Abstract
Drug development for front-line treatment of epithelial ovarian cancer (EOC) has been stagnant for almost three decades. Traditional cell culture methods for primary drug screening do not always accurately reflect clinical disease. To overcome this barrier, we grew a panel of EOC cell lines in three-dimensional (3D) cell cultures to form multicellular tumor spheroids (MCTS). We characterized these MCTS for molecular and cellular features of EOC and performed a comparative screen with cells grown using two-dimensional (2D) cell culture to identify previously unappreciated anti-cancer drugs. MCTS exhibited greater resistance to chemotherapeutic agents, showed signs of senescence and hypoxia, and expressed a number of stem cell-associated transcripts including ALDH1A and CD133. Using a library of clinically repurposed drugs, we identified candidates with preferential activity in MCTS over 2D cultured cells. One of the lead compounds, the dual COX/LOX inhibitor licofelone, reversed the stem-like properties of ovarian MCTS. Licofelone also synergized with paclitaxel in ovarian MCTS models and in a patient-derived tumor xenograft (PDX) model. Importantly, the combination of licofelone with paclitaxel prolonged the median survival of mice (>141 days) relative to paclitaxel (115 days), licofelone (37 days), or vehicle (30 days). Increased efficacy was confirmed by Mantel-Haenszel hazard ratio compared to vehicle (HR=0.037) and paclitaxel (HR=0.017). These results identify for the first time an unappreciated, anti-inflammatory drug that can reverse chemotherapeutic resistance in ovarian cancer, highlighting the need to clinically evaluate licofelone in combination with front-line chemotherapy in primary and chemotherapy-refractory EOC.
INTRODUCTION
Epithelial ovarian cancer (EOC) is the most common reproductive cancer. EOC is the 9th most prevalent cancer and the 5th most common cause of cancer-related death in women (1). The average 5- and 10-year survival rates for women diagnosed with EOC are ~44% and ~30%, respectively (2). These poor outcomes are attributed to many factors including late stage diagnosis, disease heterogeneity, and drug resistance. Front-line therapy (surgical resection followed by taxane and platinum-based chemotherapy) shows success in most patients by eliminating the detectible disease. However, many patients will ultimately develop recurrent, chemotherapy resistant tumors, and the majority of patients will succumb to the disease within one year (3). Efforts to add a third cytotoxic agent in clinical trials have shown no improvement in survival (4). There continues to be an urgent need to develop new drugs that can target mechanisms associated with resistance to chemotherapy and disease recurrence.
Recently it has become clear, stem cell properties, or “stemness”, can play a critical role in both progression and development of drug resistance in many types of cancers. Two common stem cell factors, CD133 and ALDH1A, are of interest for improving preclinical drug development models. EOC cells with expression of both CD133 and ALDH1A have been identified (5–7). CD133, also known as promomin-1, is a transmembrane glycoprotein encoded on the PROM1 gene. Aldehyde dehydrogenase-1A1 (ALDH1A) is a member of the 17 different isotypes of aldehyde dehydrogenase enzymes. CD133 expression in cell culture models, animal models, and patient tumors is associated with enhanced tumorigenicity, drug resistance, and disease recurrence (8–10). ALDH1A expression has a negative correlation to progression-free survival (PFS) in EOC patients and knockdown of ALDH1A restored chemo sensitivity in vitro (11). Co-expression of CD133 and ALDH1A subpopulations in EOC patients are associated with both decreased time-to-recurrence and patient survival (12). Front line drug screening models, that more accurately represent stem-like properties in vitro, could be useful in developing more efficacious drugs to treat EOC.
In this study, we used a model of multicellular tumor spheroids (MCTS) using three-dimensional (3D) cell culture. We observed induction of common cellular features associated with drug resistance (cellular senescence, hypoxia, and stem-like properties) in EOC cells grown in 3D. MCTS formation has been shown to induce stem like properties, including CD133 and ALDH1A expression, across many different cancer cell lines (13–15). Given these observations, the use of MCTS drug screening has been proposed to better predict patient drug response (16). Therefore, the focus of this study was to i) exploit MCTS models for drug screening in order to identify repurposed drugs with previously unappreciated anti-tumor activity in ovarian cancer, and ii) determine if viable drug hits could synergize with frontline therapies to improve response and/or overcome acquired resistance. Using various ovarian cancer MCTS models as screening tools, we identified drugs with preferential anti-proliferative/cell killing activity in spheroids compared to 2D cultures. We show that one of the top candidate drugs, licofelone, reverses stem-like properties of MCTS and enhances paclitaxel activity in both EOC cell line MCTS and EOC patient derived xenografts.
MATERIALS and METHODS
Cell culture
EOC and HIO cell lines used in these studies were previously described in detail (17–20). Cell lines used in this study were kindly provided by Dr. Thomas Hamilton (A1847, A2780, OVCAR3, OVCAR4, OVCAR5, OVCAR8, OVCAR10, PEO1, SKOV3) or were derived by Dr. Andrew Godwin (C30, CP70, UPN275) while at the Fox Chase Cancer Center (Philadelphia, PA). Cell lines were authenticated by the Clinical and Molecular Oncology Laboratory at KUMC by multiplex short tandem repeat (STR) using the Promega PowerPlex 16 System used for human identity testing run on an Applied Biosystems instrument. All of the EOC cell lines were maintained in normal growth media consisting of RPMI 1640 media supplemented with 10% fetal bovine serum (FBS) (vol/vol), insulin (7.5 µg/mL), penicillin (100 U/mL), and streptomycin (100 µg/mL) at 37°C in a humidified atmosphere with 5% CO2. HIO cells were cultured in medium 199 and MCDB 105 (1:1) supplemented with 15% FBS (vol/vol), insulin (0.25 U/mL), L-glutamine (2 mM), penicillin (100 U/mL), and streptomycin (100 µg/mL). Cells were passaged using 0.25% Trypsin with 2.21 mM EDTA and 1X sodium bicarbonate (Corning) for 5 minutes. Cell lines were kept in passage for no longer than two months. Mycoplamsa testing was performed by either Hoechst staining or the LookOut Myco PCR Test (Sigma).
Spheroids were formed using the liquid-overlay method with agarose coated 96-well plates, modified from a previous publication (21). Briefly, 1.5% agarose was dissolved in RMPI 1640 media and then used to coat the bottom of 96-well flat bottom plates with 50 µL of the agarose solution. Agarose was allowed to solidify for a minimum of 30 minutes and then 3,000 cells were plated in 50 µL using standard cell culture media. These plated cells were then allowed to form spheroids undisrupted for four days. On day 4 spheroids were visually inspected and 50 µL of fresh, standard media was supplemented to each well.
For secondary assays, spheroids were removed from each well using wide bore pipette tips and pooled together from 96 wells for each single treatment. Spheroids were centrifuged at 2,000 RPM for 5 minutes at 4°C, the media was removed, and washed with sterile PBS. Following another spin and removal of PBS, spheroids were dissociated in a 1:1 (vol/vol) combination of trypsin and Accutase (10 mL) for 30 minutes at 37°C, with mild shaking every 3 to 10 minutes. Cells were then rinsed, spun down, suspended, and plated overnight on a 10 cm cell culture dish. Following overnight seeding, viable cells were counted for subsequent assays following standard protocols.
Cell cycle analysis
Cells were stained using propidium iodide (Guava Cell Cycle reagent, EMD Millipore) following the manufacturer’s established protocol. The cell cycle assays were performed three independent times with two technical replicates for each. A Guava Easycyte HT instrument (EMD Millipore) was used to measure the changes in cell cycle.
Immunofluorescence
Sections (4 µm) from formalin-fixed, paraffin-embedded (FFPE) MCTS were made for immunofluorescence (IF) staining. After deparaffinization and rehydration, tissue sections were treated using citrate buffer (pH 6.0) for antigen retrieval. Sections were blocked with 1% normal goat serum for 1 h then incubated with Ki67 rabbit mAB (1:1,000, Cell Signaling Tech) overnight at 4 °C. Cells were washed with PBS and then incubated with secondary antibody (goat anti-rabbit IgG, DyLight 594, Thermo Scientific) for 1 h at room temperature. The coverslips containing cells were mounted onto glass slides in VECTASHIELD® mounting medium with DAPI (Vector Labs, Inc.). Pimonidazole staining was performed following the manufacture’s protocol (Hypoxyprobe). Fluorescence digital images were captured using a Nikon Eclipse 80i microscope attached with a Nikon Q-imaging camera adaptor. MetaMorph Image Analysis software (version 7.7.0.0) was used to acquire and analyze images.
Immunohistochemistry
Immunohistochemical staining was performed using the IntelliPATH FLX Automated Stainer at room temperature. Epitomics (Abcam) was used for Immunohistochemical staining according to the following procedure. Four micron paraffin sections were mounted on Superfrost (Fisher) slides and baked for 60 minutes at 60° C then deparaffinized. Epitope retrieval was performed in Biocare Decloaking Chamber, under pressure for 5 min, using pH 6.0 Citrate buffer followed by a 10 minute cool down period. Endogenous peroxidase was blocked with 3% H2O2 for 10 minutes followed by incubation with Ki-67 (M7240, ThemoFisher) (1:200) primary antibody for 30 min., followed by Envision+Mouse, Dako (Carpinteria, CA) for 30 minutes and DAB+ chromogen (Dako, Carpinteria, CA.) for 5 minutes. After washing, a light hematoxylin counterstain was performed, following which the slides were dehydrated, cleared, and mounted using permanent mounting media. Images were captured using a Nikon Eclipse 80i microscope attached with a Nikon Q-imaging camera adaptor and analysis was performed with HALO 2.0 next generation digital pathology (Indica Labs).
Immunoblot analysis
All lysates were extracted using RIPA buffer supplemented with protease inhibitors (Roche Molecular Biochemicals) and phosphatase inhibitors (Fisher Scientific). Protein concentration was measured using the DC Protein Assay (Bio Rad) following manufacture’s protocol. 30 µg of whole-cell extract was electrophoresed on a 4–20% precast gradient polyacrylamide gel (Bio-Rad) and transferred onto nitrocellulose membranes using the Trans-Blot Turbo (Bio-Rad). After blocking with 5% skim milk (Difco), membranes were incubated overnight at 4 °C with primary antibodies, HIF1-α (1:1,000, Cell Signaling), JMJD1A (1:1,000 Cell Signaling), and PARP (1:500, Cell Signaling). HRP-conjugated secondary antibody (1:10,000) was incubated at room temperature; development was carried out using chemiluminescence substrate (Pierce). To measure mitochondrial complexes, the OXPHOS array, the Total OXPHOS Rodent WB Antibody Cocktail (Abcam, Cambridge, United Kingdom) was used following manufacture’s recommendations. Lysates were heated for 5 minutes to 50 °C and run on the gel as described above. For transfer, a high pH (11) CAPS transfer buffer was used for 2 hours at 100 mA onto a pvdf membrane. Antibody treatment was performed as described above using a 1:500 dilution. Pixel densities of blot images were calculated using Image-J software (NIH). Changes in protein levels were normalized to loading controls and expressed as fold change relative to treatment controls.
RT-PCR and RNASeq analysis
RNA was isolated using Trizol and Phase Lock Gel Heavy tubes (5 Prime) followed by RNeasy Mini Kits (Qiagen) following manufacture’s protocols. RNA quality (A260:A280 ratio > 1.8) and quantity was assessed using the Infinite 200Pro (Tecan). For TaqMan qRT-PCR, 1 µg of RNA of subjected to reverse transcription using SuperScript III (ThermoFisher) following manufacture’s protocol. For amplification, 10 µL of TaqMan Gene Expression Master Mix (2x) (Applied Biosystems) was combined with 2 µL of diluted cDNA (1:2), 7 µL of sterile water, and 1 µL of TaqMan primers (PROM1, NANOG, POU5F1, SOX2, CD44, KLF4, ALDH1A, LOX, JMJD1A, VEGF, PPIA, BACTIN, and GAPDH). Amplification was performed on the Bio-Rad CFX96 (BioRad). The fluorescence threshold value was calculated using the CFX96 real-time system software Ver 2.1 (BioRad). The relative change in mRNA levels was measured by the delta-delta CT method normalizing to the geometric mean of three housekeeping genes (PPIA, BACTIN, and GAPDH).
For high-throughput PCR analyses, the Fluidigm BioMark™ HD system was used to run 96x96 dynamic arrays. Basal mRNA expression levels were measured in spheroids following 72 h treatment with vehicle, 1 µM paclitaxel, 20 µM licofelone, or 1 µM paclitaxel plus 20 µM licofelone. Total RNA was extracted as described above and three biological replicates were pooled for analysis. 500 ng of mRNA were reverse transcribed and the resulting cDNA was pre-amplified using a multiplexed specific target amplification protocol with gene specific TaqMan assays (Applied Biosystems). The Qiagen Glucose Metabolism RT2 profiler PCR array was used to measure gene transcript expression. All subsequent assays were performed following the manufacture’s recommended protocols. Absolute Ct values were determined using amplification curves and equality of amplifiable mRNA was assessed by comparing geometric means of three internal reference controls.
For transcriptome sequencing (RNA-seq), RNA was isolated from 2D cultures at 80% confluency, from 3D cultures at 7 days of spheroid growth from twelve ovarian cancer cell lines (A1847, SKOV3, OVCAR3, OVCAR8, C30, PEO1, OVCAR5, OVCAR10, OVCAR4, UPN275, A2780, and CP70). RNA was prepared for paired end sequencing on a HiSeq 2500 using a stranded library prep kit (Illumina). Initial analyses were prepared using RSEM expected gene counts. First, data were filtered to remove non/low-expressed genes. This resulted in a total of ~14,000 genes examined for differential expression between the different 2D and 3D cultures. Next, normalization factors were calculated to scale the library sizes followed by estimation of tag wise negative binomial dispersion values. The EdgeR Bioconductor package was used to conduct a differential expression analysis, comparing gene expression between 3D cultures and 2D cultures. To identify potential pathway regulators, IPA Pathway Analysis (QIAGEN) was applied to the significantly overexpressed genes in the 3D compared to 2D cultures (FDR < 0.05 and fold-change > 1.5), and the top 5 regulators based on p value were reported. Data were made publicly available through the Sequence Read Archive by uploading fasq files for all cell lines sequenced in 2D and 3D cultures (PRJNA472611).
Drug screening
Four EOC cell lines (A1847, OVCAR3, OVCAR4, and OVCAR8) were selected for drug screening. 2D cultures were plated overnight before drug treatment, while spheroids were grown for 4 days before the addition of drug. A custom 304 drug library was purchased from the 10 mM, FDA-Approved Library at Selleckchem. Drugs were diluted into 1 mM stock plates in DMSO and then twice diluted in media by the Hamilton Nimbus 96 with the final dilution of 5 µl drugs into 95 µl cell media, for a final concentration of 10 µM drug. After 72 h of drug exposure, cell viability was measured using CellTiter-Glo incubated 1:1 to culture media for 1 h at 37°C. Relative viability was established for the control (DMSO) treated cells and directly compared to the 2D and 3D cultures for each drug treatment. Secondary screening was performed at 25 µM following the same plating protocol across all 3D hits and select hits from 2D culture and hits from 2D and 3D culture. For all dosage response and function assays, licofelone powder was purchased from Santa Cruz Biotechnology and glafenine hydrochloride powder was purchased from Sigma Aldrich Inc.
DNA mutation analysis
To determine if any specific gene mutation(s) correlated with response to drug treatments, we used an in silico approach to reanalyze cell line DNA sequencing data available from our laboratory. Specifically, sequence variants found using the TruSeq Amplicon Cancer Panel (Illumina) (22) were classified as Tier 1, Tier 2, or Tier 3 using QIAGEN Clinical Insight (QCI™). Tier 1 and 2 mutants were compared to the IC50 for either licofelone or glafenine. Mutational analysis was performed through the CLIA approved and CAP accredited Clinical Molecular Oncology Laboratory (CMOL) at KUMC.
Drug combination assays
Drug combination studies were performed using the Combination Index (CI) method described by Chou and Talalay (23). Spheroids from A1847 and OVCAR8 cell lines were grown for 4 days before addition of either paclitaxel (1 µM or 250 nM) or licofelone (10 µM or 20 µM) and then 3 days later serial dilutions of licofelone were added on top of the paclitaxel treated cells or serial dilutions of paclitaxel were added on top of the licofelone treated cells. Assays were performed as biological triplicate using triplicate wells within each experiment. Cell viability was evaluated using CellTiter-Glo and the viability data were then analyzed using CalcuSyn (version 2.1, BioSoft, UK) to calculate the CI at each molar ratio evaluated. Drug combinations which yielded CI values less than 1 were considered to be synergistic.
Ovarian Cancer PDX Model
Preclinical efficacy studies were performed using patient-derived xenograft (PDX) models for EOC. De-identified fresh clinical samples were provided from the KU Cancer Center’s Biospecimen Repository Core Facility (BRCF) along with detailed clinical outcomes and phenotypes. Tissue specimens were obtained from individuals enrolled under the repository’s IRB approved protocol (HSC #5929) and following U.S. Common Rule. Once the patient provides written, informed consent in accordance with our IRB protocol, discard tumor tissue is collected in the OR by BRCF staff and placed in McCoy’s media without FBS. The sample is de-identified to the user and transported directly to the laboratory for processing and implanting. For these in vivo drug efficacy experiments, tumor from a treatment naïve patient diagnosed with a high grade serous ovarian cancer (Stage IIIc) was provided. The tissue was minced into slurry and added to a syringe in a 1:1 volume with McCoy’s media. Four to six-week-old NOD scid gamma (NSG) mice were anesthetized using gaseous isoflurane and tumor slurry was injected intraperitoneally. Once the implanted tumor grew in the recipient mice it developed bloody ascites that was collected, cleared of red blood cells using a Ficol gradient and passaged into additional NSG mice. The second generation PDX mice are evaluated weekly for overall health and body weight, once abdominal distention was pronounced, ascites was collected for further studies.
For drug efficacy studies, 21 days after intraperitoneal inoculation with tumor cells from ascites, the mice were randomized into four different treatment groups (n=7) of either i.p. delivery of vehicle, paclitaxel (12 mg/kg), licofelone (50 mg/kg), or paclitaxel plus licofelone. Mice were monitored for disease progression (development of ascites fluid) and overall survival. Median survival was compared to each group and survival curves were compared using the Mantel-Haenszel hazard ratio (HR).
Statistics
All statistics (t-test and Two-way ANOVA) were performed using GraphPad Prism 6 (Ver 6.02, GraphPad Software Inc).
RESULTS
MCTS Formation Induces Cellular Changes Associated with Drug Resistance in Solid Tumors
A three dimensional cell culture of EOC cell lines was adapted from a previous study, using the liquid overlay method on agarose coated 96-well plates (21). Tight spheroids were formed after four days in six ovarian tumor-derived cell lines (Figure 1A, Supplemental Figure 1), while three of the cell lines formed loose aggregates (green scale bar) (Supplemental Figure 1). Compact spheroids, which remained adherent following pipetting, were selected for phenotypic analysis.
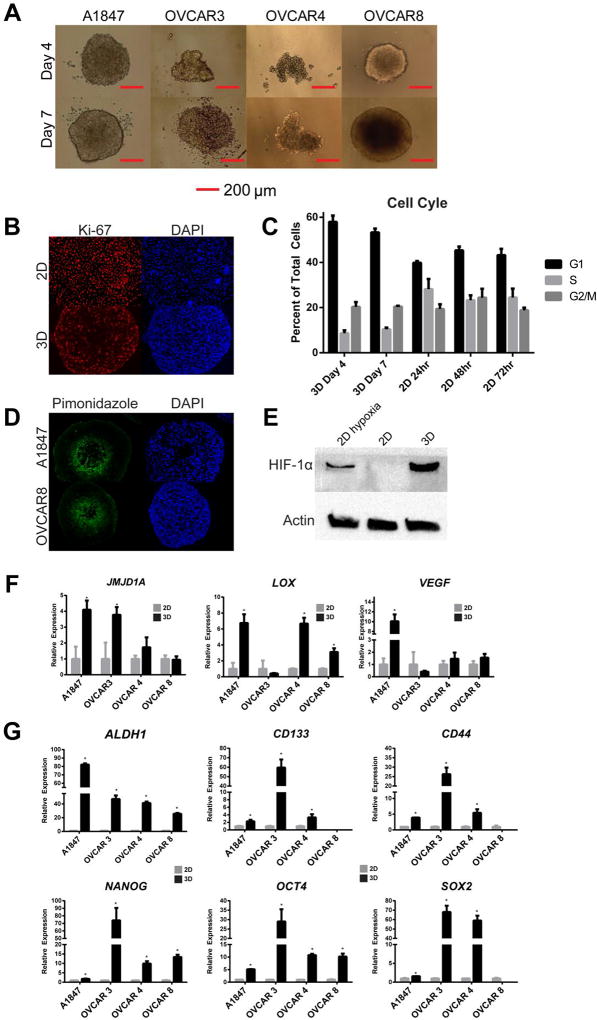
Figure 1. MCTS formation induces senescence, hypoxia and stemness in ovarian cancer cells.
(A) Light microscopy images of MCTS of EOC cell lines. (B) Immunofluorescence staining of Ki-67 in 2D A1847 cells and in a cross-section of MCTS. (C) Cell cycle analysis reveals an increase in G1 cell populations at both day 4 and day 7 in A1847-derived MCTS, compared to 2D cells. (D) Piminidozole staining in A1847 and OVCAR8 cells indicates areas of cellular hypoxia in spheroid cores at day 4. Cell nuclei were labeled with DAPI. (E) HIF-1α protein is stabilized in A1847 2D cultures under hypoxia (0.5 % oxygen) and in A1847 MCTS under atmospheric (normoxic) oxygen but not in A1847 2D cultures. (F) Expression of hypoxia related transcripts were increased in ovarian MCTS compared to parental 2D cultures under normoxia after 7 days (n=3,* = p<0.05, t-test). (G) Several stem-like genes were upregulated in in day 7 MCTS compared to parental 2D cultures (* = p<0.05, t-test). All bar graphs are represented as mean +/− SD, n=3.
Cell proliferation in MCTS was measured using immunofluorescence (IF) staining and cell cycle analysis. Staining for the proliferation marker Ki-67 showed a gradient of positive cells on the spheroid surface and Ki-67 negative cells in the spheroid center (Figure 1B). However, near ubiquitous positive staining of Ki-67 was observed in 2D cultures (Figure 1B). Cell cycle analysis showed an increased accumulation of cells in G1 phase (58% at day 4 and 53% at day 7) as compared to cells in 2D cultures (40%, 45%, and 34% at 24 h, 48 h and 72 h, respectively) (Figure 1C). In the tightly formed MCTS, the smaller ones, e.g., formed using OVCAR3 and OVCAR4 cells, still showed a similar percentage of Ki-67 positive cells (27% and 37%, respectively) to that of the larger spheroids A1847 (41%) and OVCAR8 (36%) indicating that changes in spheroid size is independent of the percent of proliferative cells across the cell line panel (Supplemental Figure 2).
Cellular hypoxia was detected using pimonidazole staining in spheroid sections. Pimonidazole staining was positive in the spheroid center at day 4 (Figure 1D). Using Western blot analysis, HIF-1α is stabilized in A1847 spheroids at day 4 under atmospheric oxygen levels and 2D cultures under hypoxic conditions, but not in 2D cultures under normoxic conditions (Figure 1E). Gene expression analysis of hypoxia regulated genes (JMJD1A, LOX, VEGF) using qRT-PCR was performed across four EOC cell lines (A1847, OVCAR3, OVCAR4 and OVCAR8) when grown under 2D and 3D culture conditions. Significant increases (p< 0.05, n=3, t-test) of the hypoxia-related genes were observed in the 3D cultured cells relative to the 2D cultured cells (Figure 1F).
In addition, expression of stem cell genes associated with drug resistance in EOC (ALDH1A, CD133, CD44) and traditional stem cell markers (NANOG, OCT4, SOX2) were measured in tumor cells from both 2D and 3D cultures. The expression of ALHD1A, NANOG, and OCT4 was significantly increased (p< 0.05, n=3, t-test) in 3D cultures compared to 2D cultures across all four ovarian cancer cell lines (Figure 1G). The other transcripts (CD133, CD44, and SOX2) were significantly increased (p< 0.05, n=3, t-test) in A1847, OVCAR3, and OVCAR4 cell lines (Figure 1G). None of these transcripts were significantly increased in 3D culture of immortalized, non-tumorigenic human ovarian surface epithelial cell line, HIO107, indicating the drug induction of stem-like properties may be specific to tumorigenic cells in 3D cultures (Supplemental Figure 3A-C). Interestingly, the protein expression of JMJD1A, a known hypoxia response element that regulates stemness in cancer (24), is overexpressed in the spheroids compared to 2D cells (Supplemental Figure 4). Taken together, these data suggest there is an increase in multiple cellular mechanisms associated with clinical drug resistance in MCTS when compared to 2D cultures in EOC cell lines.
Ovarian MCTS Paclitaxel Resistance is Associated with Increased Stemness
To measure paclitaxel response, cells grown in 2D were plated overnight before drug addition, whereas ovarian cancer cell-derived spheroids were allowed to develop for four days before drug treatment (Figure 2A). MCTS showed a robust decrease in paclitaxel response compared to 2D cultures (Figure 2B). Consequently, Ki-67 staining revealed a decrease in Ki-67 positive cells compared to control following paclitaxel treatment (41.09 % vs 23.72 %) (Figure 2C). Using qRT-PCR, we found multiple stem cell markers showed a significant, robust increase in ovarian MCTS following 1 µM paclitaxel treatment (Figure 2D, *=p< 0.05, t-test). Interestingly, in 2D cultured cells, small molecule drugs, such as dasatinib, nilotinib, ganestespib, with robust in vitro activity, but limited clinical activity, also induced extensive expression of stem-like genes (red bars) over typical cancer-associated genes (black bars) following drug treatment (Supplemental Figure 5A-C).
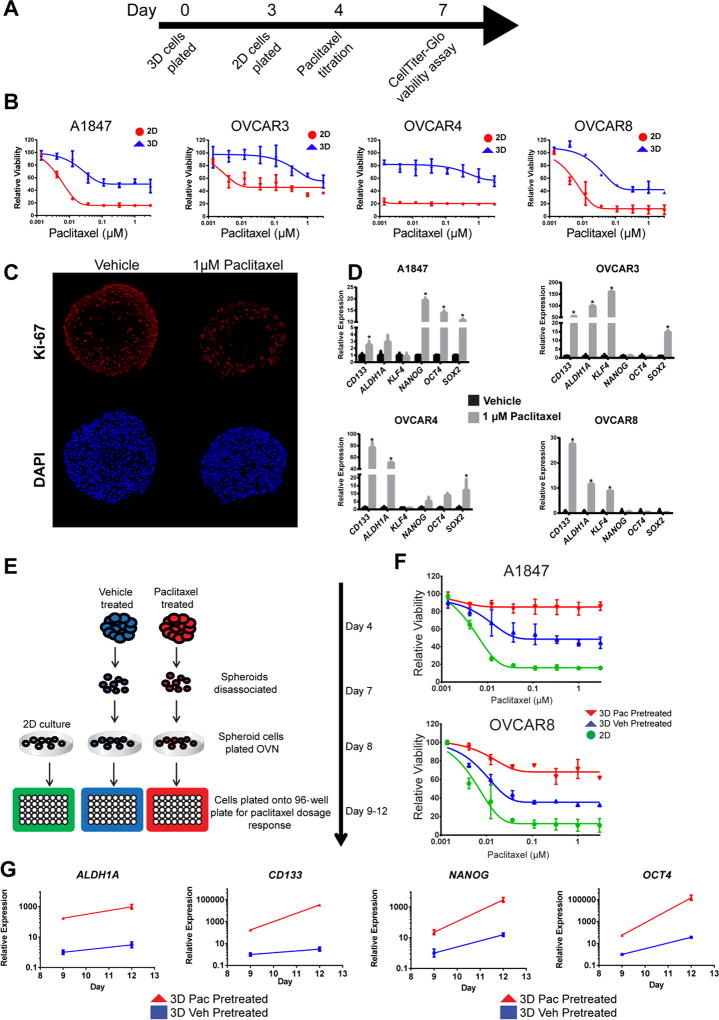
Figure 2. Paclitaxel resistance is associated with increased stem-like properties in ovarian cancer MCTS.
(A) Graphical representation of 2D and 3D culture growth for paclitaxel treatment. (B) Ovarian MCTS showed a robust decrease in paclitaxel sensitivity compared to 2D cultures. (C) Immunofluorescence detection of Ki-67, and DAPI, reveals a decrease in Ki-67 expression following paclitaxel treatment in MCTS. (D) Gene expression was increased following 1 µM paclitaxel treatment in MCTS compared to 2D cultures measured using qRT-PCR (n=3) (*= p< 0.05, t-test). (E) Graphic representation of spheroid disassociation experiments. (F) Cells grown in 2D culture derived from vehicle (blue) or paclitaxel (red) pretreated spheroids have reduced paclitaxel response compared to standard 2D cultured cells (green). (G) Transcript expression of stem-like genes is maintained in 2D cultures derived from vehicle (blue) or paclitaxel (red) pretreated spheroids for three days. All data are represented as mean +/− SD, n=3.
To test if phenotypic changes in MCTS are contributing to decreased paclitaxel sensitivity or driven by reduced drug penetration, we formed MCTS, dissociated the spheroids, and evaluated paclitaxel sensitivity of dissociated cells under 2D culture conditions (Figure 2E). Cells derived from MCTS showed reduced paclitaxel response compared to parental 2D cultures in both A1847 and OVCAR8 cells (Figure 2F, compare ● and ▲). Cells derived from paclitaxel treated MCTS showed an even greater decrease in paclitaxel response when evaluated under 2D culture conditions (Figure 2F, compare ▼ with ● and ▲). Furthermore, the expression of stem cell markers remained high in the MCTS-derived cells even after growth for 3 days under 2D conditions (Figure 2G). Interestingly, in cells derived from paclitaxel treated spheroids, stem cell-associated gene expression was greater than in those derived from vehicle treated spheroids (Figure 2G, compare red and blue lines), suggesting that the 3D phenotype is stable. These data show using a MCTS model of EOC induces a drug resistant phenotype which could be exploited for front-line drug screening.
Drug Repurposing Screening in MCTS Identifies Unappreciated Drug Candidates
To determine if phenotypic changes in MCTS lead to alter drugs response compared to 2D cultures, we screened a targeted library of FDA-approved drugs to identify drug candidates that can be repurposed in order to accelerate a path to the clinic. RNA sequencing was performed between 2D and 3D cultures (BioProject ID: PRJNA472611) to identify potential regulatory pathways altered in MCTS and in turn to develop a focused library of compounds. In cell lines grown under 3D conditions, 630 genes showed at least a 1.5-fold over expression compared to 2D cultures, with a False Discovery Rate (FDR) of less than 0.05 (Figure 3A, blue points). Likewise, 96 genes showed at least a 1.5-fold over expression with a FDR of less than 0.05 in 2D cultures when compared to 3D cultures (Figure 3A, red points). Ingenuity Pathway Analysis (IPA) of the overexpressed genes in 3D cultures showed the top upstream regulators were related to cellular hypoxia: HIF1A (p=1.34E-10), EPAS1 (p=4.55E-08), and cobalt chloride (p=1.22E-07) (Figure 3B). Interestingly, the two top drug hits predicted to reverse 3D gene expression were a nonsteroidal anti-inflammatory drug (NSAID), fluticasone (p=1.44E-09), and an anti-diabetic, rosiglitazone (p=4.55E-07). Other proteins associated with drug resistance, such as the ABC transporters, were found at the RNA level to also be overexpressed in spheroids compared to 2D cultures (Supplemental Table 1). In total, the mRNA transcripts for seven of the 72 known ABC transporters were found to be elevated, four of which were significantly overexpressed in the spheroids compared to 2D cells (logFC> 1.5-fold, FDR< 0.05). However, none of these encoded proteins were identified as top regulators of the 3D culture phenotype using IPA pathways. Based on the pathways identified from the RNA-seq analysis, a drug library (n=304) including known cancer therapeutics, and agents that target inflammation and metabolic disorders, was selected from Selleck Chemicals’ FDA-approved drug library.
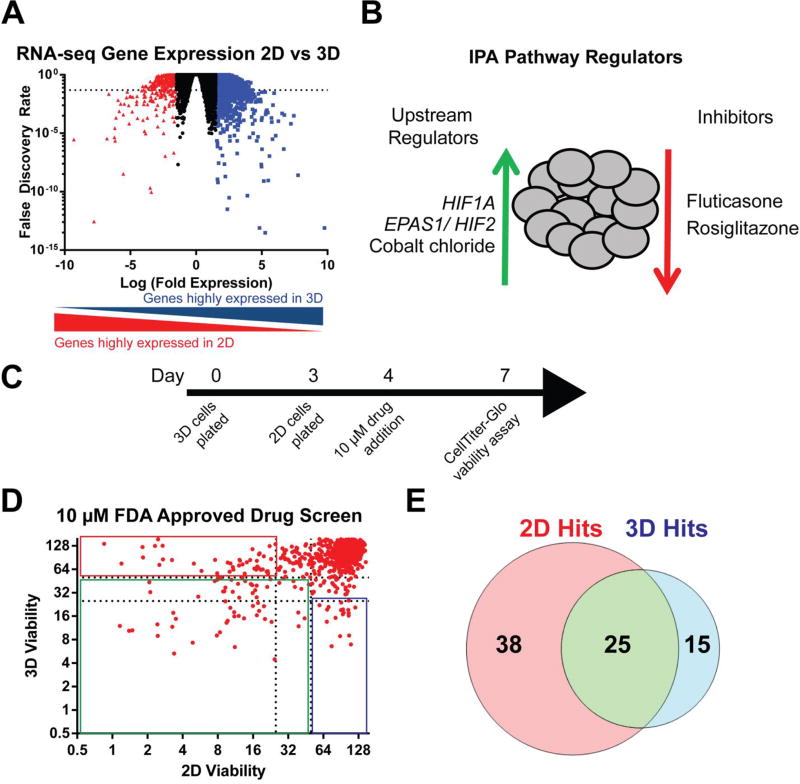
Figure 3. Drug repurposing screen in ovarian cancer MCTS identifies unappreciated drug candidates.
(A) Analysis of RNA-seq data identified genes expressed 1.5-fold or greater in 3D cultures (blue) versus 2D and genes 1.5-fold or greater in 2D cultures (red) versus 3D with a FDR less than 0.05 (dashed line). (B) IPA shows the top five regulators inferred from the genes identified to be over expressed in 3D cultures. (C) Representative time-line for drug screening in 2D and 3D cultures. (D) Relative viability compared to vehicle (DMSO) compared in both 2D and 3D cultures following 72 h treatment using 10 µM drug. (E) Venn diagram representing unique and common drug hits: 2D hits with >75% reduction viability in 2D cultures and < 50% reduction in viability in 3D cultures; 2D and 3D hits with > 75% reduction in viability for both 2D and 3D cultures; and 3D hits with > 75% reduction in viability in 3D cultures and < 50% reduction in viability in 2D cultures.
For this screening, four EOC cell lines (A1847, OVCAR3, OVCAR4, and OVCAR8) were grown in 3D culture for four days or in 2D culture overnight before 10 µM drug was added and incubated for 72 h (Figure 3C). Relative viability to DMSO controls was directly compared between 2D and 3D cultures using Cell-TiterGlo, which better measures spheroid drug response than other viability methods (Supplemental Figure 6A-C). Drugs classified as three different hits: i) preferential 2D hits (> 75% reduction in relative viability in 2D cultures and < 50% reduction in relative viability in 3D cultures); ii) 2D and 3D hits (> 50% reduction in relative viability for both 2D and 3D cultures); and iii) preferential 3D hits (>75% reduction in relative viability in 3D cultures and < 50% reduction in relative viability in 2D cultures). Overall, there were 78 drugs classified as hits, with 38 being enriched in 2D, 25 in 2D and 3D, and 15 in 3D (Figure 3 D-E). Interestingly, the preferential 3D hits were mostly comprised of drugs from the inflammatory (47%) and metabolic (26%) groups compared to anticancer drugs (20%) (Table 1). We searched these drugs on PubMed for previous studies in EOC (numerator) or all cancers (denominator) and found few positive findings (Table 1). These results confirm many of these drugs have not been identified in previous screens for cancer. Drug screening in HIO107 and HIO118 showed no 3D drug hits (Supplemental Figure 7), indicating these compounds may target the more quiescent drug resistance cells. Two drugs, licofelone and glafenine, were validated as 3D hits across at least three EOC cell lines in a secondary screen at 25 µM (Supplemental Figure 8). Interestingly the hits from each class, as defined during primary drug screening, tend to retain the same activity in secondary screening if they showed enough activity to be classified (Supplemental Figure 9A). Validation of select agents classified as 2D (afatinib) hits or 2D and 3D (bortezomib) hits confirmed the activity across multiple cells lines in a dose-dependent manor (Supplemental Figure 9B-C). These results identified licofelone and glafenine as candidate anticancer drugs with enhanced activity against ovarian MCTS that would not have been identified through traditional 2D drug screening.
Table 1. 3D Hits from Primary Screening.
List of the fifteen 3D drug hits identified from the primary drug screen. The disease indication from most drugs was from either inflammation or metabolic disease. The specific target or use for each specific drug as listed from the Selleckchem database. PubMed search results for searching each drug with either “ovarian cancer” (numerator) or “cancer” (denominator) indicates most 3D specific drugs have had limited studies in ovarian cancer.
| Drug | Indication | Target or Use | Pub Med Hits |
|---|---|---|---|
| Pizotifen malate | Inflammation | Serotonin antagonist | 0/0 |
| Risedronic acid (Actonel) | Metabolic Disease | Osteoporosis drug | 0/109 |
| Hydroxyurea (Cytodrox) | Cancer | Free radical nitroxide | 43/4234 |
| Pioglitazone hydrochloride (Actos) | Metabolic Disease | Cytochrome P450 (CYP)2C8 and CYP3A4 | 3/529 |
| Mecarbinate | Metabolic Disease | Antihypertension | 0/0 |
| Licofelone | Inflammation | COX/LOX | 0/24 |
| OSI-420 (Erlotinib) | Cancer | EGFR | 82/5077 |
| Geniposidie | Inflammation | Glucagon-like peptide-1 | 0/2 |
| Phenformin HCl | Metabolic Disease | Anti-diabetic. Glycolysis | 2/129 |
| Cinepazide maleate | Inflammation | Calcium Channel blocker | 0/0 |
| Beclomethasone dipropionate | Inflammation | Glucocorticoid | 0/60 |
| Dexamethasone acetate | Inflammation | Glucocorticoid | 302/14137 |
| Allylthiourea | Inflammation | Ammonia oxidation | 0/3 |
| Bergapten | Cancer | DNA mutagen | 0/76 |
| Glafenine | Inflammation | NSAID | 0/4 |
Licofelone and Glafenine Reduce Stemness in Ovarian MCTS
In order to confirm the preferential activities of licofelone and glafenine in 3D culture models, we performed secondary functional analyses. These assays focused on the ability of licofelone and glafenine to reverse the drug resistant signatures induced by MCTS.
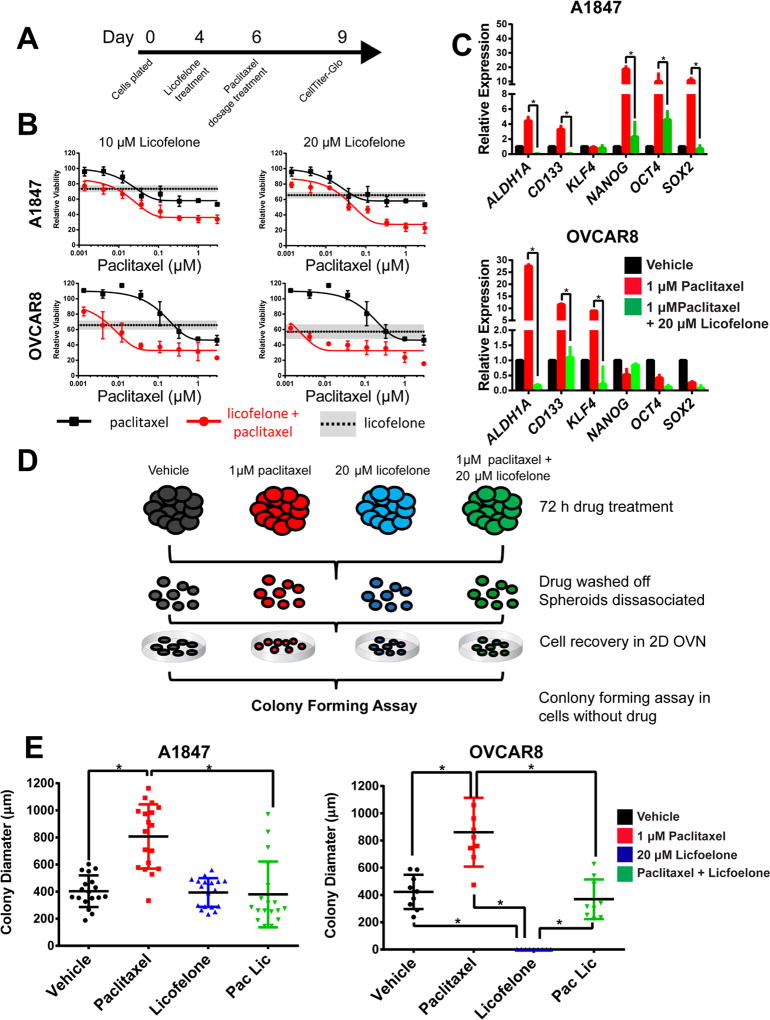
Licofelone, a dual COX/LOX inhibitor developed by Merckle and Ratiopharm (Teva Pharmaceuticals, Germany) to treat osteoarthritis (25, 26), was evaluated using a dosage response from 195 nM to 50 µM and showed preferential activity in 3D cultures compared to 2D cultures following 72 h treatments (Figure 4A). Specifically, the IC50 values for licofelone were lower in 3D cultures than in 2D cultures across A1847 (21.6 µM vs 50+ µM), OVCAR3 (13.5 µM vs 50+ µM), OVCAR4 (33.5 µM vs 50+ µM), and OVCAR8 (17.4 µM vs 44 µM) cell lines.
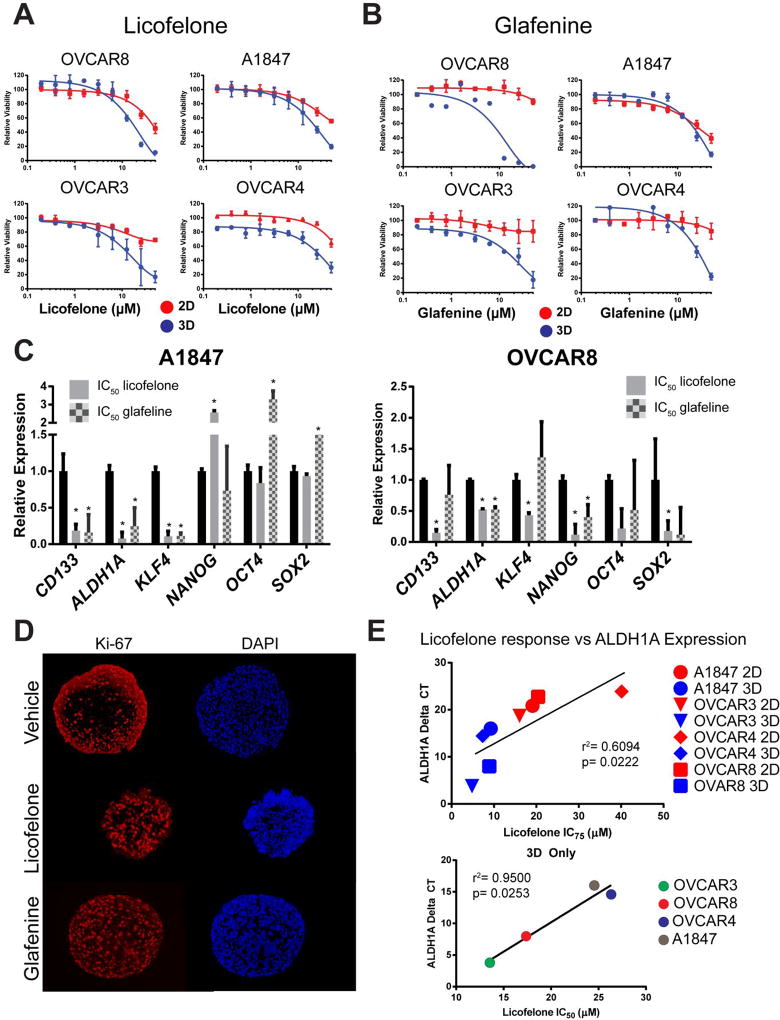
Figure 4. Licofelone and glafenine decrease stemness in ovarian cancer MCTS.
(A-B) Dosage response to the top two 3D specific drug hits, licofelone and glafeline, following 72 h treatments shows both drugs are more sensitive in ovarian MCTS than 2D cultures. (C) Expression of stem-like transcripts following 72 h, IC50 dosage treatment of both drugs in MCTS was measured using qRT-PCR. (D) Immunofluorescence detection of Ki-67 and DAPI reveals an increase in Ki-67 expression following treatment with licofelone or glafeline at their respective IC50 values in MCTS. (E) Correlation of baseline licofelone expression in 2D and 3D to either the IC75 of licofelone (top panel) or the IC50 of licofelone (bottom panel). All data are represented as mean +/− SD, n=3.
Glafenine, an NSAID derivative of anthranilic acid, was developed by Alecandria Co. (Egypt) for the relief of all types of pain. Due to a high incidence of anaphylaxis and kidney failure it has been withdrawn from the market in most countries (27–29). When evaluated using a dosage response from 195 nM to 50 µM, glafenine also showed preferential activity in 3D cultures compared to 2D cultures following 72 h treatments (Figure 4B). The IC50 values for glafenine were consistently lower in 3D cultures as compared to 2D cultures of A1847 (24 µM vs 33 uM), OVCAR3 (18.0 µM vs 50+ µM), OVCAR4 (30.3 µM vs 50+ µM), and OVCAR8 (9.1 µM vs 50+ µM) cell lines.
To test the effect of both drugs on stem-like gene expression, A1847 and OVCAR8 MCTS were treated with licofelone or glafenine for 72 h at the calculated IC50 concentrations and RNA was isolated for qRT-PCR. In A1847 spheroids, licofelone and glafenine significantly reversed the expression of CD133, ALDH1A, and KLF4 while the expression of NANOG, OCT4, and SOX2 were either not changed or actually increased (Figure 4C, * = p<0.05, t-test). In OVCAR8 MCTS licofelone treatment significantly reversed the expression of CD133, ALDH1A, KLF4, NANOG, and SOX2 while glafenine significantly reversed the expression of only ALDH1A and NANOG (Figure 4C, * = p<0.05, t-test). Immunofluorescence analysis of A1847 spheroid sections, following 72 h treatment with IC50 concentrations of licofelone or glafenine, showed an increased number of Ki-67 positive cells (63.5% and 67.9%, respectively) when compared to vehicle treated MCTS (41%) (Figure 4D).
Interestingly, we observed licofelone showed enhanced activity in 2D cultured cells, which were derived following paclitaxel treatment of spheroids, as compared to the parental 2D cultured cells (Supplemental Figure 10A-B). Furthermore, licofelone showed greater efficacy in comparison to a COX-2 specific inhibitor, celecoxib, in A1847 spheroids (Supplemental Figure 11), suggesting the dual inhibition of COX/LOX better inhibits ovarian MCTS cell growth than COX-2 inhibition alone.
Finally, licofelone response was correlated to baseline ALDH1A expression across all four cell lines in 2D and 3D cultures. Since licofelone response did not reach less than 50% cell viability in most 2D cultures, the IC75 was used across 2D (red dots) and 3D cultures (blue dots). Licofelone IC75 showed a significant correlation to baseline ALDH1A expression levels (r2= 0.6094, p= 0.0222) (Figure 4E, top panel). However, in MCTS alone, the correlation of IC50 and ALDH1A expression was much more robust (r2= 0.9500, p= 0.0253) (Figure 4E, bottom panel). Mutational analysis of the various ovarian cancer cell lines found no obvious correlation to either licofelone or glafenine activity. Specifically, Tier 1 or Tier 2 mutations found, when using the TruSeq Amplicon Cancer Panel (Illumina) and classified using QIAGEN Clinical Insight (QCI™), did not find common gene alterations, albeit from a small panel, associated with drug sensitivity to licofelone or glafenine (Supplemental Table 2).
Taken together these data demonstrate two different anti-inflammatory drugs, licofelone and glafenine, reverse stem-like properties of ovarian cells grown as MCTS. Therefore, we hypothesized that reversing stem-like properties of ovarian MCTS and inducing a more proliferative phenotype could enhance the activity of paclitaxel by circumventing drug resistance when combined with licofelone or glafenine.
Licofelone Synergistically Enhances Paclitaxel Activity and Blocks Stem-like Properties in Ovarian MCTS
To test for synergistic activity with paclitaxel, licofelone was selected for further development due to a more favorable clinical profile. MCTS from A1847 and OVCAR8 cell lines were grown for four days and pretreated for 48 h with licofelone before treatment with a serial dilution of paclitaxel for 72 h (Figure 5A). In both A1847 and OVCAR8 spheroids, there was leftward shift in dosage response, a decrease in IC50, and a decrease in the percent viability remaining at 3 µM paclitaxel (Figure 5B). To quantify the synergy between licofelone and paclitaxel, combination index (CI) values were calculated using the Chou-Talalay method. Strong synergy was measured in both A1847 and OVCAR8 spheroids (Supplemental Figure 12). Likewise, pretreatment with paclitaxel followed by treatment with licofelone showed significant synergy in A1847 and OVCAR8 spheroids (Supplemental Figure 13A-C).
Figure 5. Licofelone synergistically enhances paclitaxel activity in combination while reversing the stem-like characteristics of ovarian MCTS.
(A) Representative time-line of drug combinations in A1847 and OVCAR8 spheroids. (B) Dosage response to paclitaxel alone (black line) is lower than paclitaxel following 72 h pretreatments with 10 µM licofelone (top) or 20 µM licofelone (bottom) (red lines). Licofelone alone treatments of 10 µM or 20 µM are represented by the horizontal dashed lines (mean) and grey boxes (SD). (C) Expression of cancer stem cell-related genes following 72 h treatment of vehicle (DMSO), 1 µM paclitaxel, or 1 µM paclitaxel and 20 µM licofelone in A1847 or OVCAR8 spheroids. (D) Graphical representation of secondary colony forming assays from MCTS. (E) Colony size (µm) was significantly increased in cells derived from previously paclitaxel treated MCTS and was decreased from cells derived from paclitaxel and licofelone treatments (n>10, *=p<0.05, two-way ANOVA). Colony formation assays were carried out in the absence of drug treatment. All data are represented as mean +/− SD, n=3.
Licofelone, in combination with paclitaxel, led to a significant reduction in the expression of a number of stem-like transcripts in A1847 MCTS (ALDH1A, CD133, NANOG, OCT4, and SOX2) and OVCAR8 MCTS (ALDH1A, CD133, and KLF4) when compared to paclitaxel alone (Figure 5C, n=3, *= p<0.05, t-test). The combination therapy of 1 µM paclitaxel and 20 µM licofelone in A1847 MCTS significantly increased the expression of cleaved poly (ADP-ribose) polymerase (PARP) compared to total PARP (3-fold) compared to vehicle treatment, as measured by Western blot. Single treatments compared to vehicle only showed a trend towards increase in licofelone (1.3-fold) and paclitaxel (2.2-fold) (Supplemental Figure 14, n=3, p<.05, t-test).
Given that we found that the spheroids formed in our studies were resistant to both paclitaxel and carboplatin (Figure 2 and Supplemental Figure 15A), we also assessed whether licofelone could enhance the activity of a platinum-based drug. Interestingly, the combination of licofelone and carboplatin showed a minimal response in A1847 MCTS. However, in OVCAR8-derived spheroids, the combination of 20 µM licofelone treatment reduced the IC50 of carboplatin by ~3.8 fold (24 µM vs 92 µM) (Supplemental Figure 15B). With the existence of PARP inhibitors that already work in concert with DNA repair deficiency and platinum induced DNA damage, we chose instead to focus the remaining experiments on a drug that could more universally enhance taxol’s activity.
To test the stem-like function of MCTS cells that survived drug treatments, A1847 or OVCAR8 MCTS were treated with drug (vehicle, 1 µM paclitaxel, 20 µM licofelone, or 1 µM paclitaxel plus 20 µM licofelone) for 72 h, washed repeatedly, disassociated, then re-plated into 2D cultures, followed by colony forming assays (Figure 5D). For the A1847 cells, the size of individual colonies increased significantly in cells that survived paclitaxel treatment alone when compared to vehicle treatments (Figure 5E, n=30, p<0.05, Two-way ANOVA). Interestingly, the cells which survived the combination of licofelone and paclitaxel showed significantly reduced colony forming ability (i.e., size) when compared to paclitaxel treatment alone (Figure 5E, n=30, p<0.05, Two-way ANOVA). In comparison, and somewhat unexpected, the ability to form colonies after treatment of spheroids followed by dissociation and re-plating was completely lost in OVCAR8 cells that survived licofelone treatment (Figure 5E). Alternatively, the size of colonies formed by OVCAR8 cells that survived paclitaxel treatment was significantly increased when compared to vehicle treatments (Figure 5E, n=30, p<0.05, Two-way ANOVA). The combination of licofelone with paclitaxel significantly reduced subsequent colony size compared to paclitaxel alone (Figure 5E, n=30, p<0.05, Two-way ANOVA). These data demonstrate that licofelone can reverse the downstream stem-like function (as measured by colony diameter) that is induced by paclitaxel treatment.
Here we have found licofelone not only synergizes with paclitaxel, but it can reverse the stem-like properties observed in ovarian cancer cells grown as MCTS. Therefore, licofelone appears to be a promising agent for further preclinical development for combination therapy with paclitaxel in EOC.
Licofelone Reduces Mitochondrial Metabolism, a Potential Mechanism to Regulate Stemness in Ovarian Cancer Cells
Mitochondrial metabolism has been shown to be a critical regulator of cancer stemness (30). To measure the potential effect of licofelone alone or in combination with paclitaxel, we performed both a gene expression array of transcripts coding for enzymes within the citric acid cycle (TCA) and an immunoblot array of oxidative phosphorylation (OXPHOS) mitochondrial complexes. From the gene expression array analysis, we observed that paclitaxel induces expression (>1 log(ddCT)) of most of the TCA transcripts across both A1847 and OVCAR8 spheroids (Supplemental Figure 16A). Analysis of the OVCAR8 spheroids found that licofelone (20 µM) treatment results in reduced expression of several TCA transcripts, including IDH2, IHDH3A, MDH2, PCK2, SDHD, and SUCLG2 (Supplemental Figure 16A). Recent studies have shown that inhibiting mitochondrial complexes, and therefore inhibiting OXPHOS, could reverse stemness in cancer cells(30). In our spheroids, treatment with 20 µM licofelone reduced the expression of multiple OXPHOS complexes (I, II, and IV) in OVCAR8 spheroids, while no noticeable effect was observed in A1847 spheroids (Supplemental Figure 16B). These results suggest that licofelone could be modulating the expression of stem-like markers through regulation of mitochondrial metabolism and OXPHOS.
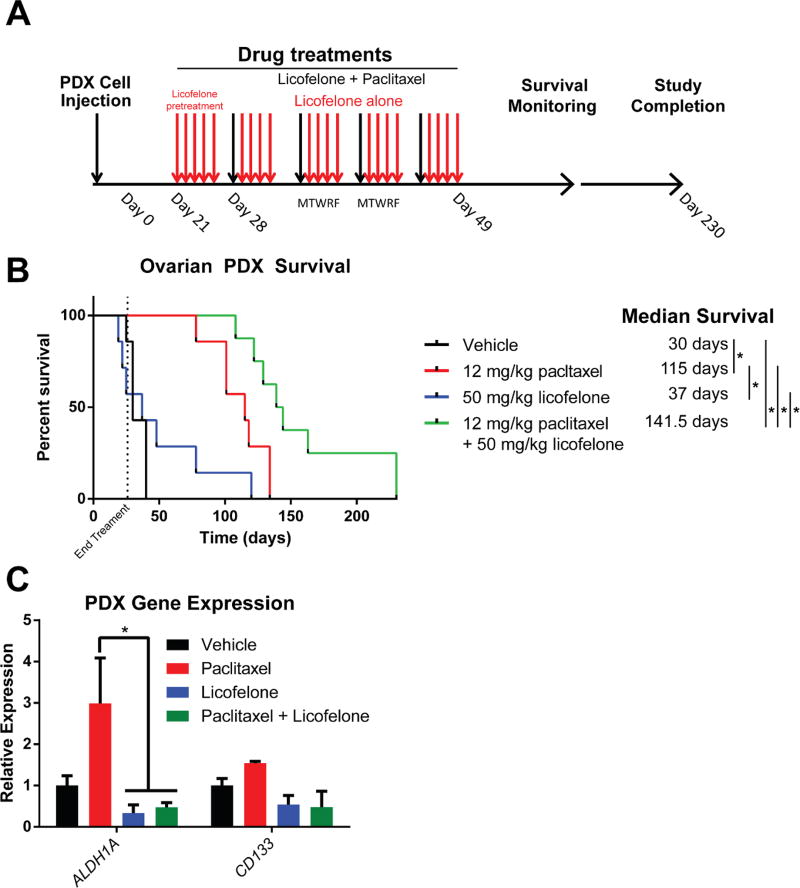
Licofelone Combination Increased the Efficacy of Paclitaxel in Ovarian Cancer PDX Mice
To support in vivo analysis of licofelone, an ovarian cancer PDX ascites model was generated from both subcutaneous (s.c.) and intraperitoneal (i.p.) passaging of ovarian tumor in NOD scid gamma (NSG) mice. De-identified fresh tumor tissue was obtained following informed consent from a treatment naïve patient diagnosed with high grade serous (Stage IIIc) ovarian cancer (see MATERIALS and METHODS section for specific details). The ovarian tumor cells from this PDX model grew within the ascites fluid. Mice were injected i.p. with tumor cells isolated from this primary PDX ascites model (passage 3). Twenty-one days after inoculation the mice were sorted into four different treatment groups (n=7) for i.p. delivery of vehicle, paclitaxel (12 mg/kg), licofelone (50 mg/kg), or paclitaxel plus licofelone. Mice were monitored for disease progression (development of ascites fluid) and overall survival (Figure 6A). Median survival was calculated for each group and survival curves were compared using the Mantel-Haenszel hazard ratio (HR). In the EOC PDX model, licofelone alone shows minimal difference in median survival (37 days) when compared to vehicle (30 days) (Figure 6B, Supplemental Table 3). As expected, paclitaxel has enhanced efficacy compared to vehicle alone (median survival 115 days, HR= 0.0452) (Figure 6B, Supplemental Table 3). Importantly, in the combination group the median survival (>141 days, some mice were tumor free past 200 days) was significantly improved compared to vehicle alone (HR = 0.0373), licofelone alone (HR = 0.046), and to paclitaxel alone (HR = 0. 172) (Figure 6B, Supplemental Table 3). Transcript expression analysis revealed that paclitaxel treatment increased the expression of CD133 and significantly increased the expression of ALDH1A (Figure 6C, p< 0.05, Two-way ANOVA). Likewise, the combination of licofelone and paclitaxel significantly reversed the expression of ALDH1A compared to paclitaxel alone (Figure 6C, p< 0.05, Two-way ANOVA). Interestingly, the percent of Ki-67 positive cells in the recurrent tumor cells isolated from the ascites was significantly reduced in the 1 µM paclitaxel group compared to vehicle (Supplemental Figure 17A-B, p< .05, t-test). However, in the licofelone treated tumor cells, the percent of Ki-67 positive cells was significantly increased compared to vehicle, while the combination of paclitaxel and licofelone showed no statistical different compared to vehicle (Supplemental Figure 17A-B, p< .05, t-test). These data confirm our previous observations that paclitaxel reduces cell proliferation, while licofelone induces proliferation alone and reverses the paclitaxel proliferative phenotype.
Figure 6. Licofelone significantly improves paclitaxel efficacy in an ovarian cancer PDX mouse model.
(A) Graphical representation of PDX development and treatment timeline. (B) Kaplan-Meyer survival curve for each arm of therapy: vehicle (black, n=7), 12 mg/kg paclitaxel (red, n=7), 50 mg/ kg licofelone (blue, n=7), and 12 mg/kg paclitaxel + 50 mg/kg licofelone (green, n=8). Median survival was significantly higher in the combination group (~141days, p< 0.05), compared to paclitaxel alone (115 days), licofelone alone (37 days), or vehicle (30 days). (C) Expression of stem-like genes (ALDH1A, CD133) is increased in paclitaxel PDX tumors at the time of sacrifice while licofelone treatment inhibits the induction (n=4, p< 0.05, Two-way ANOVA).
Taken together, these data support the notion that licofelone, a clinically repurposed drug unappreciated by traditional drug screening, can enhance the activity of the front-line chemotherapeutic agent, paclitaxel. The clinical development of licofelone for EOC could provide benefits to primary and recurrent patients with EOC, although further studies using additional models might be warranted. Outside recent activities around PARP inhibitors in EOCs with homologous recombination (HR) deficiency, e.g., BRCA mutations, these patients have not seen improvements in front-line therapy for almost three decades.
DISCUSSION
Limited clinical success of EOC drugs could be attributed to the fact that many drugs are not targeting the stem-like cells that persist after chemotherapy (8). We showed some molecular targeted agents with limited clinical efficacy produce a similar stem-like phenotype after high dose treatment. In this study, we demonstrate that 3D in vitro screening is superior to traditional screening approaches for identifying new drug leads for the treatment of women with chemotherapy refractory ovarian cancer. We extrapolate that this in vitro screening approach can be used to identify more robust drug leads for other solid tumors as well.
Studies have used MCTS to validate primary drug screens (31, 32). However, these approaches rely on traditional drug screening methods in two-dimensional cultures to identify the initial drug hits, focused primarily on cytotoxic activity. As our results show, the activity of some drugs are preferentially revealed in 3D than in 2D and therefore, would not have been identified using traditional screening approaches for cytotoxic drugs.
The use of three-dimensional cell culture in primary screening employed in this study can be used to better mimic the clinical disease, including hypoxia and stem-like properties which are driving mechanisms of drug resistance(14, 33, 34). Cellular hypoxia was not only identified as the top regulator from the IPA data, it has also been shown to drive stemness through epigenetic regulation in many different cancer models(35–38). Regulation of hypoxia-related epigenetic modifiers has been shown to reduce spheroid forming function of ovarian cancer cells (39). Likewise, JMDJ1A is overexpressed in MCTS compared to 2D cells which in itself can regulate the expression of stem-like genes and influence chemotherapy sensitivity in ovarian cancer cells (24). Taken together, the relationship between hypoxia and stemness could be a critical regulator of response to paclitaxel and is mimicked in our ovarian MCTS models used for drug repurposing screens.
To exploit these differences for drug screening, we directly compared the activity of clinically repurposed drugs across 2D and 3D cultures to identify novel candidates, unappreciated by traditional screening in 2D cultures. The drugs classified as 2D and 3D hits were predominantly from the current cancer therapeutic class that would have been identified by traditional screening and validated in in vivo xenograft tumor models. Recent studies have suggested that 3D models will be predictive of patient response(16, 40) while our study focused on drugs with activity only in the 3D model would certainly have been overlooked by a traditional screening approach. Most of these 3D hits came from metabolic or anti-inflammatory drugs that had not been extensively studied in cancers. To date, the top two 3D only hits, licofelone and glafenine, have not been evaluated in preclinical or clinical studies for EOC. Our screening results explain why these have not been studied in EOC previously.
Licofelone is a dual inhibitor of COX-2/5-LOX developed for the treatment of arthritis. COX-2, or cyclooxygenase-2, is prostaglandin synthase that converts arachidonic acid to PGH2. COX-2 is overexpressed in EOC patients and may play a role in tumor progression(41, 42). Interestingly, COX-2 has been shown to induce stem cells in breast cancer cell lines(43). 5-LOX, or 5-lipoxygenase, is associated with promoting hypoxia response and inflammation in EOC (44, 45). While the use of licofelone in cancer therapy has been limited, it has been shown to inhibit cigarette smoke-induced lung cancer (46) and block pancreatic cancer induction with a marked decrease in stem cell markers (47). While licofelone has not been evaluated clinically for cancer, including ovarian, celecoxib, a COX-2 inhibitor, enhanced the efficacy of chemotherapy in a Phase II trial (48). In our MCTS model, licofelone showed greater activity than celecoxib and is known to have fewer side effects (49). Furthermore, the enhanced cytotoxic activity of licofelone in cells correlates with higher basal expression of ALHD1A. Interestingly, in OVCAR8 MCTS, licofelone reduced the expression of cancer stem cell-related transcripts (ALDH1A and CD133) and the canonical stem cell markers (OCT4, SOX2, and NANOG). This effect corresponded to a complete loss in colony forming activity, whereas in A1847-derived MCTS, licofelone reduced the expression of only the cancer-related stem transcripts. In this study, we showed licofelone could reverse stem-cell like characteristics of ovarian MCTS and enhance sensitivity to the front-line chemotherapeutic agent, paclitaxel.
Both licofelone and celecoxib are NSAIDs which might indicate a role of mitochondrial regulation blocking stemness in our MCTS model (50). The use of NSAIDs, specifically aspirin, has been retrospectively shown to lower the risk of EOC in a dose and frequency dependent manner(51). Recent studies have shown that mitochondrial metabolism is both favored and critical for cancer stem-like cells (30). Ovarian cancer stem cells favored OXPHOS to glucose metabolism, shunted pyruvate to the Krebs cycle, and showed an increase in ROS production (52). EOC spheroid stem cells have been reported to favor the Krebs cycle to induce hypoxia resistant metabolism(53). Targeting mitochondrial function by inhibiting complex III with atovaquone showed a reduction in OXPHOS and inhibited breast cancer stem cells (54). Licofelone can induce mitochondrial apoptosis in colon cancer cells independent of arachidonic acid cascade activity(55). In our studies we showed that the expression of TCA-related genes is upregulated in response the paclitaxel therapy, and strongly downregulated in OVCAR8-derived spheroids following licofelone treatment. The level of expression of stem cell-related genes in OVCAR8 spheroids is much more sensitive to licofelone than A1847 cells, suggesting a relationship between mitochondrial metabolism and stemness in ovarian MCTS. Further studies will help provide insight into the regulation of stem-like cell expansion or induction in EOC patient tumors.
Overall, this study highlights the importance of alternative drug screening models in order to better mimic in vitro the disease state of clinical EOC. Drugs with specific activity in ovarian cancer MCTS show the ability to induce proliferation, reverse stemness, and in turn show synergistic activity with front-line chemotherapy. These results suggest the possibility to develop future clinical trials with companion diagnostics to evaluate licofelone in combination with front-line chemotherapy in women with primary and recurrent ovarian cancer.
Supplementary Material
SIGNIFICANCE.
This study highlights the use of an in vitro spheroid 3D drug screening model to identify new therapeutic approaches to reverse chemotherapy resistance in ovarian cancer.
Acknowledgments
We would like to thank Dr. Scott Weir (KUMC) for his valuable input regarding drug screening, and Stacey Hembruff (KUCC) who assisted with treatments of animals used for the PDX models. We would like to thank Ms. Tara Meyer (Biospecimen Repository Core Facility) for her assistance in preparing MCTS sections and Ms. Kelly Cannova and Ms. Kristi O’Keefe, founders of the OERRUN Ovarian Cancer Foundation for support.
GRANT SUPPORT
The work is supported by funding from the NIH (RO1 CA140323) and an ovarian cancer planning group award from the KU cancer center (P30 CA168524) to A.K. Godwin, the Madison & Lila Self Graduate Fellowship to J. Hirst, the BIRCWH K12 Career Development award to H.B. Pathak, and the OVERRUN Ovarian Cancer Foundation. A.K. Godwin is the Chancellors Distinguished Chair in Biomedical Sciences endowed Professor and a Kansas Bioscience Authority Eminent Scholar. The authors also gratefully acknowledge the support of the Biospecimen Repository Core Facility and the Biostatistics and Informatics Shared Resource funded in part by the National Cancer Institute Cancer Center Support Grant (P30 CA168524). The Ingenuity Pathways Analysis (IPA) software used in this manuscript was supported by the Biostatistics and Informatics Shared Resource (BISR), funding by the National Cancer Institute (NCI) Cancer Center Support Grant P30 CA168524, and the Kansas IDeA Network of Biomedical Research Excellence Bioinformatics Core, supported by the National Institute of General Medical Science award P20 GM103418.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICT OF INTEREST
The authors declare no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin LA, Huang B, Miller RW, Tucker T, Goodrich ST, Podzielinski I, et al. Ten-year relative survival for epithelial ovarian cancer. Obstet Gynecol. 2012;120:612–618. doi: 10.1097/AOG.0b013e318264f794. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths RW, Zee YK, Evans S, Mitchell CL, Kumaran GC, Welch RS, et al. Outcomes after multiple lines of chemotherapy for platinum-resistant epithelial cancers of the ovary, peritoneum, and fallopian tube. Int J Gynecol Cancer. 2011;21:58–65. doi: 10.1097/IGC.0b013e3182049273. [DOI] [PubMed] [Google Scholar]
- 4.Bookman MA, Brady MF, McGuire WP, Harper PG, Alberts DS, Friedlander M, et al. Evaluation of new platinum-based treatment regimens in advanced-stage ovarian cancer: a Phase III Trial of the Gynecologic Cancer Intergroup. J Clin Oncol. 2009;27:1419–1425. doi: 10.1200/JCO.2008.19.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrandina G, Bonanno G, Pierelli L, Perillo A, Procoli A, Mariotti A, et al. Expression of CD133-1 and CD133-2 in ovarian cancer. Int J Gynecol Cancer. 2008;18:506–514. doi: 10.1111/j.1525-1438.2007.01056.x. [DOI] [PubMed] [Google Scholar]
- 6.Ferrandina G, Martinelli E, Petrillo M, Prisco MG, Zannoni G, Sioletic S, et al. CD133 antigen expression in ovarian cancer. BMC Cancer. 2009;9:221. doi: 10.1186/1471-2407-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kryczek I, Liu S, Roh M, Vatan L, Szeliga W, Wei S, et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer. 2012;130:29–39. doi: 10.1002/ijc.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steg AD, Bevis KS, Katre AA, Ziebarth A, Dobbin ZC, Alvarez RD, et al. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin Cancer Res. 2012;18:869–881. doi: 10.1158/1078-0432.CCR-11-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbin ZC, Katre AA, Steg AD, Erickson BK, Shah MM, Alvarez RD, et al. Using heterogeneity of the patient-derived xenograft model to identify the chemoresistant population in ovarian cancer. Oncotarget. 2014;5:8750–8764. doi: 10.18632/oncotarget.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curley MD, Therrien VA, Cummings CL, Sergent PA, Koulouris CR, Friel AM, et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27:2875–2883. doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 11.Landen CN, Jr, Goodman B, Katre AA, Steg AD, Nick AM, Stone RL, et al. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol Cancer Ther. 2010;9:3186–3199. doi: 10.1158/1535-7163.MCT-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oktem G, Bilir A, Uslu R, Inan SV, Demiray SB, Atmaca H, et al. Expression profiling of stem cell signaling alters with spheroid formation in CD133high/CD44high prostate cancer stem cells. Oncol Lett. 2014;7:2103–2109. doi: 10.3892/ol.2014.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Hua R, Wang X, Huang M, Gan L, Wu Z, et al. Identification of stem-like cells and clinical significance of candidate stem cell markers in gastric cancer. Oncotarget. 2016;7:9815–9831. doi: 10.18632/oncotarget.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Wang L, Ma L, Xu J, Liu C, Zhang J, et al. Significantly increased expression of OCT4 and ABCG2 in spheroid body-forming cells of the human gastric cancer MKN-45 cell line. Oncol Lett. 2013;6:891–896. doi: 10.3892/ol.2013.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghavan S, Mehta P, Ward MR, Bregenzer ME, Fleck EMA, Tan L, et al. Personalized Medicine-Based Approach to Model Patterns of Chemoresistance and Tumor Recurrence Using Ovarian Cancer Stem Cell Spheroids. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-17-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pathak HB, Zhou Y, Sethi G, Hirst J, Schilder RJ, Golemis EA, et al. A Synthetic Lethality Screen Using a Focused siRNA Library to Identify Sensitizers to Dasatinib Therapy for the Treatment of Epithelial Ovarian Cancer. PLoS One. 2015;10:e0144126. doi: 10.1371/journal.pone.0144126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godwin AK, Meister A, O'Dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci U S A. 1992;89:3070–3074. doi: 10.1073/pnas.89.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao KS, Godwin AK, Johnson SW, Ozols RF, O'Dwyer PJ, Hamilton TC. Evidence for altered regulation of gamma-glutamylcysteine synthetase gene expression among cisplatin-sensitive and cisplatin-resistant human ovarian cancer cell lines. Cancer Res. 1995;55:4367–4374. [PubMed] [Google Scholar]
- 20.Godwin AK, Testa JR, Handel LM, Liu Z, Vanderveer LA, Tracey PA, et al. Spontaneous transformation of rat ovarian surface epithelial cells: association with cytogenetic changes and implications of repeated ovulation in the etiology of ovarian cancer. J Natl Cancer Inst. 1992;84:592–601. doi: 10.1093/jnci/84.8.592. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich J Seidel C Fau - Ebner R, Ebner R Fau - Kunz-Schughart LA, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat Protoc. 2009;4:309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 22.Crow J, Atay S, Banskota S, Artale B, Schmitt S, Godwin AK. Exosomes as mediators of platinum resistance in ovarian cancer. Oncotarget. 2017 doi: 10.18632/oncotarget.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 24.Ramadoss S, Sen S, Ramachandran I, Roy S, Chaudhuri G, Farias-Eisner R. Lysine-specific demethylase KDM3A regulates ovarian cancer stemness and chemoresistance. Oncogene. 2017;36:6508. doi: 10.1038/onc.2017.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laufer S, Tries S, Augustin J, Dannhardt G. Pharmacological profile of a new pyrrolizine derivative inhibiting the enzymes cyclo-oxygenase and 5-lipoxygenase. Arzneimittelforschung. 1994;44:629–636. [PubMed] [Google Scholar]
- 26.Gay RE, Neidhart M, Pataky F, Tries S, Laufer S, Gay S. Dual inhibition of 5-lipoxygenase and cyclooxygenases 1 and 2 by ML3000 reduces joint destruction in adjuvant arthritis. J Rheumatol. 2001;28:2060–2065. [PubMed] [Google Scholar]
- 27.Dechezlepretre S, Lechat P, Wickelholz M. Comparison of the biological effects in rat of high doses of two 4-amino-7-chloroquinoline derivatives: chloroquine and glafenine. Arch Toxicol. 1975;34:309–314. doi: 10.1007/BF00353850. [DOI] [PubMed] [Google Scholar]
- 28.Boeijinga JK, van der Vijgh WJ. Double blind study of the effect of glafenine (Glifanan) on oral anticoagulant therapy with phenprocoumon (Marcumar) Eur J Clin Pharmacol. 1977;12:291–296. doi: 10.1007/BF00607429. [DOI] [PubMed] [Google Scholar]
- 29.Raken G. Treatment of pain with Glifanan in chronic degenerative diseases of the locomotor system. Fortschr Med. 1972;90:405–408. [PubMed] [Google Scholar]
- 30.Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer. 2016;114:1305–1312. doi: 10.1038/bjc.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fayad W, Rickardson L, Haglund C, Olofsson MH, D'Arcy P, Larsson R, et al. Identification of agents that induce apoptosis of multicellular tumour spheroids: enrichment for mitotic inhibitors with hydrophobic properties. Chem Biol Drug Des. 2011;78:547–557. doi: 10.1111/j.1747-0285.2011.01170.x. [DOI] [PubMed] [Google Scholar]
- 32.Kenny HA, Lal-Nag M, White EA, Shen M, Chiang CY, Mitra AK, et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat Commun. 2015;6:6220. doi: 10.1038/ncomms7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenzel C, Riefke B, Grundemann S, Krebs A, Christian S, Prinz F, et al. 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res. 2014;323:131–143. doi: 10.1016/j.yexcr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 34.Rotem A, Janzer A, Izar B, Ji Z, Doench JG, Garraway LA, et al. Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc Natl Acad Sci U S A. 2015;112:5708–5713. doi: 10.1073/pnas.1505979112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu CP, Du HD, Gong HL, Li DW, Tao L, Tian J, et al. Hypoxia promotes stem-like properties of laryngeal cancer cell lines by increasing the CD133+ stem cell fraction. Int J Oncol. 2014;44:1652–1660. doi: 10.3892/ijo.2014.2307. [DOI] [PubMed] [Google Scholar]
- 36.Iriondo O, Rabano M, Domenici G, Carlevaris O, Lopez-Ruiz JA, Zabalza I, et al. Distinct breast cancer stem/progenitor cell populations require either HIF1alpha or loss of PHD3 to expand under hypoxic conditions. Oncotarget. 2015;6:31721–31739. doi: 10.18632/oncotarget.5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iida H, Suzuki M, Goitsuka R, Ueno H. Hypoxia induces CD133 expression in human lung cancer cells by up-regulation of OCT3/4 and SOX2. Int J Oncol. 2012;40:71–79. doi: 10.3892/ijo.2011.1207. [DOI] [PubMed] [Google Scholar]
- 38.van den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson C, Qiu L, Hong Y, Karnik T, Tadros G, Mau B, et al. The histone demethylase KDM4B regulates peritoneal seeding of ovarian cancer. Oncogene. 2017;36:2565–2576. doi: 10.1038/onc.2016.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JS, Choi YD, Lee JH, Nam JH, Choi C, Lee MC, et al. Expression of cyclooxygenase-2 in epithelial ovarian tumors and its relation to vascular endothelial growth factor and p53 expression. Int J Gynecol Cancer. 2006;16(Suppl 1):247–253. doi: 10.1111/j.1525-1438.2006.00477.x. [DOI] [PubMed] [Google Scholar]
- 42.Singhal PK, Spiegel G, Driscoll D, Odunsi K, Lele S, Rodabaugh KJ. Cyclooxygenase 2 expression in serous tumors of the ovary. Int J Gynecol Pathol. 2005;24:62–66. [PubMed] [Google Scholar]
- 43.Majumder M, Xin X, Liu L, Tutunea-Fatan E, Rodriguez-Torres M, Vincent K, et al. COX-2 Induces Breast Cancer Stem Cells via EP4/PI3K/AKT/NOTCH/WNT Axis. Stem Cells. 2016 doi: 10.1002/stem.2426. [DOI] [PubMed] [Google Scholar]
- 44.Wen Z, Liu H, Li M, Li B, Gao W, Shao Q, et al. Increased metabolites of 5-lipoxygenase from hypoxic ovarian cancer cells promote tumor-associated macrophage infiltration. Oncogene. 2015;34:1241–1252. doi: 10.1038/onc.2014.85. [DOI] [PubMed] [Google Scholar]
- 45.White KL, Schildkraut JM, Palmieri RT, Iversen ES, Jr, Berchuck A, Vierkant RA, et al. Ovarian cancer risk associated with inherited inflammation-related variants. Cancer Res. 2012;72:1064–1069. doi: 10.1158/0008-5472.CAN-11-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balansky R, Ganchev G, Iltcheva M, Nikolov M, Maestra SL, Micale RT, et al. Modulation by licofelone and celecoxib of experimentally induced cancer and preneoplastic lesions in mice exposed to cigarette smoke. Curr Cancer Drug Targets. 2015;15:188–195. doi: 10.2174/1568009615666150216170008. [DOI] [PubMed] [Google Scholar]
- 47.Mohammed A, Janakiram NB, Madka V, Brewer M, Ritchie RL, Lightfoot S, et al. Targeting pancreatitis blocks tumor-initiating stem cells and pancreatic cancer progression. Oncotarget. 2015;6:15524–15539. doi: 10.18632/oncotarget.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Legge F, Paglia A, D'Asta M, Fuoco G, Scambia G, Ferrandina G. Phase II study of the combination carboplatin plus celecoxib in heavily pre-treated recurrent ovarian cancer patients. BMC Cancer. 2011;11:214. doi: 10.1186/1471-2407-11-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bias P, Buchner A, Klesser B, Laufer S. The gastrointestinal tolerability of the LOX/COX inhibitor, licofelone, is similar to placebo and superior to naproxen therapy in healthy volunteers: results from a randomized, controlled trial. Am J Gastroenterol. 2004;99:611–618. doi: 10.1111/j.1572-0241.2004.04133.x. [DOI] [PubMed] [Google Scholar]
- 50.Vaish V, Tanwar L, Kaur J, Sanyal SN. Chemopreventive effects of non-steroidal anti-inflammatory drugs in early neoplasm of experimental colorectal cancer: an apoptosome study. J Gastrointest Cancer. 2011;42:195–203. doi: 10.1007/s12029-010-9188-2. [DOI] [PubMed] [Google Scholar]
- 51.Trabert B, Ness RB, Lo-Ciganic WH, Murphy MA, Goode EL, Poole EM, et al. Aspirin, nonaspirin nonsteroidal anti-inflammatory drug, and acetaminophen use and risk of invasive epithelial ovarian cancer: a pooled analysis in the Ovarian Cancer Association Consortium. J Natl Cancer Inst. 2014;106:djt431. doi: 10.1093/jnci/djt431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasto A, Bellio C, Pilotto G, Ciminale V, Silic-Benussi M, Guzzo G, et al. Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget. 2014;5:4305–4319. doi: 10.18632/oncotarget.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao J, Qian F, Tchabo N, Mhawech-Fauceglia P, Beck A, Qian Z, et al. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS One. 2014;9:e84941. doi: 10.1371/journal.pone.0084941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fiorillo M, Lamb R, Tanowitz HB, Mutti L, Krstic-Demonacos M, Cappello AR, et al. Repurposing atovaquone: targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget. 2016;7:34084–34099. doi: 10.18632/oncotarget.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavolari S, Bonafe M, Marini M, Ferreri C, Bartolini G, Brighenti E, et al. Licofelone, a dual COX/5-LOX inhibitor, induces apoptosis in HCA-7 colon cancer cells through the mitochondrial pathway independently from its ability to affect the arachidonic acid cascade. Carcinogenesis. 2008;29:371–380. doi: 10.1093/carcin/bgm265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.