Abstract
Genome wide association studies have identified many disease-associated non-coding single nucleotide polymorphisms, but cannot distinguish functional SNPs (fSNPs) from others that reside incidentally within risk loci. To address this challenge, we developed an unbiased high-throughput screen that employs type IIS enzymatic restriction to identify fSNPs that allelically modulate the binding of regulatory proteins. We coupled this approach, termed SNP-seq, with flanking restriction enhanced pulldown (FREP) to identify regulation of CD40 by 3 disease-associated fSNPs via 4 regulatory proteins, RBPJ, RSRC2, and FUBP-1/TRAP150. Applying this approach across 27 loci associated with juvenile idiopathic arthritis, we identified 148 candidate fSNPs, including two that regulate STAT4 via the regulatory proteins SATB2 and H1.2. Together, these findings establish the utility of tandem SNP-seq/FREP to bridge the gap between GWAS and disease mechanism.
Introduction
Polygenic diseases including juvenile idiopathic arthritis (JIA), rheumatoid arthritis (RA), multiple sclerosis (MS), and type 1 diabetes (T1D) arise through a complex interplay of genetic and environmental factors. Involvement of multiple alleles, each with modest impact, complicates genetic dissection of disease pathogenesis.1–3
Over the last 15 years, GWAS have enabled genetic exploration of polygenic diseases through the systematic identification of loci associated with disease risk. However, translation into mechanistic understanding has proven unexpectedly challenging. Most loci contain multiple variants, most commonly single nucleotide polymorphisms, each of which is potentially a functional SNP (fSNP). The search for fSNPs is complicated by the fact that most variants reside in non-coding regions, such as introns, 5′ and 3′ UTR, or intergenic regions, while SNPs that alter protein coding are uncommon.4–6 Bioinformatic and epigenetic analysis suggests that relatively few non-coding fSNPs alter canonical transcription factor binding sites.7 Thus proceeding from GWAS to fSNP remains a major challenge.
Recently, innovative strategies have been described to identify non-coding fSNPs. These include proteome-wide analysis of SNPs (PWAS)8; bioinformatic enrichment using genetic, expression quantitative trait locus (eQTL), and epigenetic data7,9,10; and massively parallel reporter assays (MPRAs) that test the translational impact of candidate SNPs.11–13 These approaches each face limitations. PWAS is a SNP-by-SNP method that requires SILAC labeling, restricting the screen to cells grown in culture. Epigenetic and eQTL data are specific for lineage and activation state. MPRAs reflect the transcriptional complement of model cells that can be transfected efficiently. Further, technical complexity places these methods beyond the range of most investigators. Thus there remains a need for complementary approaches that are amenable to a broad spectrum of cellular lineages while employing widely accessible methods.
Single nucleotide polymorphism-next generation sequencing (SNP-seq) employs restriction endonuclease protection, modified from the approach pioneered by Van Dyke and colleagues14, in an unbiased high-throughput experimental assay to identify fSNPs that bind regulatory proteins from an unrestricted range of input lineages. Application of flanking restriction enhanced pulldown (FREP)15 then enables efficient identification of fSNP-bound regulatory proteins. Using tandem SNP-seq/FREP, we studied the CD40 locus associated with RA, MS and systemic lupus erythematosus16–19, and then applied the approach in high-throughput manner across 608 SNPs associated with oligoarticular and seronegative polyarticular JIA20, enabling in each case the identification of previously unrecognized DNA-protein associations implicated by GWAS in human disease risk.
Results
Tandem SNP-seq/FREP
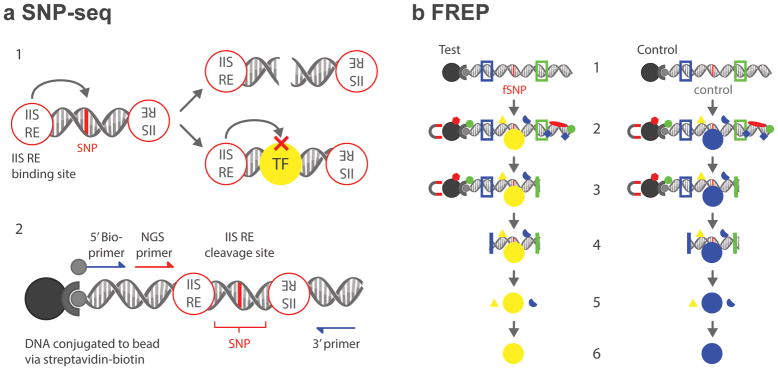
SNP-seq detects SNPs that bind regulatory proteins to control cognate gene expression, using the capacity of type IIS restriction enzymes (IIS RE, e.g. Bpm I) to cleave DNA in a sequence-independent manner at a fixed distance to one side of the binding site (Fig. 1A, upper panel). A 31bp sequence containing each SNP is introduced into a double-stranded DNA construct, with the SNP itself positioned directly at the IIS RE cleavage site (Fig. 1A, lower panel). A library of constructs is incubated with nuclear extract from a disease-related cell population as a source of regulatory proteins. If a SNP binds a regulatory protein, it will be protected from IIS RE cleavage and amplified by PCR. Otherwise, it will be cut and negatively selected.
Figure 1. Diagram of tandem SNP-seq and FREP.
A. 1. SNPs that fail to bind regulatory proteins such as transcription factors (TF) are negatively selected by PCR after type IIS restriction enzyme (IIS RE) cleavage (upper); protected fSNPs can then be enriched by PCR. A. 2. SNP-seq construct. A 31 bp SNP sequence with the SNP centered in the middle on the Bpm I cutting site is flanked with two Bpm I binding sites. A next-generation sequencing (NGS) primer is included for high throughput sequencing. The whole construct can be amplified using G5 and G3 primers. B. 1. The FREP construct with BamH I (blue) and EcoR I (green) restriction sites flanking a 31bp sequence centered on the fSNP of interest (red) and attached to a magnetic bead by streptavidin and biotin. Parallel procedures using the test fSNP and a control sequence enables identification of sequence-specific protein associations. B. 2. Incubation with nuclear extract followed by extraction of constructs from unbound nuclear proteins by magnetic bead separation. B. 3. EcoR I digestion removes 3′ DNA and proteins. B. 4. BamH I digestion removes 5′ DNA, the beads and proteins and proteins binding single stranded-DNA, which is not cut and therefore is extracted with the bead. B. 5. Protein complex identification with mass spectrometry. B. 6. Identification of associated proteins for each SNP.
Once an fSNP is confirmed experimentally, identification of fSNP-bound proteins is accomplished using FREP.15 In this method, the fSNP is employed as bait to pull down regulatory proteins from nuclear extract, reducing non-specific binding through removal of both 5′ and 3′ DNA ends of the FREP construct via sequential restriction enzyme cleavage. For the present studies, FREP was modified by performance in parallel with an irrelevant control sequence to enable identification of differential protein binding by mass spectrometry (Fig. 1B).
Identification of fSNPs at the CD40 locus
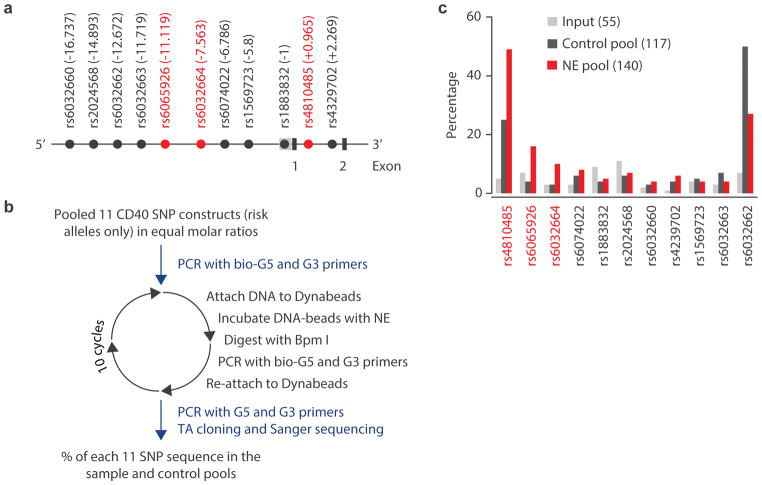
To test SNP-seq, we first studied CD40. This locus contains 11 SNPs in linkage disequilibrium (LD) R2 > 0.8, residing within ~17 kb of the 5′ promoter and within intron 1 (Fig. 2A).21 We engineered risk alleles from each SNP into independent SNP-seq constructs and incubated the 11-construct pool with nuclear extract from human BL2 B cells, followed by Bpm I digestion and PCR amplification. As a control, the construct pool was incubated without nuclear extract, repeating the procedure for 10 cycles as summarized in Fig. 2B. PCR products from both experimental and control samples were cloned into TA vectors and sequenced by Sanger sequencing. The percentage of each SNP in the input (grey bar), experimental (red bar) and control (black bar) after mutated sequences were eliminated is shown in Fig. 2C. Relatively even numbers of reads for all sequences were observed in the input sample. However, SNPs rs4810485, rs6065926 and rs6032664 showed greater than 100% enrichment in experimental over control conditions, indicating strong protection from Bpm I cutting and implicating these sites as candidate fSNPs (Fig. 2C).
Figure 2. Screening of fSNPs within the CD40 locus.
A. Partial genomic arrangement at CD40 showing the relative positions of the 11 SNPs (in LD R2> 0.8) to the transcription start site +1 and exons 1 and 2. SNPs ultimately implicated by SNP-seq are shown in red. B. The experimental procedure for SNP-seq for the CD40 locus; NE, nuclear extract. C. Screening by SNP-seq at CD40 showing the percentage of each SNP in the input (grey), control (black) and NE (red) pools. The numbers in the parenthesis represent the total numbers that were counted after Sanger sequencing. The uneven amplification reflects PCR bias towards certain sequences such as rs4810485 and rs6032662.
Validation of three fSNPs at the CD40 locus
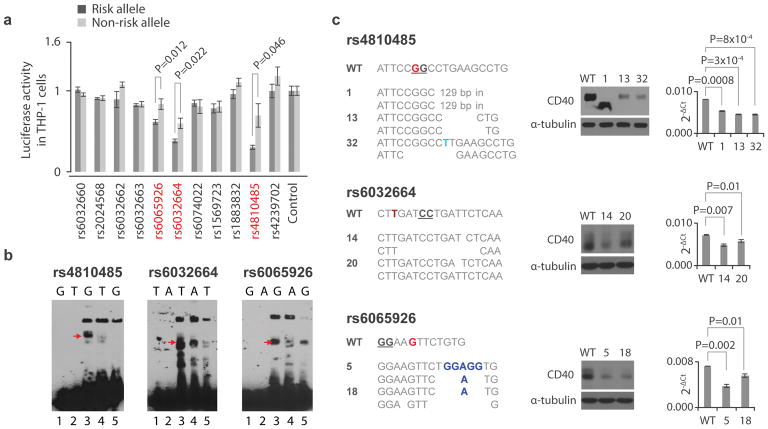
To confirm that rs4810485, rs6065926 and rs6032664 are fSNPs, we performed a luciferase reporter assay for all 11 SNPs. We cloned a 41bp SNP-centered fragment containing either risk or non-risk allele into the pGL3 vector (Promega) and transfected each reporter construct into THP-1, a CD40-expressing human monocyte cell line22, together with control vector pRL (Promega). While 8 of 11 SNPs showed no allele-imbalanced luciferase activity, SNPs rs4810485, rs6065926 and rs6032664 exhibited a significant difference between alleles (Fig. 3A). These data confirm that the variants identified by SNP-seq are functional. In each case luciferase activity was lower for the risk allele, suggesting a negative regulatory role.
Figure 3. Validation of fSNPs rs4810485, rs6032664, rs6065926, rs1883832, and rs6074022 as CD40 fSNPs.
A. Reporter assay showing relative luciferase activity in human THP-1 cells between the risk (black) and non-risk (grey) alleles of 11 CD40 SNPs. Candidate fSNPs identified by SNP-seq highlighted in red (mean +/− SD, n=3 biological replicates, t-test with 2 tails without correction for multiple hypothesis testing). B. EMSA showing allele-specific gel shifting (arrows, n=3 independent biological replicates with similar results). rs4810485, G: risk/major allele, T: non-risk/minor allele; rs6032664, T: risk/major allele, A: non-risk/minor; rs6065926: G; risk/major, A: non-risk/minor; rs1883832: C: risk/major allele, T: non-risk/minor allele; and rs6074022, C: risk/major allele, T: non-risk/minor allele; Red arrow, allele-specific shift. Lane 1: risk allele probe only; lane 2: non-risk allele probe only; lane 3:risk allele probe with nuclear extract; lane 4: non-risk allele probe with nuclear extract; lane 5: risk allele probe with nuclear extract and excess unlabeled probe as cold competitor. C. CRISPR/Cas9 targeting rs4810485 (upper), rs6032664 (middle) and rs6065926 (lower) in the human B cell line BL2. Left: sequences showing mutations at the three SNP sites; Middle: Western blots (n=3 independent replicates with similar results); and Right: qPCR showing CD40 expression in all mutants (mean +/− SD, n=3 biological replicates, t-test with 2 tails). Red nucleotides (nts) represent the fSNPs. The genotype of WT BL2 cells at the three SNPs is homozygous. Blue nts reflect insertion; underlined nts indicate the NGG PAM for CRISPR/Cas9 targeting; in: insertion. WT: control for CRISPR/Cas9. Numbers indicate mutant clone designations.
We next performed electrophoretic mobility shift assay (EMSA) to assess allele-imbalanced nuclear protein binding, using 31bp biotinylated DNA fragments centered upon each SNP. These experiments confirmed allele-imbalanced gel shifting between the major/risk allele (lane 3) and minor/non-risk allele (lane 4) for all three candidate fSNPs, including specific competition using an unlabeled risk allele competitor (lane 5) (Fig. 3B, red arrows).
To support the contention that these SNPs participate in the regulation of CD40, we employed CRISPR/Cas9 to perturb the associated sequences in BL2 cells. Three mutant clones were generated for rs4810485 (clones 1, 13 and 32) (Fig. 3C, upper) and two clones each for rs6032664 (clones 14 and 20) (Fig. 3C, middle) and rs6065926 (clones 5 and 18) (Fig. 3C, lower). WT and mutant sequences are listed in Fig. 3C (left). One clone at rs4810485 (clone 1) represented a homozygous mutation with the deletion of 1 nt and a 129bp insertion, resulting in a CD40 truncation confirmed by Western blot (Fig. 3C, middle). All mutants corresponding to the three fSNP sites showed significantly decreased CD40 protein and mRNA (Fig. 3C, middle and right). Together, these data validate the identification of rs4810485, rs6065926 and rs6032664 as non-coding fSNPs for CD40.
We compared SNP-seq findings with in silico analysis using HaploReg 4.1, a web-based tool for epigenetic and functional annotation of genetic variants.23 We scored each of the 11 CD40 SNPs on a scale of 0 to 5 to reflect the number of positive annotations for histone methylation (2 markers), DNase hypersensitivity, predicted protein binding, and predicted alteration in binding motifs: rs4810485 scored 5, rs6032664 scored 3, and rs6065926 scored 2, showing that SNP-seq identifies fSNPs not predicted by this method (Supplementary Table 1). Two SNPs in addition to rs4810485 scored 5 on the HaploReg scale and yet were not identified by SNP-seq: rs6074022 and rs1883832. Both have been implicated genetically in MS24,25 and were slightly enriched in the SNP-seq pool (Fig. 2C); rs1883832 also exhibited a trend toward greater luciferase activity (Fig. 3A). We performed EMSA on these 2 SNPs, finding allele-imbalanced gel shifting for rs1883832 (Supplementary Fig. 1). SNP-seq as applied to CD40 was therefore able to identify most but not all candidate non-coding fSNPs as determined by EMSA.
Identification of CD40 fSNP regulatory proteins by FREP
To understand how the CD40 fSNPs identified by SNP-seq modulate gene expression, we performed FREP to define associated regulatory proteins. Pulldown using BL2 nuclear extract revealed one protein associated specifically with rs4810485, 8 with rs6032664, and 15 with rs6065926 (Supplementary Table 2). Based on peptide spectrum count, we chose for further functional analysis RBPJ for rs4810485 (4 peptides in test vs. 0 in control), RSRC2 for rs6032664 (7 vs. 0), and FUBP1+TRAP150 for rs6056926 (each 5 vs. 0).
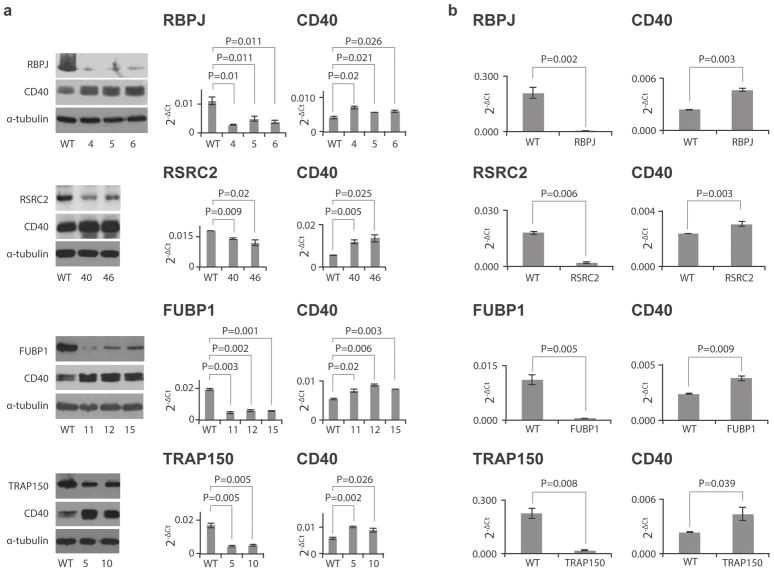
To confirm a functional role for these proteins, we performed knockdown in BL2 cells. Using shRNA in the pRNAi-hU6-puro vector system (Biosettia), we identified 3 stable clones for RBPJ (clones 4, 5 and 6) and FUBP1 (clones 11, 12 and 15) and 2 for RSRC2 (clones 40, 46) and TRAP150 (clones 5 and 10) that showed reduced expression of their respective genes (Fig. 4A) via Western blot (Fig. 4A, left) and real time PCR (Fig. 4A, middle and right). Knocking down each protein resulted in a significant increase in CD40, consistent with our luciferase data indicating a negative regulatory role for the three fSNPs.
Figure 4. Expression of CD40 in RNAi knockdown human B cells and human synovial fibroblasts.
A. Expression of CD40 in human BL2 clones with down-regulation of RBPJ, RSRC2, FUBP1 and TRAP150 (from top to bottom) by stable shRNA targeting. Left: Western blots showing expression of targeted protein and CD40; Middle: qPCR showing the expression of RBPJ, RSRC2, FUBP1 and TRAP150 in knockdown cells (mean +/− SD, n=3 biological replicates, t-test with 2 tails); and Right: qPCR showing CD40 expression in the same cells (mean +/− SD, n=3 biological replicates, t-test with 2 tails). B. qPCR showing the expression of CD40 in human synovial fibroblasts (right) (mean +/− SD, n=3 biological replicates, t-test with 2 tails) with down-regulation of RBPJ, RSRC2, FUBP1 and TRAP150 (from top to bottom at left) by transient siRNA targeting (mean +/− SD, n=3 biological replicates, t-test with 2 tails). Western blots reflect 3 independent replicates with similar results.
To assess lineage specificity, we performed RNAi knockdown in human synovial fibroblasts, a lineage that also expresses CD40.26 To ensure that observed effects did not reflect incidental impact on the housekeeping reference gene, we employed siRNAs targeting sequences distinct from those in the BL2 studies, as well as a different transfection system, transient transfection via RNAi Max (Life Technologies). Consistent with the BL2 findings, knockdown of each gene (Fig. 4B, left) up-regulated CD40 expression in fibroblasts (Fig. 4B, right). Together, these results establish regulation of CD40 via three disease-associated fSNPs.
RBPJ and TRAP150 bind their associated fSNPs
To confirm that RBPJ and TRAP150 regulate CD40 expression via their corresponding SNPs, rs4810485 and rs6065926 respectively, we performed gel super-shift. Antibodies to RBPJ and TRAP150 yielded super-shifted bands for RBPJ at rs4810485 and TRAP150 at rs6065926 (Fig. 5A, red arrows). Multiple supershifted bands suggest binding to several distinct protein complexes containing RBPJ or TRAP150. Although we were unable to detect super-shifted bands for RSRC2 and FUBP1, we did observe enhanced binding of these proteins to both rs6032664 and rs6065926, suggesting stabilization of the interaction of these proteins with their corresponding fSNPs (Supplementary Fig. 2).
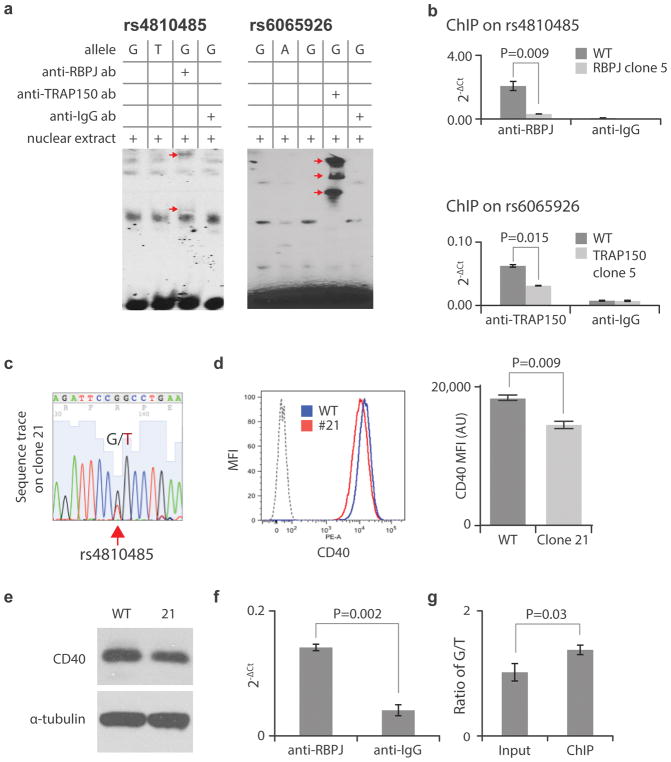
Figure 5. Demonstration of the binding of RBPJ to rs4810485 and TRAP150 to rs6065926.
A Gel super-shifting showing the binding of RBPJ to rs4810485 and TRAP150 to rs6065926. Arrows indicate the super-shifted bands in lane 3 containing the relevant antibody for risk alleles at rs4810485 and rs6065926. G and T: risk and non-risk allele for rs4810485; G and A: risk and non-risk allele for rs6065923. ab: antibody. Data reflective of 3 independent replicate experiments with similar results. B. ChIP showing endogenous binding of RBPJ to rs4810485 (upper) and TRAP150 to rs6065926 (lower); (mean +/− SD, n=3 biological replicates, t-test with 2 tails). C. Sequencing trace showing the heterozygous genotype G/T on rs4810485 from mutant 21. D. and E. Flow cytometry (representative histogram from triplicate biological repeats) (mean +/− SD, n=3 biological replicates, t-test with 2 tails) and Western blot showing reduced expression of CD40 in mutant 21 versus WT control (data reflect 3 independent biological replicates with similar results). F. ChIP of mutant 21 with an anti-RBPJ antibody showing the specific binding of RBPJ to rs4810485 site by comparison with an anti-IgG antibody (mean +/− SD, n=3 biological replicates, t-test with 2 tails). G. The ratio of risk allele G versus non-risk allele T at rs4810485 in input and ChIP DNA showing a significant enrichment of the G allele in the ChIP sample (mean +/− SD, n=3 biological replicates, t-test with 2 tails).
To validate endogenous binding of RBPJ to rs4810485 and TRAP150 to rs6065926, we performed chromatin immunoprecipitation (ChIP) with WT BL2 cells and shRNA knockdown clones (clone 5 for RBPJ and clone 5 for TRAP150 as shown in Fig. 4A). For both RBPJ (upper) and TRAP150 (lower), we noted significant enrichment with RBPJ- or TRAP150-specific antibodies compared with control anti-IgG antibody, as well as between WT and shRNA knockdown BL2 cells (Fig. 5B). Together, these data demonstrate exogenous and endogenous binding of RBPJ to rs4810485 and TRAP150 to rs6065926.
In the process of generating mutations by CRISPR/Cas9 with a homologous sequence as a repair guide, we identified a mutant clone heterozygous for G/T at rs4810485, instead of G/G in WT BL2 cells (Fig. 5C). Both flow cytometry and Western blot showed down-regulation of CD40, as predicted since the T is a non-risk allele (Fig. 5D, E). We performed ChIP on this clone using anti-RBPJ and observed enrichment of rs4810485 compared to control anti-IgG (Fig. 5F). To confirm allele-imbalanced binding of RBPJ to the risk allele (G) versus the non-risk allele (T), we sequenced the rs4810485 DNA fragments from both input and ChIP samples by cloning into TA vectors. As predicted, a modest but statistically significant enrichment of the G allele was observed (Fig. 5G), supporting the allele specificity of the rs4810485 fSNP.
High-throughput screen with SNP-seq across 27 JIA loci
We next employed SNP-seq to screen 608 SNPs in LD (R2 > 0.8) with 27 loci associated with JIA.20 In total, we screened 1,223 SNP-seq constructs representing both risk and non-risk alleles, including 7 SNPs with 3 alleles each. We generated the library through parallel oligonucleotide library synthesis (LC Sciences) and performed 10 cycles of SNP-seq using nuclear extract from healthy donor peripheral blood mononuclear cells (PBMC) as the source of regulatory proteins. Each cycle of SNP-seq was performed in duplicate, together with a control that consisted of constructs exposed to IIS RE restriction in the absence of nuclear extract, to normalize for uneven input and for bias in PCR amplification. SNP sequences were quantitated after 4, 7 and 10 cycles of enrichment using barcoded NGS.27
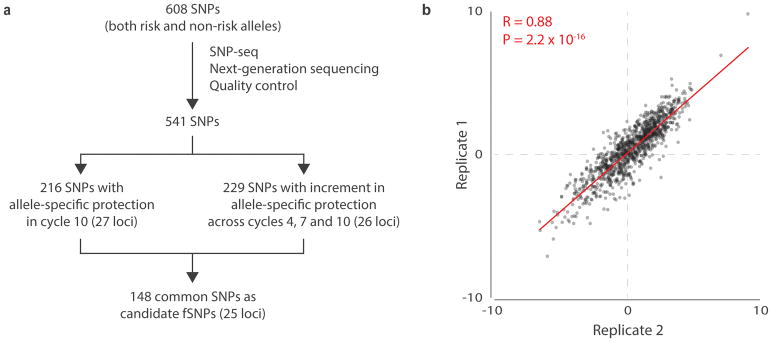
The resulting sequence data were analyzed as outlined (Fig. 6A). For quality control, we first selected only sequences containing 3′ Bpm I binding sites with sequence 5′-CTCCAG-3′. The 5′ Bpm I binding site is embedded within in the NGS primer. This strategy ensures that we include only constructs that should have been cleaved by Bpm I unless protected by binding proteins. Second, since transcription factors typically recognize 6 to 12 bp degenerate DNA sequences28, we selected only sequences in which 12bp on either side of the target SNP corresponded accurately to the input SNP sequences, eliminating mutants arising during PCR amplification. Third, we eliminated SNPs for which complete sequence data were not available across cycles. We thereby ended up with a collection of 541 SNPs across all 27 loci. We normalized the read count at each allele by dividing the number of sequence reads for each replicate by the number of reads for that allele in the control (no nuclear extract), as shown diagrammatically in Supplementary Fig. 3 and Supplementary Fig. 4. Reproducibility was assessed by plotting these normalized values across replicates at cycle 10, finding a Pearson correlation coefficient r = 0.88 (p = 2.2 × 10−16) (Fig. 6B).
Figure 6. SNP-seq high-throughput screening of 608 JIA-associated SNPs.
A. Diagram the data analysis procedure for SNP-seq. The arms are described in detail in Figs. S2 and S3. B. Correlation of the normalized sequence counts (count from sample treated with nuclear extract divided by count from control without nuclear extract) across 2 SNP-seq replicates (n=2) at cycle 10 in 541 SNPs, plotted in log2 transformation using R (See URLs https://cran.r-project.org, version 3.4.1) function “cor” and “cor.test” with the default setting using two-sided P value for Pearson correlation.
To identify SNPs with allele-specific protection, we removed SNPs for which fewer sequence reads were detected at cycle 10 for each allele with nuclear extract than control, indicating lack of protection; SNPs eliminated at this stage included the 7 SNPs with 3 alleles. We thereafter employed two parallel analytical approaches. First (Fig. 6A, left; Supplementary Fig. 3), we removed SNPs for which the difference in protection between the two alleles at cycle 10 was less than 20%, a proportion selected empirically from our experience screening the CD40 locus. We also eliminated SNPs for which replicates showed inconsistent results, defined as a ratio between replicates outside the range of 0.5 to 2.5, a range selected to recover SNPs from all loci. This curation left a set of 216 SNPs across 27 JIA loci. Second (Fig. 6A, right; Supplementary Fig. 4), we identified SNPs for which allele-specific protection increased with cycle number, indicating progressive enrichment. For each SNP, and for each of the two replicates, we calculated the ratio of the normalized sequence count between alleles and fitted a linear model of the ratio across the three cycle points (cycle 4, 7, and 10) for each replicate. The slope of change with cycle number (β in the linear model) was termed the cycle point coefficient, and a P value was calculated comparing the observed coefficient to the null hypothesis of no change in allele ratio across cycles, without correction for multiple hypothesis testing. We retained only SNPs with an absolute value of cycle point coefficient > 0.03 and P values < 0.3 in both replicates, using R as per Methods. These coefficient and P cutoffs were determined by the elbow points in the empirical distributions (the point at which the change in slope was greatest). We eliminated SNPs for which replicate results were inconsistent, defined again for consistency as a ratio of cycle point coefficients outside the range of 0.5 to 2.5. In this way, we collected a second set of 229 SNPs across 26 JIA loci. We then merged the two sets of SNPs together to identify 148 candidate fSNPs across 25 JIA loci (Supplementary Table 3).
Characterization of fSNPs at the JIA-associated STAT4 locus
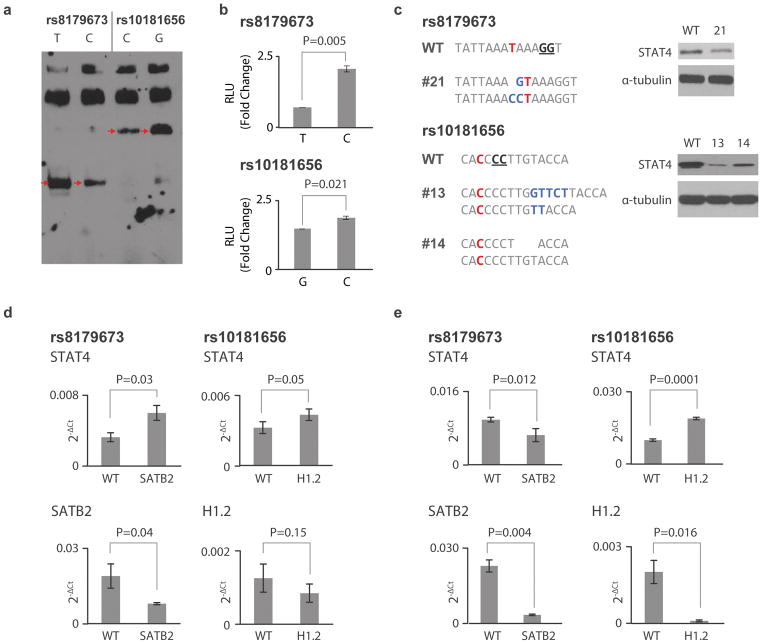
To test our high-throughput screen, we selected one locus for more detailed characterization. SNP-seq identified 4 candidate fSNPs in the STAT4 locus, a region also implicated in RA and T1D: rs11889341, rs4853459, rs8179673 and rs10181656 (Supplementary Table 3). Using EMSA with nuclear extract from Jurkat T cells, we confirmed allelic binding at rs8179673 and rs10181656 (Fig. 7A). Both SNPs, 502bp apart, are in intron 8 of STAT4, approximately 68 kb downstream of the STAT4 transcriptional start site. Neither SNP registered prominently by HaploReg 4.1 annotation (each 2 out of 5, in the mid-range of all SNPs within the locus, Supplementary Table 4). Luciferase reporter assay confirmed allele-dependent regulatory activity for both SNPs (Fig. 7B). We then performed CRISPR/Cas9 to generate site mutations in Jurkat T cells. We obtained one mutant (clone 21) at rs8179673 and two (clones 13 and 14) at rs10181656 (Fig. 7C, left). Western blot showed that these mutations significantly reduced STAT4 expression (Fig. 7C, right).
Figure 7.
Characterization of fSNPs at the STAT4 locus. A. EMSA showing allele-specific gel shifting (arrows, n=3 independent biological replicates with similar results). rs8179673, T: risk/major allele, C: non-risk/minor; rs10181656, C: risk/major allele and G: non-risk/minor allele. B. Luciferase reporter assay showing allele-imbalanced reporter activity between the two alleles of rs8179673 and rs10181656 (mean +/− SD, n=4 biological repeats, t-test with 2 tails). C. Sequences showing mutations at SNPs rs8179673 (clone 21) and rs10181656 (clone 13 and 14) in human Jurkat T cells targeted with CRISPR/Cas9 (left). Red nts represent the fSNPs. The genotype of WT Jurkat T cells at the two SNPs is homozygous. Blue nts represent insertion; underlined nts indicate the NGG PAM for CRISPR/Cas9 targeting. Western blots showing expression of STAT4 in the targeted clones (right, reflective of 3 independent biological replicates with similar results.). D. and E. qPCR showing expression of STAT4 (upper) in SATB2 (right) and H1.2 (left) knockdown human Jurkat T cells (D) and human synovial fibroblasts (E) (mean +/− SD, n=3 biological repeats, t-test with 2 tails). WT: WT control transfected with either an empty CRISPR/Cas9 vector (Addgene) or control lentivirus (Santa Cruz, Cat#: sc-108080) for RNAi knockdown.
We then performed FREP using Jurkat T cell nuclear extract, comparing each candidate fSNP with an irrelevant control (Supplementary Table 5). On the basis of peptide spectral count, we selected for further analysis H1.2 for rs8179673 (73 peptides vs. 0 in control) and SATB2 for rs10181656 (102 vs. 0). We then employed RNAi knockdown in Jurkat T cells using lentiviral particles (Santa Cruz Cat#: sc-76456-V for SATB2 and sc-37970-V for H1.2). Knockdown of SATB2 induced STAT4 expression in Jurkat T cells, and H1.2 knockdown exhibited a similar trend (Fig. 7D). We then performed RNAi knockdown in human synovial fibroblasts stimulated with Th17 and TNFα.29 Compared with control RNAi, we observed down-regulation of STAT4 in SATB2 knockdown cells (Fig. 7E, left) but increased expression of STAT4 in H1.2 knockdown cells (Fig. 7E, right), showing lineage-specific regulation of STAT4 via these two fSNPs.
To assess the overall accuracy of SNP-seq, we selected 14 candidate fSNPs at random from 9 additional JIA-associated loci. By EMSA, 11 of these showed allele-imbalanced gel shifting (Supplementary Fig. 5). Together with the STAT4 locus (2 of 4 positive), these data indicate a true-positive rate greater than 70% (76% including the CD40 SNP-seq, a screen conducted using risk alleles only). We then performed EMSA on 7 SNPs at the STAT4 locus and 6 at the FAS locus that were not enriched by SNP-seq. We observed two clear positives at FAS, in addition to two borderline SNPs (one at each locus), a false negative rate of 15–31% (14–24% including CD40) (Supplementary Fig. 6).
Discussion
GWAS launched a new era in the study of complex diseases by identifying numerous loci associated with disease risk. Most of these loci do not contain genetic variants that affect protein coding, implicating regulatory variants. Since many non-coding variants reside in LD with each tagging SNP, GWAS by itself cannot define which variants are functional. We developed SNP-seq as a high-throughput experimental approach to identify polymorphisms that modulate the binding of regulatory proteins. Coupled with FREP15, SNP-seq enabled us to identify three fSNPs regulating CD40 via four regulatory proteins. Applying this approach genome-wide in JIA, SNP-seq identified 148 candidate fSNPs, including two that modulate STAT4 via previously unrecognized DNA-protein interactions. These observations suggest that tandem SNP-seq and FREP represents a useful strategy to identify novel pathways of gene regulation, potentially opening the associated proteins to therapeutic targeting.
SNP-seq has both strengths and weaknesses. It is an unbiased high-throughput screen that can be performed across many SNPs simultaneously, testing risk and non-risk alleles in the same reaction to provide SNP-specific internal controls. As an experimental tool, SNP-seq avoids the probabilistic nature of in silico methods, while requiring in vitro techniques no more challenging than PCR. Because SNP-seq can be performed with any nuclear extract, it can be used to study primary cells, circumventing the lineage limitations intrinsic to SILAC proteomics and reporter assays based on cells maintained in culture. However, SNP-seq will detect only variants that modulate the binding of regulatory proteins, not those that alter protein coding or affect gene expression through splicing or regulatory RNA. Some protein-DNA interactions rely on the 3D structure of the DNA in addition to the recognition motif.30,31 Whereas SNP-seq employs only a short synthetic DNA sequence, it will not detect fSNPs for which protein binding requires higher-order structures or longer recognition sequences.
Importantly, SNP-seq will identify fSNPs only if the related regulatory proteins are in the nuclear extract tested. Results will therefore vary with lineage and with developmental or activation state. This limitation is counterbalanced by the ability to use any lineage or combination of lineages as the nuclear protein source. In many cases, the effect sizes of fSNPs and their associated proteins will be modest, as observed here, because the odds ratios for most GWAS loci are small (e.g. CD40 0.87, STAT4 1.29).16,20 Although SNP-seq can screen many loci at once, the number of SNPs that can be tested at once is limited by the number of quality reads that can be obtained across cycles. While we expect that the method can screen more than the 27 loci and 1,223 alleles tested simultaneously here, the maximum number that can be evaluated is unknown.
Like other screening tools, SNP-seq is not completely accurate. Across 21 SNPs that identified as candidate fSNPs and characterized here by EMSA, more than 75% were positive. However, some fSNPs were missed, including rs1883832 at CD40 that had been implicated by HaploReg data and genetic association.24,25 Thus, while SNP-seq enables identification of candidate fSNPs, complementary approaches will be helpful, and detailed fSNP-by-fSNP validation remains essential.
Using SNP-seq and FREP, we identified RBPJ, RSRC2, and FUBP1/TRAP150 as regulatory proteins that control CD40. RBPJ is the major nuclear transducer of Notch signaling.32 Its DNA binding consensus sequence is 5′-a/tgTTCCCACgg/ct-3′ 33, resembling the rs4810485 site 5′-AGATTCC[G/T]GCC-3′ and suggesting that RBPJ may directly recognize this fSNP. Interestingly, RBPJ has itself been implicated directly in RA and T1D by GWAS, and may be an autoantigen in MS.34–36 Less is known about the other proteins, although available data are intriguing. FUBP1 is a ssDNA binding protein that activates the far-upstream element (FUSE) to stimulate c-myc expression.37 TRAP150 is a transcriptional co-activator.38 RSRC2 is implicated in cell proliferation.39 Proteins engaging rs1883832, a candidate fSNP positive by EMSA but not identified by SNP-seq, were not sought here.
We reported previously that RA risk is associated with higher CD40 expression.21 This conclusion is supported by our CRISPR/Cas9 data showing down-regulation of CD40 with mutations at or around these fSNPs. Unexpectedly, luciferase studies showed lower activity for risk alleles at all three fSNPs, suggesting that they bind negative regulators. Similarly, RNAi knockdown of RBPJ, RSRC2, and FUBP1/TRAP150 enhanced CD40 expression in both BL2 cells and fibroblasts. This apparent paradox could reflect the known biology of RBPJ and FUBP1. In isolation, RBPJ is a transcriptional repressor. However, when bound to a co-activator such as NICD (Notch intracellular domain) released from Notch1 upon activation, it becomes a transcriptional activator.32 Similarly, FUBP1 can either activate or repress transcription, modulated by proteins such as PUF60.40 Lack of co-activators and/or expression of co-repressors could explain how regulatory proteins enhance target expression in some contexts and reduce it in others. To test this hypothesis, we performed RNAi in fibroblasts to down-regulate either Notch1 or PUF60. While no effect of Notch1 knockdown was noted, PUF60 knockdown significantly decreased CD40 expression (Supplementary Fig. 6). Further, chromosomal conformation capture (3C) data suggested the presence of a DNA-protein complex that encompasses rs4810485, rs6032664, and rs6056926 (Supplementary Fig. 7). How these fSNPs and their associated proteins regulate CD40 will be an important subject for future study.
JIA is a family of inflammatory arthritides that affects approximately one per thousand children.41 SNP-seq identified 148 candidate fSNPs in 25 of 27 loci associated with JIA.20 More stringent analytical conditions could have reduced the number of candidate fSNPs, at the expense of fewer loci covered. Validation studies at STAT4, a mediator of IL-12 and IL-23 signaling, confirmed two fSNPs. FREP identified a novel interaction between rs10181656 and SATB2, a MAR binding protein.42 The observed interaction is supported by the presence within rs8179673 of the MAR binding motif AAA[C/T]AAA.43 SATB2 is a chromatin organizer that has been shown to bind and activate the immunoglobulin heavy chain enhancer.44 For rs8179673, FREP identified an interaction with H1.2, a regulator of higher-order chromatin structure mutated in germinal center B cell lymphomas.45 Both fSNPs are 68 kb away from the transcriptional start site of STAT4, suggesting long-range chromatin effects. How SATB2 and H1.2 act via JIA-associated fSNPs to modulate arthritis risk remains to be established.
Collectively, these data show that SNP-seq and FREP represent a promising approach to translating GWAS data into molecular insight. Both CD40 and STAT4 are modulated by more than one fSNP, highlighting the complexity of genetic regulation. Areas for future exploration include target enrichment with complementary bioinformatic strategies and use of distinct nuclear extracts to explore the lineage specificity of GWAS risk loci. Together with other approaches, tandem SNP-seq and FREP could accelerate the understanding of human disease pathogenesis through population-level genetics.
Online Methods
Cells and culture
Human THP-1 and Jurkat T cells were purchased from ATCC and the human B cell line BL2 was from DSMZ. Human synovial fibroblasts were isolated from tissues discarded after synovectomy or joint replacement surgery and were used experimentally between passage 5 and 8. THP-1, Jurkat T and BL2 cells were cultured in RPMI1640 medium and fibroblasts in DMEM. All medium was supplemented with 10% fetal bovine serum.
Primers and antibodies
All primers were purchased from IDT as listed in Supplementary Table 6 except for the primers for qPCR of RBPJ, RSRC2, FUBP1, TRAP150, CD40 and GAPDH. These primers were purchased from Genecopoeia with Cat#: HQP021663 for FUBP1, HQP022955 for human CD40, HQP017342 for RSRC2, HQP023418 for THRAP3 and HQP006940 for GAPDH. Anti-human antibodies were purchased and used as listed in Supplementary Table 7.
SNP-seq
SNP-seq constructs were built according to Fig. 1A, 2. The SNP sequence is 31 bp long centered upon the SNP of interest. The sequences for PCR amplification primers bio-G5 and G3 and for the library oligonucleotides are listed in Supplementary Table 6. For fSNP screening of CD40, 100 ng of pooled DNA was amplified by PCR with bio-G5 and G3 for 15 cycles with AccuPrime Taq (Thermo Fisher Scientific) at 95C for 90 s; 58C for 90 s and 72C for 40 s. After gel purification, 10 ng of biotinylated DNA was attached to 4 μl streptavidin-Dynabeads (Invitrogen) according to the manufacturer’s protocol. The DNA-beads were then incubated with 100 μg nuclear extract for 1 hr at room temperature in LightShift Chemiluminescent EMSA Kit reaction buffer (Thermo Fisher Scientific). After washing and separation, the DNA-beads were digested with 2 μl Bpm I (NEB) for 30 min at 37C. After another wash and separation, the DNA was amplified again with bio-G5 and G3, and re-attached to the Dynabeads for the next SNP-seq cycle. 10 cycles were performed in total. DNA from cycle 10 was cloned into pGEM-T easy vector (Promega) for Sanger sequencing since no next generation sequencing site was included in the SNP-seq construct employed for CD40. For the high-throughput screen in JIA, a sequencing library was prepared with DNA from different cycles according to published protocol.27 Cycle point coefficients (β in the linear model) and P values for change in allele ratio across cycles were calculated by the R function “lm” with the default setting and considering three time points (cycle 4, 7, and 10) as number 1, 2 and 3 in the model.
FREP
FREP was performed as described in15, modified as follows. Instead of using one SNP sequence plus a “cold competitor” as the control, we performed parallel FREP assays using either a non-specific sequence for the CD40 studies (CCR6DNP/TG):/5Biosg/AATGATACGGCGACCACCGAGGATCCGTGGCTGCTGCAGAATGGGGGGTGCTGGTGAATTCTCGTATGCCGTCTTCTGCTTG15 or a risk allele from another SNP as the control for the STAT4 studies, comparing the peptide spectrums of the eluted protein complexes. All proteins identified in test samples but absent in controls are listed in Supplementary Table 2 and Supplementary Table 5.
Luciferase reporter assay
Luciferase reporter assay was performed exactly according to the manufacture’s manual (Promega, pGL3 Luciferase Reporter Vectors Technical Manual). 0.1 μg of pGL3 expression vector was co-transfected with the control vector pRL by TransIT-2020 (Muris) into 4.5×104 THP-1 cells in a well of a 96 well plate. After 48 hr incubation, luciferase activity was measured with the Dual-Glo luciferase assay system (Promega).
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific) according to manufacturer instructions. For probe, a 31 bp SNP fragment with the SNP centered in the middle was made by annealing two biotinylated oligos. Nuclear proteins were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) per manufacturer instructions from BL2 cells for CD40 locus SNPs and from Jurkat T cells for STAT4 locus SNPs. For gel super-shifting, 10 μg of antibody was added before an additional 10 min incubation.
CRISPR/Cas9
CRISPR/Cas9 was done using the LentiCRISPR v2 vector system (Addgene) exactly as per manufacturer instructions. 50 μg CRISPR/cas9 vectors were transfected into human BL2 cells and cloned in 96 well plates as described previously.21 Single puromycin-resistant clones were harvested for genomic DNA isolation. DNA fragments crossing each fSNP were amplified and sequenced. Cells positive for mutations, except for the homozygous mutations, were subcloned and the same DNA fragments were cloned into TA vectors for sequencing of both alleles.
Western blots
Whole cell proteins were isolated with RIPA buffer (Sigma, Cat#: R0278) according to manufacturer instructions. Western blots were performed as described previously.21
Flow Cytometry
The CD40 levels in BL2 cells were measured by FACS analysis with PE anti-hCD40/TNFRSF5 antibody (R&D; Cat#: FAB6321P). As a negative control, an PE anti-IgG2B-PE (R&D; Cat #IC0041P) was used. In brief, 0.5×106 cells were incubated with 5 ul antibody in PBS/0.02%FBS at 4C for 30 min. After washing, FACS was performed as previously described21 and analyzed using FlowJo version 6.
qPCR
Total RNAs were isolated with RNeasy Mini kit (Qiagen, Cat#: 74106). cDNA was synthesized with SuperScript III Reverse Transcriptase (Invitrogen, Cat#: 18080093) after the RNA sample was treated with DNase I (Invitrogen, Cat#: 18068015). All the procedures were performed following the manufacture’s protocols. qPCR was done with StepOne real time PCR system according to the protocol for power SYBR green PCR master mix (A&B Applied Biosystems).
RNAi knockdown
For shRNA stable knockdown in human BL2 cells, the pRNAi-hU6-puro system was used as per manufacturer instructions (Biosettia, Cat#: Sort-02A). 20 μg DNA was transfected into 1×107 BL2 cells as described previously.21 Cells were cloned and subcloned with limiting dilution and selected in 1 μg/ml puromycin.
For siRNA transient knockdown in human synovial fibroblasts, cells were transfected with siRNA by reverse transfection at 30 nM siRNA using the RNAi Max reagent (Life Technologies) in 10% FBS-containing media on day 0. Cells were then switched to low serum media containing 1% FBS on day 1 and pre-stimulated with 1 μg IL-17 and 1 μg TNFα on day 2. RNA samples were collected on day 332. Target siRNA was purchased from Invitrogen with siRNA ID: s223923 for RBPJ, s16968 for FUBP1, s19359 for TRAP150, and s35220 for RSRC2.
ChIP
ChIP was performed as described previously.46 The primers that were used are listed in Supplementary Table 6 and the antibodies are listed in Supplementary Table 7.
3C assay
3C was done exactly following the protocol described at described (see URLs). Briefly, 1×107 BL2 cells were fixed with 1% formaldehyde at RT for 10 min. Nuclei were isolated and cut with 30 μl BamH I overnight at 37°C, 950 rpm. Digested nuclei were diluted to 7 ml and ligated with and without 1600U T4 DNA ligase for 4 hours in a 16°C water bath and for 30 minutes at RT. DNA was then isolated and purified. PCR was performed with primers described in Supplementary Table 6 and resolved by 1% gel. Expected fragments were cloned into TA vector for sequencing with T7 primer.
Statistical analysis
P values were calculated using Student’s T test with 2 tails without correction for multiple hypothesis testing. Error bars represent standard deviation. For EMSA, DNA sequencing and Western blot, the data in each case represent three biological replicates. For qPCR, ChIP and luciferase reporter assays, individual data points represent biological replicates.
Data Availability Statement
The next generation sequencing data from SNP-seq are available at http://diwulab.web.unc.edu/files/2018/02/SequencingData608SNP-v2-GL.xlsx. The other data supporting this study are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
We thank Drs. Pui Y. Lee, I-Cheng Ho, Peter Libby and Robert M. Plenge for scientific discussions concerning this work. This work was supported by grants from the Arthritis National Research Foundation, National Multiple Sclerosis Society, NIH R21 NS096443 and NIH R21 AR070378 (GL), and from the Rheumatology Research Foundation, NIH R01 AR065538, NIH P30 AR070253, and the Fundación Bechara (PAN).
Footnotes
URLs
3C protocol: http://www.epigenesys.eu/images/stories/protocols/pdf/20111025155834_p31.pdf.
Figure 6D: https://cran.r-project.org, version 3.4.1
Author Contributions
GL developed SNP-seq and FREP, designed the study, performed all the experiments and analyzed the data in the laboratory of PAN. GL and PAN drafted the manuscript. MM-B performed experiments and analysis and revised the manuscript. DW and JC performed sequencing data analyses. YY assisted with experiments. PC and AL assisted with analysis of CD40 expression by FACS. MB assisted with the 3C assay. HNN performed siRNA on human synovial fibroblasts and assisted with the ChIP assays in the laboratory of MBB. EAH assisted with the CRISPR/Cas9 experiments. YO, MM-B and SR assisted with the fine-mapping analysis at the CD40 and STAT4 locus. H-JW assisted with figure formatting and data analysis. SR, EAH, and RLM assisted with data analysis and revised the manuscript.
References for main text
- 1.Bogdanos DP, et al. Twin studies in autoimmune disease: genetics, gender and environment. J Autoimmun. 2012;38:J156–69. doi: 10.1016/j.jaut.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Stahl EA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44:483–9. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas CL, Lenardo MJ. Identifying genetic determinants of autoimmunity and immune dysregulation. Curr Opin Immunol. 2015;37:28–33. doi: 10.1016/j.coi.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little boxes. Nat Genet. 2014;46:659. doi: 10.1038/ng.3028. [DOI] [PubMed] [Google Scholar]
- 5.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deplancke B, Alpern D, Gardeux V. The Genetics of Transcription Factor DNA Binding Variation. Cell. 2016;166:538–554. doi: 10.1016/j.cell.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2014 doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butter F, et al. Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet. 2012;8:e1002982. doi: 10.1371/journal.pgen.1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trynka G, et al. Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-coding Variants within Complex-Trait Loci. Am J Hum Genet. 2015;97:139–52. doi: 10.1016/j.ajhg.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue F, Ahituv N. Decoding enhancers using massively parallel reporter assays. Genomics. 2015;106:159–64. doi: 10.1016/j.ygeno.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulirsch JC, et al. Systematic Functional Dissection of Common Genetic Variation Affecting Red Blood Cell Traits. Cell. 2016;165:1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tewhey R, et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardenbol P, Van Dyke MW. Sequence specificity of triplex DNA formation: Analysis by a combinatorial approach, restriction endonuclease protection selection and amplification. Proc Natl Acad Sci U S A. 1996;93:2811–6. doi: 10.1073/pnas.93.7.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, et al. The Rheumatoid Arthritis Risk Variant CCR6DNP Regulates CCR6 via PARP-1. PLoS Genet. 2016;12:e1006292. doi: 10.1371/journal.pgen.1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raychaudhuri S, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–23. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Multiple Sclerosis Genetics C et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Multiple Sclerosis Genetics C et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazgiourakis VM, et al. A common SNP in the CD40 region is associated with systemic lupus erythematosus and correlates with altered CD40 expression: implications for the pathogenesis. Ann Rheum Dis. 2011;70:2184–90. doi: 10.1136/ard.2010.146530. [DOI] [PubMed] [Google Scholar]
- 20.Hinks A, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet. 2013;45:664–9. doi: 10.1038/ng.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G, et al. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 2013;9:e1003487. doi: 10.1371/journal.pgen.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson LL, Castle BE, Kehry MR. CD40-mediated signaling in monocytic cells: up-regulation of tumor necrosis factor receptor-associated factor mRNAs and activation of mitogen-activated protein kinase signaling pathways. Int Immunol. 2001;13:273–83. doi: 10.1093/intimm/13.3.273. [DOI] [PubMed] [Google Scholar]
- 23.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokolova EA, et al. Association of SNPs of CD40 gene with multiple sclerosis in Russians. PLoS One. 2013;8:e61032. doi: 10.1371/journal.pone.0061032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson EM, Concepcion E, Oashi T, Tomer Y. A Graves’ disease-associated Kozak sequence single-nucleotide polymorphism enhances the efficiency of CD40 gene translation: a case for translational pathophysiology. Endocrinology. 2005;146:2684–91. doi: 10.1210/en.2004-1617. [DOI] [PubMed] [Google Scholar]
- 26.Fries KM, et al. CD40 expression by human fibroblasts. Clin Immunol Immunopathol. 1995;77:42–51. doi: 10.1016/0090-1229(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 27.Larman HB, et al. PhIP-Seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J Autoimmun. 2013;43:1–9. doi: 10.1016/j.jaut.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levo M, Segal E. In pursuit of design principles of regulatory sequences. Nat Rev Genet. 2014;15:453–68. doi: 10.1038/nrg3684. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen HN, et al. Autocrine Loop Involving IL-6 Family Member LIF, LIF Receptor, and STAT4 Drives Sustained Fibroblast Production of Inflammatory Mediators. Immunity. 2017;46:220–232. doi: 10.1016/j.immuni.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 31.Fordyce PM, et al. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat Biotechnol. 2010;28:970–5. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–46. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tun T, et al. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965–71. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Querol L, et al. Protein array-based profiling of CSF identifies RBPJ as an autoantigen in multiple sclerosis. Neurology. 2013;81:956–63. doi: 10.1212/WNL.0b013e3182a43b48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsiao HH, et al. Quantitative characterization of the interactions among c-myc transcriptional regulators FUSE, FBP, and FIR. Biochemistry. 2010;49:4620–34. doi: 10.1021/bi9021445. [DOI] [PubMed] [Google Scholar]
- 38.Choi JH, et al. Thrap3 docks on phosphoserine 273 of PPARgamma and controls diabetic gene programming. Genes Dev. 2014;28:2361–9. doi: 10.1101/gad.249367.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurehara H, et al. A novel gene, RSRC2, inhibits cell proliferation and affects survival in esophageal cancer patients. Int J Oncol. 2007;30:421–8. [PubMed] [Google Scholar]
- 40.Liu J, et al. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell. 2000;5:331–41. doi: 10.1016/s1097-2765(00)80428-1. [DOI] [PubMed] [Google Scholar]
- 41.Nigrovic PA, Raychaudhuri S, Thompson SD. Genetics and the classification of arthritis in adults and children. Arthritis Rheumatol. 2018;70:7–17. doi: 10.1002/art.40350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez JD, et al. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–35. [PMC free article] [PubMed] [Google Scholar]
- 43.Boulikas T. Chromatin domains and prediction of MAR sequences. Int Rev Cytol. 1995;162A:279–388. doi: 10.1016/s0074-7696(08)61234-6. [DOI] [PubMed] [Google Scholar]
- 44.Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17:3048–61. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lunning MA, Green MR. Mutation of chromatin modifiers; an emerging hallmark of germinal center B-cell lymphomas. Blood Cancer J. 2015;5:e361. doi: 10.1038/bcj.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noss EH, Nguyen HN, Chang SK, Watts GF, Brenner MB. Genetic polymorphism directs IL-6 expression in fibroblasts but not selected other cell types. Proc Natl Acad Sci U S A. 2015;112:14948–53. doi: 10.1073/pnas.1520861112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The next generation sequencing data from SNP-seq are available at http://diwulab.web.unc.edu/files/2018/02/SequencingData608SNP-v2-GL.xlsx. The other data supporting this study are available from the corresponding authors upon reasonable request.