.76, 130.92, 135.33, 153.05, 162.26, 180.28. MS: m/z: 341 (M+ + 1). Anal. calcd. for C20H11F3O2: C, 70.59; H, 3.26; F, 16.75; found: C, 70.49; H, 3.35; F, 16.65.
3-(2-(Trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-28): yield: 81%; mp 105–108 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.06 (1H, m, ArH), 8.11–8.14 (3H, m, ArH), 8.00–8.04 (2H, m, ArH), 7.88–7.91 (1H, m, ArH), 7.21–7.33 (3H, m, ArH), 6.97 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.45, 119.94, 122.63, 122.85, 123.76, 123.96, 124.05, 124.64, 128.13, 128.62, 129.71, 129.92, 130.43, 130.74, 130.95, 135.36, 153.07, 162.28, 180.29. MS: m/z: 341 (M+ + 1). Anal. calcd. for C20H11F3O2: C, 70.59; H, 3.26; F, 16.75; found: C, 70.48; H, 3.35; F, 16.38.
3-(3,5-Bis(trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-29): yield 80%, mp 96–101 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.09 (1H, m, ArH), 8.13–8.18 (3H, m, ArH), 7.97–8.02 (2H, m, ArH), 7.84 (1H, s, ArH), 7.41–7.53 (3H, m, ArH), 6.93 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.48, 119.98, 122.67, 122.86, 123.75, 123.94, 124.03, 124.62, 128.12, 128.61, 129.74, 129.95, 130.46, 130.77, 130.94, 135.33, 153.06, 162.23, 180.25. MS: m/z: 409 (M+ + 1). Anal. calcd. for C21H10F6O2: C, 61.78; H, 2.47; F, 27.92; found: C, 61.68; H, 2.55; F, 27.83.
3-(4-Fluoro-2-(trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-30): yield 81%, mp 155–159 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.06 (1H, d, J = 8.6 Hz, ArH), 8.13–8.16 (3H, m, ArH), 8.06 (1H, m, ArH), 8.01 (1H, m, ArH), 7.64–7.66 (1H, m, ArH), 7.44–7.47 (1H, m, ArH), 7.23 (1H, m, ArH), 6.86 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.65, 110.99, 114.75, 117.47, 119.99, 121.45, 122.69, 123.74, 123.96, 125.94, 126.63, 128.17, 128.67, 129.94, 130.47, 135.34, 153.05, 162.25, 164.55, 180.42. MS: m/z: 359 (M+ + 1). Anal. calcd. for C20H10F4O2: C, 67.05; H, 2.81; F, 21.21; found: C, 67.11; H, 2.79; F, 21.33.
3-(2-Fluoro-4-(trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-31): yield 82%, mp 151–155 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.08 (1H, d, J = 8.6 Hz, ArH), 8.14–8.17 (3H, m, ArH), 8.03–8.06 (2H, m, ArH), 7.22–7.43 (3H, m, ArH), 6.85 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.23, 110.67, 114.74, 117.45, 119.67, 121.65, 122.76, 123.34, 123.78, 125.45, 126.67, 128.45, 128.89, 129.45, 130.67, 135.67, 153.65, 162.67, 164.56, 180.56. MS: m/z: 359 (M+ + 1). Anal. calcd. for C20H10F4O2: C, 67.05; H, 2.81; F, 21.21; found: C, 67.12; H, 2.76; F, 21.34.
3-(2-Fluoro-6-(trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-32): yield 73%, mp 122–127 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.07 (1H, d, J = 8.6 Hz, ArH), 8.12–8.14 (3H, m, ArH), 8.02–8.06 (2H, m, ArH), 7.19–7.28 (3H, m, ArH), 6.86 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.64, 110.93, 114.74, 117.45, 119.93, 121.45, 122.66, 123.74, 123.93, 125.92, 126.65, 128.14, 128.64, 129.93, 130.44, 135.36, 153.04, 162.27, 164.54, 180.47. MS: m/z: 359 (M+ + 1). Anal. calcd. for C20H10F4O2: C, 67.05; H, 2.81; F, 21.21; found: C, 67.22; H, 2.76; F, 21.32.
3-(5-Fluoro-2-(trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-33): yield 74%, mp 144–147 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.05 (1H, m, ArH), 8.10–8.13 (3H, m, ArH), 8.02–8.05 (2H, m, ArH), 7.74–7.78 (1H, m, ArH), 7.11–7.23 (2H, m, ArH), 6.85 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.62, 110.92, 114.73, 117.49, 119.98, 121.48, 122.69, 123.78, 123.97, 125.96, 126.68, 128.16, 128.67, 129.95, 130.46, 135.34, 153.07, 162.25, 164.56, 180.43. MS: m/z: 359 (M+ + 1). Anal. calcd. for C20H10F4O2: C, 67.05; H, 2.81; F, 21.21; found: C, 67.25; H, 2.65; F, 21.54.
3-(3-Fluoro-5-(trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-34): yield 69%, mp 110–115 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.11 (1H, m, ArH), 8.14–8.19 (3H, m, ArH), 8.02–8.00 (2H, m, ArH), 7.66 (1H, s, ArH), 7.21 (2H, m, ArH), 6.94 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.67, 110.95, 114.45, 117.78, 119.45, 121.98, 122.45, 123.67, 123.76, 125.54, 126.67, 128.54, 128.89, 129.43, 130.54, 135.55, 153.43, 162.54, 164.34, 180.23. MS: m/z: 359 (M+ + 1). Anal. calcd. for C20H10F4O2: C, 67.05; H, 2.81; F, 21.21; found: C, 67.14; H, 2.74; F, 21.45.
3-(4-Fluoro-3-(trifluoromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-35): yield 81%, mp 158–164 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.10 (1H, m, ArH), 8.16–8.20 (3H, m, ArH), 8.04–8.08 (2H, m, ArH), 7.69 (1H, s, ArH), 7.44 (1H, d, J = 8.9 Hz, ArH), 7.23–7.25 (1H, m, ArH), 6.96 (1H, s, –CH–). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 110.64, 110.93, 114.74, 117.45, 119.93, 121.45, 122.66, 123.74, 123.93, 125.92, 126.65, 128.14, 128.64, 129.93, 130.44, 135.36, 153.04, 162.27, 164.54, 180.47. MS: m/z: 359 (M+ + 1). Anal. calcd. for C20H10F4O2: C, 67.05; H, 2.81; F, 21.21; found: C, 67.21; H, 2.66; F, 21.29.
3-(4-(Chloromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-36): yield 74%, mp 116–120 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.05 (1H, m, ArH), 8.11–8.15 (3H, m, ArH), 8.05–8.07 (2H, m, ArH), 7.66 (2H, d, J = 8.6 Hz, ArH), 7.29 (2H, d, J = 8.6 Hz, ArH), 6.98 (1H, s, –CH–), 4.64 (2H, s, CH2Cl). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 48.54, 110.56, 119.87, 122.45, 123.34, 123.78, 125.56, 128.45, 128.76, 128.99, 129.45, 130.43, 130.55, 135.67, 137.34, 153.56, 162.56, 180.34. MS: m/z: 321 (M+ + 1). Anal. calcd. for C20H13ClO2: C, 74.89; H, 4.08; Cl, 11.05; found: C, 74.79; H, 4.18; Cl, 11.25.
3-(3-(Chloromethyl)phenyl)-1H-benzo[f]chromen-1-one (B-37): yield 77%, mp 101–105 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.11 (1H, m, ArH), 8.14–8.17 (3H, m, ArH), 8.06–8.09 (2H, m, ArH), 7.33–7.49 (3H, m, ArH), 6.98 (1H, s, –CH–), 4.69 (2H, s, CH2Cl). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 48.23, 110.23, 119.45, 122.45, 123.65, 123.45, 125.65, 128.54, 128.34, 128.34, 129.45, 130.56, 130.45, 135.23, 137.56, 153.34, 162.23, 180.45. MS: m/z: 321 (M+ + 1). Anal. calcd. for C20H13ClO2: C, 74.89; H, 4.08; Cl, 11.05; found: C, 74.78; H, 4.24; Cl, 10.96.
3-(4-(Dimethylamino)phenyl)-1H-benzo[f]chromen-1-one (B-38): yield 74%, mp 100–105 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.04 (1H, m, ArH), 8.14–8.17 (3H, m, ArH), 8.05–8.07 (2H, m, ArH), 7.41 (2H, d, J = 8.7 Hz, ArH), 7.22–7.25 (2H, m, ArH), 6.99 (1H, s, –CH–), 2.89 (3H, s, N(CH3)2), 2.83 (3H, s, N(CH3)2). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 42.56, 42.78, 110.56, 114.34, 119.54, 119.89, 122.65, 123.45, 123.67, 127.45, 128.34, 128.69, 129.56, 130.45, 135.67, 147.54, 153.56, 162.24, 180.56. MS: m/z: 316 (M+ + 1). Anal. calcd. for C21H17NO2: C, 79.98; H, 5.43; N, 4.44; found: C, 79.86; H, 5.66; N, 4.34.
3-(3-(Acetoxy)phenyl)-1H-benzo[f]chromen-1-one (B-39): yield 78%, mp 160–165 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.11 (1H, m, ArH), 8.14–8.18 (3H, m, ArH), 8.03–8.05 (2H, m, ArH), 7.77–7.82 (2H, m, ArH), 7.33–7.35 (2H, m, ArH), 6.94 (1H, s, –CH–), 2.16 (3H, s, OCOCH3). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 23.34, 110.65, 119.45, 121.46, 121.84, 122.65, 123.74, 123.92, 126.76, 127.56, 128.16, 128.66, 129.97, 130.47, 135.37, 153.08, 162.28, 168.75, 180.32. MS: m/z: 331 (M+ + 1). Anal. calcd. for C21H14O4: C, 76.35; H, 4.27; found: C, 76.29; H, 4.34.
3-(4-(Acetoxy)phenyl)-1H-benzo[f]chromen-1-one (B-40): yield 81%, mp 159–163 °C. 1H NMR (CDCl3, 300 MHz, δ, TMS = 0): 10.10 (1H, m, ArH), 8.14–8.18 (3H, m, ArH), 8.02–8.04 (2H, m, ArH), 7.66 (2H, d, J = 8.5 Hz, ArH), 7.13–7.15 (2H, m, ArH), 6.96 (1H, s, –CH–), 2.14 (3H, s, OCOCH3). 13C NMR (CDCl3, 75 MHz, δ, TMS = 0): 23.54, 110.25, 119.96, 121.48, 121.86, 122.67, 123.74, 123.95, 126.73, 127.66, 128.14, 128.62, 129.93, 130.42, 135.35, 153.02, 162.22, 168.75, 179.99. MS: m/z: 331 (M+ + 1). Anal. calcd. for C21H14O4: C, 76.35; H, 4.27; found: C, 76.42 H, 4.18.
In vitro cholesterol esterase assay
Porcine cholesterol esterase inhibition was assayed spectrophotometrically at 405 nm at 25 °C. The assay buffer was 100 mM sodium phosphate and 100 mM NaCl, at pH 7.0. A stock solution of CEase (200 μg mL–1) was prepared in 100 mM sodium phosphate buffer, at pH 7.0, and kept at 0 °C. A 1:200 dilution was done with the same buffer immediately before starting the measurement. Sodium taurocholate (12 mM) was dissolved in assay buffer and kept at 25 °C. A stock solution of para-nitrophenyl butyrate (20 mM) was prepared in acetonitrile. The final concentration of acetonitrile was 3%, that of the substrate para-nitrophenyl butyrate was 20 μM, and that of sodium taurocholate was 6 mM. Assays were performed with a final concentration of 10 ng mL–1 of CEase. Into a cuvette containing 430 μL assay buffer, 500 μL of the sodium taurocholate solution, 20 μL acetonitrile, 10 μL of the para-nitrophenyl butyrate solution, and 30 μL of an inhibitor solution in DMSO were added and thoroughly mixed. After incubation for 5 min at 25 °C, the reaction was initiated by adding 10 μL of the enzyme solution with concentration 1 μg mL–1. All the experiments were performed in triplicate and values were expressed as means of three experiments.8
Enzyme kinetics study
Synthesized compounds were further investigated to determine the type of inhibition and enzyme kinetics studies were carried out. The Lineweaver–Burk plot was established from which we can calculate the Km, the Vmax of the slope of the inhibitor and the value of α (a constant that defines the degree to which inhibitor binding affects the affinity of the enzyme for substrate).19
Docking study
To study the binding mode of the synthesized compounds, we docked the most potent compound B-16 at the catalytic site of lipase. The X-ray coordinates of the catalytic domain of human bile salt activated lipase (hBAL, hCEase) was obtained from the protein data bank (PDB entry: ; 1F6W; resolution 2.3 Å).20 The docking study was carried out using GOLD v5.3 software.26 GOLD performs genetic algorithm based ligand docking to optimize the conformation of the ligand at the receptor binding site. The GoldScore fitness function was used to evaluate the various conformations of the ligand at the binding site. GoldScore comprises four components: protein–ligand hydrogen bond energy, protein–ligand van der Waals (vdw) energy, ligand internal vdw energy and ligand torsional strain energy.26B-16 was docked ten times and the conformation associated with the highest scoring value was considered to analyze various drug–receptor interactions. The structure of B-16 was drawn in ChemDraw Ultra (2010) and subjected to energy minimization using the MM2 force field as implemented in the Chem 3D Ultra software.
Conflicts of interest
The authors declare no competing interests.
Supplementary Material
Acknowledgments
The authors are grateful to the University Grants Commission for providing funds under the University with Potential for Excellence (UPE) Scheme and Rajiv Gandhi National Fellowship (RGNF) to carry out the research work. The authors are also thankful to Dr. Sahil Sharma, Post-doctoral research Fellow at the Memorial Sloan Kettering Cancer Center, New York, USA for his valuable support and guidance.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7md00565b
‡This research article is dedicated to Dr. Sahil Sharma.
References
- Mahalwar R., Khanna D. Eur. J. Pharmacol. 2013;711:57–62. doi: 10.1016/j.ejphar.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Heng S., Tieu W., Hautmann S., Kuan K., Pedersen D. S., Pietsch M., Gutschow M., Abell A. D. Bioorg. Med. Chem. 2011;19:7453–7463. doi: 10.1016/j.bmc.2011.10.042. [DOI] [PubMed] [Google Scholar]
- Pietsch M., Gutschow M., J. Med. Chem., 2005, 48 , 8270 –8288 , (and the references therein) . [DOI] [PubMed] [Google Scholar]
- (a) Quistad G. B., Liang S. N., Fisher K. J., Nomura D. K., Casida J. E. Toxicol. Sci. 2006;91:166–175. doi: 10.1093/toxsci/kfj124. [DOI] [PubMed] [Google Scholar]; (b) Schmidinger H., Birner-Gruenberger R., Riesenhuber G., Saf R., Susani E. H., Hermetter A. ChemBioChem. 2005;6:1776–1781. doi: 10.1002/cbic.200500013. [DOI] [PubMed] [Google Scholar]
- (a) Lin M. C., Lin G. Z., Hwang C. I., Jian S. Y., Lin J., Shen Y. F., Lin G. Protein Sci. 2012;21:1344–1357. doi: 10.1002/pro.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lin M. C., Yeh S. J., Chen I. R., Lin G. Protein J. 2011;30:220–227. doi: 10.1007/s10930-011-9323-3. [DOI] [PubMed] [Google Scholar]; (c) Chiou S. Y., Lai C. Y., Lin L. Y., Lin G. BMC Biochem. 2005;6:17. doi: 10.1186/1471-2091-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lin G., Liao W. C., Chiou S. Y. Bioorg. Med. Chem. 2000;8:2601–2607. doi: 10.1016/s0968-0896(00)00196-6. [DOI] [PubMed] [Google Scholar]; (e) Lin G., Shieh C. T., Tsai Y. C., Hwang C. I., Lu C. P., Cheng G. H. Biochim. Biophys. Acta. 1999;1431:500–511. doi: 10.1016/s0167-4838(99)00073-4. [DOI] [PubMed] [Google Scholar]; (f) Feaster S. R., Lee K., Baker N., Hui D. Y., Quinn D. M. Biochemistry. 1996;35:16723–16734. doi: 10.1021/bi961677v. [DOI] [PubMed] [Google Scholar]
- Heynekamp J. J., Hunsaker L. A., Vander J. T. A., Royer R. E., Deck L. M., Vander J. D. L. Bioorg. Med. Chem. 2008;16:5285–5294. doi: 10.1016/j.bmc.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard H. M., Brown W. M., Deck J. A., Hunsaker L. A., Deck L. M., Vander J. D. L. Biochim. Biophys. Acta. 2002;596:381–391. doi: 10.1016/s0167-4838(01)00304-1. [DOI] [PubMed] [Google Scholar]
- Muscia G. C., Hautmann S., Buldain G. Y., Gutschow M. Bioorg. Med. Chem. Lett. 2014;24:1545–1549. doi: 10.1016/j.bmcl.2014.01.081. [DOI] [PubMed] [Google Scholar]
- Point V., Benarouche A., Zarillo J., Guy A., Magnez R., Fonseca L., Raux B., Leclaire J., Buono G., Fotiadu F., Durand T., Carriere F., Vaysse C., Couedelo L., Cavalier J. F. Eur. J. Med. Chem. 2012;58:452–463. [Google Scholar]
- Li B., Zhou B., Lu H., Ma L., Peng A. Y. Eur. J. Med. Chem. 2010;45:1955–1963. doi: 10.1016/j.ejmech.2010.01.038. [DOI] [PubMed] [Google Scholar]
- Wei Y., Peng A. Y., Wang B., Ma L., Peng G., Du Y., Tang J. Eur. J. Med. Chem. 2014;74:751–758. doi: 10.1016/j.ejmech.2013.03.025. [DOI] [PubMed] [Google Scholar]
- Pietsch M., Gutschow M. J. Biol. Chem. 2002;277:24006–24013. doi: 10.1074/jbc.M112252200. [DOI] [PubMed] [Google Scholar]
- Deck L. M., Baca M. L., Salas S. L., Hunsaker L. A., Vander-Jagt D. L. J. Med. Chem. 1999;42:4250–4256. doi: 10.1021/jm990309x. [DOI] [PubMed] [Google Scholar]
- Lin G., Chiou S. Y., Hwu B. C., Hsieh C. W. Protein J. 2006;25:33–43. doi: 10.1007/s10930-006-0013-5. [DOI] [PubMed] [Google Scholar]
- Lin G., Shieh C. T., Ho H. C., Chouhwang J. Y., Lin W. Y., Lu C. P. Biochemistry. 1999;38:9971–9981. doi: 10.1021/bi982775e. [DOI] [PubMed] [Google Scholar]
- Lin G., Tsai Y., Liu H. C., Liao W. C., Chang C. H. Biochim. Biophys. Acta. 1998;1388:161–174. doi: 10.1016/s0167-4838(98)00184-8. [DOI] [PubMed] [Google Scholar]
- Cygler M., Schrag J. D., Sussman J. L., Harel M., Silman I., Gentry M. K., Doctor B. P. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Singh J. V., Gupta M. K., Singh P., Sharma S., Nepali K., Bedi P. M. S. Bioorg. Med. Chem. Lett. 2017;27:850–854. doi: 10.1016/j.bmcl.2017.01.020. [DOI] [PubMed] [Google Scholar]
- Singh M., Kaur M., Silakari O. Eur. J. Med. Chem. 2014;84:206–239. doi: 10.1016/j.ejmech.2014.07.013. [DOI] [PubMed] [Google Scholar]
- Terzyan S., Wang C. S., Downs D., Hunter B., Zhang X. C. Protein Sci. 2000;9:1783–1790. doi: 10.1110/ps.9.9.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Thangapandian S., Lee K. W. J. J. Biomol. Struct. Dyn. 2012;29:921–936. doi: 10.1080/07391102.2012.10507419. [DOI] [PubMed] [Google Scholar]
- Juvale K., Stefan K., Wiese M. Eur. J. Med. Chem. 2013;67:115–126. doi: 10.1016/j.ejmech.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Yonehara M., Tetsuhashi M., Noguchi-Yachide T., Hashimoto Y., Ishikawa M. Bioorg. Med. Chem. 2010;18:1194–1203. doi: 10.1016/j.bmc.2009.12.036. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Yonehara M., Kitahara K., Shimokawa J., Hashimoto Y., Ishikawa M. Heterocycles. 2011;83:2563–2575. [Google Scholar]
- Hwang D., Jo G., Hyun J., Lee S. D., Kohc D., Lima Y. Magn. Reson. Chem. 2012;50:62–67. doi: 10.1002/mrc.3790. [DOI] [PubMed] [Google Scholar]
- GOLD v5.3, Cambridge Crystallographic Data Centre, Cambridge, U.K., 2015.
- Wallace A. C., Laskowski R. A., Thornton J. M. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.