Abstract
Up to 30% of patients with metastatic breast cancer eventually develop brain metastasis, yet the pathological mechanism behind this development remains poorly understood. Here we profiled long non-coding RNAs in brain metastatic tumors from breast cancer patients and found that the X-inactive specific transcript (XIST) was significantly downregulated in these tissues. XIST expression levels inversely correlated with brain metastasis, but not with bone metastasis in patients. Silencing of XIST preferentially promoted brain metastatic growth of XISThigh cells in our xenograft models. Moreover, knockout of XIST in mice mammary glands accelerated primary tumor growth as well as metastases in the brain. Decreased expression of XIST stimulated epithelial-mesenchymal-transition (EMT) and activated c-Met via MSN-mediated protein stabilization, which resulted in the promotion of stemness in the tumor cells. Loss of XIST also augmented secretion of exosomal microRNA-503, which triggered M1-M2 polarization of microglia. This M1-M2 conversion upregulated immune suppressive cytokines in microglia which suppressed T-cell proliferation. Furthermore, we screened an FDA-approved drug library and identified fludarabine as a synthetic lethal drug for XISTlow breast tumor cells and found that fludarabine blocked brain metastasis in our animal model. Our results indicate that XIST plays a critical role in brain metastasis in breast cancer by affecting both tumor cells and the tumor microenvironment, and that the XIST-mediated pathway may serve as an effective target for treating brain metastasis.
INTRODUCTION
Around 30% of patients with metastatic breast cancer will eventually develop brain metastasis, which profoundly affects the cognitive and sensory function as well as morbidity of the patients(1). The development of brain metastasis is a complex, multi-step process, including invasion of tumor cells through the blood brain barrier (BBB) to reach the brain parenchyma, adaptation to the brain microenvironment to acquire growth factors and signaling that are critical for their survival and sustained growth. Adaptation is mainly achieved by reprograming of the cells within the brain metastatic niche that consists of astrocytes, microglia, and various immune cells (2). Therefore, only a small population of disseminated tumor cells, that have the appropriate genetic profile, will eventually outgrow in the brain (3,4). Although many genes such as COX2, Cx43, and CTSS have been shown to play roles in brain metastasis by affecting some of these steps, it is not clear whether there exists a single master regulatory oncogene or tumor suppressor whose expression controls the entire process of brain metastasis(3,5). Our group has previously shown the importance of non-coding RNAs, especially microRNAs (miRNAs) in breast cancer brain metastasis, through their targeting of multiple oncogenes that regulate the invasive ability of cancer cells, as well as cancer stem cell-like populations(6,7). Another group of non-coding RNAs called long non-coding RNAs (lncRNAs) have recently drawn much attention due to their potential roles in tumor progression(8). LncRNAs are more complex in structure, and their mechanisms of gene modulation include chromatin remolding, as well as transcriptional and post-transcriptional regulation(9). Therefore, it is likely that this group of genes also play critical roles in the key steps of brain metastasis, including stem cell growth and reprograming tumor microenvironment.
Microglia are the major innate immune cells found in the brain that become activated under many pathological conditions including infection, injury, and cancer of the central nervous system (CNS)(10). Activated microglia are abundant source of inflammatory molecules that can affect the development of neurodegenerative diseases as well as tumor progression (11). Therefore, microglia are key components of the tumor microenvironment in brain metastasis. Similar to macrophages, activated microglia have both tumor suppressive (M1) and tumor promoting (M2) roles depending on the activation of specific signal pathways (12). However, it is not yet clear how metastatic tumor cells evade the cytotoxic effect of M1 microglia, while at the same time inducing the M1-M2 phenotypic change that supports their growth. In addition to microglia, tumor infiltrating lymphocytes (TILs), which are known to be able to efficiently eliminate tumor cells by triggering a series of anti-tumor responses, are frequently found at tumor sites in CNS cancer and metastatic brain tumors, even though a normal brain is considered to be an immune privileged site(13). However, how tumor cells evade the anti-tumor effect of TILs is yet to be clarified. In this study, we show that the loss of lncRNA X-inactive specific transcript (XIST) promotes breast cancer brain metastasis by enhancing both stemness and aggressiveness of tumor cells through induction of EMT- and MSN-mediated up-regulation of c-Met. We also show that loss of XIST in tumor cells causes local immune suppression by converting the microglia to the M2 phenotype through the transport of exosomal miR-503 from the tumor cells.
MATERIALS AND METHODS
Cell culture and reagents
Human breast carcinoma cell lines, MCF7, ZR75-1, SKBR3 and MDA-MB231 (MDA231), were purchased from American Type Culture Collection. SIM-A9 was purchased from Kumi Nagamoto-Combs, through Kerafast.com. MDA-MB231BrM2a (231BrM) was a kind gift from Dr. Massague (Memorial Sloan-Kettering Cancer Center)(4). SKBrM3 and MDA-MB231BrM2a are brain metastatic cell lines derived from parental SKBR3 and MDA-MB-231 cells through three rounds of in vivo selections(14). SKBR3, SKBrM3, MCF7, MDA231 and 231BrM were cultured in DMEM medium supplemented with 10% FBS, streptomycin (100 mg/ml) and penicillin (100 units/ml). ZR75-1 was cultured in RPMI medium with 10%FBS. SIM-A9 was cultured in DMEM/F12 medium with 5% FBS. T-cells were isolated from mouse spleen by using the pan T-cell isolation kit (Miltenyi Biotec) and cultured in RPMI with 10% FBS. All cells were grown at 37 °C in a 5% CO2 atmosphere. All cell lines were obtained between 2010 and 2016, and they were authenticated by qRT-PCR analysis for the expression of 20 signature genes.
Plasmids and reagents
Lentiviral vectors expressing shRNA for XIST and MSN were obtained from abm Inc, and Dharmacon, respectively. siRNA targeting MPP1, MSN and MID1 were purchased from Dharmacon. The plasmids expressing dCAS9 (lenti dCAS-VP64_Blast) and MS2 (lenti MS2-P65-HSF1_Hygro), and gRNA backbone plasmid (lenti sgRNA (MS2)_zeo backbone) were obtained from Addgene. The gRNAs targeting XIST promoter were cloned into the backbone plasmid as described previously(15). gRNA-11 and gRNA-8 target 5′TGTCCGGCTTTCAATCTTCT3′ and 5′GCAGCGCTTTAAGAACTGAA3′, respectively. DNA oligomers were synthesized by Invitrogen. The plasmid expressing miR503 was purchased from System Biology Inc (SBI).
Quantitative real-time PCR
Total RNAs were isolated from cells and reverse transcribed as described previously(16). The cDNA was then amplified with a pair of forward and reverse primers for the following genes:
XIST-F 5′ GTGATCAGCACCCCAGCTAT3′
XIST-R 5′ TCTTTCTTTTCCTCCCAGCA3′
c-Met-F 5′GGTCCTTTGGCGTCGTCCTC3′
c-Met-R 5′ CTCATCATCAGCGTTATCTTC3′
MSN-F 5′ ACCGGGAAGCAGCTATTTGA3′
MSN-R 5′ GAACTTGGCACGGAACTTAA3′
MID1-F 5′ CCTGTCAACATGTTGAAGTC3′
MID1-R 5′ GCAATCTGCTGAGCCAGTTT3′
MPP1-F 5′ATTGAATACTGTGACCGAGG3′
MPP1-R 5′ TTCTAGGATCTCATCCCCCA3′
E-Cad-F 5′ GACCAGGACTATGACTACTTGAAACG3′
E-Cad-R 5′ ATCTGCAAGGTGCTGGGTGAACCTT3′
Vim-F 5′ ATTCCACTTTGCGTTCAAGG3′
Vim-R 5′ CTTCAGAGAGAGGAAGCCGA3′
mT-antigen-F 5′ GGAAGCAAGTACTTCACAAGGG3′
mT-antigen-R 5′ GGAAAGTCACTAGGAGCAGGG3′
Mouse-CD86-F 5′TTACGGAAGCACCCACGATG3′
Mouse-CD86-R 5′ CCTGTTACATTCTGAGCCAGT3′
Mouse-CD204-F 5′TTCAATGACAGCATCCCTTCC3′
Mouse-CD204-R 5′ GCTTTCGATTCTCTCCTCCAT3′
Mouse-Arg1-F 5′ TCCTTAGAGATTATCGGAGCG3′
Mouse-Arg1-R 5′ GTCTTTGGCAGATATGCAGG3′
Mouse-CD74-F 5′ AGAGCCAGAAAGGTGCAGC3′
Mouse-CD74-R 5′ GATGCATCACATGGTCCTGG3′
Mouse-CD80-F 5′TACACCACTCCTCAAGTTTCC3′
Mouse-CD80-R 5′CAGGTAATCCTTTTAGTGTCTG3′
Mouse-Arg2-F 5′ CAAATTCCTTGCGTCCTGACG3′
Mouse-Arg2-R 5′GGTACCTATTGCCAGGCTGT3′
Mouse-IL10-F 5′ AGCAGAGTGAAGACTTTCTTTC3′
Mouse-IL10-R 5′ CCTTGCTCTTGTTTTCACAGG3′
PCR reactions were performed using CFX Connect (Bio-Rad) and iTaq Universal SYBR Green Supermix (Bio-Rad). The thermal cycling conditions composed of an initial denaturation step at 95°C for 5 min followed by 40 cycles of PCR using the following profile: 94 °C, 30 s; 60 °C, 30 s; and 72 °C, 30 s. Taqman realtime PCR reagent and primer were purchased from Invitrogen. TaqMan PCR was performed as described by the manufacture.
Human tissue samples
Human primary breast cancer specimens were obtained from surgical pathology archives of the, Wake Forest Baptist Comprehensive Cancer Center (WFBCCC), and Cooperative Human Tissue Network (CHTN). Human breast cancer brain metastasis samples were obtained from CHTN and Tumor Tissue and Pathology Shared Resource at WFBCCC. All tissue sections were obtained by surgical resection.
Immunocytochemistry and immunofluorescence
Cells or frozen section tissues were fixed with methanol, washed with PBS and blocked with 2% BSA for 1hr. The sections were then washed with PBS and incubated with anti-c-Met (1/500: R&D), anti-E-Cad (1/500, Cell signaling Technology), anti-Vimentin (1/500; Cell signaling Technology), anti-mT-antigen-FITC (1/200; Novus), anti-CK14 (1:200, Abcam), anti-CK8/18 (1:200, Abcam), anti-IBA-1 (1:200, Abcam) and anti-CD163 (1/500; Cell signaling Technology) antibodies for 12 hrs at 4 °C. Samples were then incubated with anti-rabbit IgG Alexa Fluor 555 molecular probe (Cell Signaling Technology) for 1hr at room temperature if primary antibody is not conjugated with florescence. Fluorescence images were taken by a fluorescent microscope (Olympus IX71).
Gene-expression microarray profiling
RNAs were extracted, labeled and hybridized to Human Gene 1.0 ST array (Affymetrix) using the manufacturer’s protocol. Normalization of the data was performed using the RMA algorithm. Existing breast cancer cohort data were analyzed as described previously. For the cancer cohort data analysis, we compiled a microarray dataset of 710 patients from GEO (accession numbers: GSE12276, GSE2034, GSE2603, GSE5327, and GSE14020) as we published previously(14). These datasets were all normalized using MAS5.0, and each microarray was centered to the median of all probes. For each patient, brain-metastasis free survival was defined as the time interval between the surgery and the diagnosis of metastasis.
GSEA
The Gene MatriX file (.gmx) was generated as described previously(14) by combining the top 200 down-regulated genes when MCF7 cells were treated with pathway-specific inhibitors (GSE31912) and also by adopting oncogenic signatures that were deposited in Molecular Signatures Database (MsigDB). Gene Cluster Text file (.gct) was generated from TCGA mRNA data of invasive breast cancer patients. Top 300 patients with highest XIST expression were defined as XIST-high patients, and top 300 patients with lowest XIST expression were defined as XISTlow patients. The number of permutations was set to 1000.
Animal experiments
All animal experiments were done in accordance with a protocol approved by the Wake Forest Institutional Animal Care and Use Committee. For experimental metastasis assay, nude mice (7–8 weeks) were injected with luciferase-labeled tumor cells in PBS into left cardiac ventricle in a total volume of 100 μl. To confirm a successful injection, the photon flux from whole body of the mice was immediately measured using IVIS Xenogen bioimager (Caliper). The brain metastasis progression was monitored and the luminescence was quantified at the indicated time points. At the endpoint of the study, whole brain was removed, incubated in RPMI-1640 medium with 0.6 mg/ml luciferin for 5 min and photon flux was measured. Intracranial injection was performed as previously described. In brief, luciferase-labeled cancer cells in PBS were injected into mouse brain. Photon flux of mice was measured using IVIS xenogeny bioimager weekly. XISTlox/lox mouse strain is a kind gift of Dr. Rudolf Jaenisch(17) and was obtained from the MMRRC repository. Tg(MMTV-cre)4Mam/J (MMTV-Cre/Line D) mouse strain was purchased from Jackson laboratory. B6.FVB-Tg(MMTV-PyVT)634Mul/LellJ (MMTV-PyMT) mouse strain was purchased from Jackson laboratory. The MMTV-PyMT and XISTlox/lox were back crossed to B6129F1 for 5 generations before cross breeding. XISTlox/lox and MMTV-Cre were crossbred to generate MMTV-CRE/XISTlox/lox. MMTV-CRE/XISTlox/lox was further crossbred with MMTV-PyMT to generate XISTlox/lox/MMTV-PyMT or MMTV-CRE/XISTlox/lox/MMTV-PyMT. Siblings were used as a control for each experiment. Mammary gland whole mount slides were generated as described previously(18). The early stage (4 weeks) hyperplastic nodules were counted under microscope. Tumor areas at age of 8 weeks were measured by ImageJ. Tumor length, height and width were measured by caliper, and tumor size was calculated using the formula, 1/2 x L x W x H. The degree of lung metastases was measured by counting the number of metastatic lesions on the lungs at the end point (24 weeks). Micrometastases in the brain were defined as mT-antigen-positive clusters, and the number of lesions was counted under fluorescent microscope.
Exosome isolation
Exosomes were isolated by sequential ultracentrifugation(19). Briefly, CM harvested from cell culture was centrifuged at 300 × g for 10 minutes to remove cells. Exosomes were collected by centrifugation at 2,000 × g for 20 minutes. The supernatant was again centrifuged at 16,500 × g for 20 minutes to remove microvesicles. The supernatant was then passed through a 0.2μm filter (Sarstedt, Nümbrecht, GERMANY) to remove particles larger than 200 nm. Exosomes were then collected by ultracentrifugation at 120,000 × g for 70 minutes. Pellets were washed in PBS and centrifuged 70 minutes at 120,000 × g for 70 min and finally suspended in PBS.
Plasma exosomes were isolated as described previously(19). In brief, 1ml of serum was loaded onto the qEV size exclusion columns (Izon), and exosomes were elution with PBS. Isolated exosomes were concentrated by centrifugation using the Amicon® Ultra-4 10 kDa column.
The size distribution of isolated exosomes was analyzed by nano-particle tracking analysis using Nanosight NS50 and by eletronmicroscopy.
Exosome uptake analysis
Exosomes were isolated from cells infected with lentivirus expressing PalmGFP (a kind gift from Dr. Breakefield(20). Human microglial cells were stained with PKH26 according to product manual and seeded onto cell culture slide. Cells were treated with 50 μg/ml exosomes for 24hr and they were fixed and stained with DAPI (Invitrogen). The exosome uptake was observed by Olympus FV1200 spectral laser scanning confocal microscope.
CFSE assay
T cells were isolated from the spleen of BALB/C mouse by using Pan T Cell Isolation Kit II (Miltenyi Biotec) followed by incubation with Dynabeads™ Mouse T-Activator (Thermo Fisher) and 2U/ml IL2 for 2 days. 106 activated T cells were labeled by CellTrace™ CFSE dye (Thermo Fisher) and co-cultured with same amount of microglia for 5 days followed by FACS analysis.
Analysis of H3K4me3 ChIP-seq data
We acquired the publicly available ChIP-seq data from Sequence Read Archive (SRA) of NCBI (https://www.ncbi.nlm.nih.gov/sra). The H3K4me3 ChIP-seq data were derived from the following projects: PRJNA266312(21), PRJNA142885(22), and PRJNA363069. The raw fastq files were derived from SRA, by the fastq-dump tool. We checked the data quality of the fastq files by using Fastqc software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and identified all of the fastq files passed the quality control. Then we aligned the raw fastq files to hg19 genome by bowtie2. The peak calling was enabled by MACS2 software using default parameters. The visulization is enabled by IGV software from Broad Institute (http://software.broadinstitute.org/software/igv/).
Statistical analysis
All analysis was calculated by GraphPad Prism. Data are presented as mean ±SD. The p-value is calculated by an unpaired Student’s t-test. Gene correlation analysis was calculated by liner regression analysis. The Kaplan-Meier survival analysis was calculated by log-rank (Mantel Cox) test. Significance between each groups were represented as *p<0.05, **p<0.01.
RESULTS
XIST expression is down regulated in breast cancer brain metastasis
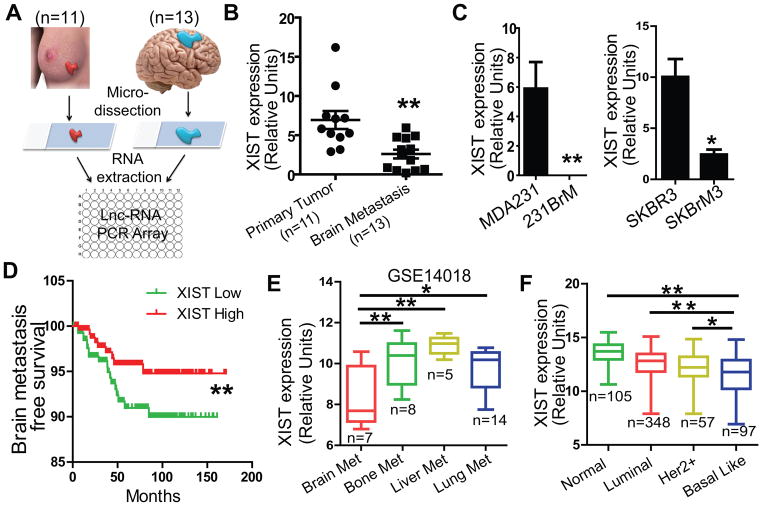
To identify key lncRNAs in brain metastasis of breast cancer, we isolated RNA from micro-dissected primary tumors and brain metastatic tumors (Fig. 1A). The expression profiles of lncRNAs were examined in pooled samples using a quantitative real-time PCR (qRT-PCR) based lncRNA array which we recently developed. The cutoff threshold was set to over 10-fold difference between the two groups followed by validation of selected lncRNAs in individual sample. We found that XIST was the most highly differentially expressed lncRNA between the two groups (Fig. 1B; Supplementary Fig. S1A and B). XIST gene is located on the X chromosome and is known to be involved in X chromosome inactivation by suppressing the genes on this chromosome in a cis-acting manner(23). We also examined the expression of XIST in two paired brain tropic tumor cell lines and found that XIST expression was also significantly lower in these cells compared to their parental cells (Fig. 1C). The results of Kaplan-Meier analyses of 710 breast cancer patients using combined cohort data (14) indicated that low levels of XIST was associated with a poor brain metastasis-free survival, but not bone-metastasis free survival (Fig. 1D; Supplementary Fig. S1C). Furthermore, XIST expression was found significantly down-regulated in brain metastatic lesions compared to other metastatic tumors based on an organ-specific cohort analysis (Fig. 1E). We also examined XIST expression in various subtypes of breast cancer using the TCGA database and found that XIST expression levels were significantly decreased in basal-type breast tumors (Fig. 1F), which are the most common types of breast cancer that known to metastasize to the brain. Interestingly, our analysis of TCGA ChIP-seq database for various breast cancer cell lines revealed that XIST expression was strongly correlated to the histone modification of H3K4me3 at the promoter region of XIST gene (Supplementary Fig. S1D). These results strongly indicate that the loss of XIST expression plays a key role in brain metastasis.
Fig. 1. Down-regulation of XIST on the X chromosome positively correlates with brain metastasis.
(A) Schematic diagram of sample collection and lncRNA analysis. Tumors from the primary sites (n=11) and the brain metastatic sites (n=13) were sectioned, and tumor cells were micro-dissected followed by extraction of RNAs. They were then subjected to the PCR-based lncRNA analysis for the panel of 92 key cancer-related lncRNAs. (B) TaqMan PCR analyses of XIST expression in clinical samples used for LncRNA array. (C) XIST expression was examined by qRT-PCR in two pairs of isogenic cell lines that preferentially metastasize to the brain. (D) Kaplan-Meier analysis for brain metastasis-free survival using a combined GEO cohort database which includes a total of 710 breast cancer patients. (E) XIST expression was examined in metastatic tumors from breast cancer patients using the GSE14018 cohort data. (F) XIST expression was examined for the patients with breast cancer in each subtype using the TCGA database.
XIST preferentially suppresses brain metastasis in vivo
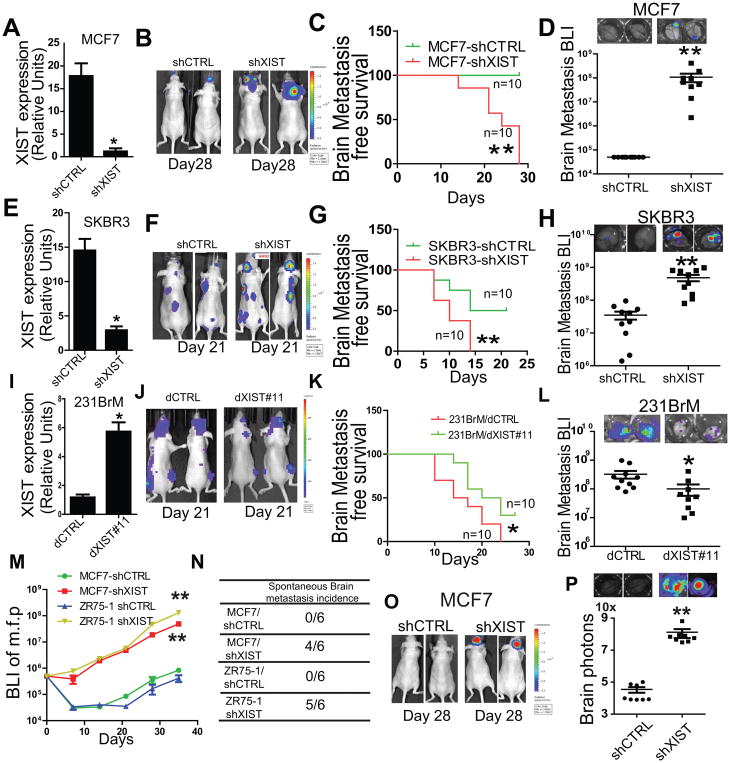
To address the question how downregulation of XIST is involved in brain metastasis, we first established cell lines expressing small hairpin RNAs (shRNAs) that targeted four different locations in the XIST gene. The knockdown effects of shXIST in MCF7 cells which have high endogenous levels of XIST were confirmed by qRT-PCR (Fig. 2A). To examine the effects of XIST on breast cancer metastasis in vivo, we inoculated luciferase labeled MCF7 cells with or without knocking down of XIST into nude mice by intracardiac injection followed by monitoring the tumor growth by measuring the amount of bioluminescence (BLI). We found that the knockdown of XIST in both cell lines significantly promoted their brain metastatic abilities (Fig. 2B–D). This metastasis suppressive function of XIST was also verified with another two XIST high breast cancer cell line, SKBR3 and ZR75-1 in vivo (Fig. 2E–H; Supplementary Fig. S2 A–C). To further examine whether gain of function of XIST suppresses metastasis, we used the dCas9 system (15) to activate and restored XIST expression in 231BrM cells using gRNAs that target key transcriptional activation regions. We have designed 20 gRNAs based on the prediction by the Benchling website and successfully restored the expression of XIST by gRNA dXIST#11(Fig. 2I). We found that the over expression of XIST in 231BrM cell significantly attenuated its metastatic ability to the brain but not to the bone (Fig. 2J–L; Supplementary Fig. S2 D and E). These in vivo results suggest that knockdown and over expression of XIST preferentially promotes and suppresses brain metastasis, respectively (Fig. S2F). Notably, knock down of XIST in MCF7 and ZR75-1 significantly promoted their growth in vivo (Fig. 2M). We also observed a high spontaneous brain metastasis incidence in shXIST groups, suggesting that knockdown of XIST enhances tumor invasion and extravasation at the primary site (Fig. 2N; Supplementary Fig. S2G). To further test the ability of XIST-knockdown cells to colonize the brain, we transplanted MCF7-shCTRL or MCF7-shXIST cells into nude mice by intracranial injection followed by monitoring tumor growth by bioluminescence imaging. Knockdown of XIST in MCF7 cells significantly promoted tumor growth in the brain (Fig. 2O and P). These results suggest that XIST contributes to multiple metastatic events which are crucial for colonization and outgrowth of tumor cells preferentially in the brain.
Fig. 2. Knockdown of XIST preferentially promotes brain metastasis in vivo.
(A–H) Intracardiac injection of MCF7 and SKBR3 with or without knock down of XIST. MCF7 and SKBR3 cells were infected with lentivirus expressing shXIST (5 target sites in combination) to establish knockdown cell lines, and the knockdown of XIST was confirmed by qRT-PCR (A and E). MCF7-shCTRL, MCF7-shXIST, SKBR3-shCTRL and SKBR3-shXIST cells (5×105) were implanted into nude mice by intracardiac injection, and tumor growth was monitored by measuring luciferase signals twice a week. The mice were sacrificed on Day 28, and brains were removed and ex vivo signal of luciferase were measured. Representative photos of BLI were shown (B and F). The Kaplan-Meier analysis of brain-metastasis free survival was performed (C and G). Ex vivo signals in the brain at the end point was examined (D and H). (I) XIST expression in 231BrM-dXIST #11 was validated by qRT-PCR. (J–L) 2×105 231BrM/dCTRL and 231BrM/dXIST#11 cells were inoculated into nude mice by intracardiac injection followed by monitoring tumor growth. Representative photos of BLI were shown (J). The Kaplan-Meier analysis of brain-metastasis free survival was performed (K). Ex vivo signals in the brain at the end point was examined (L). (M) 5×105 MCF7 and ZR75-1 with or without knock down of XIST were orthotopically implanted into mammary fat pad, and the growth of primary tumors were measured by BLI. (N) Brain metastasis incidence was examined after 5 weeks of implantation. (O) 50,000 cells of MCF7-shCTRL and MCF7-shXIST were implanted into nude mouse brain by intra-cranial injection, and the tumor growth was monitored by IVIS twice a week. On day 28, brains were removed and luciferase signals were measured using the IVIS imager. (P) Quantification and representative photos of ex vivo images of the brain from tumor bearing mice.
Mammary specific knockout of XIST enhances brain metastasis
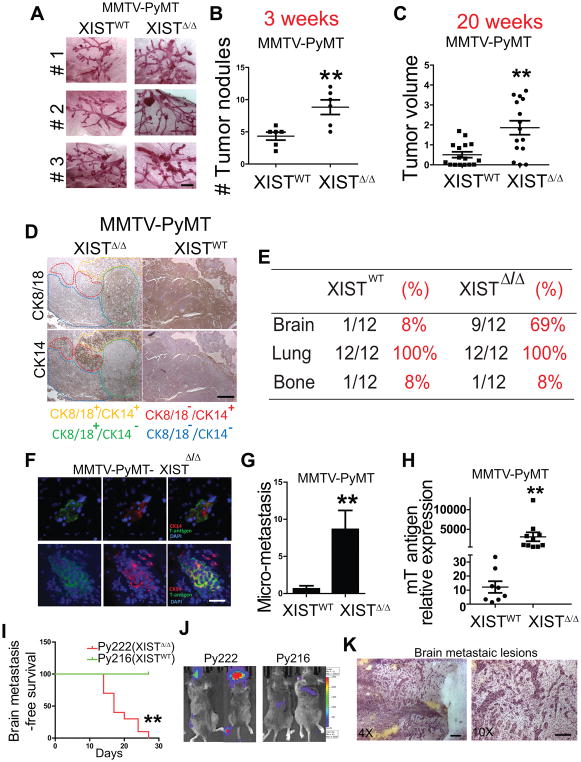
To gain a further insight into the role of XIST in tumorigenesis and metastasis in immunocompetent animals, we generated a mammary-specific XIST knockout mouse by crossbreeding XISTlox/lox and MMTV-Cre mouse. However, the knockout of XIST was insufficient to induce tumorigenesis even at 12 months of age. Therefore, we further crossbred the hybrid mouse with the MMTV-PyMT mouse, which is known to spontaneously develop lung metastasis but not brain metastasis(24). The resultant PyMT-XISTΔ/Δ mouse developed mammary tumors at a significantly accelerated rate compared to the PyMT-XISTWT mouse. As early as 3 weeks after birth when the mammary gland starts to develop, hyperplastic nodules appeared in the mice of both genotypes. However, the XIST knockout mice had a significantly higher number of nodules compared to the control mice, suggesting an early onset of tumor development and a higher frequency of tumor initiation in the XIST knockout mouse (Fig. 3A and B). At 8 weeks, by which time the mammary glands have developed and the end buds have started to branch, XIST knockout mice developed significantly larger tumors compared to control mice in their respective mammary glands (Supplementary Fig. S3A and B). At 20 weeks, the volumes of palpable tumors in XIST knockout mice were significantly larger than that of control mice (Fig. 3C). The MMTV-PyMT mouse model represents luminal breast cancer, and we have shown that decreased XIST expression was more prominent in triple negative breast cancer patients with a basal-like phenotype. Therefore, we speculated that the loss of XIST promotes the phenotypic switch from luminal-like to basal-like tumor in MMTV-PyMT model. Interestingly, we found a mixture of luminal- and basal-type tumor cell populations in the XIST knockout tumors stained by anti-CK18 (luminal) and anti-CK-14 (basal) antibodies, while the majority of tumors in control mice were CK18+ luminal type (Fig. 3D). Importantly, 69% of the XIST knockout mice developed micrometastasis in the brain at 24 weeks, which could be detected by anti-mT-antigen antibody and majority of them were CK14 positive (Fig. 3E–G). This observation was further verified by qRT-PCR analysis of mouse brains using mT antigen-specific primers (Fig. 3H). We also established two syngenic cell lines isolated from XIST WT (Py216) and XISTΔ/Δ (Py222) tumors and verified the XIST expression by qRT-PCR (Supplementary Fig. S3C). We then tested the metastatic ability of these cells in wild type mice by intracardiac injection and found that the knockout of XIST preferentially promoted brain metastasis but not bone metastasis (Fig. 3I and J; Supplementary Fig. S3D). H&E analysis also demonstrated a clear perivascular growth pattern of tumor cells in the brain metastatic lesions which was consistent with other established brain metastatic models (Fig. 3K). Our data strongly suggest that the knockout of XIST drastically promoted brain metastasis in both MMTV-PyMT transgenic and syngenic models which were consistent with the xenograft data. These results further support our notion that the loss of XIST plays a critical role in brain metastasis.
Fig. 3. Knockout of XIST enhances brain metastases in MMTV-PyMT model.
(A) Representative photos of mammary gland whole mount of PyMT-XISTWT and PyMT-XISTΔ/Δ mice at age of 3 weeks. Scale bar represents 2 mm. (B) Number of tumor nodules in the gland shown in (A) was counted. (C) Tumor volumes were measured at age of 20 weeks, using the formula, V = (L x W x W)/2. (D) CK14 and CK8/18 expression was examined by immunohistochemistry in primary tumor from PyMT mice with or without knock out of XIST. Scale bar represents 500 μm. (E) Distant metastasis rates of MMTV-PyMT mice with or without knock out of XIST. (F) At age of 24-28 weeks, brains were removed from these mice, sectioned and subjected to immunohistochemical analysis for CK14 and mT-antigen to examine micro-metastases. Scale bar represents 50 μm. (G) The number of micro-metastatic lesions in the brain was counted in 10 slides in each mouse. (H) The expression of mT antigen in mouse brain was examined by q-PCR. (I) Kaplan-Meier analysis of brain-metastasis free survival of C57/B6 mice inoculated with 5×105 Py216 and Py222 cells. (J) Representative image of end point. (K) H&E staining of brain metastatic lesions from a Py222 inoculated mouse. Scale bar represents 200 μm (left) and 100μm(right).
Knockdown of XIST promotes EMT and stemness
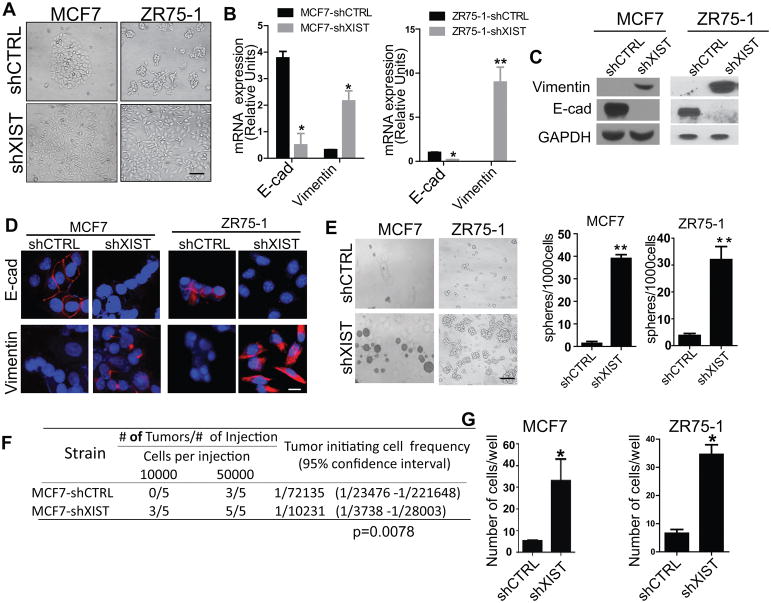
Our in vivo data demonstrated that loss of XIST promotes malignancy of tumor cells which enable them metastasize to the brain. However, the molecular mechanism underneath is not clear. During the establishment of XIST knock down cell lines from MCF7 and ZR75-1, we observed a morphological change of those cells with the acquisition of a mesenchymal phenotype (Fig. 4A), suggesting that XIST may play a role in the epithelial to mesenchymal transition (EMT). Tumor cells are known to gain invasiveness, migratory ability, and stemness following the EMT(25). We therefore examined the expression of EMT markers on these cells and found that E-cadherin and vimentin were significantly suppressed and upregulated, respectively, when XIST was knocked down (Fig. 4B–D; Supplementary Fig. S4A and B). We also observed the same results in Py216 and Py222 cell lines (Supplementary Fig. S4C and D). Because the EMT is closely associated with the properties of cancer stem-like cells that are known to be involved in tumor progression, we performed mammosphere assays to test the self-renewal ability of cells with or without knockdown of XIST. As shown in Fig. 4E and Supplementary Fig. S4E, knockdown of XIST significantly increased their sphere forming ability. In addition, an in vivo limiting dilution assay indicated that knock down of XIST significantly enhanced the tumor initiating ability of these cells (Fig. 4F). Furthermore, because cancer stem cells have characteristic of intrinsic therapy resistance, we performed clonogenic assays in cells with or without knockdown of XIST followed by radiation and found that knockdown of XIST significantly increased their survival after radiation (Supplementary Fig. S4F). Knockdown of XIST also promoted cell invasion and migration, as shown using a transmigration assay that mimics the blood-brain barrier (Fig. 4G). These results strongly indicate that a loss of XIST enhances the EMT and tumor stemness, and hence, promotes metastatic progression.
Fig. 4. Knockdown of XIST promotes EMT and stemness.
(A) Representative photos showing morphological changes in MCF7-shXIST and ZR75-1-shXIST. Scale bar represents 50 μm. (B–E) Expressions of EMT markers (E-cadherin and vimentin) in these cells were examined by qRT-PCR (B), by western blot (C), and by immunocytochemistry (D). Scale bar represents 50 μm. (E) MCF7 and ZR75-1 with or without knockdown of XIST were cultured in the mammosphere medium for 7 days. Representative photos are shown. Scale bar represents 100 μm. (left panel). The number of spheres was counted under a microscope (right panel). (F) A limiting dilution assay was performed for MCF7-shCTRL and MCF7-shXIST to examine their tumor initiating abilities by injecting the indicated number of cells into mammary fat pad of nude mice followed by measuring tumor formation. p=0.0076 (G) Transmigration assay of MCF7 and ZR75-1 with or without knock down of XIST.
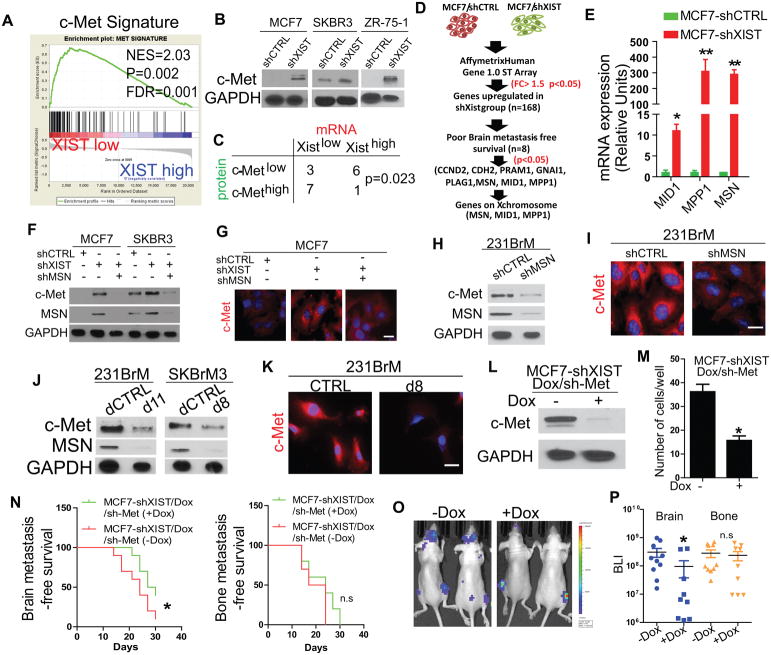
Knockdown of XIST activates the c-Met pathway by upregulating MSN
To identify key downstream effectors of XIST in breast cancer patients that affect brain metastasis, we performed a pathway screening based on Gene Set Enrichment Analysis (GSEA) by using the TCGA database of 937 patients and 119 breast specific oncogenic signatures established previously(14). This approach identifies up and down regulated pathways based on the enrichment of signature genes in patients with two phenotypes, e.g. patients with high or low expression of XIST. Therefore, the result of the screening is not dependent on the expression of a single gene and rather represents overall activation of a pathway which is a consequence of both transcriptional and post-transcriptional regulation. Out of those pathways tested, four of them, c-Met, EIF4E, Myc, and CCND1 pathways, were significantly enriched in patients with low XIST levels (Fig. 5A; Supplementary Fig. S5A). Among these four pathways, the c-Met pathway showed the highest enrichment score (NES = 2.1), and it was the only pathway significantly correlated with brain metastasis-free survival in breast cancer patients (p = 0.004, Fig. S5A). Western blot analysis indicated that there was a strong upregulation of total c-Met in three pairs of XIST knockdown cell lines (Fig. 5B). Furthermore, knockdown of XIST strongly increased c-Met phosphorylation even without HGF stimulation, indicating that c-Met signaling is constitutively activated in XISTlow cells (Supplementary Fig. S5B). We also found a negative correlation between the RNA levels of XIST and the protein expression of c-Met in brain metastasis samples from patients (Fig. 5C). These data suggest that loss of XIST upregulates c-Met and its downstream signaling pathway. To understand how XIST regulates c-Met, we performed a gene expression array analysis on MCF7 cells with or without knockdown of XIST and found that 168 genes were significantly upregulated in MCF7-shXIST cells (>2-fold change, p < 0.05, false discovery rate [FDR] < 0.05) (Fig. 5D). Interestingly, several EMT drivers and mesenchymal markers were among these genes (Supplementary Fig. S5C), which is consistent with our finding in Fig. 4. Notably, out of the 168 genes, eight genes were found to be significantly correlated with poor brain metastasis-free survival (Fig. 5D). Because XIST functions only in a cis-acting manner on the X chromosome, from among the eight genes, we chose to further examine MID1, MPP1, and MSN which are located on the X- chromosome (Supplementary Fig. S5D). We then confirmed upregulation of these genes after knocking down of XIST in MCF7 (Fig. 5E). Among those genes, MSN has been previously reported to indirectly bind c-Met through CD44, and mediates c-Met internalization(26). Upon internalization, c-Met could sort into early endosomes for degradation or traffic to the non-degradative peri-nuclear compartment(27). Therefore, we explored the possibility that XIST-mediated MSN expression contributes to c-Met upregulation by mediating its accumulation in the peri-nuclear compartment. Notably, c-Met expression was drastically decreased by knockdown of MSN but not by MPP1 or MID1in MCF7-shXIST cells (Fig. 5F; Supplementary Fig. S5E). In addition, gene correlation analyses in breast cancer cell lines demonstrated that MSN is the only gene whose expression is inversely correlated with XIST in triple negative tumor cell lines, suggesting a distinct role of MSN in triple negative tumors which are known to express high amounts of c-Met (Supplementary Fig. S5F). Our immunocytochemical analysis indicated that c-Met was localized to the perinuclear region in MCF7-shXIST cells (Fig. 5G). In addition, knockdown of MSN also suppressed c-Met expression and reduced its accumulation in the perinuclear compartment in 231BrM cells (Fig. 5H and I). These results indicate that down regulation of XIST enhances c-Met expression by upregulating MSN, which then facilitates perinuclear localization and stabilization of the c-Met protein. We also generated SKBrM3 cell subline that overexpressed XIST using the dCas9 approach and found that the expression of both MSN and c-Met were strongly suppressed in the XIST overexpressing cells (Fig. 5J; Supplementary Fig. S5G). Moreover, the restoration of XIST expression suppressed c-Met expression by diminishing the perinuclear accumulation of the c-Met protein (Fig. 5K). To further validate the notion that c-Met is a crucial downstream of XIST to enhance brain metastasis, we suppressed c-Met in MCF7-shXIST, 231BrM and SKBrM3 cells by doxycycline-inducible c-Met shRNA. Loss of c-Met in these cells significantly decreases their transmigration abilities (Fig. 5L and M; Supplementary Fig. S5, H and I). We also found that knock down of c-Met in MCF7-shXIST cells significantly decreased their metastatic abilities on brain but not on bone in vivo (Fig. 5N–P). IHC analysis also showed strong c-Met expression in tumors from XIST-knockout mice (Supplementary Fig. S5J). These results further support our notion that induction of c-Met in XISTlow cells is mediated by MSN.
Fig. 5. XIST modulates c-Met expression through MSN.
(A) Breast cancer patients in TCGA were grouped in “XIST-high (top 30%)” and “XIST-low (bottom 30%)” followed by performing Gene Set Enrichment Analysis (GSEA) for 119 oncogenic pathways. The HGF/c-Met pathway showed the highest enrichment score among these pathways, and the result of GSEA for this pathway is shown. (B) c-Met expression was examined by western blot in 3 different breast cancer cell lines with or without knockdown of XIST. (C) XIST mRNA expression was examined by TaqMan qPCR, and c-Met protein expression was examined by IHC in brain metastatic samples of patients. The correlation of these expression levels was calculated by Chi-square test. (D) Flow chart of expression profile analysis of MCF7-shCTRL and MCF7-shXIST cells. (E) The expression of MSN, MID1 and MPP1was examined in MCF7-shCTRL and MCF7-shXIST cells by qRT-PCR. (F) MSN was knocked-down by shRNA in MCF7-shXIST and SKBR3-shXIST cells, and the expressions of MSN and c-Met were examined by western blot. (G) Immunocytochemical analysis of c-Met expression in MCF7 cells with or without knockdown of XIST or MSN. Scale bar represents 10 μm. (H) MSN was knocked down by shRNA in 231BrM cells and the expression of MSN and c-Met was examined by western blot. (I) Immunocytochemical analysis of c-Met expression in 231BrM with or without knockdown of MSN. Scale bar represents10 μm. (J) c-Met and MSN expression was examined by western blot in 231BrM and SKBrM3 cells with or without expression of XIST by dCas9. (K) Immunocytochemical analysis of c-Met expression in 231BrM cells with or without expressing XIST by dCas9. Scale bar represents 10 μm. (L) Western blot analysis of c-Met in MCF7-shXIST/Dox/sh-Met cells with or without addition of doxycycline. (M) Transmigration assay of MCF7-shXIST/Dox/sh-Met cells with or without treatment of doxycycline. (N) Mice were intracardiacally inoculated with 5×105 MCF7-shXIST/Dox/sh-Met cells followed by doxycycline treatment (100μg/ml) in drinking water. Brain (left panel) and bone (right panel) metastasis-free survival were analyzed. (O) Representative BLI image of mice at end point. (P) Quantification of ex vivo BLI from the brain and bone of tumor bearing mice.
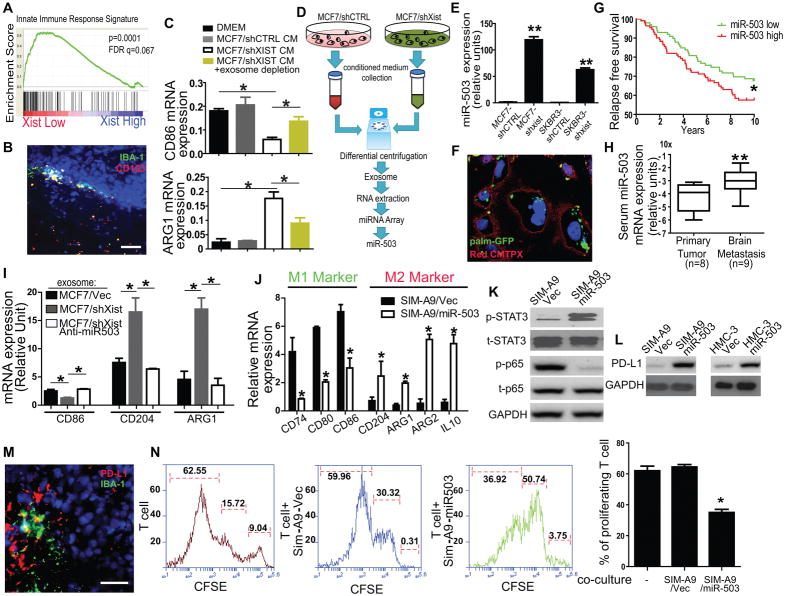
XISTlow cells secrete exosomal miR-503 to reprogram microglia in the brain
The effect of the loss of XIST on metastasis is more profound in the brain compared to other distant organs in the clinical settings and this selectivity was also evident in our xenograft model. These observations suggest that the tumor microenvironment may affect the colonization ability of cells with low levels of XIST expression in the brain. Because microglia are one of the major components of the brain microenvironment, and they have been previously shown to suppress brain metastasis in lung cancer, it is plausible that XISTlow cells reprogram microglia towards a pro-metastatic phenotype through cell-cell communication (28). Interestingly, when we examined the relationship between XIST expression and 95 immune-related signaling pathways in MSigDB by GSEA analysis, the innate cell immune signature was highly enriched in XISTlow patients (Fig. 6A). Immunohistochemical analysis of brain metastatic lesions in breast cancer patients revealed a strong localization of M2 microglia (both IBA-1 and CD163 positive) around the tumor invasion front (Fig. 6B). Metastatic cells are known to educate the tumor microenvironment by various factors including cytokines and exosomes(29). We found that conditioned medium of MCF7-shXIST cells significantly promotes the M1-M2 conversion of microglia as shown by the expression of M1 gene CD86 and M2 gene ARG1. However, depletion of exosomes by ultracentrifuge significantly attenuated such effect, indicating that exosomes are major contributor to the M1-M2 conversion of microglia (Fig. 6C). Although it is not clear how M2 microglia is generated in the tumor areas by exosomes, recent studies showed that microRNAs carried by exosomes play critical roles in the generation of metastatic niche even before the presence of tumor cells(30). Therefore, we hypothesized that exosomes secreted from tumor cells mediate the generation of M2 microglia in the tumor lesions. To test this hypothesis, we first prepared exosomes from MCF7 and MCF7-shXIST cells followed by size validation by the nanoparticle tracking analysis and electron microscopy (Supplementary Fig. S6A and B). RNAs were extracted from the exosomes and they were subjected to microRNA expression profiling (Fig. 6D). Our results revealed that miR-503 was among the most highly expressed microRNAs in the exosomes from MCF7-shXIST cells, and it ranked as the most highly expressed microRNA encoded by the X chromosome. We further validated the induction of miR-503 in cells with knockdown of XIST (Fig. 6E). As shown in Fig. 6F, exosomes were robustly uptake by microglials at a physiological relevant concentration (10mg/ml). We also examined the relationship between miR-503 expression and relapse-free survival in breast cancer patients using the GSE22220 database and found that a higher expression of miR-503 was negatively correlated with the relapse-free survival (Fig. 6G), suggesting that miR-503 plays a role in metastatic progression. Furthermore, we isolated exosomes from the serum of breast cancer patients with or without brain metastasis followed by examine the miR-503 levels. We detected a significantly higher level of miR-503 in the serum-derived exosomes from patients with brain metastasis than in those from other patients, suggesting that exosomal miR-503 is a potential biomarker for brain metastasis (Fig. 6H). To test the effect of exosomal miR-503 on microglia, exosomes were prepared from (i) MCF7 cells, (ii) MCF7-shXIST cells, or (iii) MCF7-shXIST cells expressing anti-miR-503, and then incubated with SIM-A9, a mouse microglia cell line. We examined the expression of the M1 and M2 markers in these treated microglia, and found that the treatment of microglia with exosomes from MCF7-shXIST cells significantly up- and down-regulated the M2 and M1 markers, respectively. This effect of the exosomes was rescued by suppressing miR-503 expression in the MCF7-shXIST cells (Fig. 6I). In addition, CM derived from brain tropic tumor cells (231BrM and SKBRM3) significantly promoted the microglial M1-M2 transition (Supplementary Fig. S6C). When miR-503 was ectopically expressed in SIM-A9 cells, M2 markers were significantly up-regulated while the expression of M1 markers was suppressed (Fig. 6J). In addition, the ectopic expression of miR-503 suppressed ROS in SIM-A9 cells, which is also indicative of the M1/M2 conversion. (Supplementary Fig. S6D). These results strongly suggest that exosomal miR-503 is capable of converting microglia from the M1 to the M2 phenotype. STAT3 and NF-κB are two major signaling pathways that known to play critical roles in the M1-M2 conversion of macrophage(31). We found that miR-503 strongly increased phosphorylation of STAT3 and decreased phosphorylation of the p65 subunit of NF-κB, suggesting a systematic reprogramming of M1 and M2 pathways by miR-503 (Fig. 6K). Treatment of microglia with CM from brain tropic breast cancer cells (231BrM and SKBrM3) further validated our hypothesis (Supplementary Fig. S6E). Furthermore, this miR-503-mediated M1-M2 conversion was significantly suppressed by the STAT3 inhibitor, STATTIC (Supplementary Fig. S6F and G). These results indicate that exosomal miR-503 promotes the M1-M2 conversion through manipulating STAT3 and NF-κB pathways. Similar to M2 macrophages, M2 microglia have been reported to promote progression of brain tumors by releasing cytokines and growth factors (28,32). Because tumor-associated M2 macrophages are also known to suppress T-cell immunity by expressing PD-L1(33,34). Indeed, Ectopic expression of miR-503 in both human and mouse microglial cells increased the expression of PD-L1 (Fig. 6L). Immunohistochemical analysis of brain metastatic lesions from breast cancer patients also revealed that PD-L1 expression was co-localized with microglial cells adjacent to the tumor, but not with astrocytes (Fig. 6M; Supplementary Fig. S6H). Importantly, when mouse T cells were co-cultured with SIM-A9 expressing miR-503, the proliferation rate of T cells was significantly suppressed (Fig. 6N). These results strongly suggest that exosomal miR-503 derived from tumor cells promotes M1-M2 conversion of microglia followed by enhancing their PD-L1 expression to suppress local immunity and thereby enhance tumor growth.
Fig. 6. Phynotypic conversion of microglia from M1 to M2 by exosomal miR503 secreted from XISTlow cells.
(A) XIST-high and XIST-low patients were defined as in Fig. 5A. Ninety five immune-related signaling pathways from MSigDB were tested by GSEA. The innate immune pathway showed the highest enrichment score and the result is shown. (B) A metastatic tumor in the brain of breast cancer patient was subjected to immunohistochemical analysis for the M2 microglia markers, IBA-1 and CD163. Scale bar represents 100 μm. (C) Mouse microglial cell line, SIM-A9, was treated with conditioned medium with or without exosome depletion by ultra-centrifuge. The expressions of CD86 and ARG1 were examined by qRT-PCR after 24 hours (D) Flow chart of exosomal microRNA array analysis. Total exosomes from MCF7-shCTRL and MCF7-shXIST cells were isolated by differential centrifugation, and they were verified for the size by the nano-particle tracking analysis and by electron microscopy (see Fig. S6A and B). Small RNAs were extracted from the exosomes and subjected to microRNA array analysis (Affymetrix; Mir2.0 array). (E) The expressions of miR-503 in two paired breast cell lines with or without knockdown of XIST were examined by TaqMan-qPCR. (F) Exosomes were isolated from conditioned medium of MCF-shXIST cells and then labeled with lentivirus expressing PalmGFP. Mouse microglial cell, SIM-A9, was treated with the exosomes for 24 hours. The result of immunocytochemical analysis shows that the exosomes were taken up by the microglial cells. Scale bar represents 10 μm. (G) Relapse-free survival analysis of miR-503 was examined in breast cancer patients using GSE22220. (H) Exosomes were prepared from the serum of patients with or without brain metastasis, using Exoquick (SystemBio). RNAs were prepared from the exosomes, and the level of miR-503 was examined by TaqMan qPCR. (I) Exosomes were prepared from conditioned medium prepared from MCF7-shCTRL, MCF7-shXIST and MCF7-shXIST/Anti-miR503 cells. SIM-A9 cells (50,000 cells) were treated with exosomes (50 μg of particles) for 48 hrs. RNAs were then extracted, and M1 (CD86) and M2 (CD204, ARG1) markers were examined by qRT-PCR. (J) Expressions of M1 (CD74, CD80, CD86) and M2 (CD204, ARG1/2, IL10) markers in SIM-A9 cells with or without expressing miR-503 were measured by qRT-PCR. (K) Western blot analyses for p-STAT3, STAT3, p-p65, and p65 in SIM-A9 cells with or without expressing miR-503 was performed. (L) The expression of PD-L1 in SIM-A9 and human microglial cell (HMC-3) with or without the expression of miR-503 was examined by western blot. (M) Brain metastatic tumor of a breast cancer patient was subjected to immunohistochemical analysis using anti-PD-L1 and anti-IBA-1 antibodies. Scale bar represents 50 μm. (N) SIM-A9 cells with or without miR-503 expression were co-cultured with primary T-cells isolated from mouse spleen for 5 days, and T-cell proliferation was measured by the CFSE assay. The left three panels show FACS analysis. The right panel represents the results of T-cell proliferation.
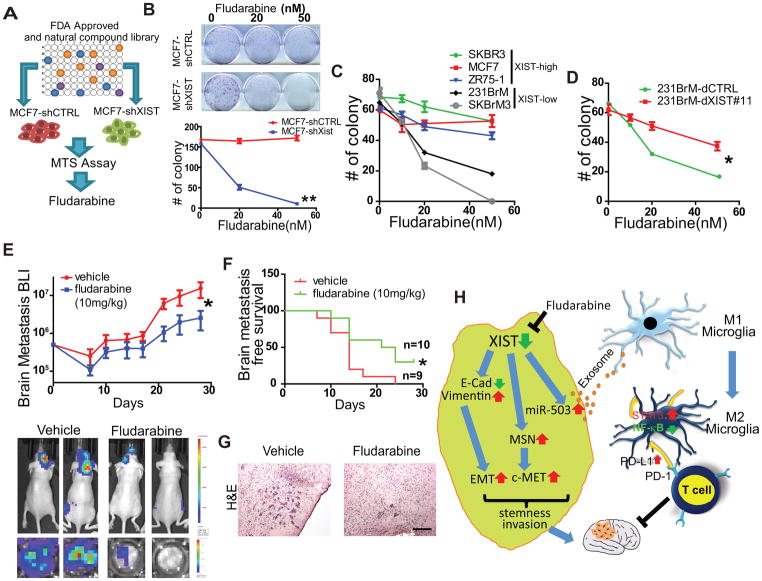
Fludarabine selectively inhibits the growth of XISTlow tumor cells
We have shown that cells with low levels of XIST are highly aggressive and metastasize to the brain. Therefore, identifying a drug that specifically targets these cells would be a promising therapeutic approach to treat brain metastasis. To accomplish this goal, we used a FDA-approved drug library (n = 1250) and a natural compound library (n = 140) to screen compounds that selectively suppress the growth of XIST low cells followed by MTS assay (Fig. 7A). Seven drugs were picked up by the first round of screening (Supplementary Fig. S7A). Among those seven drugs, we found that fludarabine was the most effective drug that kills MCF7-shXIST cells compared to MCF7-shCTRL (Fig. 7B). In addition, fludarabine was found to be highly selective for cells with low levels of XIST expression as it showed selective inhibition of 231BrM and SKBrM3 cells, compared to SKBR3, MCF7, and ZR75-1 cells which have high levels of XIST (Fig. 7C). This inhibitory effect was rescued by restoring the expression of XIST by dCas9 in 231BrM cells (Fig. 7D). This selective cytotoxic effect on XISTlow cells was not observed for cisplatin and paclitaxel, two chemotherapy drugs commonly used for treating breast cancer patients (Supplementary Fig. S7 B). It should be noted that the IC50 for fludarabine on XISTlow cells was around 20 nM, which is more than 10-fold lower than the effective dose for leukemic cells, for which fludarabine is currently in clinical use(35). The in vitro toxicity of fludarabine on primary neuronal cells was minimal (Supplementary Fig. S7C). Because fludarabine is known to be BBB permeable(36), we tested the efficacy of this drug in vivo on tumors with low levels of XIST expression by transplanting MCF7-shXIST cells into nude mice by intracardiac injection followed by administration of fludarabine intraperitoneally (10 mg/kg) every 2 days for 28 days. We found that fludarabine significantly delayed the onset of brain metastasis, and suppressed the growth of tumor cells in the brain (Fig. 7E–G) without notable toxicity (Supplementary Fig. S7D). Although the specific mechanism of action of this drug on cells with low levels of XIST expression is not yet clear, our results suggest a promising utility of this drug for the patients with XISTlow breast cancer cells that spread to the brain.
Fig. 7. Fludarabine selectively suppresses the growth of breast cells with low expression of XIST.
(A) FDA-approved drug library and natural compound library that contain 1,250 and 140 compounds, respectively, were screened by MTS assays in MCF7 cells with or without knockdown of XIST. (B) Clonogenic assay of MCF7-shCTRL cells and MCF7-shXIST cells treated with various doses of fludarabine. (C) Clonogenic assay of cancer cell lines with differential expression of XIST that were treated with fludarabine. (D) Clonogenic assay of 231BrM with or without expressing dCas9 of XIST (E) 5×105 MCF7-shXIST cells were inoculated into nude mice by intracardiac injection followed by treating mice with or without fludarabine (10 mg/kg, every other day by i.p. injection). The tumor growth was monitored by IVIS twice a week (upper panel). Lower panel shows representative IVIS images of whole bodies and ex vivo image of the brains. (F) Kaplan-Meier analysis for brain metastases-free survival of the mice in (E). (G) Representative photos of H&E staining of the brain metastatic lesions of mice treated with or without fludarabine. Scale bar represents 100 μm. (H) The proposed downstream effect of XIST in brain metastasis. Decreased XIST in cancer cells promotes EMT, upregulates c-Met through MSN, and upregulates miR-503 which is secreted through exosome. Exosomal miR-503 activates microglia and triggers M1 to M2 conversion, which suppresses T cell activities. Fludarabine selectively kills XISTlow cancer cells.
DISCUSSION
Brain metastasis is a highly complex and selective process due to the unique microenvironment of this specialized organ. In this study, we identified XIST as a key lncRNA which functions as a metastasis suppressor gene in the brain. Down-regulation of XIST activates three distinct pathways, EMT, MSN/c-Met, and release of exosomal miR-503 (Fig. 7H). Together they promote the expression of metastatic traits in tumor stem cells and trigger the reprogramming of the microenvironment through exosomal-mediated communication. The result of our analysis of the TCGA database indicates that XIST expression is decreased in 48% of basal like breast cancer, while it is down regulated in 28% of Her2+ and 19% of luminal A subtype cancers. It should be noted that most of the brain metastases of breast cancer belong to basal-like and Her2+ subtypes (37). Importantly, XIST was found to be down regulated in 78% of tumor tissues from patients with brain metastases, compared to 32% in patients with bone metastases and 41% with lung metastasis. These observations are consistent with the results of our study and further support our notion that XIST exerts a metastasis suppressive function preferentially in the brain.
XIST silences X chromosome genes in a cis-acting manner, mainly through PRC2 complex-mediated H3K27me3 methylation(38). How this lncRNA functions only on the X chromosome in a cis-acting manner is not well understood. However, when XIST is artificially introduced to autosomes, a similar cis-acting effect of chromosome inactivation was observed, indicating that autosomal chromatin is fully competent to be inactivated by XIST (39). Aberrant expression of XIST has been implicated to inactivate microRNAs by a “sponge” effect and also to dysragulate the AKT signal(40). In this study, we showed that both MSN and miR-503 are up-regulated following knockdown of XIST. Importantly, both genes are encoded by the X chromosome, and hence, they are under the direct epigenetic control of XIST. Therefore, the XIST axis is considered to be a gender-specific oncogenic pathway.
We previously demonstrated that metastatic growth of breast cancer in the brain is strongly associated with the self-renewal abilities of cancer stem cell (41,42). Moreover, the link between the EMT and the properties of CSCs has been well established in breast cancer(43). Therefore, the increased stemness of cells with low XIST expression can in part be attributed to the XIST-mediated EMT. Upon knockdown of XIST, we observed a systematic up-regulation of multiple transcription factors that target the E-box (ZEB1, ZEB2, and TWIST2), as well as components of the Notch and Wnt pathways (HEY1, TCF4, and WNT5A) that are known to be potent EMT promoters(44). However, the up regulation of EMT genes is not directly mediated either by MSN or miR-503 on the X chromosome (data not shown). Therefore, we speculate that the EMT phenotype is caused by a net effect of XIST down regulation of the X chromosome, which is consistent with the observation that XIST is decreased in many of the basal-like breast cancer samples.
We have shown that the loss of XIST up regulates c-Met expression through activation of MSN on the X chromosome. It is noteworthy that high levels of c-Met signaling in pancreatic cancer and glioblastoma have also been reported to be associated with an increased expression of stem cell-related genes(45). Moreover, we previously showed that c-Met signaling specifically promotes brain metastasis by inducing angiogenesis through CXCL1 and IL8 secretion, suggesting that activation of this pathway also modifies the tumor microenvironment(14). Therefore, c-Met signaling is likely to contribute to brain metastasis at multiple-levels that affect the malignancy of tumor cells as well as in the local microenvironment. MSN encodes moesin, which is one of the three components of the ERM complex (ezrin/radixin/moesin). In addition to c-Met internalization, ERM can also serve as a signal transducer which binds to Grb2 and SOS to further amplify c-Met signaling(46). Moreover, perinuclear c-Met has been shown to be able to activate another key oncogenic signaling, STAT3 pathway, by direct phosphorylation and facilitates STAT3 to enter into the nucleus(47).
Successful colonization of brain metastatic cells requires reciprocal communications with microenvironmental cells and local immune cells. We have previously shown that a group of inflammatory cytokines that activate astrocytes and brain endothelial cells are highly secreted by brain metastatic cells(14). In addition to soluble factors, tumor cell-secreted miRNAs in exosomes are considered to be important mediators between the tumor and the host microenvironment(30). Here, we have shown that the loss of XIST enhances the secretion of exosomal miR-503, which is also encoded by the X chromosome, and that exosomal miR-503 is capable of reprogramming microglia from M1 to M2 type by modulating the STAT3 and NF-κB pathways. This M1-M2 conversion is reported to occur through activation of signaling pathways by either direct interaction with tumor cells or through soluble factors(48). However, when exosomes were depleted in our M1-M2 conversion assay system, this phenotypic change was drastically diminished, indicating that exosomal miR-503, rather than a soluble factor, plays a major role in the microglial M1-M2 transition in XIST-mediated brain metastasis.
Normal brain is considered to be “immune-privileged,” and normally cytotoxic T cells are unable to penetrate the BBB. However, recent mounting evidence has indicated that both T and B cells are frequently found in brain tumors, including glioma and metastatic tumors(13). The amount of T-cells and the level of expression of immune check point genes in the brain tumor areas have been associated with prognosis in patients with brain metastases(13). We have shown here that immune suppression is one of the mechanisms by which M2 microglia promote tumor progression, and this effect is achieved by the up-regulation of PD-L1 in the microglia. Although several studies have shown that many growth factors that directly promote the survival and migration of tumor cells are also secreted from M2 microglia, it is likely that both mechanisms contribute to tumor growth in the brain and that these two mechanisms are not mutually exclusive. Nevertheless, our data provide a promising clue of how to exploit immunotherapy for the treatment of brain metastasis. A recent study has shown that there is a synergetic effect between immune checkpoint inhibitors and an oncolytic virus which increases intratumoral M1-like macrophages in a mouse GBM model(49), suggesting a potential immune therapy to treat brain metastasis could be developed by boosting T cell activities by pharmacologically manipulating the population of these microglial subtypes in brain metastatic sites.
Several groups, including ours, have shown that there is a strong correlation between the amount of a particular exosomal miRNA in circulation and the prognosis of cancer patients(50). In this study, we have demonstrated that exosomal miR-503 levels are positively correlated with brain metastasis status. Exosomes secreted from cancer cells can be taken up by non-tumor cells in metastatic niches even before the arrival of tumor cells(51). Although the effect of exosomes is likely to be maximum at the local site, it is possible that exosomal miR-503, shed by the primary tumor, can also be taken up by microglia to generate a pre-metastatic niche at an early stage of tumor progression. Furthermore, exosomal miR-503 present in patient serum may serve as a promising biomarker, with the level of miR-503 in circulation helping to predict the risk of future brain metastasis.
We have shown that fludarabine acts as a synthetic lethal drug in tumor cells with low XIST levels. Importantly, the IC50 of fludarabine against tumor cells having low XIST levels is at least ten times lower than the clinical dose used in treating leukemia, and furthermore fludarabine showed significant efficacy in our animal model of brain metastasis without any obvious adverse effect. Therefore, it is possible that the synthetic lethality of fludarabine to XISTlow cells, which are mainly triple-negative, is in part due to a synergistic effect with ROS production. Fludarabine is a BBB-permeable compound whose neurotoxicity is tolerable even at a relatively high dose(52) and it is already approved by the FDA. Therefore, fludarabine could be used as a potential therapeutic agent for the treatment of breast cancer patients with brain metastases that have low levels of XIST expression. In summary, we have shown that the lncRNA, XIST, plays a critical role in brain metastasis in breast cancer in a gender specific manner by affecting both tumor cells and the tumor microenvironment, and that the XIST-mediated pathway may serve as an effective target for the treatment of brain metastasis.
Supplementary Material
SIGNIFICANCE.
Findings describe mechanisms of how loss of the lncRNA XIST promotes brain metastasis in breast cancer and identify fludarabine as a potential therapeutic agent that specifically eliminates XISTlow tumor cells in the brain.
Acknowledgments
We thank Dr. Joan Massagué for providing the brain metastatic cell lines. The XISTlox/lox mouse is kind gift from Dr. Rudolf Jaenisch. This work was supported by NIH grant R01CA173499, R01CA185650 and R01CA205067 (to K. Watabe), a pilot grant from Comprehensive Cancer Center of Wake Forest University (to F. Xing). Y. Liu is supported by F31CA200286. The Tumor Tissue and Pathology Shared Resources, and Biostatistics/Bioinformatics Shared Resource are supported by the Comprehensive Cancer Center of Wake Forest University NCI, National Institutes of Health Grant (P30CA012197).
References
- 1.Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 2.Lowery FJ, Yu D. Brain metastasis: Unique challenges and open opportunities. Biochim Biophys Acta. 2017;1867(1):49–57. doi: 10.1016/j.bbcan.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–84. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 4.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533(7604):493–98. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73(4):1434–44. doi: 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing F, Sharma S, Liu Y, Mo YY, Wu K, Zhang YY, et al. miR-509 suppresses brain metastasis of breast cancer cells by modulating RhoC and TNF-alpha. Oncogene. 2015;34(37):4890–900. doi: 10.1038/onc.2014.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Lull ME, Block ML. Microglial activation and chronic neurodegeneration. Neurotherapeutics. 2010;7(4):354–65. doi: 10.1016/j.nurt.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat Med. 2017;23(9):1018–27. doi: 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- 12.Hu X, Leak RK, Shi Y, Suenaga J, Gao Y, Zheng P, et al. Microglial and macrophage polarization-new prospects for brain repair. Nat Rev Neurol. 2015;11(1):56–64. doi: 10.1038/nrneurol.2014.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berghoff AS, Fuchs E, Ricken G, Mlecnik B, Bindea G, Spanberger T, et al. Density of tumor-infiltrating lymphocytes correlates with extent of brain edema and overall survival time in patients with brain metastases. Oncoimmunology. 2016;5(1):e1057388. doi: 10.1080/2162402X.2015.1057388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing F, Liu Y, Sharma S, Wu K, Chan MD, Lo HW, et al. Activation of the c-Met Pathway Mobilizes an Inflammatory Network in the Brain Microenvironment to Promote Brain Metastasis of Breast Cancer. Cancer Res. 2016;76(17):4970–80. doi: 10.1158/0008-5472.CAN-15-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandyopadhyay S, Pai SK, Gross SC, Hirota S, Hosobe S, Miura K, et al. The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer Res. 2003;63(8):1731–6. [PubMed] [Google Scholar]
- 17.Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22(4):323–4. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 18.Plante I, Stewart MK, Laird DW. Evaluation of mammary gland development and function in mouse models. J Vis Exp. 2011;(53) doi: 10.3791/2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6:7029. doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaligne R, Popova T, Mendoza-Parra MA, Saleem MA, Gentien D, Ban K, et al. The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 2015;25(4):488–503. doi: 10.1101/gr.185926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22(2):246–58. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–78. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 24.Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8(4):212. doi: 10.1186/bcr1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 26.Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 2002;16(23):3074–86. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11(12):834–48. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 28.He BP, Wang JJ, Zhang X, Wu Y, Wang M, Bay BH, et al. Differential reactions of microglia to brain metastasis of lung cancer. Mol Med. 2006;12(7–8):161–70. doi: 10.2119/2006-00033.He. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sleeman JP. The metastatic niche and stromal progression. Cancer Metastasis Rev. 2012;31(3–4):429–40. doi: 10.1007/s10555-012-9373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–7. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prima V, Kaliberova LN, Kaliberov S, Curiel DT, Kusmartsev S. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc Natl Acad Sci U S A. 2017;114(5):1117–22. doi: 10.1073/pnas.1612920114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottlieb CE, Mills AM, Cross JV, Ring KL. Tumor-associated macrophage expression of PD-L1 in implants of high grade serous ovarian carcinoma: A comparison of matched primary and metastatic tumors. Gynecol Oncol. 2017;144(3):607–12. doi: 10.1016/j.ygyno.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 35.Plunkett W, Gandhi V, Huang P, Robertson LE, Yang LY, Gregoire V, et al. Fludarabine: pharmacokinetics, mechanisms of action, and rationales for combination therapies. Semin Oncol. 1993;20(5 Suppl 7):2–12. [PubMed] [Google Scholar]
- 36.Jensen K, Johnson LA, Jacobson PA, Kachler S, Kirstein MN, Lamba J, et al. Cytotoxic purine nucleoside analogues bind to A1, A2A, and A3 adenosine receptors. Naunyn Schmiedebergs Arch Pharmacol. 2012;385(5):519–25. doi: 10.1007/s00210-011-0719-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niwinska A, Murawska M, Pogoda K. Breast cancer brain metastases: differences in survival depending on biological subtype, RPA RTOG prognostic class and systemic treatment after whole-brain radiotherapy (WBRT) Ann Oncol. 2010;21(5):942–8. doi: 10.1093/annonc/mdp407. [DOI] [PubMed] [Google Scholar]
- 38.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19(4):429–42. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500(7462):296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35(1):142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xing F, Kobayashi A, Okuda H, Watabe M, Pai SK, Pandey PR, et al. Reactive astrocytes promote the metastatic growth of breast cancer stem-like cells by activating Notch signalling in brain. EMBO Mol Med. 2013;5(3):384–96. doi: 10.1002/emmm.201201623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuda H, Kobayashi A, Xia B, Watabe M, Pai SK, Hirota S, et al. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem-like cells with macrophages and stromal cells. Cancer Res. 2012;72(2):537–47. doi: 10.1158/0008-5472.CAN-11-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22(5–6):396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Wu JJ, Hynes M, Dosch J, Sarkar B, Welling TH, et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141(6):2218–27 e5. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Sperka T, Geissler KJ, Merkel U, Scholl I, Rubio I, Herrlich P, et al. Activation of Ras requires the ERM-dependent link of actin to the plasma membrane. PLoS ONE. 2011;6(11):e27511. doi: 10.1371/journal.pone.0027511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kermorgant S, Parker PJ. Receptor trafficking controls weak signal delivery: a strategy used by c-Met for STAT3 nuclear accumulation. J Cell Biol. 2008;182(5):855–63. doi: 10.1083/jcb.200806076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Y, Le W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol Neurobiol. 2016;53(2):1181–94. doi: 10.1007/s12035-014-9070-5. [DOI] [PubMed] [Google Scholar]
- 49.Saha D, Martuza RL, Rabkin SD. Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer Cell. 2017;32(2):253–67 e5. doi: 10.1016/j.ccell.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, Ma YY, Wang J, Zeng XF, Li R, Kang W, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016;9:139–48. doi: 10.2147/OTT.S95565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, et al. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell. 2016;30(2):243–56. doi: 10.1016/j.ccell.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 52.Lee MS, McKinney AM, Brace JR, Santacruz K. Clinical and imaging features of fludarabine neurotoxicity. J Neuroophthalmol. 2010;30(1):37–41. doi: 10.1097/WNO.0b013e3181ce8087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.