 Suppressing tumor cell metabolism is an attractive strategy for treating cancer. We identified a 2,3-didithiocarbamate-substituted naphthoquinone 3i that inhibited the proliferation of tumor cells by disturbing their metabolism.
Suppressing tumor cell metabolism is an attractive strategy for treating cancer. We identified a 2,3-didithiocarbamate-substituted naphthoquinone 3i that inhibited the proliferation of tumor cells by disturbing their metabolism.
Abstract
Tumor cells reprogram their cellular metabolism by switching from oxidative phosphorylation to aerobic glycolysis to support aberrant cell proliferation. Suppressing tumor cell metabolism has become an attractive strategy for treating cancer patients. In this study, we identified a 2,3-didithiocarbamate-substituted naphthoquinone 3i that inhibited the proliferation of tumor cells by disturbing their metabolism. Compound 3i reduced cancer cell viability with IC50 values from 50 nM to 150 nM against HCT116, MCF7, MDA-MB231, HeLa, H1299 and B16 cells. Further, compound 3i was found to suppress ATP production in cultured cancer cells, inhibit the M2 isoform of pyruvate kinase (PKM2) which is a rate-limiting enzyme in the glycolytic pathway and block the subsequent transcription of the downstream genes GLUT1, LDH and CCND1. In addition, exposure to compound 3i significantly suppressed tumor growth in a B16 melanoma transplantation mouse model and a spontaneous breast carcinoma mouse model in vivo. The identification of compound 3i as a tumor metabolic suppressor not only offers a candidate compound for cancer therapy, but also provides a tool for an in-depth study of tumor metabolism.
1. Introduction
Cancer cells differ from most normal cells in metabolism. Normal cells rely generally on mitochondrial oxidative phosphorylation to generate energy from glucose, whereas cancer cells instead rely on glycolysis.1–4 This difference suggests that targeting tumor metabolism could be a selective approach to suppress tumor growth.5–7 Glucose goes through a series of biochemical transformations with production of ATP in the process of glycolysis, and each step is catalyzed by specific enzymes.5 Influencing these enzymes to reduce or reverse the abnormal reprogrammed metabolism of cancer cells is an attractive therapeutic strategy for cancer patients. Pyruvate kinase (PK) is a rate-limiting enzyme that regulates the final step in glycolysis and catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to adenosine diphosphate (ADP) to yield pyruvate and adenosine triphosphate (ATP).8,9 There are four isoforms of PK (M1, M2, L and R) in mammalian cells: the M1 isoform (PKM1) is expressed in many differentiated tissues, PKM2 is expressed during embryonic development and overexpressed in tumor tissues, PKL and PKR are expressed in liver and erythrocytes, respectively.10–12 Many studies have shown that tumorigenesis is connected with the re-expression of PKM2 together with downregulation of the expression of PKM1 and other isozymes.9,13 In addition, PKM2 activates β-catenin to induce CCDN1 and c-Myc expression and upregulate GLUT1 and lactate dehydrogenase A (LDHA).14–17 Upregulation of these glycolysis genes increases glucose intake, consumption and lactate production to promote tumorigenesis.1
We previously synthesized a series of dithiocarbamate-substituted naphthoquinone derivatives and evaluated their anti-proliferative effects at the cellular level and PKM2 inhibition activity at the enzyme level.18,19 To further improve the activity, we continue to optimize the structure. In this study, we synthesized a new 2,3-didithiocarbamate-substituted naphthoquinone compound 3i (Fig. 1A), which exhibited a more potent anti-proliferative effect than previously synthesized compounds. It reduced cancer cell viability with IC50 values from 50 nM to 100 nM against HCT116, MCF7, MDA-MB231, HeLa, H1299 and B16 cells. We also found that compound 3i suppressed tumor cell metabolism by reducing ATP production in cancer cells, inhibiting PKM2 activity and blocking the subsequent transcription of the downstream genes GLUT1, LDH and CCND1. In addition, exposure to compound 3i significantly suppressed tumor growth in a B16 melanoma transplantation mouse model and a spontaneous breast carcinoma mouse model in vivo.
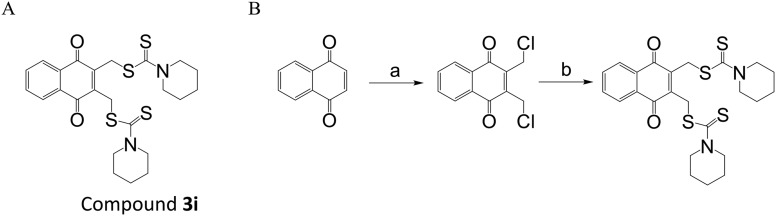
Fig. 1. The structure of and synthetic route toward compound 3i. (A) The structure of compound 3i. (B) The synthetic route toward compound 3i. Reagents and conditions: (a) formaldehyde, HCl, HAc, H2O, 0 °C, 68.9%; (b) CS2, piperidine, CH3CN, rt, 83.5%.
2. Materials and methods
2.1. Synthesis of compound 3i
2.1.1. Procedure for preparation of 2,3-bis-chloromethyl-[1,4]naphthoquinone
The 1,4-naphthoquinone (1 g, 6.3 mmol) in glacial acetic acid (20 mL) was placed in a 100 mL round-bottomed flask, and 36% aqueous formaldehyde (6 mL) was added. The reaction solution was cooled in ice-water. Dry hydrogen chloride was passed in for 2 h. The solution became red, then was kept at room temperature for 48 h. The reaction mixture was poured onto ice and extracted with ethyl acetate. The combined organic fractions were washed with brine, dried (Na2SO4), and concentrated under reduced pressure. Purification of the crude residue by column chromatography (petroleum ether/ethyl acetate) afforded the title compound (yellow solid). The yield of this reaction was 68.9%. 1H NMR (400 MHz, CDCl3) δ 8.18–8.20 (m, 2H, ArH), 7.81–7.83 (m, 2H, ArH), 4.72 (s, 4H, 2CH2Cl).
2.1.2. Procedure for preparation of dipiperidine-dithiocarbamic acid 3-dipiperidinethiocarbamoylsulfanylmethyl-1,4-dioxo-1,4-dihydronaphthalen-2-ylmethyl ester (3i)
Carbon disulfide (180 μL, 3 mmol) and piperidine (297 μL, 3 mmol) were added to CH3CN (5 mL) and the resulting solution was stirred for 30 minutes. 2,3-Bis-chloromethyl-[1,4]naphthoquinone (254 mg, 1 mmol) was added in portions at frequent intervals. Then, the reaction mixture was kept at room temperature for 48 h. The reaction mixture was concentrated in vacuo, diluted with H2O, and extracted with CH2Cl2. The combined organic fractions were washed with brine, dried (Na2SO4), and concentrated under reduced pressure. Purification of the crude residue by column chromatography (petroleum ether/CH2Cl2) afforded compound 3i (yellow solid). The yield of this reaction was 83.5%. Mp 142–143 °C. 1H NMR (400 MHz, CDCl3) δ 8.11–8.13 (m, 2H, ArH), 7.73–7.75 (m, 2H, ArH), 4.86 (s, 4H, 2CH2S), 4.29 (q, 4H, 2NCH2), 3.87 (q, 4H, 2NCH2), 1.70 (m, 12H, 2CH2CH2CH2). 13C NMR (100 MHz, CDCl3) δ 194.21, 183.95, 143.97, 133.84, 132.05, 126.65, 34.11, 24.25. HR-MS (ESI+) m/z: 505.1112 [M + H]+, 527.0931 [M + Na]+. Found: 505.1100 [M + H]+, 527.0903 [M + Na]+.
2.2. Cell culture
Cell lines were grown with routine culture techniques in RPMI 1640 supplemented with 9% fetal bovine serum at 37 °C in 5% CO2.
2.3. MTS cell proliferation assay
Cells were plated in 96-well plates at a density of 5000–10 000 cells per well. 12 h after seeding, the cells were treated with various concentrations of test compounds for 48 h. Cell viability was assessed with the MTS assay (Promega) according to the manufacturer's instructions.
2.4. Measurement of ATP
HCT116 cells were seeded into 6-well plates at a density of 5 × 105 cells per well. 24 h after seeding, the cells were treated with 10 μM or 20 μM compound 3i for 6 h. ATP levels were measured using the Cell Titer-Glo Luminescent Cell Viability Assay (Promega).
2.5. PKM2 activity assay
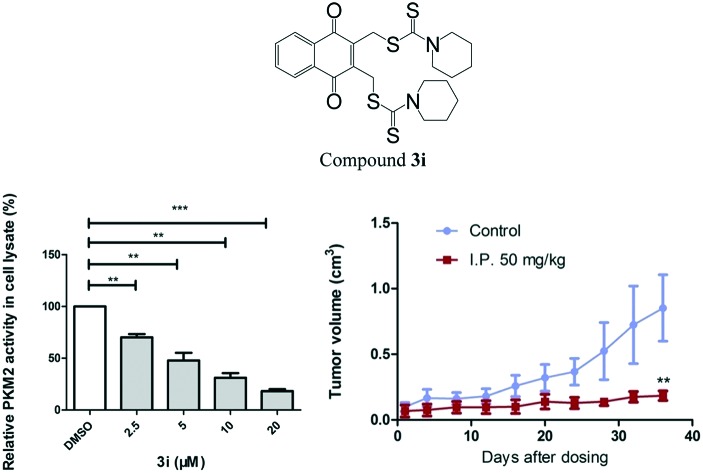
Pyruvate kinase activity was measured with a fluorescent pyruvate kinase–lactate dehydrogenase coupled assay as previously described.20 To evaluate PKM2 activity in cell lysates, HCT116 cells were treated with various concentrations of compound 3i for 8 h and lysed in NP40 lysis buffer immediately before measuring the pyruvate kinase activity as described previously.21
2.6. Quantitative real-time PCR (RT-qPCR)
HCT116 cells were seeded into 6-well plates at a density of 5 × 105 cells per well. 24 h after seeding, the cells were treated with 20 μM compound 3i for 8 h. Total RNA was extracted using TRIzol (Invitrogen). cDNA synthesis was carried out using a cDNA synthesis kit (Transgene). qPCR was then carried out using a SYBR Green Master Mix (Transgene) in a Bio-rad real-time PCR machine. The PCR primer sequences used are listed in ESI† Table S1.
2.7. B16 melanoma transplantation mouse model
Female C57BL/6 mice were injected with 1 × 106 B16 cells subcutaneously in the armpits. Approximately 6 days later, B16 tumors appeared, and the mice were paired (N = 7) and injected with compound 3i (25 mg kg–1 and 50 mg kg–1) or the vehicle. Compound 3i was dissolved in 5% (v/v) DMAC (dimethylacetamide) and added to olive oil. Intraperitoneal injection was performed in the mice. The animals were injected once every two days. Tumor volume was calculated using the following equation: V = L(S2)π/6, where L is the longer and S is the shorter of the two tumor dimensions.
2.8. Spontaneous breast carcinoma mouse model
C57BL/6 spontaneous breast carcinoma mice22 were paired (N = 3) and injected with compound 3i (50 mg kg–1) or the vehicle, when spontaneous tumor volumes reached 0.1 cm3. Intraperitoneal injection was performed in the mice. The animals were injected once every two days.
All experiments were performed in compliance with the relevant laws and institutional guidelines of the Institute Research Ethics Committee of Peking University Health Science Center, and the committee had approved the experiments.
2.9. Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0. Data are presented as mean ± SD (n = 3).
3. Results and discussion
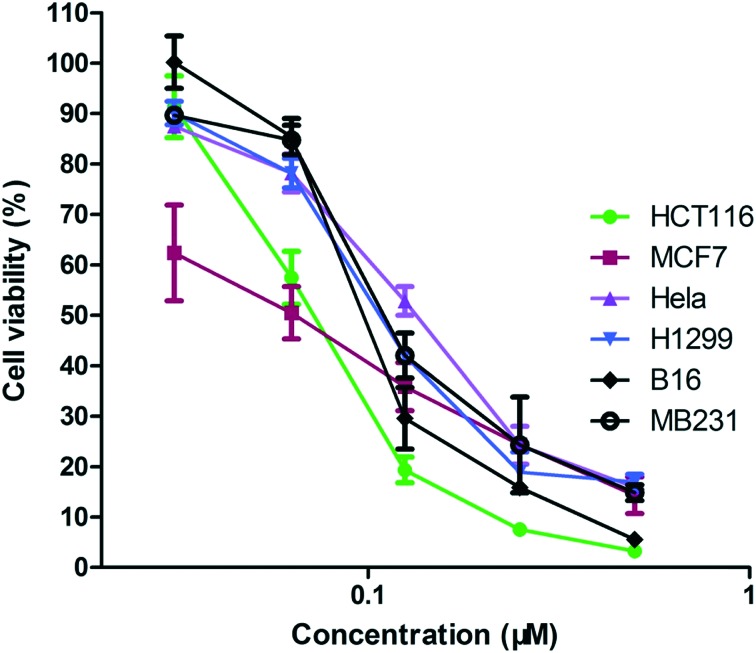
We prepared compound 3i by the synthetic route shown in Fig. 1B. To determine the efficiency of compound 3i as an anti-tumor agent, we assessed the in vitro cytotoxicity of 3i using several different tumor cell lines derived from human colon cancer (HCT116), breast cancer (MCF7), breast cancer (MDA-MB231), cervical cancer (HeLa) and lung cancer (H1299), and mouse melanoma (B16). The results are presented in Table 1. Compound 3i reduced cancer cell viability at nanomolar concentrations with IC50 values against HCT116, MCF7, MDA-MB231, HeLa, H1299 and B16 cells from 50 nM to 150 nM in MTS reduction assays. In particular, compound 3i exhibited a dose-dependent cytotoxicity (Fig. 2). To further explore the selectivity of the target compound against cancer cells, we tested its cytotoxicity in BEAS-2B cells derived from normal human bronchial epithelial cells. As seen in Table 1, compound 3i showed higher selectivity for H1299 cancer cells than BEAS-2B normal cells, which indicated that compound 3i probably has low toxicity to normal cells.
Table 1. In vitro cytotoxicity of compound 3i.
| IC50 ± SD (μM) | |
| HCT116 | 0.077 ± 0.011 |
| MCF7 | 0.061 ± 0.030 |
| MDA-MB231 | 0.135 ± 0.010 |
| HeLa | 0.124 ± 0.010 |
| H1299 | 0.109 ± 0.015 |
| B16 | 0.104 ± 0.007 |
| BEAS-2B | 32.61 ± 2.04 |
Fig. 2. Compound 3i reduces cancer cell viability. Cells were treated with increasing concentrations of compound 3i. Cell viability was measured using MTS.
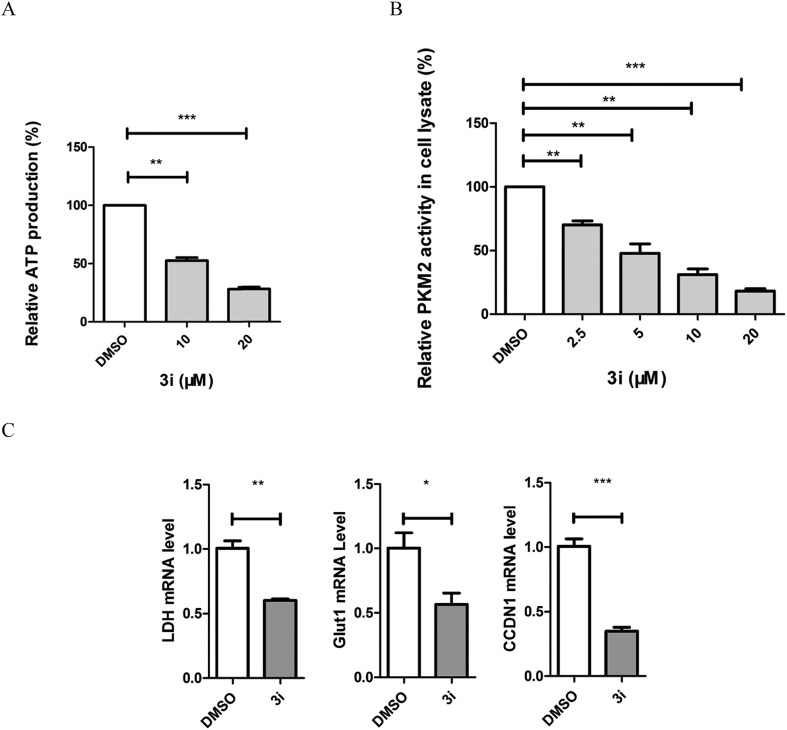
The direct consequence of inhibiting tumor cell metabolism is the decrease of ATP production in cells. We treated HCT116 cells with 10 μM and 20 μM compound 3i and tested ATP production with an ATP assay kit. The results showed that compound 3i significantly influenced the metabolic function by impairing the cellular ATP production (Fig. 3A). Compound 3i has a similar naphthoquinone skeleton to shikonin23 which was reported to affect the metabolism of tumor cells by inhibiting PKM2 activity, therefore, we tested the influence of compound 3i on PKM2 activity using a fluorescence PK–LDH coupled assay according to a previously reported method.20 Shikonin was used as the positive control. As shown in Table 2, compound 3i (IC50 = 0.88 ± 0.37) displayed higher inhibitory activity than shikonin (IC50 = 8.82 ± 2.62). Moreover, compound 3i showed inhibition of PKM2 with lower inhibition of PKM1 and PKL. To investigate whether compound 3i is able to inhibit PKM2 in cells, we treated cells in which PKM2 is highly expressed with 2.5 μM, 5 μM, 10 μM and 20 μM compound 3i and assayed PKM2 activity in the corresponding cell lysates. We found that compound 3i inhibited PKM2 activity in cells in a dose-dependent manner (Fig. 3B). In addition, there is extensive evidence that PKM2 coactivates β-catenin to induce its downstream gene CCND1 and c-Myc transcription, resulting in upregulation of GLUT1 and LDHA. We evaluated the transcription of these genes. Cells were treated with 20 μM compound 3i, and RT-PCR showed a significant reduction of GLUT1, LDH and CCND1 (Fig. 3C). These data suggest that compound 3i probably interferes with the energy metabolism of cancer cells by inhibiting PKM2 activity.
Fig. 3. Compound 3i can regulate the metabolism of tumor cells. (A) Compound 3i inhibited cellular ATP production. HCT116 cells were treated with 10 μM and 20 μM compound 3i for 6 h and ATP production was tested with an ATP assay kit. (B) Compound 3i inhibited PKM2 activity in cells in a dose-dependent manner. HCT116 cells were treated with 2.5 μM, 5 μM, 10 μM and 20 μM compound 3i for 8 h and PKM2 activity was tested in the corresponding cell lysates using a fluorescence PK–LDH coupled assay. (C) Compound 3i inhibited the transcription of PKM2-regulated glycolytic genes GLUT1, LDH and CCND1. HCT116 cells were treated with 20 μM compound 3i for 8 h and qPCR was performed.
Table 2. In vitro inhibitory activity (IC50) of 3i and shikonin on different PKM2 isoforms.
| PKM2 (μM) | PKM1 (μM) | PKL (μM) | IC50(PKM1)/IC50(PKM2) | IC50(PKL)/IC50(PKM2) | |
| 3i | 0.88 ± 0.37 | 5.07 ± 0.13 | 2.90 ± 0.64 | 5.8 | 3.3 |
| Shikonin | 8.82 ± 2.62 | 12.96 ± 3.37 | 39.25 ± 6.53 | 1.5 | 4.5 |
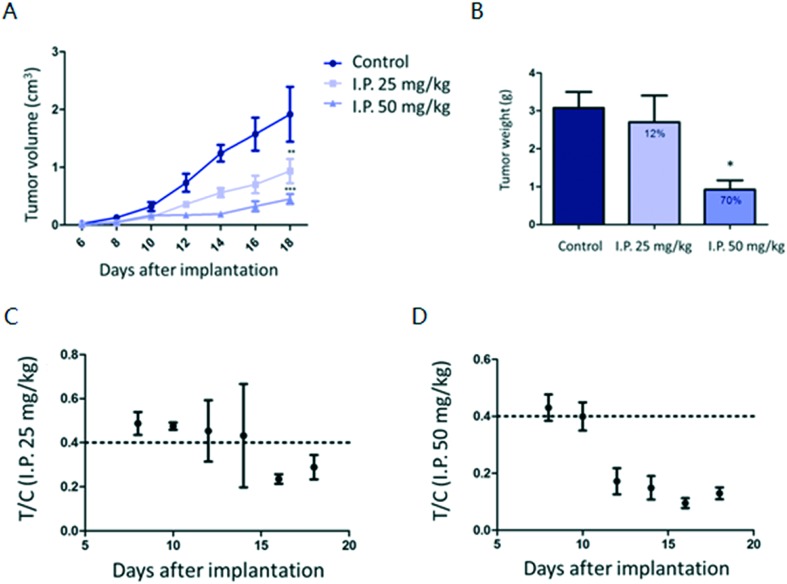
Based on the potent inhibitory effect of compound 3i on tumor cells in vitro, we next assessed its inhibitory efficiency in mouse models. The in vivo anti-tumor effect of compound 3i was first evaluated in a B16 transplantation mouse model. As shown in Fig. 4A, intraperitoneal injection of compound 3i at 25 and 50 mg kg–1 every two days for a period of 12 days demonstrated significant dose-dependent inhibition of tumor growth over the course of the treatment. The mice treated with compound 3i at 50 mg kg–1 showed tumors of 30% tumor weight compared to the vehicle-treated (control) group (Fig. 4B). The T/C values (relative tumor volume growth rate) of the 50 mg kg–1 treatment group were close to or less than 40% at each time point, which is consistent with the high efficiency of compound 3i (Fig. 4D). In addition, compound 3i had no significant effect on body weight over the course of this experiment, suggesting that it was well tolerated in vivo.
Fig. 4. Compound 3i inhibited B16 tumor growth in a dose-dependent manner. (A) Mice were treated with 25 mg kg–1 or 50 mg kg–1 compound 3i or vehicle. Tumor volume was measured once every two days. (B) The weight of individual tumors was measured. (C and D) The T/C values of the 25 mg kg–1 and 50 mg kg–1 treatment groups at selected time points.
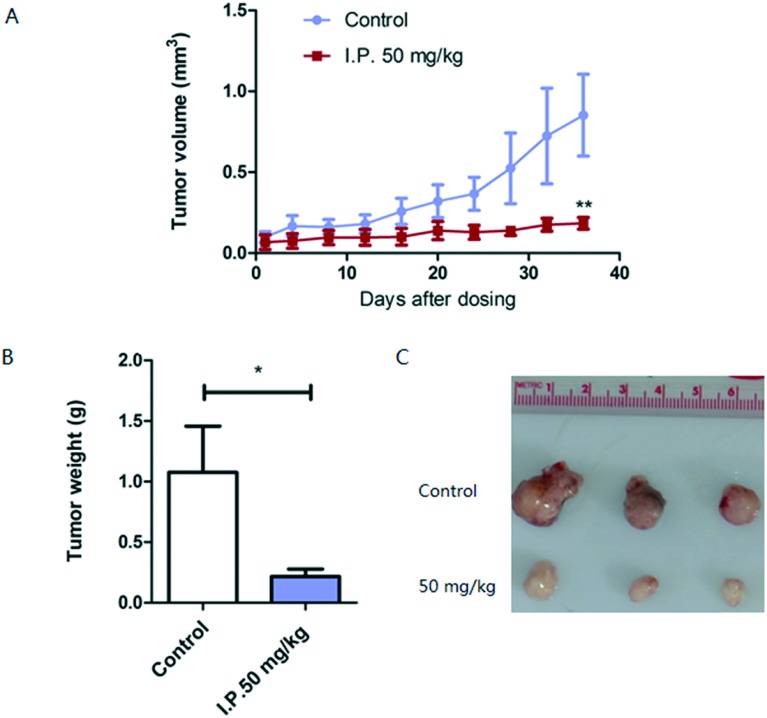
To further assess the therapeutic efficiency of this compound, we used a PTEN deletion-mediated mouse spontaneous breast tumor model.22 Mice were divided into two groups when tumors reached a volume of 0.1 cm3, and compound 3i at 50 mg kg–1 or the vehicle was injected intraperitoneally every two days for 36 days. As shown in Fig. 5A, tumor growth was significantly suppressed over this course of treatment. The mouse tumor weight after treatment with compound 3i was approximately 20% of that of the control (Fig. 5B and C).
Fig. 5. Compound 3i significantly inhibited mouse spontaneous breast tumor growth. (A) Mice were treated with compound 3i or the vehicle. Tumor volume was measured once every four days. (B and C) Tumors were removed after sacrificing the mice. The weight of individual tumors was measured.
4. Conclusions
Modulation of metabolism is a key characteristic of highly proliferative cancer cells which allows both rapid ATP generation and access to metabolites needed as cellular building blocks. Here, we describe a metabolic suppressor compound 3i which is a previously unreported compound. Compound 3i reduced cancer cell viability in a high response with IC50 values in nanomolar concentrations and suppressed ATP production in cancer cells. Meanwhile, compound 3i inhibited a rate-limiting enzyme PKM2 in the glycolytic pathway. However, the cytotoxicity of compound 3i was much higher than the PKM2 inhibitory activity, which suggested that compound 3i had other mechanisms to influence tumor cell metabolism and suppress cell proliferation. The 1,4-naphthoquinone moiety of compound 3i has been reported to influence other proteins, such as DT-diaphorase24 and the P2X7 receptor,25 which may lead to the poor activity relationship at the enzyme and cellular levels. However, there is no doubt that compound 3i targets PKM2 and affects tumor cell metabolism. In future studies, we will focus on investigating other mechanisms of compound 3i.
In this study, we also found that compound 3i significantly inhibited PTEN loss-mediated mouse spontaneous breast tumor growth. Previous studies have indicated that PTEN-negative human hepatocellular carcinoma cell lines show upregulation of PKM2 expression, which is advantageous for cell proliferation and anchorage-independent growth.26 This probably accounts for the fact that compound 3i is very sensitive to PTEN loss in the spontaneous tumor model. The mechanism will be investigated in our further work. In addition, we will also continue the optimization of the structure of 2,3-didithiocarbamate-substituted naphthoquinones and prepare suitable formulation for identifying new compounds with better physico-chemical properties and higher efficiency.
Conflicts of interest
The authors declare no competing interest.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Key grants #81430056, #81372491 and #81402777) and the China Postdoctoral Science Foundation (#2014M560026 and #2015T80028).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8md00062j
References
- Yang W., Lu Z. Cancer Lett. 2013;339:153–158. doi: 10.1016/j.canlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi A. T., Gomperts B. N. Clin. Cancer Res. 2015;21:2440–2444. doi: 10.1158/1078-0432.CCR-14-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkert J., Ripperger T., Schieck M., Schlegelberger B., Steinemann D., Illig T. Oncotarget. 2016;7:67626–67649. doi: 10.18632/oncotarget.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Butler E. B., Tan M. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoedo N. D., Obre E., Rossignol R. Biochim. Biophys. Acta. 2017;1858:674–685. doi: 10.1016/j.bbabio.2017.02.005. [DOI] [PubMed] [Google Scholar]
- Scharping N. E., Delgoffe G. M. Vaccines. 2016;4:46. doi: 10.3390/vaccines4040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N., De Melo J., Tang D. Int. J. Cell Biol. 2013;2013:242513. doi: 10.1155/2013/242513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Bamezai R. N. Protein Sci. 2010;19:2031–2044. doi: 10.1002/pro.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Semenza G. L. Trends Endocrinol. Metab. 2012;23:560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Inoue H., Tanaka T. J. Biol. Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- Yang W., Lu Z. J. Cell Sci. 2015;128:1655–1660. doi: 10.1242/jcs.166629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W., Gao X., Aldape K., Lu Z. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton T. L., Jacks T., Vander Heiden M. G. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Wang H., Yang J. J., Liu X., Liu Z. R. Mol. Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Wang H., Yang J. J., Chen J., Jie J., Li L., Zhang Y., Liu Z. R. J. Biol. Chem. 2013;288:15971–15979. doi: 10.1074/jbc.M112.448753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning X., Qi H., Li R., Li Y., Jin Y., McNutt M. A., Liu J., Yin Y. Eur. J. Med. Chem. 2017;138:343–352. doi: 10.1016/j.ejmech.2017.06.064. [DOI] [PubMed] [Google Scholar]
- Ning X., Qi H., Li R., Jin Y., McNutt M. A., Yin Y. J. Enzyme Inhib. Med. Chem. 2018;33:126–129. doi: 10.1080/14756366.2017.1404591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M. G., Christofk H. R., Schuman E., Subtelny A. O., Sharfi H., Harlow E. E., Xian J., Cantley L. C. Biochem. Pharmacol. 2010;79:1118–1124. doi: 10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk H. R., Vander Heiden M. G., Wu N., Asara J. M., Cantley L. C. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A., Pesce B., Cordon-Cardo C., Pandolfi P. P. Nat. Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Chen J., Xie J., Jiang Z., Wang B., Wang Y., Hu X. Oncogene. 2011;30:4297–4306. doi: 10.1038/onc.2011.137. [DOI] [PubMed] [Google Scholar]
- Flader C., Liu J., Borch R. F. J. Med. Chem. 2000;43:3157–3167. doi: 10.1021/jm000179o. [DOI] [PubMed] [Google Scholar]
- Faria R. X., Oliveira F. H., Salles J. P., Oliveira A. S., von Ranke N. L., Bello M. L., Rodrigues C. R., Castro H. C., Louvis A. R., Martins D. L., Ferreira V. F. Eur. J. Med. Chem. 2018;143:1361–1372. doi: 10.1016/j.ejmech.2017.10.033. [DOI] [PubMed] [Google Scholar]
- Nemazanyy I., Espeillac C., Pende M., Panasyuk G. Biochem. Soc. Trans. 2013;41:917–922. doi: 10.1042/BST20130034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.