Abstract
Genome-wide identification and characterization of long non-coding RNAs (lncRNAs) in individual immune cell lineages helps us better understand the driving mechanisms behind melanoma and advance personalized patient treatment. To elucidate the transcriptional landscape in diverse immune cell types of peripheral blood cells (PBC) in stage IV melanoma, we used whole transcriptome RNA sequencing to profile lncRNAs in CD4+, CD8+, and CD14+ PBC from 132 patient samples. Our integrative computational approach identified 27,625 expressed lncRNAs, 2,744 of which were novel. Both T cells (i.e., CD4+ and CD8+ PBC) and monocytes (i.e., CD14+ PBC) exhibited differential transcriptional expression profiles between melanoma patients and healthy subjects. Cis- and trans-level co-expression analysis suggested that lncRNAs are potentially involved in many important immune-related pathways and the programmed cell death receptor 1 (PD-1) checkpoint pathways. We also identified 9 gene co-expression modules significantly associated with melanoma status, all of which were significantly enriched for three mRNA translation processes. Age and melanoma traits closely correlated with each other, implying that melanoma contains age-associated immune changes. Our computational prediction analysis suggests that many cis- and trans-regulatory lncRNAs could interact with multiple transcriptional and post-transcriptional regulatory elements in CD4+, CD8+, and CD14+ PBC, respectively. These results provide novel insights into the regulatory mechanisms involving lncRNAs in individual immune cell types in melanoma and can help expedite cell type-specific immunotherapy treatments for such diseases.
Keywords: lncRNA, cell type-specific expression, stage IV melanoma, immune cell lineage, RNA-seq, peripheral blood cell
Introduction
Melanoma is responsible for the majority of skin cancer related death with limited effective treatment available for stage IV patients (1). Despite recent treatment successes of immunotherapy and targeted agents, the majority of patients develop resistance to treatment for reasons that are still largely unknown. To better understand the underlying the disease mechanism, instead of traditional approaches focusing on antibody and cytokine production or the expression of selected cell-associated molecules, more comprehensive approaches to investigate the full spectrum of the transcriptome of individual cell types in peripheral blood cells (PBCs) are desired (2).
LncRNAs are highly heterogeneous molecules with functional versatility due to their diverse structures and interactions with other molecules (3). Recently, deregulation of lncRNAs has been reported to be greatly involved in various human diseases such as cancer (4–6) and heart disease (7,8). LncRNAs are also widely expressed in immune cells serving as important regulators of gene expression throughout the immune system (9). However, the contribution of lncRNA-related biological activities in various immune compartments in response to disease treatment still remains little known. Integrative analysis of lncRNA expression profiling in diverse immune cell types (e.g., CD4+, CD8+, and CD14+ cells) will greatly expedite the discovery of lncRNA regulatory mechanisms and shed light on new treatment options for melanoma patients.
Previous melanoma studies mostly focused on melanoma located in the skin tissue and the peripheral blood (10), while few have investigated genome-wide transcriptome profiling of individual immune cell types in PBCs of melanoma patients (11). Furthermore, many studies only focused on protein-coding genes (12,13), overlooking the vast landscape of noncoding genes such as lncRNAs. Hence, an in-depth investigation of lncRNA roles in individual immune cell types of PBCs will provide a much richer understanding of molecular regulatory mechanisms in melanoma patients.
In this study, we developed an integrative computational functional genomic approach to interrogate the lncRNA expression profiles and their potential regulatory roles in CD4+, CD8+, and CD14+ PBCs of stage IV melanoma patients. Our study presented a most comprehensive landscape of lncRNA profiling in CD4+, CD8+, and CD14+ PBCs of melanoma patients, which holds great potential to provide clues for future cell type-specific immunotherapy treatment.
Materials and Methods
Sample collection and isolation of various immune cell types
Participant recruitment and blood sample collection were conducted using protocols approved by the Mayo Clinic Institutional Review Board (IRB#12-002580 and IRB#13-002293). Twenty-three normal healthy subjects were selected from a Southeastern Minnesota community cohort of adults self-identifying as having no autoimmunity, allergy, immunodeficiency, immunosuppressive treatments, or cancer. Eleven patients with a diagnosis of stage IV melanoma to receive immune therapy were selected. Of these donors, 2 normal healthy subjects and 10 melanoma patients commonly contain CD4+, CD8+, and CD14+ PBC samples (Supplementary Table S1).
Isolation of CD4+, CD8+, and CD14+ PBCs
Blood was drawn into sodium heparin tubes (APP Pharmaceuticals, NDC 63323-540-11). CD4+, CD8+, and CD14+ cells were isolated from whole blood using anti-CD4/CD8/CD14 microbeads (130-090-877/130-090-878/130-090-879, Miltenyi Biotec) and Automatics purification as per the manufacturer’s instructions. A small portion of the cell isolate was set aside for flow cytometry using anti-CD3 APC-H7, anti-CD4 PE-Cy7, anti-CD11b-APC, anti-CD8 PE, anti CD14 – FITC (Miltenyi Biotec) to assess the purity. All antibodies were purchased from BD Pharmingen unless otherwise indicated. Isolated cells were re-suspended at 1 million/ml in RPMI containing 10% FBS and either immediately lysed in 0.7 ml QIAzol lysis reagent (Qiagen) or incubated for 4 h at 37 °C with 25 μl/ml anti-CD3/antiCD28 Human T-Activator Dynabeads (111.32D, Invitrogen) for CD4 and CD8 cells or 1 μg/ml pI:C (528906, Calbiochem), LPS (L439, Sigma) CPG ODN2006, Invivogen) and PGN (Sigma, 77140) for CD14 cells. Lysates were stored at −80 °C until processing for RNAseq.
RNA and cDNA library preparation
RNA was prepared using the Qiagen miRNeasy kit according to the manufacturer’s directions. RNA samples with integrity values of ≥ 6.0 by Agilent Bioanalyzer (Santa Clara, CA) were processed into TruSeq libraries according to the manufacturer’s instructions (RNA Prep Kit v2, Illumina, San Diego, CA) by the Mayo Clinic Medical Genome Facility Gene Expression Core. Paired-end DNA adaptors (Illumina) with a single “T” base overhang at the 3’end were immediately ligated to the “A-tailed” cDNA population. Unique indexes, included in the standard TruSeq Kits (12-Set A and 12-Set B), were incorporated at the adaptor ligation step for multiplex sample loading on the flow cells. Libraries (8–10 pM) were loaded onto paired-end flow cells to generate cluster densities of 700,000/mm2 following Illumina’s standard protocol for cBot and cBot paired-end cluster kit version 3. The flow cells were sequenced as 51 X 2 paired-end reads on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit version 3 and HCS v2.0.12 data collection software. Base-calling was performed using RTA version 1.17.21.3.
RNA-seq data analysis
Raw RNA-seq reads generated from each RNA-seq library were assessed for duplication rate and gene coverage using FastQC. The reads were aligned to the human reference genome (GRCh38) with the GENCODE annotation (gencode.v23.annotation.gtf) by TopHat (Version 2.0.12). The transcriptomes were then reconstructed by the Cufflinks tool (Version 2.2.1).
Identification of known and novel lncRNAs
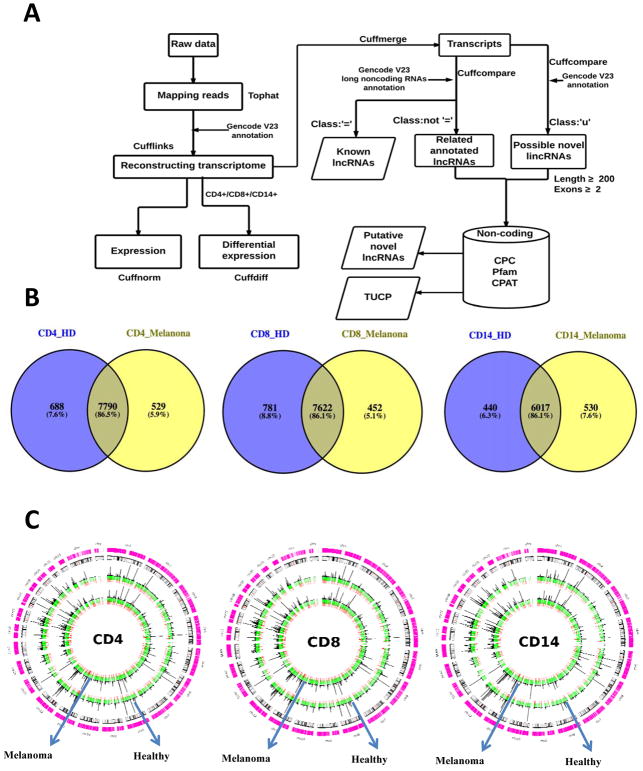
We compared our reconstructed transcriptome with the GENCODE annotation V23 using the Cuffcompare script. Based on the comparison with the GENCODE annotation, we kept the transcripts in the ‘u’ category (i.e., unknown intergenic transcripts), which were considered as candidates of novel long intergenic noncoding RNAs (lincRNAs). Based on the comparison with the GENCODE lncRNA annotation, the transcripts in the ‘=’ category were kept as known lncRNAs, and other transcripts in the non ‘=’ category (i.e., the ‘i’, ‘j’, ‘o’, ‘p’, and ‘x’ categories) were considered as candidate novel lncRNAs. These candidate novel lncRNAs were then filtered based on 1) the lncRNA features (i.e., sequence length larger than 200 nt and exon number greater than 2), and 2) the coding potential using the Coding Potential Calculator (CPC) algorithm (14), the Pfam database (15), and the Cording Potential Assessing Tool (CPAT) (16). The putative novel lncRNAs were then obtained (Fig. 1A).
Fig. 1.
Identification and characterization of lncRNAs in CD4+, CD8+, and CD14+ PBCs. (A) The computational bioinformatics workflow to assemble the transcriptome and identify lncRNAs. (B) The overlap of the expressed lncRNAs between stage IV melanoma patients and healthy donors in CD4+, CD8+, and CD14+ PBCs, respectively. (C) The expression and distribution of lncRNAs in CD4+, CD8+, and CD14+ PBCs (putative novel lncRNAs, red; annotated lncRNAs, green; the FPKM value (FPKM < 50) for histogram).
Identification of expressed lncRNAs, uniquely expressed lncRNAs, and differentially expressed lncRNAs
For each PBC type, one lncRNA was considered expressed if its mean Fragments Per Kilobase of transcript per Million mapped reads (FPKM) value was greater than 0.1 in either healthy subject group or stage IV melanoma patient group; one lncRNA was considered uniquely expressed if it expressed only in stage IV melanoma patients or normal healthy subjects; one lncRNA was considered differentially expressed if its expression level had greater than a 1.5-fold change and a false discovery rate (FDR) less than 0.1 between two groups using the Cuffdiff script.
MiTranscriptome database
Differentially expressed lncRNAs of melanoma in the skin tissues were downloaded from the MiTranscriptome database (17). We used the liftover tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver) to convert the gene coordinates of these lncRNAs from the GRCh37 to the GRCh38 annotation.
Statistical analysis
The principle component analysis (PCA) was conducted using the prcomp function in the R software. Hierarchical clustering was performed using the pvclust R package(18) with the multiscale bootstrap resampling (i.e., 1000 time), and p-values were computed for each of the clusters (e.g., AU, approximately unbiased; BP, bootstrap probability).
The permutation test was used to evaluate the statistical significance of the epigenetic marks associated with lncRNAs. The p values of the permutation test were calculated by P value = (E + 1)/(R + 1), where R is the number of permutations (equal to 1000 in our study), and E is the number of permutation test statistics (i.e., number of lncRNAs associated with selected epigenetic marks) that are greater than or equal to the observed test statistic (i.e., number of identified lncRNAs associated with epigenetic marks).
Pathway and network enrichment analysis
The Ingenuity Pathway Analysis tool (IPA, QIAGEN) was used to annotate the genes of interest, map and generate putative biological processes/functions, networks, and pathways based on the manually curated knowledge database of molecular interactions extracted from the public literature. The enriched pathways and interaction networks were generated using both direct and indirect known relationships/connectivity. These pathways and networks were ranked by the enrichment score, which measures the probability that the genes of interest were included in a network by chance.
Weighted gene co-expression network construction and gene module detection
The R package “WGCNA” was used to construct the weighted gene co-expression network (19). The transcripts with the mean FPKM value great than 0.1 in CD4+, CD8+, and CD14+ PBCs of stage IV melanoma patients or normal healthy subjects were used for the analysis. After transforming the FPKM value by log2(1+FPKM), weighted gene co-expression analysis was performed for CD4+, CD8+, and CD14+ PBCs subgroups, respectively. To minimize the noise and spurious associations, transcripts with similar expression pattern(s) were clustered using the module detection function “blockwiseModules” with the parameters networkType = “signed” and TOMType = “signed”. The expression profile of a given module was represented by its first principal component (i.e., module eigengene), which can explain the most variation of the module expression levels. Modules with highly correlated module eigengenes (correlation > 0.75) were merged together. The module membership (kME) of each gene was calculated by correlating the gene expression profile with the module eigengene, representing the extent of a gene close to a given module. The function “overlapTableUsingKME” was used to assess whether two modules were preserved based on a hypergeometric test of module kME.
ChIP-seq data analysis
We used the publicly available ChIP-seq datasets (CD4, CD8, and CD14 primary cells) from the Human Epigenome Atlas Release 9 (http://www.epigenomeatlas.org) generated by the Epigenomics Roadmap project (20). The histone mark peaks (i.e., H3K4me3 and H3K36me3) were detected using the MACS2 (version 2.1.1) (21) with default parameters (FDR<0.05). The liftover tool was used to convert the genome coordinates of the significant peaks from the GRCh37 to the GRCh38 annotation. We then identified the lncRNAs with H3K36me3 or H3K4me3 mark peaks in the region of +/− 3 kb transcription start sites of lncRNAs. We downloaded the super-enhancer datasets of the CD8 and CD14 PBCs from the dbSUPER database and identified the super-enhancer-associated lncRNAs as the ones whose transcription start sites were assigned to super-enhancers within a 50 kb window.
Transcription factor binding analysis
The human transcription factor binding motifs were retrieved from the ENCODE database (http://compbio.mit.edu/encode-motifs/). Using the FIMO tool (22), we searched transcription factor binding motifs (P < 0.0001) within the regulatory regions of lncRNAs (i.e., 1 kb upstream and 1 kb downstream from the transcription start sites of lncRNAs).
Prediction of lncRNA interactions
We used the RNAplex software (23) to identify potential lncRNA-lncRNA interactions based on the RNA duplex energy prediction. The RNAplex parameter was set as -e-20.
To investigate potential interactions between miRNAs and lncRNAs, we downloaded all human miRNAs in the miRBase v21 and predicted potential target lncRNAs for each miRNA using the miRanda software (24) with default parameters.
To identify potential protein-RNAs interactions, we employed the catRAPID algorithm (25) to predict potential protein-lncRNA interactions based on the information of protein and RNA domains involved in the macromolecular recognition. The protein-lncRNA interactions with the highly ranking distribution (i.e., the stringent parameters: the star rating > 2, RNA-binding motifs, and domains) were considered as significant hits.
Data availability
The sequencing data set has been deposited in the NCBI Gene Expression Omnibus (GEO) database (GEO accession number GSE104744).
Results
Systematic identification and characterization of lncRNAs in diverse immune cell types
To obtain a comprehensive lncRNA catalog of individual immune cell types, we developed an integrative transcriptome assembly pipeline designed to identify expressed lncRNAs in our dataset (Fig. 1A). In total, we reconstructed 345,417 transcripts from 65,247 loci across all 132 samples (Supplementary Table S1) using our transcriptome assembly pipeline. Of these transcripts, 24,881 were known lncRNAs (Supplementary Table S2), and 2,744 were putative novel lncRNAs (Supplementary Table S3; https://sites.google.com/view/yujizhanglab/data-scripts?authuser=0). Meanwhile, 19,476 protein-coding genes (98.7%) in the GENCODE annotation (V23) were also recovered in our assembled transcriptome. The high degree of overlap provides an independent measure of the comprehensive coverage in our dataset.
We observed that stage IV melanoma patients and healthy subjects shared thousands of expressed lncRNAs in CD4+, CD8+, and CD14+ PBCs respectively (Fig. 1B), i.e., the percentage of lncRNAs expressed in both groups is more than 86%. These lncRNAs were widely distributed across all the chromosomes in the human genome (Fig. 1C). We further investigated the differential expression profiling between stage IV melanoma patients and healthy subjects. In CD4+ PBC, we identified 781 differentially expressed genes (including 98 lncRNA genes) and 445 differentially expressed transcripts (including 10 lncRNA). In CD8+ PBC, we identified 789 differentially expressed genes (including 88 lncRNA genes) and 292 differentially expressed transcripts (including 10 lncRNA). In CD14+ PBC, we identified 186 differentially expressed genes (including 22 lncRNA genes) and 32 differentially expressed transcripts (including 2 lncRNAs).
The cell type-specific expression patterns of lncRNAs
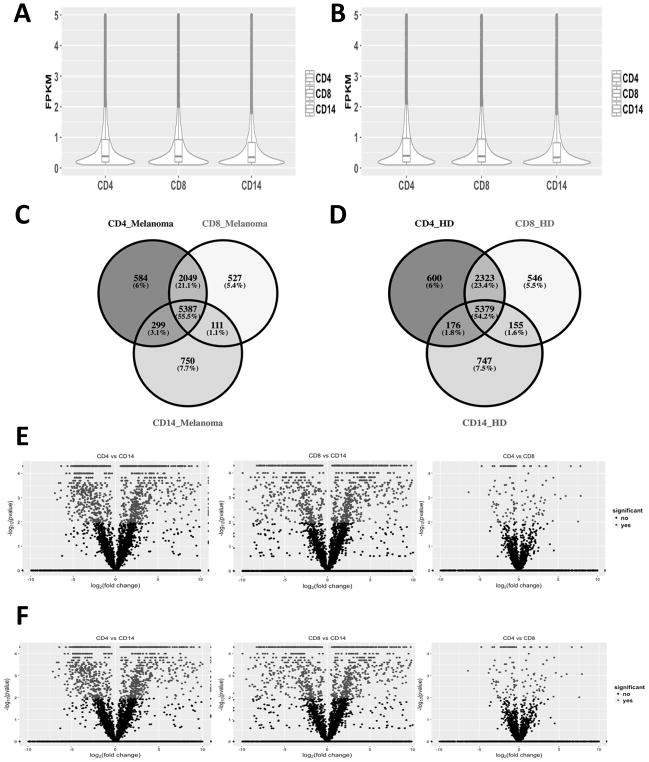
To investigate the cell-specific expression profilings of lncRNAs in CD4+, CD8+, and CD14+ PBCs, we compared their expression profiles between melanoma patients and healthy subjects. Overall, lncRNAs were expressed at relatively lower levels in comparison with mRNAs in all three immune cell types. Approximately 75% lncRNAs showed low expression level (Mean FPKM value < 1) in both stage IV melanoma patients (Fig. 2A) and healthy subjects (Fig. 2B). We also observed that hundreds of lncRNAs were uniquely expressed in CD4+, CD8+, or CD14+ PBCs of melanoma patients (Fig. 2C) and healthy subjects (Fig. 2D), respectively. In addition, there were many differentially expressed lncRNAs between CD4+, CD8+, and CD14+ PBCs (Fig. 2E and 2F), particularly between the T cell subsets (CD4+ or CD8+ PBCs) and monocytes (CD14+ PBC).
Fig. 2.
Comparison of the lncRNAs between stage IV melanoma patients and healthy subjects in CD4+, CD8+, and CD14+ PBCs, including (1) The density distribution of expressed lncRNAs in stage IV melanoma patients (A) and healthy donors (B) of CD4+, CD8+, and CD14+ PBCs; (2) The number of expressed lncRNAs and their overlaps across CD4+, CD8+, and CD14+ PBCs in stage IV melanoma patients (C) and healthy donors (D); and (3) the differentially expressed lncRNAs by pairwise comparison between CD4+, CD8+, and CD14+ PBCs in stage IV melanoma patients (E) and healthy donors (F).
To evaluate the cell type- and tissue-specific expression patterns of lncRNAs, we explored the expression of some known lncRNAs associated with melanoma in our dataset. First, we examined the expression profiles of lncRNAs differentially expressed in the skin tissue of melanoma. Among 426 differentially expressed lncRNAs in the skin tissue of melanoma cataloged in the MiTranscriptome database, 63 were also expressed in our dataset. However, they were not differentially expressed between stage IV melanoma patients and healthy subjects in CD4+, CD8+, or CD14+ PBCs (Supplementary Fig. S1A). Second, we examined the expression of 7 key immune-associated lncRNAs from one previous report (26) in our datasets (Supplementary Fig. S1B), all of which showed no significant expression differences between stage IV melanoma patients and healthy subjects in CD4+, CD8+, or CD14+ PBCs. Third, we examined the expression of 8 key melanoma-associated lncRNAs from the experimentally supported Lnc2Cancer database (27) and one previous study (28) in our dataset (Supplementary Fig. S1C). Out of 8 lncRNAs, MALAT1 and SPRY4-IT1 were expressed in stage IV melanoma patients (Mean FPKM value > 0.1) and showed differential expression patterns between stage IV melanoma patients and healthy subjects in each of three cell types. Taken together, lncRNAs exhibited tight cell-/tissue-specific expression patterns between melanoma patients and healthy subjects in all three immune cell types.
lncRNAs cis-regulate their neighboring protein-coding transcripts enriched in important immune-associated pathways
To investigate potential cis-regulatory roles of lncRNAs on protein-coding transcripts, we computed the pairwise expression correlations between lncRNAs and their neighboring protein-coding transcripts (i.e., a neighboring protein-coding transcript needs to be within a 10kb distance to a lncRNA). Overall, we identified 205, 236, and 228 highly correlated lncRNA-mRNA pairs (i.e., Spearman correlation coefficient |r| >0.7, and p-value < 1E-07) in CD4+, CD8+, and CD14+ PBCs, respectively (Fig. 3A; Supplementary Table S4). Most of these lncRNAs are known lncRNAs, of which 7, 12, and 12 are novel lncRNAs in CD4+, CD8+, and CD14+ PBCs, respectively. Specifically, 192 lncRNA-mRNA pairs (94%) in CD4+ PBC, 223 lncRNA-mRNA pairs (95%) in CD8+ PBC, and 222 lncRNA-mRNA pairs (97%) in CD14+ PBC contained immune-associated genes based on the immunologic signatures in the MSigDB database (29).
Fig. 3.
LncRNAs potentially cis-regulate the protein-coding transcripts associated with the immune biological processes. (A) The heatmap of lncRNAs (i.e., Mean FPKM value) from the significant lncRNA-mRNA pairs (i.e., Spearman correlation coefficient |r| >0.7, P value < 1E-07 in at least one PBC) in CD4+, CD8+, and CD14+ PBCs. (B) IPA functional enrichment analysis of protein-coding transcripts from the lncRNA-mRNA pairs in CD4+, CD8+, and CD14+ PBCs. (C) The number of positively and negatively correlated lncRNAs from the lncRNA-mRNA pairs in CD4+, CD8+, and CD14+ PBCs (N_cor, negatively correlated lncRNAs; P_cor, positively correlated lncRNAs), respectively. (D) The number of lncRNAs including the same and opposite transcriptional direction with the protein-coding transcripts in CD4+, CD8+, and CD14+ PBCs (O_dir, the opposite direction; S_dir, the same direction), respectively.
The functional enrichment analysis by IPA revealed that the lncRNA-mRNA pairs in CD4+, CD8+, and CD14+ PBCs were significantly enriched in immune-related pathways (Fig. 3B; Supplementary Table S4). In CD4+ PBC, 3 top upstream regulators including TCF7 (p-value =1.40E-04), IL27 (p-value =1.93E-04), and EBI3 (p-value =2.60E-04) were associated with the immune response pathways. In CD8+ PBC, the top upstream regulator TGFB1 (p-value =1.57E-04) belongs to the wnt/β-catenin signaling pathway (−log(P-value) =2.07) which plays a critical role in T-cell immunity. In CD14+ PBC, the top upstream regulator MAPK8 (p-value = 1.43E-05) was associated with the immune response pathway.
We also investigated the relationship of the correlation and the transcription directions between lncRNAs and their neighboring mRNAs. Over 95% of lncRNA-mRNA pairs were positively correlated (Fig. 3C), consistent with the previous report on cis-acting noncoding RNAs (30). Over 70% of lncRNA-mRNA pairs had opposite transcriptional directions in CD4+, CD8+, or CD14+ PBCs (Fig. 3D). These findings are consistent with previous studies suggesting that lncRNAs have potential cis-regulatory roles on their neighboring protein-coding genes (31,32).
lncRNAs trans-regulate protein-coding transcripts enriched in immune and mRNA translation pathways
To investigate potential trans-regulatory roles of lncRNAs on protein-coding transcripts, we performed a weighted gene co-expression network analysis (WGCNA) (19) to identify groups of coexpressed transcripts (i.e., modules). We adopted the coding–to–non-coding strategy to identify the lncRNA-mRNA coexpression associations in a module (33): for a module of interest, we identified statistically significant correlations between each lncRNA and mRNAs that are important in specific known functional processes. The functions of each lncRNA were then inferred by the “guilty-by-association” approach (34).
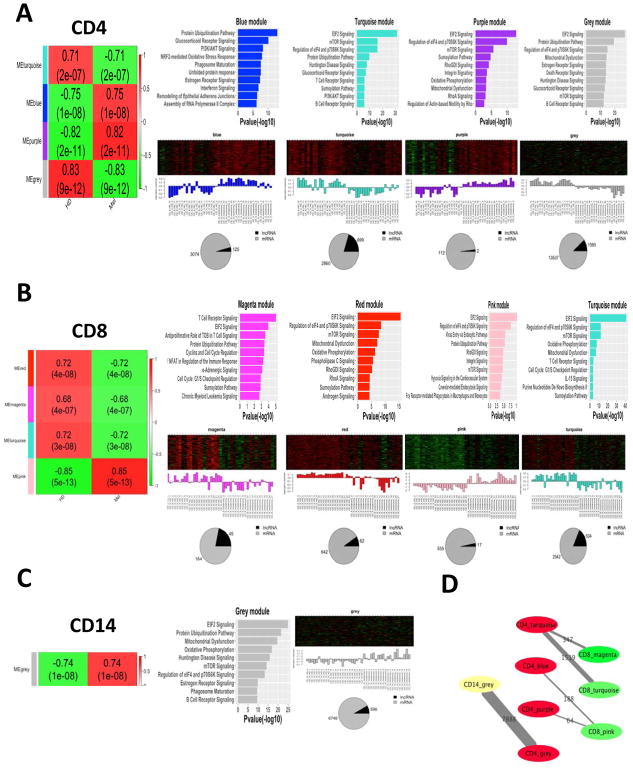
Overall, we identified 13, 17, and 17 coexpression modules in CD4+, CD8+, and CD14+ PBCs, respectively (Supplementary Fig. 2A). Notably, 4 modules (|correlation coefficient| > 0.7, p-value <1E–6) in CD4+ PBC, 4 modules (|correlation coefficient| > 0.68, p-value <1E–6) in CD8+ PBC, and 1 module (correlation = 0.74, p-value =10–8) in CD14+ PBC were statistically correlated with the melanoma status (Fig. 4A, 4B, and 4C). Interestingly, these modules were also significantly correlated with age (|correlation coefficient| > 0.57, P-value <2E–5, Supplementary Fig. S2A), while none of them were associated with the patient status (M1b – metastasis to the lung; M1c – other metastasis), disease response (Stable Disease (SD), Partial Response (PR), and Progressive Disease (PD); assessed at 3-months post initiation of ipilimumab therapy), or sex.
Fig. 4.
The weighted gene co-expression network analysis (WGCNA) in CD4+, CD8+, and CD14+ PBCs. (A) In CD4+ PBC, left of the panel represents the co-expression modules high correlated with melanoma (HD, normal healthy subjects; Mel, stage IV melanoma patients): numbers of each square represent the correlation of module and the melanoma trait (correlation > 0.7, p-value <10–6); color of each square corresponds to correlation: positive correlation (Red) and negative correlation (Green). The upper right of the panel shows the biological functions statistically enriched in these modules, in which the length of bars indicate the significance by IPA analyses (i.e., −log10(P-Value)). The middle right of the panel represents the heatmaps of the expression patterns of all transcripts in this module across all samples (red, increased expression; black, neutral expression; green, decreased expression), in which the barplots show the corresponding module eigengene expression value, the pie chart shows the ratio of mRNAs and lncRNAs in the module, and the numbers of mRNAs and lncRNAs in each module are shown next to the pie chart. Similar results are shown for CD8+ PBC (B) and CD14+ PBC (C) respectively. (D) The remarkably conservative modules in these 9 modules across CD4+, CD8+, and CD14+ PBCs (P-value <1E-10).
We also explored lncRNAs associated with important immune processes in these 9 co-expression modules (trans-regulatory lncRNAs). Specifically, we identified 361, 151, and 3 trans-regulatory lncRNAs in CD4+, CD8+, and CD14+ PBCs significantly associated with immune responses pathways respectively (−log(p-value) > 2; Supplementary Table S5, S6, and S7). In addition, these 9 coexpression modules were also all enriched in 3 mRNA translation pathways, including EIF2 signaling, mTOR signaling, and regulation of eIF4 and p70S6K signaling pathway (−log(p-value) > 2; Fig. 4A, 4B, and 4C). In total, we identified 228, 158, and 8 trans-regulatory lncRNAs associated with these three pathways in CD4+, CD8+, and CD14+ PBCs respectively (Supplementary Tables S5, S6, and S7). Of these trans-regulatory lncRNAs, 184 (81%) and 134 (85%) trans-regulatory lncRNAs were in turquoise modules of CD4+ and CD8+ PBCs (Supplementary Tables S5 and S6), suggesting that these modules contain major molecular regulators in these pathways.
Among these 9 coexpression modules correlated with the melanoma status, the transcripts of 6 modules were significantly overlapped between CD4+ and CD8+ PBCs (p-value <1E–10), the transcripts of 2 modules were significantly overlapped between CD4+ and CD14+ PBCs (p-value <1E–10), and no transcript was significantly overlapped between CD8+ and CD14+ PBCs (Fig. 4D and Supplementary Fig. S2B). These results further confirmed that the CD4+ and CD8+ T cells shared more similar expression profiles than the CD14+ monocyte.
The lncRNAs associated with programmed cell death receptor 1 checkpoint pathways
Using the immune checkpoint blockers to block the interactions between programmed cell death receptor 1 (PD-1) and its ligands is a widely acknowledged approach for cancer immunotherapy. To investigate the impact of PD-1 checkpoint in diverse immune cell types, we explored PD-1 expression and related pathways, including PI3K/PTEN/Akt/mTOR signaling pathway and the RAS/RAF/MEK/ERK signaling pathway (35). We observed that the PD-1/PDCD1 gene was up-regulated in T cell subsets (CD4+ or CD8+ PBCs) but not monocytes (CD14+ PBC) of melanoma patients compared with healthy subjects, dominantly expressed by the PDCD1-201 transcript (Supplementary Fig. S3A and S3B). We also identified 11 lncRNAs coexpressed with the PDCD1-201 transcript in CD8 PBC. In 8 coexpression modules of T cells correlated with the melanoma status, 435 and 167 trans-regulatory lncRNAs associated with PD-1 checkpoint pathways in CD4 and CD8 PBCs, respectively (|correlation coefficient| > 0.7, p-value <1E–6; Supplementary Table S8). Intriguingly, all these modules contain the mTOR signaling pathway (Supplementary Fig. S3C and S3D). The fact that 335 (77%) and 133 (80%) trans-regulatory lncRNAs were in turquoise modules of CD4+ and CD8+ PBCs suggested that the lncRNAs in these modules were potentially associated with PD-1 checkpoint pathways.
The potential regulation of lncRNA expression at the transcriptional level
To investigate potential regulatory mechanisms mediating lncRNA expression at the transcriptional level, we explored the association between the lncRNA regions and various biological annotations including histone modification sites, super enhancers, transcription factor binding sites, and single nucleotide polymorphisms (SNPs).
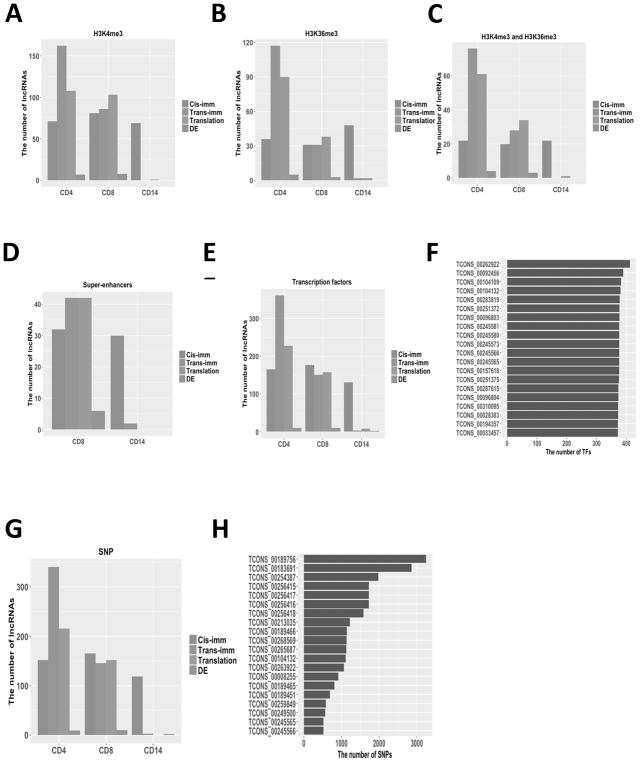
The histone H3 lysine 4 tri-methylation (H3K4me3) is a hallmark of actively transcribed promoters, and the histone H3 lysine 36 tri-methylation (H3K36me3) is enriched not only within actively transcribed gene bodies but also active enhancers (36). To explore the potential epigenetic modification on lncRNA expression, we classified lncRNAs into the H3K4me3 and H3K36me3 marked groups based on the chromatin status of putative regulatory regions of lncRNAs in the ChIP-seq datasets generated by the Epigenomics Roadmap project (20). By focusing on the biologically significant lncRNAs in our dataset (i.e., immune-associated, translation-associated, and differentially expressed lncRNAs) in CD4+, CD8+, and CD14+ PBCs, we found more lncRNAs marked with H3K4me3 than those marked with H3K36me3 (Fig. 5A and 5B). CD4+ PBC had the largest number of lncRNAs commonly marked with H3K4me3 and H3K36me3 (Fig. 5C). Our permutation test suggested that our lncRNAs were indeed enriched in epigenetic marks (P value < 0.001 based on 10,000 permutations; Supplementary Fig. S4 and Table S9). The fact that many lncRNAs were marked with the epigenetic modifications (e.g. H3K4me3 and H3K36me3), indicated that these lncRNAs were transcribed in CD4+, CD8+, and CD14+ PBCs.
Fig. 5.
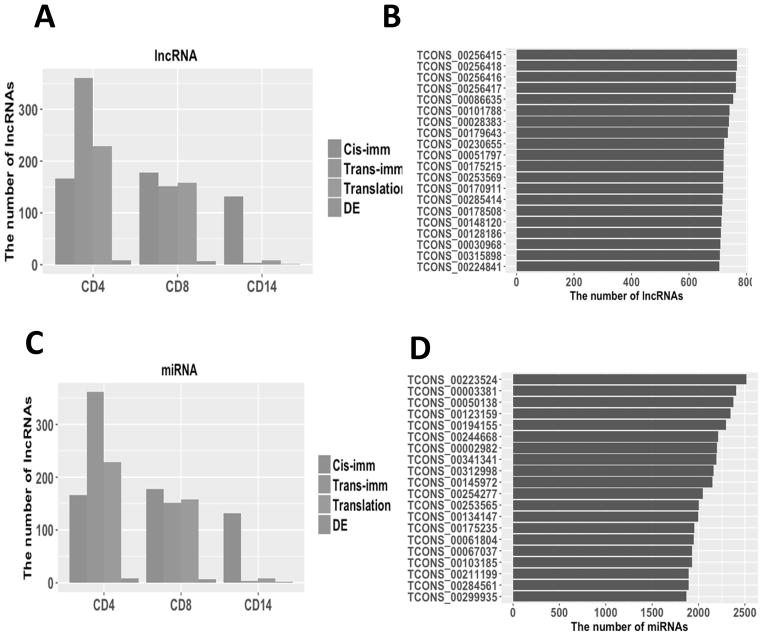
The impact of transcriptional regulatory elements for the immune-associated, translation-associated, and differently expressed lncRNAs in CD4+, CD8+, and CD14+ PBCs. (A–D) The impact of the epigenetic modifications: (A) The number of lncRNAs marked with H3K4me3; (B) The number of lncRNAs marked with H3K36me3; (C) The number of lncRNAs marked with H3K4me3 and H3K36me3; (D) The number of lncRNAs marked with the super-enhancer in CD8+ and CD14+ PBCs. (E, F) The impact of the transcription factors: (E) The number of lncRNAs including TF motifs; (F) Top 20 lncRNAs with the largest number of potential transcription factor. (G, H) The impact of the SNPs: (G) The number of lncRNAs containing at least one cancer risk-associated SNP; (H) Top 20 lncRNAs potentially contained the largest number of cancer risk-associated SNPs.
Super-enhancers are large clusters of transcriptional enhancers which regulate expression of genes associated with specific diseases and cell identity (37). Within the biologically significant lncRNAs, we identified 105 lncRNAs annotated by the super-enhancers dbSUPER database (38) in CD8+ and CD14+ PBCs (Fig. 5D), suggesting that these lncRNAs may play important transcriptional regulatory roles in immune cells of stage IV melanoma patients.
To investigate whether lncRNAs can be potentially regulated by transcription factors (TFs), we searched the regulatory regions of the biologically significant lncRNAs (i.e., upstream 1kb and downstream 1kb to the start sites of lncRNAs) in CD4+, CD8+, and CD14+ PBCs for potential transcription factor binding motifs based on the annotation of the ENCODE database (Fig. 5E). We found that all except 3 of these lncRNAs can be potentially bound by more than 200 TFs (Fig. 5F). The fact that these lncRNAs have many TFs in CD4+, CD8+, and CD14+ PBCs suggests that they can be similarly regulated by TFs as mRNAs do.
The Genome Wide Association Studies (GWAS) have identified thousands of SNPs associated with cancer susceptibility, over 90% of which are located in non-coding regions of the human reference genome (39). Using the non-coding somatic mutation annotation (CosmicNCV, release v80) from the COSMIC database (40), we explored whether the biologically significant lncRNAs contain any cancer risk-associated SNPs in CD4+, CD8+, or CD14+ PBCs (Fig. 5G). In total, 743 lncRNAs (93%) contained at least one cancer risk-associated SNP except the translation-associated lncRNAs in CD14+ PBC, 96% of which contain more than 2 cancer risk-associated SNPs and 14 contain more than 1000 cancer risk-associated SNPs (Fig. 5H). These results suggested that most of these lncRNAs are potentially involved in cancer risk-associated diseases.
The potential regulation of lncRNA expression at the post-transcriptional level
To investigate potential post-transcriptional regulation through molecular interaction mechanisms, we explored the possibilities of RNA-RNA interactions, miRNA-lncRNA interactions, and protein-lncRNA interactions involving our lncRNAs.
First, we investigated the lncRNA-lncRNA interactions for the biologically significant lncRNAs in CD4+, CD8+, and CD14+ PBCs using a RNA duplex energy prediction approach (23). We found that these lncRNAs can potentially interact with each other, except 2 differentially expressed lncRNAs in CD4+ PBCs, 3 in CD8+ PBCs, and 1 in CD14+ PBCs (Fig. 6A), each of which has more than 20 potentially interacting lncRNAs and over 97% have more than 100 potentially interacting lncRNAs (Fig. 6B). Their potential interactions with each other suggest that some lncRNAs can potentially regulate the expressions of other lncRNAs as well as mRNAs.
Fig. 6.
The impact of post-transcriptional regulatory elements for the immune-associated, translation-associated, and differently expressed lncRNAs in CD4+, CD8+, and CD14+ PBCs. (A, B) The lncRNA–lncRNA interactions: (A) The interacted lncRNAs; (B) Top 20 lncRNAs with multiple interacted lncRNAs. (C, D) The miRNA-lncRNA interactions: (C) miRNA targeted lncRNAs; (D) Top 20 lncRNAs binding multiple miRNAs.
The miRNA-lncRNA interactions can regulate lncRNAs either as inhibitory decoys or as regulatory targets of miRNAs (41). We investigated potential miRNA-lncRNA interactions between 2,588 annotated human microRNAs (miRBase v21) and our biologically significant lncRNAs in CD4+, CD8+, and CD14+ PBCs using the miRanda approach (24). We found that all the immune-associated and translation-associated lncRNAs have potential miRNA binding sites, except 2 differentially expressed lncRNAs in CD4+ PBCs, 3 in CD8+ PBCs, and 1 in CD14+ PBCs (Fig. 6C). Of these lncRNAs, each lncRNA had potential interaction with more than 80 miRNAs, over 98% of which have potential interactions with more than 100 miRNAs (Fig. 6D). These findings suggested that lncRNA expression can be potentially regulated by many miRNAs at the post-transcriptional level.
The protein–lncRNA interactions also play important roles in post-transcriptional regulation of the immune system (42). We employed the catRAPID algorithm (25) to investigate whether there are any potential such interactions involving 16 most highly expressed lncRNAs (i.e., mean FPKM value >100 in melanoma patients).The results showed that all 16 lncRNAs were potentially bound by multiple RNA-binding proteins (Supplementary Table S10). Of these lncRNAs, TCONS_00050166, TCONS_00212088, and TCONS_00050136 could potentially interact with 2204, 1802, and 414 RNA-binding proteins, respectively. These findings suggested that the protein-lncRNA interactions could serve as one important regulatory role for these lncRNAs.
Discussions
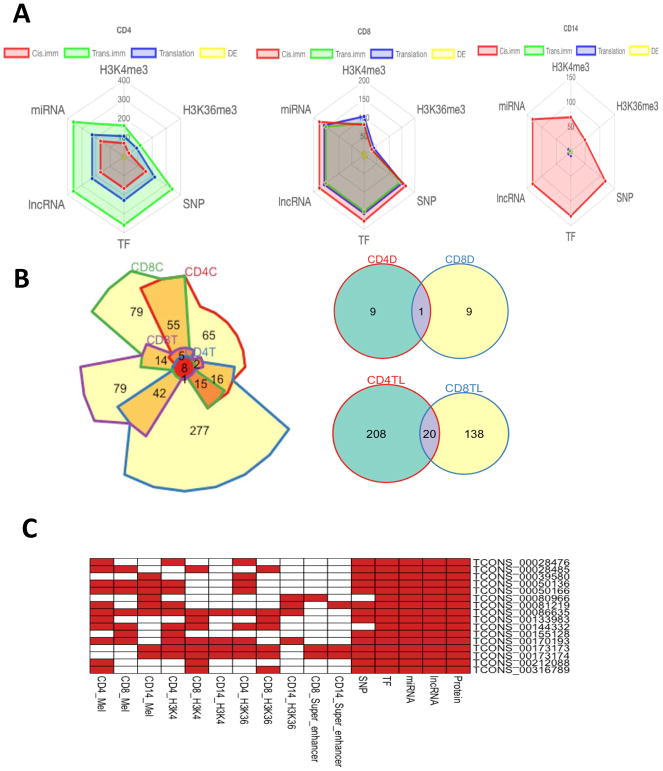
Many stage IV melanoma patients develop resistance upon their routine treatments such as immunotherapy and targeted therapy. The underlying mechanisms leading to resistance remain largely unknown. During the last decade, lncRNAs have been recognized as one of the largest regulatory RNA classes encoded in eukaryotic genomes. Our premise is that genome-wide analysis of lncRNA expression profiling in individual immune cell types in PBCs will expedite the understanding of the immune cell type-specific transcriptional regulatory mechanism in melanoma patients, thus providing resources to investigate potential biomarkers or therapeutic targets for melanoma patients in a noninvasive clinical setting in the future. As melanoma is often characterized by immune infiltration disease, we explored lncRNA expression patterns in CD4+, CD8+, and CD14+ PBCs of stage IV melanoma patients. Overall, lncRNAs exhibited widespread expression patterns in the human genome and tight cell-type-specificity in CD4+, CD8+, and CD14+ PBCs. Our analysis suggested that lncRNAs can not only mediate the expression of protein-coding transcripts in a cis- or trans-regulatory manner in the immune system (26), but also be regulated by similar regulatory mechanisms as mRNAs do. For instance, almost biologically significant lncRNAs can be regulated by both transcriptional regulatory elements and post-transcriptional regulatory elements (e.g., miRNAs and lncRNAs (Fig. 7A). By comparing CD4+ and CD8+ PBCs, we identified 29 overlapping lncRNAs in T cells, including 8 immune-associated lncRNAs, 20 translation-associated lncRNAs, and 1 differentially expressed lncRNA (Fig. 7B). TCONS_00060410, TCONS_00227034, and TCONS_00325476 were identified as both the immune-associated and translation-associated lncRNAs. Sixteen most highly expressed lncRNAs in CD4+, CD8+, and CD14+ PBCs were potentially regulated by multiple transcriptional elements and post-transcriptional elements (Fig. 7C). Through the functional enrichment analysis, we found that our lncRNAs were associated with several immune-related pathways in CD4+, CD8+, and CD14+ PBCs. Interestingly, most of the trans-regulatory lncRNAs involved in immune response pathways, PD-1 checkpoint pathways, and three mRNA translation processes were grouped in turquoise modules of CD4+ and CD8+ PBCs. Further functional investigation of these lncRNAs will be greatly appreciated to understand their biological roles in these pathways.
Fig. 7.
The comprehensive analysis of lncRNAs across CD4+, CD8+, and CD14+ PBCs. (A) Left of the panel, radar plots showing the number of lncRNAs that are mediated by multiple regulatory elements. The center of the plot is 0, and a colored dot on the respective axis indicates the number of lncRNAs that are mediated by different regulatory elements. Lines connecting the number of lncRNAs to the origin of the plot are added to improve visualization. (B) Chow-Ruskey diagrams of immune-associated cis-regulatory and trans-regulatory lncRNAs in CD4+ and CD8+ PBCs (CD4C, cis-regulatory lncRNAs in CD4 PBC; CD4T, trans-regulatory lncRNAs in CD4 PBC; CD8C, cis-regulatory lncRNAs in CD8 PBC; CD8T, trans-regulatory lncRNAs in CD8 PBC). Color of the borders around each intersection corresponds to the overlapping lncRNAs. The red circle in the middle represents the overlap of all immune-associated cis-regulatory and trans-regulatory lncRNAs. Lighter shades of red, orange, and yellow represent the overlap of fewer immune-associated cis-regulatory and trans-regulatory lncRNAs. Area of each intersection is proportional to number of lncRNAs within the intersection. Upper right of the panel, the overlapping of differentially expressed lncRNAs in CD4+ and CD8+ PBCs (CD4D, differentially expressed lncRNAs in CD4 PBC; CD8D, differentially expressed lncRNAs in CD8 PBC). Bottom right of the panel, the overlapping of translation-associated lncRNAs in CD4+ and CD8+ PBCs (CD4TL, translation-associated lncRNAs in CD4 PBC; CD8TL, translation-associated lncRNAs in CD8 PBC). (C) 16 most highly expressed lncRNAs interacted with multiple regulatory elements. Red box indicates the lncRNA interacted with regulatory element.
T cells (i.e., CD4+ and CD8+ PBCs) and monocyte (i.e., CD14+ PBC) exhibited differentially transcriptional expression profiles between melanoma patients and healthy subjects (Supplementary Fig. S5 and S6). Specifically, the expression profiles of protein-coding transcripts in CD4+ and CD8+ T cells were able to distinguish stage IV melanoma patients and healthy subjects. Furthermore, in 9 coexpression modules significantly associated with the melanoma trait, CD4+ and CD8+ T cells share more overlapping modules than CD14+ monocytes.
Age is closely related to melanoma status in diverse immune cells. Using the coexpression network analysis, we found that age and melanoma traits were both significantly correlated in the 9 coexpression modules in CD4+, CD8+, and CD14+ PBCs. Patient age has been reported as an independent prognostic factor for melanoma (43). With the increasing age, the immune system exhibits age-related changes (44), mainly due to the dysregulation of T cell function resulting in T cell immunosenescence. In T cells (CD4+ and CD8+ PBCs), the mTOR signaling pathway was enriched all 8 coexpression modules. PD-1 is an immune-checkpoint receptor which mainly expresses in T cells negatively regulating human immune response (45). Inhibition of mTOR signaling pathway can extend the life span in many different species (46) and delay the onset of age-related diseases in mice (47). Because of potential age-related changes in immune cells (48), melanoma patients in different age groups may need more personalized immunotherapeutic treatments.
There are some limitations in our study. First, CD4+, CD8+, and CD14+ PBCs primarily contain CD4 T cell, CD8 T cell, and CD14 monocyte, respectively. However, they are still heterogeneous cell types which may have distinct transcriptional expression profiles. Second, our RNA-seq dataset only contains lncRNAs with ploy-A tails, which account for a partial lncRNA transcriptome. Third, the prediction software in our analysis could introduce some computational errors. Although the multi-omics integrative analysis provides a wide understanding of the original cause of disease (genetic, environmental, or developmental) (49), the limited sample size and expensive experimental cost prevent such multi-omics experiments to be conducted in individual diseases. As the cost of single-cell sequencing technology becomes more affordable (50), we expect that an integrative analysis of diverse immune cell types at single cell level will promote a much deeper understanding of the immune response mechanism and more personalized treatment to melanoma patients.
Supplementary Material
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Deeks ED. Nivolumab: A Review of Its Use in Patients with Malignant Melanoma. Drugs. 2014;74:1233–9. doi: 10.1007/s40265-014-0234-4. [DOI] [PubMed] [Google Scholar]
- 2.Richtig G, Ehall B, Richtig E, Aigelsreiter A, Gutschner T, Pichler M. Function and Clinical Implications of Long Non-Coding RNAs in Melanoma. International journal of molecular sciences. 2017:18. doi: 10.3390/ijms18040715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–11. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer research. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nature medicine. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 7.Peters T, Schroen B. Missing links in cardiology: long non-coding RNAs enter the arena. Pflugers Archiv : European journal of physiology. 2014;466:1177–87. doi: 10.1007/s00424-014-1479-1. [DOI] [PubMed] [Google Scholar]
- 8.Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. European heart journal. 2015;36:353–68a. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends in immunology. 2014;35:408–19. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rambow F, Job B, Petit V, Gesbert F, Delmas V, Seberg H, et al. New functional signatures for understanding melanoma biology from tumor cell lineage-specific analysis. Cell reports. 2015;13:840–53. doi: 10.1016/j.celrep.2015.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felts SJ, Van Keulen VP, Scheid AD, Allen KS, Bradshaw RK, Jen J, et al. Gene expression patterns in CD4+ peripheral blood cells in healthy subjects and stage IV melanoma patients. Cancer Immunology, Immunotherapy. 2015;64:1437–47. doi: 10.1007/s00262-015-1745-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nature Reviews Cancer. 2012;12:349–61. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 13.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic acids research. 2007;35:W345–9. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic acids research. 2004;32:D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic acids research. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH roadmap epigenomics mapping consortium. Nature biotechnology. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature protocols. 2012;7:1728–40. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tafer H, Hofacker IL. RNAplex: a fast tool for RNA–RNA interaction search. Bioinformatics. 2008;24:2657–63. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 24.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome biology. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. cat RAPID omics: a web server for large-scale prediction of protein–RNA interactions. Bioinformatics. 2013;29:2928–30. doi: 10.1093/bioinformatics/btt495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends in molecular medicine. 2014;20:623–31. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ning S, Zhang J, Wang P, Zhi H, Wang J, Liu Y, et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic acids research. 2016;44:D980–5. doi: 10.1093/nar/gkv1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–22. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 29.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3. 0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nature structural & molecular biology. 2012;19:1068–75. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 31.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome research. 2012;22:577–91. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, et al. LncRNA profiling of human lymphoid progenitors reveals transcriptional divergence of B and T lineages. Nature immunology. 2015;16:1282. doi: 10.1038/ni.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. New England Journal of Medicine. 2016;375:1767–78. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome research. 2011;21:1273–83. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan A, Zhang X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic acids research. 2016;44:D164–D71. doi: 10.1093/nar/gkv1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium GP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. [Google Scholar]
- 40.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2010;39:D945–D50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic acids research. 2016;44:D231–D8. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nature immunology. 2014;15:484–91. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 43.Lasithiotakis K, Leiter U, Meier F, Eigentler T, Metzler G, Moehrle M, et al. Age and gender are significant independent predictors of survival in primary cutaneous melanoma. Cancer. 2008;112:1795–804. doi: 10.1002/cncr.23359. [DOI] [PubMed] [Google Scholar]
- 44.Pawelec G, Barnett Y, Forsey R, Frasca D, Globerson A, McLeod J, et al. T cells and aging, January 2002 update. Front Biosci. 2002;7:d1056–d183. doi: 10.2741/a831. [DOI] [PubMed] [Google Scholar]
- 45.Berger KN, Pu JJ. PD-1 pathway and its clinical application: a 20year journey after discovery of the complete human PD-1 gene. Gene. 2017 doi: 10.1016/j.gene.2017.09.050. [DOI] [PubMed] [Google Scholar]
- 46.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–45. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mannick JB, Del Giudice G, Lattanzi M, Valiante NM, Praestgaard J, Huang B, et al. mTOR inhibition improves immune function in the elderly. Science translational medicine. 2014;6:268ra179–268ra179. doi: 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Jimenez CP, Eling N, Chen H-C, Vallejos CA, Kolodziejczyk AA, Connor F, et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science. 2017;355:1433–6. doi: 10.1126/science.aah4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome biology. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giladi A, Amit I. Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries. Cell. 2018;172:14–21. doi: 10.1016/j.cell.2017.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data set has been deposited in the NCBI Gene Expression Omnibus (GEO) database (GEO accession number GSE104744).