Abstract
Although Bacillus cereus is of particular concern in food safety and public health, the role of other Bacillus species was overlooked. Therefore, we investigated the presence of eight enterotoxigenic genes, a hemolytic gene and phenotypic antibiotic resistance profiles of Bacillus species in retail meat samples. From 255 samples, 124 Bacillus isolates were recovered, 27 belonged to B. cereus and 97 were non-B. cereus species. Interestingly, the non-B. cereus isolates carried the virulence genes and exhibited phenotypic virulence characteristics as the B. cereus. However, correlation matrix analysis revealed the B. cereus group positively correlates with the presence of the genes hblA, hblC, and plc, and the detection of hemolysis (p < 0.05), while the other Bacillus sp. groups are negatively correlated. Tests for antimicrobial resistance against ten antibiotics revealed extensive drug and multi-drug resistant isolates. Statistical analyses didn’t support a correlation of antibiotic resistance to tested virulence factors suggesting independence of these phenotypic markers and virulence genes. Of special interest was the isolation of Paenibacillus alvei and Geobacillus stearothermophilus from the imported meat samples being the first recorded. The isolation of non-B. cereus species carrying enterotoxigenic genes in meat within Egypt, suggests their impact on food safety and public health and should therefore not be minimised, posing an area that requires further research.
Introduction
The diverse genus Bacillus includes harmless environmental and pathogenic species. The B. cereus group are closely related including B. anthracis, B. thuringiensis, B. mycoides, and B. cereus which are known pathogens or opportunistic pathogens to humans1. The B. subtilis group, including B. mojavensis, B. pumilus, B. fusiformis, B. licheniformis and B. subtilis, are common to soil1. The B. cereus and B. subtilis groups have been implicated in food poisoning as a result of intoxication1, either by the consumption of food containing pre-formed toxin or toxins produced by these bacteria in the human gut2–5. Bacillus circulans, B. lentus, B. amyloliquefaciens, B. simplex, B. firmus, and B. megaterium, have been rated as insignificant and disregarded in food poisoning episodes, however their presence and consequent production of enterotoxins and emetic toxins has been increasingly documented and confirmed by cellular assays6. Other aerobic spore-forming bacteria including the genera Sporosarcina, Paenisporosarcina, Brevibacillus, Paenibacillus, and Geobacillus have the potentiality to form biofilms within pipes and stainless steel equipment and to the resist industrial pasteurization posing a hazard for the food industry.
The presence of environmental isolates of Bacillus spp. harboring one or more enterotoxin gene holds crucial importance for the food safety, however, evaluation of toxin gene presence and toxin activity in Bacillus spp. other than B. cereus has not been thoroughly investigated. The diarrheal enterotoxins, Cytotoxin K (encoded by the cytK gene) and enterotoxin FM (encoded by the entFM genes), hemolysin BL (encoded by the hbl operon) and non-hemolytic enterotoxin (encoded by the nhe genes) are regarded as the main enterotoxins involved in foodborne illness studied in B. cereus7,8. There is a lack of information on the presence of these enterotoxins in the non-B. cereus groups.
As Egypt depends on imported beef meat, which reached 200,200 metric tons in the year 20169, to complement the animal protein demands, it is crucial to assess the safety of both imported meat and local products. There is increasing concern about imported meat and question whether Egypt’s food safety system can protect them from tainted foreign products. Compared to the U.S.A., EU or Australia, the food safety standards in Egypt and other underdeveloped countries are not as high. The aim of this study was to determine if the presence of enterotoxins and other putative virulence factors are contributing to the total cytotoxicity of the enterotoxic Bacillus spp. isolated from retail meat in the Egyptian market. Specifically, we have determined incidence and level of contamination with B. cereus and non-B. cereus groups in retail chicken and local and imported beef. We performed a comparative overview on the observed hemolysin BL production, cytotoxic activity, swarming, ability to form biofilms, phenotypic antibiotic resistance profile and enterotoxin gene profile (NHE, HBL and CytK)10–12.
There is an intricate link between metabolism, swarming, biofilm production, antibiotic resistance and virulence. Two soluble virulence factors, lecithinase and amylase production, were noted as phenotypic virulence markers13,14. Swarming motility was tested because it is generally considered a precursor step to biofilm formation and a major factor in pathogenesis by many human pathogens15. The presence of swarming generally increases virulence as movement over surfaces enable bacteria to migrate from sites of infection, offers protection from macrophages as swarming cells were shown to have enhanced resistance to engulfment, and become resistant to a broad range of antibiotics during swarming16–18. Biofilms are considered to promote adhesion and protect cells from antimicrobials and other external insults19. Amyloid-like fibrils have been shown to increase the biofilms’ structural integrity and allow staining with certain dyes such as Congo red20–24. However, results depending on phenotypic markers alone lack reliability in determining the virulence of Bacillus strains.
In this study, we carried out a m-PCR to detect the genes for three pore-forming enterotoxins, responsible for the diarrheal type of food poisoning: hemolysin BL (HBL), non-hemolytic enterotoxin (NHE), cytotoxin K (CytK) and enterotoxin FM (EntFM), and the hemolytic gene encoding phospholipase (plc)1,10–12 in B. cereus (n = 24) and non-B. cereus isolates (n = 97) collected from retail chicken local and imported beef meat. In addition, we calculated the level of antibiotic resistance and the multiple-antibiotic resistance indices of isolates present in meat samples.
Results
Prevalence
The 255 meat samples yielded 124 Bacillus spp. isolates containing 66 B. cereus group isolates (53.2%; B. cereus, B. thuringiensis, and B. mycoides,), 23 B. subtilis group isolates (18.5%; B. licheniformis and B. pumilus) and 35 other Bacillus spp. (28.2%; B. coagulans, B. megaterium, B. sphaericus, B. brevis, G. stearothermophilus, and P. alvei) (Fig. 1). The prevalence of the B. cereus group was 50% (33/66) of isolates within retail chicken, 66.7% (20/30) of isolates from local beef, and 46.4% (13/28) of imported meat isolates (Fig. 1). The B. subtilis group represented 21.2% (14/66), 13.3% (4/30), and 17.9% (5/28), of isolates from retail chicken, local, and imported beef, respectively. Other Bacillus spp. were isolated from chicken, local beef, and imported beef as 28.8% (19/66), 20.0% (6/30), and 35.7% (10/28) of the isolates, respectively. Out of the 124 Bacillus isolates, 66 isolates were obtained from 156 chicken meats samples and 58 isolates were recovered from the 99 beef meat. There was no significant difference between source of isolates (chicken, local and imported beef meat) in relation to the different species isolated (p > 0.06), except for G. stearothermophilus and P. alvei which were signifcantly greater in imported beef meat compared to chicken meat (p = 0.014) (Fig. 2).
Figure 1.
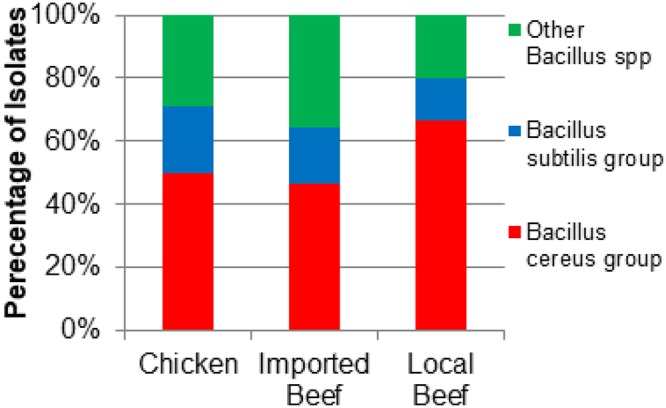
Distribution of isolates from the different sources of meat within the different Bacillus spp. groups. (Chicken, n = 66; Imported Beef, n = 28, Local Beef, n = 30).
Figure 2.
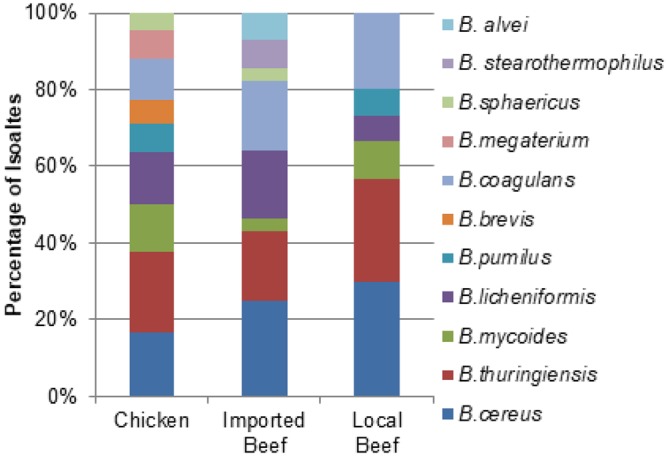
Distribution of isolates of different Bacillus species isolated from the different sources of meat. (Chicken, n = 66; Imported Beef, n = 28; Local Beef, n = 30).
Phenotypic assessment of virulence factors
Biofilm production was observed in 105/124 isolates (84.5%) (Table 1). Among the biofilm producers, the abundance was 89.4% in the chicken isolates, 83.3% in the local beef meat, and 75% in the isolates from imported frozen meat.
Table 1.
Phenotypes virulence factors distribution throughout the B. cereus and non-B. cereus strains isolated from chicken and beef meat samples.
| Source of samples | Bacillus groups | Bacillus species | Virulence phenotypes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Congo red agar test | Biofilm production | Hemolysis | Cytotoxicity | |||||||
| Weak | Moderate | Strong | α- | β- | γ- | |||||
| Chicken (n = 66) | ||||||||||
| Bacillus cereus group | cereus (n = 11) | 7 | 2 | 3 | 4 | — | 11 | — | 10 | |
| thuringiensis (n = 14) | 10 | 3 | 7 | 2 | — | 14 | — | 10 | ||
| mycoides (n = 8) | 6 | 1 | 4 | 3 | 1 | 7 | — | 7 | ||
| Bacillus subtilis group | licheniformis (n = 9) | 6 | 3 | 3 | 2 | 1 | 8 | — | 4 | |
| pumilus (n = 5) | 2 | 2 | — | 2 | — | 5 | — | 2 | ||
| Other Bacillus spp | coagulans (n = 7) | 4 | 3 | 3 | 1 | 3 | 4 | — | 4 | |
| megaterium (n = 5) | 3 | 2 | 2 | 1 | 1 | 4 | — | 3 | ||
| sphaericus (n = 3) | 3 | — | 1 | 2 | — | — | 3 | 3 | ||
| Bervibacillus | brevis (n = 4) | 3 | 1 | 2 | — | — | 4 | — | 3 | |
| Total | 42 (63.6%) | 17 (25.6%) | 25 (37.9%) | 17 (25.6%) | 6 (9.1%) | 57 (86.4%) | 3 (4.6%) | 46 (69.7%) | ||
| Local beef meat (n = 30) | ||||||||||
| Bacillus cereus group | cereus (n = 9) | 6 | 2 | 4 | 3 | 1 | 8 | — | 9 | |
| thuringiensis (n = 8) | 5 | 1 | 2 | 2 | — | 8 | — | 6 | ||
| mycoides (n = 3) | 2 | 1 | 2 | — | 3 | — | — | 3 | ||
| Bacillus subtilis group | licheniformis (n = 2) | 2 | — | — | 2 | — | 2 | — | 2 | |
| pumilus (n = 2) | 2 | — | — | 1 | 1 | 1 | — | 1 | ||
| Other Bacillus spp | coagulans (n = 6) | 3 | 2 | 1 | 2 | 1 | 3 | 2 | 4 | |
| Total | 20 (66.7%) | 6 (20%) | 9 (30%) | 10 (33.3%) | 6 (20%) | 22 (73.3%) | 2 (6.7%) | 25 (83.3%) | ||
| Frozen beef meat (n = 28) | ||||||||||
| Bacillus cereus group | cereus (n = 7) | 3 | 4 | 1 | — | 1 | 6 | — | 6 | |
| thuringiensis (n = 5) | 2 | 2 | — | 1 | 1 | 3 | 1 | 2 | ||
| mycoides (n = 1) | — | 1 | — | — | — | 1 | — | 1 | ||
| Bacillus subtilis group | licheniformis (n = 5) | 4 | 1 | 3 | 1 | 1 | 4 | — | 4 | |
| Other Bacillus spp | coagulans (n = 5) | 4 | 1 | 1 | 2 | 3 | 1 | 1 | 3 | |
| sphaericus (n = 1) | 1 | 1 | — | — | — | 1 | 1 | |||
| Geobacillus | stearothermophilus (n = 2) | 1 | 1 | — | 1 | 1 | 1 | — | 2 | |
| Paenibacillus | alvei (n = 2) | — | — | — | — | 1 | 1 | — | 2 | |
| Total | 15 (53.6%) | 10 (35.7%) | 6 (21.4%) | 5 (17.9%) | 8 (28.6%) | 17 (60.7%) | 3 (10.7%) | 21 (75%) | ||
| Total (n = 124) | 77 (62.1%) | 33 (26.6%) | 40 (32.3%) | 32 (25.8%) | 20 (16.1%) | 96 (77.4%) | 8 (6.5%) | 92 (74.2%) | ||
The other virulence factors as indicated in Table 1, reveals that 92/124 (74.2%) of the total Bacillus species isolated were cytotoxic to the Vero cells. In addition, β-hemolysis activity was observed on sheep blood agar in 77.4% of isolates, α-hemolysis activity in 16.1%, and the γ-hemolysis activity in 6.5% of the isolates.
Distribution of virulence genes among the B. cereus and non-B.cereus isolates
Eight virulence genes encoding enterotoxins were targeted in the Bacillus isolates based on PCR detection (Table S1). PCR detection included genes encoding hemolytic (hblA, hblC, and hblD) and non-hemolytic (nheA, nheB, and nheC) enterotoxin complexes, cytotoxin K (cytK), enterotoxin FM (entFM) and one hemolytic gene encoding phospholipase (plc) (Fig. 3). At least one of the nine genes were detected in all the isolates except one B. sphaericus isolated from the frozen imported beef meat. The isolated Bacillus commonly possessed cytk (58.9%) as the most prevalent toxin gene followed by hblA (45.2%), plc (40.3%) and entFM (35.5%) genes while the nheB gene was the least present (12.9%).
Figure 3.
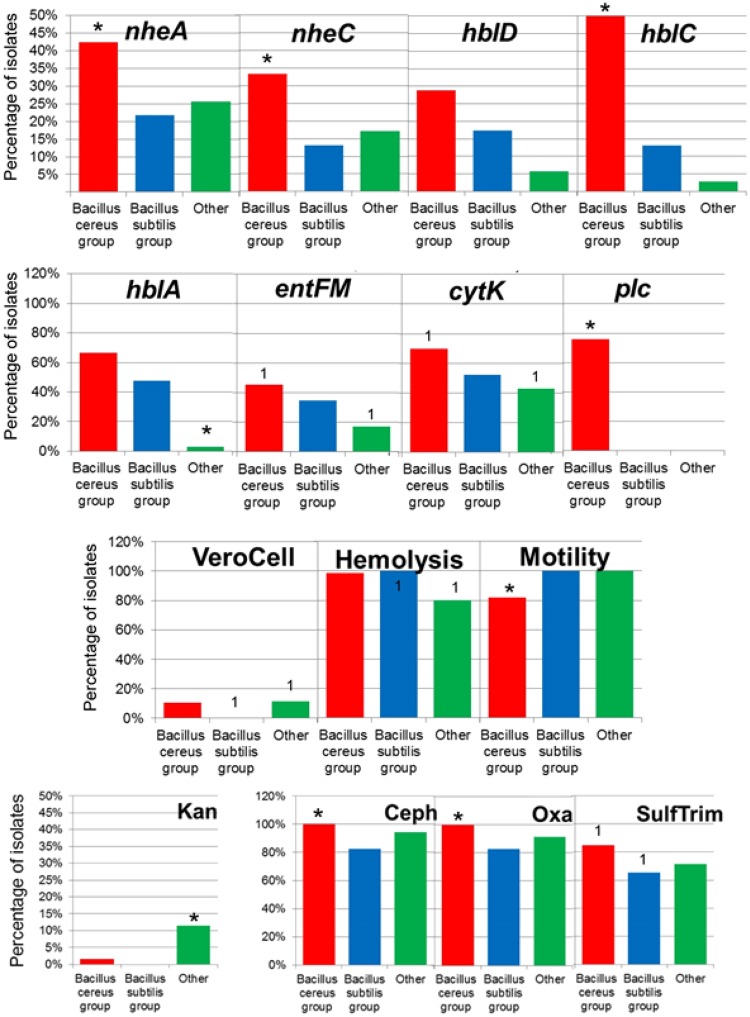
Percentage of isolates showing virulence genes, Vero cell, hemolysis, motility and antibiotic resistance (Kanamycin, KAM; Cephalothin, CEPH; Oxacillin, OXA; Sulfamethazole/Trimethoprim, SulfTrim) between Bacillus groups. Asterisks represent a group significantly different from the other two groups (p < 0.05) and numbers (example: “1”) represent groups that are significantly different from each other (p < 0.05). (B. cereus group, n = 66; B. subtilis group, n = 23; Other Bacillus group, n = 35).
The non-hemolytic genes nheA and nheC, and the hemolytic enterotoxin HBL complex gene hblC were significantly more prevalent in the B. cereus group isolates compared to the B. subtilis group, and the other Bacillus spp. isolates (p < 0.05). There was no significant difference between the number of isolates carrying the hblA hemolytic enterotoxin gene in the B. cereus group compared to B. subtilis group (p = 0.055). The other Bacillus spp. isolates showed a significantly lower abundance of the hblA gene compared to the B. cereus and B. subtilis groups (p < 0.05). There was no significant difference in the presence of the nheB gene between the groups (p > 0.13). In regard to the source of isolated Bacillus spp., there were significantly fewer incidences of the enterotoxin FM genes (entFM) within the isolates from raw chicken compared to fresh local or imported frozen meat (p < 0.05).
There were no significant difference in slime (CR) and biofilm (CV) production of the isolates in the different Bacillus spp. groups (p > 0.05). More isolates from the B. cereus group had a gene encoding phospholipase (plc), lecithinase activity, amylase activity, and significantly fewer isolates showing swarming (Fig. 3; p < 0.05).
Antimicrobial resistance prevalence
The isolates were tested for susceptibility against 10 antibiotics representing 8 classes (Table S2). The classes were glycopeptides (vancomycin), β-lactams (penicillin, oxacillin, cephalothin), quinoline (nalidixic acid), sulphonamides (sulfamethoxazole/trimepthoprim), phenicols (chloramphenicol), tetracyclines (tetracycline), aminoglycosides (kanamycin) and macrolids (erythromycin). The 124 isolates displayed a very high level of antibiotic resistances. The highest resistance was displayed to penicillin G (124/124, 100%) while the lowest resistance was recorded against the clinically important antibiotic vancomycin (2/124, 1.6%). A resistance >90% was then recorded to cephalothin and oxacillin. The 124 isolates were resistant to sulfamethazole/trimethoprim, nalidixic acid, erythromycin and tetracycline at 77.4%, 62.9%, 20.2% and 17.74%, respectively. Resistance to kanamycin was lower (4%). Resistance against chloramphenicol, an antibiotic still in use in clinic in Egypt was evidently low, with a total of 3.2% isolates being resistant (3/66 isolates from the chicken and 1/30 isolates from local beef meat).
As for the differences between the Bacillus groups, prevalence of antibiotic resistance in P. alvei and G. stearothermophilus are notably lower compared to other species. Resistance prevalence in P. alvei, G. stearothermophilus did not significantly differ compared with B. cereus, B. thuringiensis, B. mycoides, B. licheniformis, and B. coagulans, although this observation may be due to the low number of these species isolated. Resistance to the antibiotics oxacillin, cephalothin, and the antibiotic combination sulfamethazole/trimethoprim was significant more prevalent within isolates of the Bacillus cereus group compared to the B. subtilis group and other Bacillus spp. isolates (Fig. 4, p < 0.05). There was no significant difference in the percentage of isolates with antibiotic resistance to vancomycin, nalidixic acid, chloramphenicol, tetracycline, and erythromycin between the Bacillus spp. groups (p > 0.05). Kanamycin resistance was significantly greater in the other Bacillus group compared to the B. cereus and B. subtilis groups (p < 0.05).
Figure 4.
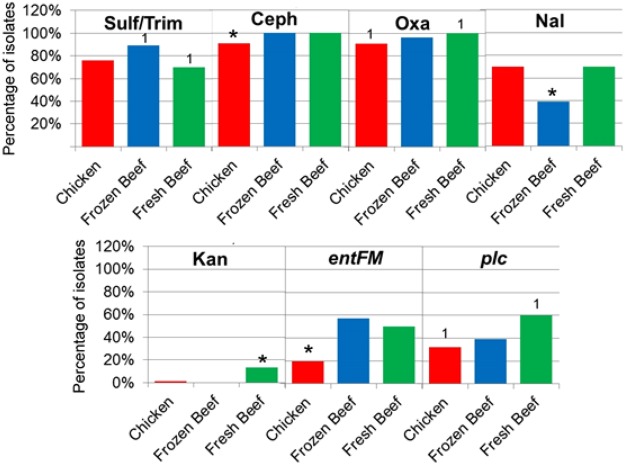
Percentage of isolates within the different meat sources demonstrating antibiotic resistance (Kanamycin, KAM; Cephalothin, CEPH; Oxacillin, OXA; Nalidixic acid, NAL; Sulfamethazole/Trimethoprim, SulfTrim) and virulence genes. Asterisks represent a group significantly different from the other two groups (p < 0.05) and numbers (example: “1”) represent groups that are significantly different from each other (p < 0.05). (Chicken, n = 66; Imported Beef, n = 28; Local Beef, n = 30).
Abundance of Antibiotic resistant Bacillus isolates from different meat sources
Bacillus spp. antimicrobial susceptibility results were stratified by isolate, source (raw chicken, local beef meat and imported beef meat) and Bacillus spp. group (Table S3).
The different source of meat showed few statistically significant variations in abundance of isolates resistant to the 10 antibiotics tested (Fig. 4). The Bacillus spp. isolates from raw retail chicken showed significantly fewer incidences of resistance to the antibiotic cephalothin (p < 0.05) compared to the isolates from local beef and imported beef. Isolated Bacillus spp. from local beef meat showed significantly more isolates resistant to oxacillin (p < 0.05) compared to isolates from raw chicken and no significant difference compared to imported beef (p > 0.15). The Bacillus spp. isolated from imported beef showed significantly fewer isolates with nalidixic acid resistance compared to isolates from raw chicken and local beef meat (p < 0.01). Isolates from local beef meat showed significantly more isolates with kanamycin resistance compared to isolates from raw chicken and imported beef meat (p < 0.02). There were significantly more isolates (p = 0.04) from imported beef with resistance to the drug combination sulfamethazole/trimethoprim compared to isolates from local beef, but there was no significant difference compared with raw chicken (p = 0.07).
Bacillus spp. resistance profile
Multiple drug resistance (MDR) for Bacillus spp. was defined as a strain non-susceptible to ≥1 antibiotic in ≥3 antibiotic classes, extensive drug-resistance (XDR) was defined as a Bacillus spp. strain non-susceptible to ≥1 antibiotic in all but ≤2 antibiotic classes and pan drug resistance (PDR) for those Bacillus spp. resistant to representatives of the 8 classes of antibiotics tested. (The antibiotics and their classes are listed in Table S2). In general, the strains were resistant to 1–7 antibiotics represented in 1–5 classes. Overall, the least number of classes was one represented by penicillin and the highest number of classes was five. The total number of MDR isolates were 85 (68.5%) and no XDR nor PDR were detected (Table S3). The MDR profiles of the Bacillus spp. collected from chicken and beef (local and imported) meat samples, resistant to the ten tested antibiotics are summarized in Table S3.
MARindex
To survey the relative predominance of resistant Bacillus isolates from raw chicken, raw local beef meat and imported frozen raw beef meat, MAR (multidrug antibiotic resistance) indices were calculated. For the raw chicken samples, the MAR index range () for chicken was 0.1–0.7 (0.4), raw local beef meat 0.3–0.7 (0.5), and imported frozen raw beef meat 0.4–0.7 (0.5). The relatively high MAR indices indicate a high-risk level of food contamination with antibiotic resistant virulent Bacillus strains.
Correlation analysis of measured variables: virulence, antibiotic resistance and meat source
Permutational MANOVA analysis using the isolated Bacillus spp. groups, isolate meat source, and the interaction term as terms within the model indicated significant difference in all three terms (p = 0.001, 0.019, and 0.002, respectively). Pairwise comparisons showed no significant difference between sources of isolates (p > 0.081) indicating little variation between meat sources. Comparisons between groups showed significant difference between all three groups (p ≤ 0.023) suggesting variation between isolates related to phenotypic and biochemical testing were related to the Bacillus group.
Associations of variables were identified within the hierarchical clustering of the heatmap (Fig. 5), PCA analysis (Fig. 6), and correlation matrix (Fig. 7). The correlation matrix indicated a stronger association of antibiotic resistance with one another than other variables such as hemolysis genes and biochemical tests. The lack of correlations between antibiotic resistance and other phenotypic traits or virulent genes suggest independence of antibiotic resistance to virulence.
Figure 5.
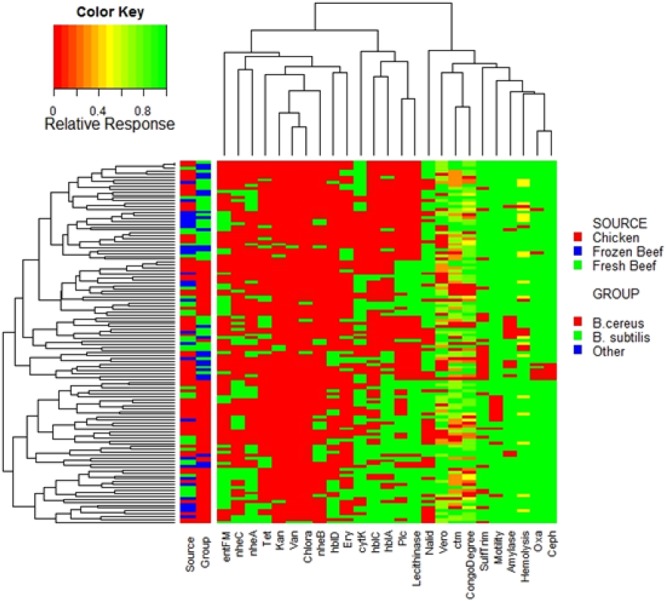
Heatmap of Individual isolates showing hierarchical clustering of isolates and factors. Binary factors (such as antibiotics or genes) indicating presence as green (relative response 1) or absence as red (relative response 0). Factors containing greater ranges were adjusted to range from 0 to 1 as indicated by color key.
Figure 6.
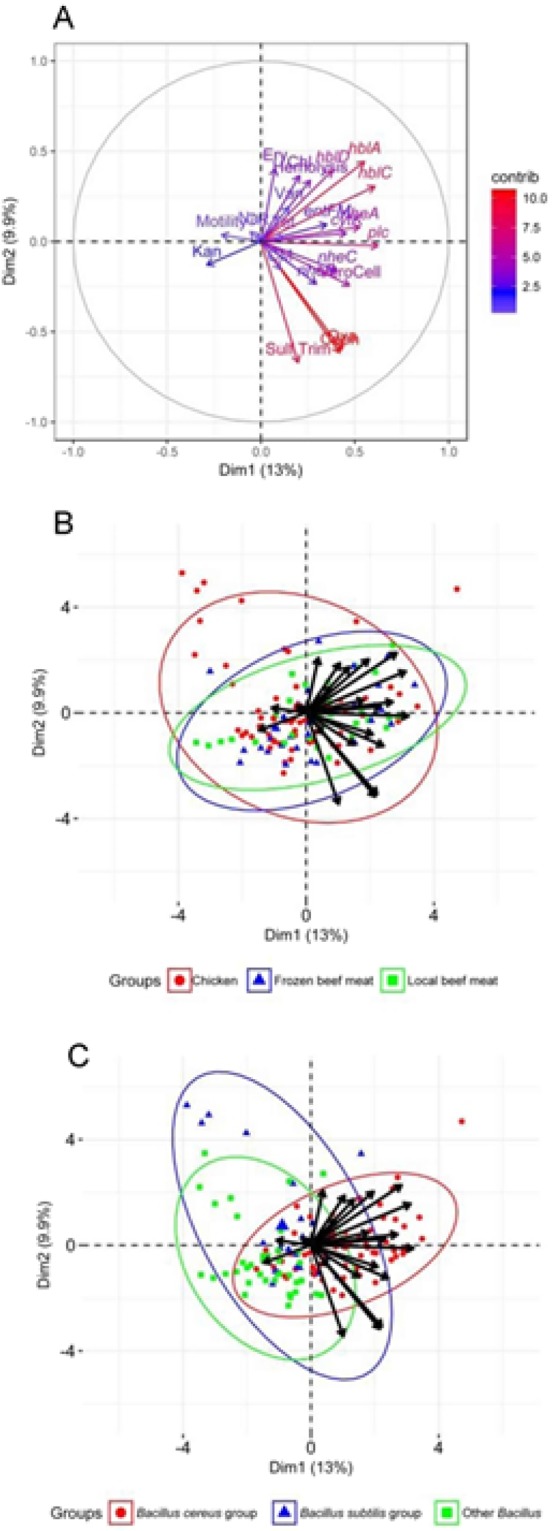
Principle component analysis of factor contribution (A) and relationship with groups (B) and meat source. (C) Ellipses represent 95% confidence intervals.
Figure 7.
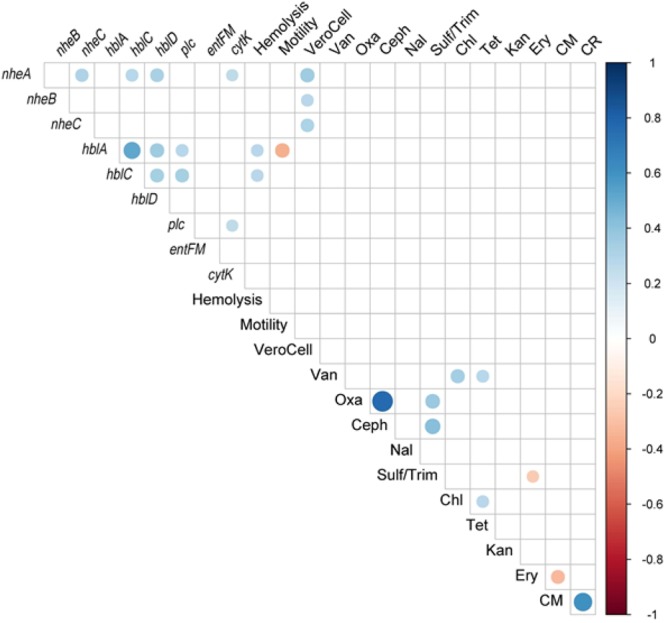
Spearman correlation matrix of phenotypic variables (antibiotic resistance, hemolytic genes, biochemical activity, and biofilm formation). Correlation matrix shows only significant (p < 0.05) correlations.
The PCA analysis (Fig. 6) also revealed a low association of antibiotic resistance to virulence factors. The variables were also unable to separate the isolates of the Bacillus spp. groups into very distinct groups indicating no clear delineation of trait to a specific group. However, increased number of isolates from groups and sources may yield greater separation of the groups.
The correlation matrices (Fig. 7) indicate the B. cereus group positively correlated with the presence of the genes hblA, hblC, and plc, and the detection of hemolysis (p < 0.05), while the other Bacillus sp. group were negatively correlated (p < 0.05). Vero cell toxicity was significantly positively correlated with the presence of nheA, nheB, and nheC genes (p < 0.05) suggesting these gene products contributed to the toxicity observed. The presence of the cytK gene was positively correlated with the presence of nheA and plc genes (p < 0.05). Hemolysis was also positively correlated with the presence of the hblA and hblC genes. Additionally, the B. cereus group was positively correlated with oxacillin resistance and negatively correlated with swarming motility. The isolates of the B. subtilis group were negatively correlated with the presence of the plc gene and cephalothin resistance (p < 0.05). The correlation matrix (Fig. 7) did not identify significant correlations between antibiotic resistances and other variables with the exception of a negative correlation of erythromycin resistance and the Casein-Mannitol (CM) methods for biofilm assessment. Chloramphenicol resistance positively correlated with vancomycin, tetracycline, and erythromycin resistance (p < 0.05). There was a strong significant positive correlation of oxacillin resistance and cephalothin resistance (p < 0.05) which was evident in the MDR strains (Table S2). The CM biofilm assessment method correlated significantly with CR as expected as both are associated with degrees of biofilm formation.
Discussion
The widespread detection of Bacillus spp. carrying enterotoxin-enconding genes in both chicken and beef implicates the possible hazardous impact on public health of these bacteria. We show for the first time non-B. cereus strains bearing an inventory of enterotoxigenic genes similar to B. cereus strains that could be the cause of diarrheal illness related to non-B. cereus strains as previously reported13,25 and thus should be considered as a risk for consumers. Further in-depth study of the sequences of the enteroxigenic genes distributed in the Bacillus genus may yield an explanation of the origin of the genes in non-B. cereus strains and the possibility of horizontal gene transfer between strains and species.
The Bacillus cereus sensu lato group are known to be involved in food poisoning26 although not always a notifiable disease in the majority of the international locations; therefore, incidence statistics is restrained even though fatal incidences were pronounced. Food poisoning caused by Bacillus cereus occurs year-round diffusely over a geographic area27. Bacillus cereus induced 0.7% of foodborne disease of the 31 primary pathogens within the US28 while in the Netherlands B. cereus was reported as a major causative agent in 5.4% of the foodborne outbreaks in 2006 and 32% of foodborne outbreaks in Norway in 200028. B. cereus is reported as the fourth and second major cause of notified FoodBorne Organisms in the European Union and France, respectively26. Meat dishes were the second most commonly implicated foods in B. cereus outbreaks29 and 24% of B. cereus outbreaks were associated with meat or poultry dishes27,28,30.
In addition to the potential risk of non-B. cereus strains to consumers the specific isolation of Paenibacillus alvei (formerly Bacillus alvei) from the imported frozen meat samples is of concern. P. alvei has been one of 22 Paenibacillus species to be recognized as responsible for transitory or authentic infections in human clinical samples31. Our study showed a prevalence at 25.8% of B. cereus group and 48.6% Bacillus spp. of 255 meat samples consistent with earlier studies reporting rates of incidences in raw beef and chicken in different countries between 23.5% and 80%25,30,32–40. This variation might be due to differences in the hygienic practices executed in the meat shops in different countries and even in the same country. It should also be emphasized that, a critical control point for microbial contamination is the storage of food at ambient temperature (room temperature) of about 5–6 h and improper cooking of food before consumption which favors endospore germination producing an increase in the total Bacillus spp. count39,40. B. cereus survives not only at room temperature41, but it is also found in heat treated meat33. The B. cereus spores are resistant to heat used in cooking leading to inadequate meat preparation allowing these pathogens to germinate, replicate, and potentially produce heat stable toxins39,42. The initial contamination of the meat might have occurred in the environment as Bacillus spp. are ubiquitous bacteria in soil, in intestinal tracts of animals, and in a variety of foods and ingredients39. Bacterial contamination of meat can also occur during processing at the slaughter house, from the water, air, soil, the workers and equipment involved or the carcass itself43–45.
The virulence factors responsible for enterotoxins production in B. cereus are primarily HBL, NHE and cytotoxin K11,12. Diarrhea can be caused by the production of one or more enterotoxin by vegetative cells in the small intestine1. High detection rates between 40 and 70% in B. cereus isolated from food origin of the HBL gene complex have been reported46–49. Similarly, high prevalence among B. cereus isolates from food and environmental sources49–53, as well as in some reference strains54 have previously been reported for the nheABC and entFM gene complexes. All isolates of the B. cereus group carried one or more enterotoxin gene of the nine investigated in this study indicating all B. cereus group isolates could cause diarrheal illness. A similar results was obtained by Smith et al.36 and Yang et al.51. Of the non-B. cereus group there was a 72% detection of one or more enterotoxin gene(s) indicating that this group may also participate in the cause of diarrheal illness1. One or more gene of the NHE complex (nheABC) was detected in 22 (81.5%) of the B. cereus isolates indicate the presence of NHE enterotoxin. A previous study recorded that, almost all isolates contained at least one gene of the NHE complex55. The isolates in the current study indicated the presence of at least one gene of the HBL complex (hblDAC) in 19 isolates (70.4%), and similarly 73% presence of HBL complex was reported in food-poisoning strains50, whereas others found HBL complex in 65.5% and 55.2% in B. cereus isolates30,56. In the present study, fewer isolates contained HBL complex genes compared to NHE complex genes, which is consistent with previous findings30. Although Ngamwongsatit et al.57 and Vyletelova and Banyko58 indicated that the three genes of HBL and NHE complexes form an operon, the present and previous report by Tewari et al.30 indicate that the structural organization of these genes can be different. Interestingly, 32.3% of isolates did not show the presence of any of the HBL complex genes and produced positive β-hemolysis on blood agar. The observed hemolytic activity might be produced by other toxins such as hemolysin I (Cereolysin O)59, hemolysin II60, hemolysin III61 or cytK62. The enterotoxin cytK gene was found in 81.5% of the B. cereus isolates which is similar with the results reported by Ngamwongsatit et al.57, whereas others detected it in smaller percent of their isolates63,64. On the other hand, the entFM gene was found in 35.5% of our Bacillus spp. isolates which is far different from the 100% recorded by Ngamwongsatit et al.57 and the 93% in their B. cereus tested isolates. Smith et al.36 isolated 27 B. cereus strains out of 60 poultry samples, which contained the gene(s) for at least one of the toxins (bceT, nheABC, hblACD), although none of the strains contained the cytK gene. This was also consistent with our results with the exception with the cytK as previously recorded.
The hazardous impact on the food production lines as a consequence of biofilm formation comes from the fact that the biofilms are dynamic structures that can release planktonic cells able to spread and colonize new areas of the production line and equipment65–70. The members of Bacillaceae family65,66 have the property to attach to biotic (meat surfaces) and abiotic (meat processing equipment such as conveyor belts, tables, knives) surfaces67, thus the removal of biofilms in the food processing environment is critical65–70. In the present study, 84.7% of the Bacillus isolates analyzed, were able to form biofilms indicating a high potential for attachment to food processing surfaces and equipment which may lead to increase spread and incidence of illness. These results agree with previous reports that foodborne B. cereus and B. thuringiensis isolates are variable (from no or weak to strong) biofilm producers65. The current study highlighted that the non-B. cereus strains isolated from different meat sources were also variable biofilm producers. To our knowledge, the capacity of these species to form biofilm have not been previously studied. The disruption of biofilm formation is a key control point in eliminating the spread of Bacillus to food and decrease illness.
Antibiotic resistance, precipitated by the overuse of antimicrobials, may rise up from a variety of mechanisms, in particular horizontal gene transfer of virulence and antibiotic resistance genes, that is frequently facilitated with the aid of biofilm formation71–73. Antimicrobial resistance is a growing problem around the world and is associated with increasing mortality and medical costs73. Determining the resistance of B. cereus to antimicrobial agents is critical for treatment during outbreaks. The isolates from beef and chicken in this study showed resistance profile highly consistent with previous reports49,74. While β-lactams have become ineffective, chloramphenicol, an antibiotic in use in Egyptian clinics, would still be effective against the isolates in this study. MAR indices calculated in this study for meat are similar to values of E. coli from poultry farms75 but are approximately double the values for Enterococcus isolates76. The high variability in MAR index and the multi-antibiotic resistance profiles of the isolates indicate a variability in effective treatment and the importance on early determination of antibiotic resistance during infection77. The introduction of resistant B. cereus into the raw meat likely indicates contamination inputs from livestock and poultry operations, during transport of the meat or meat handling at the distribution outputs and poor hygienic practices in meat shops and restaurants29,30,43,44. Because there are no criteria for MARindex for B. cereus, it is difficult to assess human health risks due to presence of antimicrobial resistant B. cereus in the meat.
The deficit of correlation or association of virulence factors and antibiotic resistances tested within the Bacillus spp. isolated in this study are consistent with observations in other species examining the correlation of virulence and antibiotic resistance78,79. The negative correlation of biofilm formation and resistance to the macrolide erythromycin might be selected for in part due to increased resistance to antimicrobial within the biofilm80. The biofilm formation in vivo maybe sufficient for resistance to macrolides at concentrations the isolates are exposed to, but not during our in vitro assay where biofilm formation is unlikely. Interestingly, He et al.81 showed that some strains of Staphylococcus epidermidis when exposed to sub-inhibitory concentrations of erythromycin showed increased expression of the resistance gene ermC with decreased biofilm formation. A similar complex relationship of biofilm formation and antibiotic resistance maybe present in the isolates in this study. The variability of virulence and antibiotic resistance profiles within the isolates are usually associated with genomic plasticity79,82 and Bacillus spp. are considered to have a plastic genome83,84. While plasmids may contain both virulence and antibiotic resistance genes85,86, the majority of plasmids currently sequenced and described within Bacillus spp. do not87–90. This lack of co-location on plasmids of virulence and antibiotic resistance genes may explain the lack of association of these factors within the isolates. Increased number of isolates and tested food products is needed to fully understand the relationship between virulence and antimicrobial resistance in Bacillus spp. and other pathogenic bacteria and further gene expression studies could also contribute to better understand the relationship between virulence and resistance.
Conclusion
Considering food safety issues, this provides confirmation that B. cereus and non-B. cereus must be considered important food-borne pathogens and underlines the need to improve monitoring. The present investigation highlights for the first time an initial finding that the non-B. cereus group poses a public health risk to consumers as a consequence to their carriage of the enterotoxigenic virulence genes and exhibiting phenotypic virulence characteristics (cytotoxicity and haemolytic activity, as well as MDR and biofilm formation) as the B. cereus. In addition, the importation of meat involves a degree of disease risk to the importing country through the introduction of pathogens previously uncommon in Egypt91. Further research is needed to better assess the risk pose to public health by non-B. cereus species.
Materials and Methods
Isolation and identification of Bacillus spp
A total of 255 retail meat samples, comprising 156 raw chicken meat and 99 beef meat (59 local and 40 imported) were purchased from different retail outlets in the local markets of Cairo and collected and transported to the laboratory following aseptic and safety precautions.
A stomacher was used to homogenize 10 g of each sample in 90 mL of buffered peptone water (BPW) for 2 min. Heat treatment of all samples at 70 °C for 15 min to was used to eliminate vegetative cells and allow the isolation of spores92. The pasteurized samples were immediately placed in ice to prevent spore germination. An amount of 100 μl was spread on Mannitol–Egg Yolk–Polymyxin (MYP) agar plates and incubated at 37 °C for 24-h both aerobically and anaerobically. The plates were examined and presumptive Bacillus spp. were confirmed based on microscopy of Gram-stained preparations and biochemical tests93. A number of 15–20 colonies were randomly selected and analyzed by cell morphology, Gram staining, ability to form endospores, growth in the presence of sodium chloride, anaerobic growth, catalase and oxidase activity, Voges-Proskauer test and growth at pH 5.792,93. The ability to ferment carbohydrates, starch hydrolysis, use of citrate as a carbon source, lecithinase activity, and growth inhibition by lysozyme were implemented according to methods described previously92,93. Reference strains for the phenotypic tests were Bacillus cereus ATCC 11778 and B. cereus ATCC 14579.
Assessment of antimicrobial resistance phenotypic profile
The Kirby–Bauer disk diffusion method94 was used to analyze the antibiotic susceptibility patterns of the Bacillus spp. isolates with antibiotic discs representing the following groups/mechanisms: Group I (inhibitors to cell wall synthesis): vancomycin (30 μg), penicillin G (10 U), oxacillin (1 μg), cephalothin (30 µg); Group II (inhibitors to nucleic acid synthesis): nalidixic acid (30 µg), sulfamethazole/trimethoprim (0.5/9.5 μg); Group III (inhibitors to protein synthesis): chloramphenicol (30 μg), tetracycline (30 μg), kanamycin (30 µg) and erythromycin (15 μg). CLSI guidelines were used to designate Bacillus spp. isolates as susceptible, intermediate, or resistant to an antibiotic95. Recent standard definitions were implemented for the phenotypic antibiotic resistance stratification of Bacillus isolates96,97.
Detection of phenotypic virulence factors and toxin encoding genes
Biofilm production
Biofilm production was assessed by the Congo red agar test and Microtiter Plate method as previously described in detail98,99. Staphlococcus epidermidis ATCC 35983, a strong slime producer, was used for positive control.
Congo red agar test
The 124 B. cereus and non-B. cereus isolates were cultured on brain heart infusion (BHI) agar augmented with 36 μg/ml of glucose and 0.8 μg/ml of Congo red. The plates were incubated at 37 °C for 24-h. After an additional 12-h at room temperature, slime production was assessed by colony color. Black colonies are strong slime producers, while red colonies lack slime production.
Microtitre Plate method
A 200 µl of the Bacillus isolates suspension equivalent to 0.8 McFarland (yielding 105 cfu/ml) in tryptone soy broth (TSB) was transferred to a 96-well polystyrene microtiter plate. The bacterial cell suspension was incubated for 18-h at 37 °C. The plates were decanted to remove well contents, washed with running tap water, allowed to dry for 30 min and stained with crystal violet (CV; 25%) for 5 min at room temperature before decantation washing and drying for 30 min. The plates were incubated for 1 min with 200 μl HCl 25% in each well. The absorbance at 570 nm of the CV stain were read for each well. Uninoculated wells subjected to the same procedures were used as negative controls.
Swarming activity
Swarming motility was observed as previously described100,101. Briefly, an overnight culture (2 × 108 cells/ml) was spotted (0.5 μl) onto the center of TrM plates (1% tryptone, 0.5%, NaCl, 0.25% agar). After 6–8 h incubation at 37 °C in a humidified chamber, the diameter of halos generated by growth were measured.
Detection of Hemolysin BL by blood agar plates
Hemolysin BL (HBL), a primary virulence factor, is a three-component enterotoxin consisting of hblA, hblD and hblC genes encoding a binding component B and two lytic components L1 and L2, respectively. The production of hemolysin BL enterotoxin of B. cereus isolates was demonstrated by discontinuous double hemolysis pattern on blood agar plates102. Overnight cultures of Bacillus isolates in BHI broth with 0.1% glucose (BHIG) were used to inoculate blood agar plates (Columbia agar +5% sheep-blood, Oxoid) by spot inoculation. The sheep blood agar plates were incubated at 24 °C and were frequently observed between 12 and 72 h. B. cereus ATCC 14579 and B. cereus INRA C15 were used as positive controls. B. cereus NC 1291 was used as a negative control.
Vero cell cytotoxicity assay
Bacillus spp. isolates were inoculated into 10 ml BHIG and incubated at 30 °C for 24-h at 150 rpm on a rotary shaker. Following incubation, 100 µl of culture was diluted into fresh 10 ml BHIG and re-incubated. The supernatant was harvested by centrifugation at 5000 g for 5 minutes and filtered with a 0.22 μm syringe filter (Millipore). Vero cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS) at 37 °C. Trypsinized Vero cells were diluted to 106 cells/ml in normal growth medium and 100 µl was placed in the wells of a Falcon 96-well flat bottom cell-culture plates. Plates were incubated until a confluent monolayer of cells was formed, approximately 24-h, at 37 °C103. Isolate-free supernatant was added (100 μl) to the first column of the 96-well plate and serially diluted by 2-fold across the columns of the plate. The plates were incubated for 18-h at 37 °C. Reactions were considered positive if greater than 50% of Vero cells showed detachment from the plate when examined under light microscopy.
PCR detection of virulence genes for enterotoxins in B. cereus
DNA isolation
The 124 isolates of Bacillus spp. isolates were grown in 5 mL nutrient broth with shaking for 18 h at 30 °C and harvested at 5,000 g for 5 min. QIAamp DNA Mini Kit was used for genomic DNA extraction and purification. The concentration and purity of genomic DNA was measured using an Ultraspec 3000 spectrophotometer at the absorbance 260 and 280 nm. PCR was performed to detect eight enterotoxigenic encoding endotoxins genes (hblA, hblC, hblD, nheA, nheB, nheC, cytK and entFM) and one hemolytic gene encoding phospholipases (plc). A positive reference strain of B. cereus ATCC 14579 and sterile MilliQ water as a negative control was used in PCR analysis104,105. Table S4 provides details about the primers used.
Conditions for PCR amplification
PCR reactions were comprised of 25 ng genomic DNA, 10 mM Tris-HCl (pH 8.3), 10 mM potassium chloride, 2.5 mM magnesium chloride, 0.8 mM dNTPs, 1 μM each primer, and 0.5 U of Taq DNA polymerase (Promega Corporation, WI, USA). Ultrapure sterile water was used as non-template DNA control and for the PCR component preparation. A PTC-100 Programmable Thermal Controller was used for PCR amplification. The optimized multiplexPCR utilized a standard 3-step cycling: 3 min at 95 °C; 35 cycles of (1) 94 °C for 30 sec, (2) 54 °C for 45 sec, and (3) 72 °C for 1.5 min; and a final extension at 72 °C for 5 min. PCR reaction were performed in triplicate. After amplification, gel electrophoresis was used to analyze PCR fragments for presence and correct size compared to positive control. PCR runs where a negative control showed amplification or positive control did not amplify were ignored and repeated.
Statistical analyses
Numerical coding was used for antibiotic resistance phenotypic, biochemical results, and gene presence. Detection or absence of a specific gene (eg. hblA) was denoted as 1 and 0, respectively. Hemolysis, Vero Cell, Congo red (CR), and CM analysis results were exchanged with numerical values matching the degree of observed activity with 0 with absence followed by sequential integers (1, 2, etc.). For antibiotic resistance, antibiotic sensitive was denoted as 0 and resistance as 1. The open statistical program R was used for statistical analysis106. Multivariant statistical analysis were carried out using functions in the ‘vegan’ package107. Binomial similarity matrices were calculated for the isolate profiles using ‘vegdist’ function and used in permutational multivariate ANOVA (MANOVA) analyses using the ‘adonis’ function. Multiple pairwise comparisons were conducted using the ‘pairwise.perm.manova’ within the ‘RVAideMemoire’ R package108. A permutational ordianation method was used to determine variables statistical significant variables in further analysis utilizing ‘ordiR2step’ function. All variables, not related to species identification, were signification (p < 0.01). Principal component analysis (PCA) was performed and visualized with the R packages ‘FactoMineR’109 and ‘factoextra’110. The ‘cor’ function was used to calculate correlations and ‘cor.test’ function was used to determine significance between variables. The ‘corrplot’ function from the ‘corrplot’ package was used to visualize significant correlations111. For multiple comparisons, False Discovery Rate was used to adjust p-values112. The function ‘heatmap.3’ in the ‘GMD’ package was used to generate heatmap representations a113. Significant difference between data shown as percentages such as biochemical tests or antibiotic resistance by source or group was determined by proportional Z test.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Electronic supplementary material
Acknowledgements
The authors extend their appreciation and indebtedness to the Deanship of Scientific Research at King Saud University for funding the work through the research group project No. RGP-162.
Author Contributions
K.M.O., A.O. and H.M.Y.Y. performed sampling, biochemical test and isolate characterization. A.D.K. performed statistical analysis and helped in the preparation of the manuscript including the bulk of the final corrections required by the reviewers and Editorial Board. K.M.O. provided resource support. K.R.H., K.S.A.M., A.S.M., T.M.D., H.A.H., I.M.I.M. and A.M.H. helped in the preparation of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Logan NA. Bacillus and relatives in foodborne illness. J. Appl. Microbiol. 2011;112:417–429. doi: 10.1111/j.1365-2672.2011.05204.x. [DOI] [PubMed] [Google Scholar]
- 2.McKillip JL. Prevalence and expression of enterotoxins in Bacillus cereus and other Bacillus spp., a literature review. Antonie Van Leeuwenhoek. 2000;77:393–399. doi: 10.1023/A:1002706906154. [DOI] [PubMed] [Google Scholar]
- 3.From C, Pukall R, Schumann P, Hormazábal V, Granum PE. Toxin-Producing Ability among Bacillus spp. Outside the Bacillus cereus Group. Appl. Environ. Microbiol. 2005;71:1178–1183. doi: 10.1128/AEM.71.3.1178-1183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.PHE. Public Health England. Health protection Infectious diseases Bacillus species (food poisoning). Wellington House 133–155 Waterloo Road London SE1 8UG (2008).
- 5.USDFA. U.S. Food and Drug Administration Bad Bug Book: Foodborne Pathogenic Microorganisms and Natural Toxins Handbook Bacillus cereus and other Bacillus spp. published by the Center for Food Safety and Applied Nutrition (CFSAN) of the Food and Drug Administration (FDA), U.S. Department of Health and Human Services. 10903 New Hampshire Avenue Silver Spring, MD 20993 1-888-INFO-FDA (1-888-463-6332) Page Last Updated: 12/16/2014.
- 6.Gopal N, et al. The Prevalence and Control of Bacillus and Related Spore-Forming Bacteria in the Dairy Industry. Front. Microbiol. 2015;6:1418. doi: 10.3389/fmicb.2015.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granum, P. & Lindbäck, T. Bacillus cereus, p 491–502. In Doyle, M & Buchanan, R (ed), Food Microbiology. ASM Press, Washington, DC, 10.1128/9781555818463.ch19 (2013).
- 8.Walker-York-Moore L, Moore SC, Fox EM. Characterization of Enterotoxigenic Bacillus cereus sensu lato and Staphylococcus aureus Isolates and Associated Enterotoxin Production Dynamics in Milk or Meat-Based Broth. Toxins. 2017;9:225. doi: 10.3390/toxins9070225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MALR. Ministry of Agriculture and Land Reclamation’s (MALR) General Organization for Veterinary Services. Egypt, Livestock and Products Annual, Government Becomes Top Meat Importer in an Effort to Curb Prices and Ensure Availability to More Citizens, Global Agricultural Information Network, USDA Foreign Agricultural Service (2016).
- 10.Stenfors-Arnesen LP, Fagerlund A, Granum PE. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008;32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- 11.Fagerlund A, Lindbäck T, Granum PE. Bacillus cereus cytotoxins Hbl, Nhe and CytK are secreted via the Sec translocation pathway. BMC Microbiology. 2010;10:304. doi: 10.1186/1471-2180-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caro-Astorga, J., Pérez-García, A., de Vicente, A. & Romero, D. A genomic region involved in the formation of adhesin fibers in Bacillus cereus biofilms. Front. Microbiol. 5, Article 745 (2015). [DOI] [PMC free article] [PubMed]
- 13.Phelps RJ, McKillip JL. Enterotoxin Production in Natural Isolates of Bacillaceae outside the Bacillus cereus Group. Appl. Environ. Microbiol. 2002;68:3147–3151. doi: 10.1128/AEM.68.6.3147-3151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgescu M, et al. Virulence and resistance features of Pseudomonas aeruginosa strains isolated from chronic leg ulcers. BMC Infect. Dis. 2016;16(Suppl 1):92. doi: 10.1186/s12879-016-1396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burall LS, et al. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: Identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 2004;72:2922–2938. doi: 10.1128/IAI.72.5.2922-2938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 17.Shrout JD, et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa. biofilm formation is nutritionally conditional. Mol. Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 18.Flemming HC, Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 19.Kearns DB. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Partridge JD, Harshey RM. Swarming: Flexible Roaming Plans. J. Bacteriol. 2013;195:909–918. doi: 10.1128/JB.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cairns LS, Hobley L, Stanley-Wall NR. Biofilm formation by Bacillus subtilis: New insights into regulatory strategies and assembly mechanisms. Mol. Microbiol. 2014;93:587–598. doi: 10.1111/mmi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dragoš A, Kovács AT, Claessen D. The Role of Functional Amyloids in Multicellular Growth and Development of Gram-Positive Bacteria. Biomolecules. 2017;7:60. doi: 10.3390/biom7030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randrianjatovo-Gbalou I, Rouquette P, Lefebvre D, Girbal-Neuhauser E, Marcato-Romain CE. In situ analysis of Bacillus licheniformis biofilms: amyloid-like polymers and eDNA are involved in the adherence and aggregation of the extracellular matrix. J. Appl. Microbiol. 2017;122:1262–1274. doi: 10.1111/jam.13423. [DOI] [PubMed] [Google Scholar]
- 25.EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) Scientific opinion on the risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA J. 2016;14(4524):93. [Google Scholar]
- 26.Glasset, B. et al. Bacillus cereus-induced food-borne outbreaks in France, 2007 to 2014: epidemiology and genetic characterisation. Euro Surveill. 21(48), 30413, 10.2807/1560-7917.ES.2016.21.48.30413 (2016). [DOI] [PMC free article] [PubMed]
- 27.Tewari A, Abdullah S. Bacillus cereus food poisoning: international and Indian perspective. J. Food Sci. Technol. 2015;52:2500–2511. doi: 10.1007/s13197-014-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bintsis T. Foodborne pathogens. AIMS Microbiology. 2017;3:529–563. doi: 10.3934/microbiol.2017.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett SD, Walsh KA, Gould LH. Foodborne Disease Outbreaks Caused by Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus—United States, 1998–2008. Clin. Infect. Dis. 2013;57:425–433. doi: 10.1093/cid/cit244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tewari A, Singh SP, Singh R. Incidence and enterotoxigenic profile of Bacillus cereus in meat and meat products of Uttarakhand, India. J. Food Sci. Technol. 2015;52:1796–1801. doi: 10.1007/s13197-013-1162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sáez-Nieto JA, et al. Paenibacillus spp. isolated from human and environmental samples in Spain: detection of 11 new species. New Microbe and New Infect. 2017;19:19–27. doi: 10.1016/j.nmni.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachhil VN, Negi SK. Bacillus cereus in meat and meat products: public health implications and control. Indian J. Public Health. 1984;28:68–69. [Google Scholar]
- 33.Schlegelova J, Brychta J, Klimova E, Napravnikova E, Babak V. The prevalence of and resistance to antimicrobial agents of Bacillus cereus isolates from foodstuffs. Vet. Med. 2003;48:331–338. doi: 10.17221/5787-VETMED. [DOI] [Google Scholar]
- 34.Sharma CS, Sharma DK, Gill JPS, Aulakh RS, Sharma J. Bacillus cereus from foods of animal origin in India and its public health significance. Acta Vet. Scand. 2003;44:P118. doi: 10.1186/1751-0147-44-S1-P118. [DOI] [Google Scholar]
- 35.Bedi SK, Sharma CS, Gill JPS, Aulakh RS, Sharma JK. B. cereus in meat and meat products: isolation, enumeration and enterotoxigenicity. J. Vet. Public Health. 2004;2:7–10. [Google Scholar]
- 36.Smith DP, Berrang ME, Feldner PW, Phillips RW, Meinersmann RJ. Detection of Bacillus cereus on selected retail chicken products. J. Food Prot. 2004;67:1770–1773. doi: 10.4315/0362-028X-67.8.1770. [DOI] [PubMed] [Google Scholar]
- 37.Guven K, Mutlu MB, Avci O. Incidence and characterization of B. cereus in meat and meat products consumed in turkey. J. Food Saf. 2006;26:30–40. doi: 10.1111/j.1745-4565.2005.00031.x. [DOI] [Google Scholar]
- 38.Mira EKI, Abuzied SMA. Prevalence of B. cereus and its enterotoxin in some cooked and half cooked chicken products. Assiut Vet. Med. J. 2006;52:70–78. [Google Scholar]
- 39.Ceuppens S, et al. Regulation of toxin production by Bacillus cereus and its food safety implications. Cri. Rev. Microbiol. 2011;37:188–213. doi: 10.3109/1040841X.2011.558832. [DOI] [PubMed] [Google Scholar]
- 40.Rao VS, Kumar RN, Kashinath L, Bhaskar V, Polasa K. Microbiological hazard identification and exposure assessment of poultry products sold in various localities of Hyderabad, India. Sci. World J. 2012;2012:7. doi: 10.1100/2012/286494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolstad I. Food poisoning caused by B. cereus—chicken. Nor. Vet. 1990;102:39. [Google Scholar]
- 42.Brown KL. Control of bacterial spores. Br. Med. Bull. 2000;56:158–171. doi: 10.1258/0007142001902860. [DOI] [PubMed] [Google Scholar]
- 43.Ceylan, Z. G. et al. Safe processing and packaging of foods. In: Microbial contaminants & contamination routes in food industry 1st open seminar arranged by SAFOODNET. Food safety and hygiene NETWORKING within new member states and associated candidate countries; FP6-022808-2006 ESPOO, Finland, January 22.23, Edited by Gun, Wirtanen & Satu, Salo Vtt Technical Research Centre of Finland. Julkaisija. Utgivare. Publisher (2007).
- 44.Raaska, L. Microbial ecology in manufacturing paper-based packaging materials for use in food industry. In: Microbial contaminants & contamination routes in food industry 1st open seminar arranged by SAFOODNET. Food safety and hygiene NETWORKING within new member states and associated candidate countries; FP6-022808-2006 ESPOO, Finland, January 22.23, Edited by Gun, Wirtanen & Satu, Salo Vtt Technical Research Centre of Finland. Julkaisija. Utgivare. Publisher (2007).
- 45.Stellato G, et al. Overlap of spoilage microbiota between meat and meat processing 1 environment in small-scale vs large-scale retail distribution. Appl. Environ. Microbiol. 2016;82:4045–4054. doi: 10.1128/AEM.00793-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aragon-Alegro LC, et al. Enterotoxigenic and genetic profiles of Bacillus cereus strains of food origin in Brazil. J. Food Prot. 2008;71:2115–2118. doi: 10.4315/0362-028X-71.10.2115. [DOI] [PubMed] [Google Scholar]
- 47.Rather MA, Aulakh RS, Gill JPS, Rao TS, Hassan MN. Direct detection of Bacillus cereus and its enterotoxigenic genes in meat and meat products by polymerase Chain reaction. J. Adv. Vet. Res. 2011;1:99–104. [Google Scholar]
- 48.Reis AL, Montanhini M, Bittencourt JV, Destro MT, Bersot LS. Gene detection and toxin production evaluation of hemolysin BL of Bacillus cereus isolated from milk and dairy products marketed in Brazil. Braz. J. Microbiol. 2013;44:1195–1198. doi: 10.1590/S1517-83822013000400024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owusu-Kwarteng J, Wuni A, Akabanda F, Tano-Debrah K, Jespersen L. Prevalence, virulence factor genes and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol. 2017;17:65. doi: 10.1186/s12866-017-0975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guinebretière MH, Broussolle V. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 2002;40:3053–3056. doi: 10.1128/JCM.40.8.3053-3056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang I, et al. Establishment of a novel multiplex PCR assay and detection of toxigenic strains of the species in the Bacillus cereus group. J. Food Prot. 2005;68:2123–2130. doi: 10.4315/0362-028X-68.10.2123. [DOI] [PubMed] [Google Scholar]
- 52.Batchoun R, Al-Sha’er AI, Khabour OF. Molecular Characterization of Bacillus cereus Toxigenic Strains Isolated from Different Food Matrices in Jordan. Foodborne Pathog. Dis. 2011;8:1153–1158. doi: 10.1089/fpd.2011.0853. [DOI] [PubMed] [Google Scholar]
- 53.Chaves JQ, Pires ES, Vivoni AM. Genetic diversity, antimicrobial resistance and toxigenic profiles of Bacillus cereus isolated from food in Brazil over three decades. Int. J. Food Microbiol. 2011;147:12–16. doi: 10.1016/j.ijfoodmicro.2011.02.029. [DOI] [PubMed] [Google Scholar]
- 54.Kim MJ, et al. Various enterotoxin and other virulence factor genes widespread among Bacillus cereus and Bacillus thuringiensis strains. J. Microb. Biotechnol. 2015;25:872–879. doi: 10.4014/jmb.1502.02003. [DOI] [PubMed] [Google Scholar]
- 55.Lotte PSA, Annette F, Per EG. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev. 2008;32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- 56.Thaenthanee S, Wong ACL, Panbangred W. Phenotypic and genotypic comparisons reveal a broad distribution and heterogeneity of hemolysin BL genes among Bacillus cereus isolates. Int. J. Food Microbiol. 2005;105:203–212. doi: 10.1016/j.ijfoodmicro.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Ngamwongsatit P, et al. Broad distribution of enterotoxins genes (hblCDA, nheABC, cytK and entFm) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int. J. Food Microbiol. 2008;21:352–356. doi: 10.1016/j.ijfoodmicro.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 58.Vyletelova, V. & Banyko, J. Detection of HBL and NHE enterotoxin genes in Bacillus cereus using multiplex PCR. Paper presented at 12th International Symposium on Microbial Ecology (ISME12). Cairns, Australia (2008).
- 59.Coolbaugh JC, Williams RP. Production and characterization of two haemolysis of Bacillus cereus. Can. J. Microbiol. 1978;3:255–273. doi: 10.1139/m78-209. [DOI] [PubMed] [Google Scholar]
- 60.Miles G, Bayley H, Cheley S. Properties of Bacillus cereus haemolysin II: a heptameric transmembrane pore. Protein Sci. 2002;11:1813–1824. doi: 10.1110/ps.0204002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baida GE, Kuzmin NP. Mechanism of action of hemolysin III from Bacillus cereus. Biochim. Biophys. Acta. 1996;1284:122–124. doi: 10.1016/S0005-2736(96)00168-X. [DOI] [PubMed] [Google Scholar]
- 62.Lund T, De Buyser ML, Granum PE. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 2000;38:254–61. doi: 10.1046/j.1365-2958.2000.02147.x. [DOI] [PubMed] [Google Scholar]
- 63.Wijnands LM, Dufrenne JB, Rombouts FM, Veld PH, Leusden FM. Prevalence of potentially pathogenic Bacillus cereus in food commodities in The Netherlands. J. Food Prot. 2006;69:2587–2594. doi: 10.4315/0362-028X-69.11.2587. [DOI] [PubMed] [Google Scholar]
- 64.Rather MA, Aulakh RS, Gill JPS, Ghatak S. Enterotoxin gene profile and antibiogram of Bacillus cereus strains isolated from raw meats and meat products. J. Food Saf. 2012;32:22–28. doi: 10.1111/j.1745-4565.2011.00340.x. [DOI] [Google Scholar]
- 65.Majed R, Faille C, Kallassy M, Gohar M. Bacillus cereus Biofilms—Same, Only Different. Front. Microbiol. 2016;7:1054. doi: 10.3389/fmicb.2016.01054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cihan AC, Karaca B, Ozel BP, Kilic T. Determination of the biofilm production capacities and characteristics of members belonging to Bacillaceae family. World J. Microbiol. Biotechnol. 2017;33:118. doi: 10.1007/s11274-017-2271-0. [DOI] [PubMed] [Google Scholar]
- 67.Giaouris E, et al. Attachment and biofilm formation by foodborne bacteria in meat processing environments: Causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci. 2014;97:298–309. doi: 10.1016/j.meatsci.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 68.Winkelströter LK, Teixeira FB, Silva EP, Alves VF, de Martinis EC. Unraveling microbial biofilms of importance for food microbiology. Microb. Ecol. 2014;68:35–46. doi: 10.1007/s00248-013-0347-4. [DOI] [PubMed] [Google Scholar]
- 69.Kuroki R, et al. Nosocomial bacteremia caused by biofilm-forming Bacillus cereus and Bacillus thuringiensis. Intern. Med. 2009;48:791–796. doi: 10.2169/internalmedicine.48.1885. [DOI] [PubMed] [Google Scholar]
- 70.Schmidt, R. H. & Erickson, D. J. Sanitary Design and Construction of Food Equipment. FSHN0409, one of a series of the Food Science and Human Nutrition Department, UF/IFAS Extension. Original publication date May 2005. Reviewed February 2017. Visit the EDIS website at, http://edis.ifas.ufl.edu.
- 71.Beceiro A, Tomás M, Bou G. Antimicrobial Resistance and Virulence: a Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013;26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schroeder M, Brooks BD, Brooks AE. The Complex Relationship between Virulence and Antibiotic Resistance. Genes. 2017;8:39. doi: 10.3390/genes8010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carruth L, Roess AA, Terefe Y, Hosh FM, Salman MD. Antimicrobial resistance and food safety in Africa. Lancet Infect. Dis. 2017;17:575–576. doi: 10.1016/S1473-3099(17)30273-6. [DOI] [PubMed] [Google Scholar]
- 74.Tahmasebi H, Talebi R, Zarif BR. Isolated of Bacillus Cereus in Chicken Meat and Investigation β-Lactamase Antibiotic-Resistant in Bacillus Cereus from Chicken Meat. Adv. Life Sci. 2014;4:200–206. [Google Scholar]
- 75.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Furtula V, et al. Antimicrobial Resistance in Enterococcus spp. Isolated from Environmental Samples in an Area of Intensive Poultry Production. Int. J. Environ. Res. Public Health. 2013;10:1020–1036. doi: 10.3390/ijerph10031020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Turnbull, P. C. B. MICs of Selected Antibiotics for Bacillus anthracis, Bacillus cereus, Bacillus thuringiensis, and Bacillus mycoides from a Range of Clinical and Environmental Sources as Determined by the Etest. J. Clin. Microbiol. 42, 3626–3634, 10.1128/10.1128/JCM.42.8.3626-3634.2004 (2004). [DOI] [PMC free article] [PubMed]
- 78.Osman KM, Amer AM, Badr JM, Saad AS. Prevalence and antimicrobial resistance profile of Staphylococcus species in chicken and beef raw meat in Egypt. Foodborne Pathog. Dis. 2015;12:406–413. doi: 10.1089/fpd.2014.1882. [DOI] [PubMed] [Google Scholar]
- 79.Hennequin C, Robin F. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 80.Gabrani, R., Sharma, G., Dang, S. & Gupta S. Interplay Among Bacterial Resistance, Biofilm Formation and Oxidative Stress for Nosocomial Infections. In: Rani V. & Yadav U. (eds) Free Radicals in Human Health and Disease. Springer, New Delhi (2015).
- 81.He H-J, et al. Erythromycin resistance features and biofilm formation affected by subinhibitory erythromycin in clinical isolates of Staphylococcus epidermidis. J. Microbiol. Immunol. Infect. 2016;49:33–40. doi: 10.1016/j.jmii.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 82.Vidana R, Rashid MU, Özenci V, Weintraub A, Lund B. The origin of endodontic Enterococcus faecalis explored by comparison of virulence factor patterns and antibiotic resistance to that of isolates from stool samples, blood cultures and food. Int. Endod. J. 2016;49:343–351. doi: 10.1111/iej.12464. [DOI] [PubMed] [Google Scholar]
- 83.Rasko DA, Altherr MR, Han CS, Ravel J. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 2005;29:303–329. doi: 10.1016/j.femsre.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 84.Earl AM, Losick R, Kolter R. Bacillus subtilis genome diversity. J. Bacteriol. 2007;189:1163–1170. doi: 10.1128/JB.01343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guerra B, Soto S, Helmuth R, Mendoza MC. Characterization of a self-transferable plasmid from Salmonella enterica serotype Typhimurium clinical isolates carrying two integron-borne gene cassettes together with virulence and drug resistance genes. Antimicrob. Agents Chemother. 2002;46:2977–2981. doi: 10.1128/AAC.46.9.2977-2981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Venturini C, Beatson SA, Djordjevic SP, Walker MJ. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 2010;24:1160–1166. doi: 10.1096/fj.09-144972. [DOI] [PubMed] [Google Scholar]
- 87.Okinaka RT, et al. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 1999;181:6509–6515. doi: 10.1128/jb.181.20.6509-6515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pannucci J, et al. DNA sequence conservation between the Bacillus anthracis pXO2 plasmid and genomic sequence from closely related bacteria. BMC Genomics. 2002;3:34. doi: 10.1186/1471-2164-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van der Auwera GA, Timmery S, Mahillon J. Self-transfer and mobilisation capabilities of the pXO2-like plasmid pBT9727 from Bacillus thuringiensis subsp. konkukian 97–27. Plasmid. 2008;59:134–138. doi: 10.1016/j.plasmid.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 90.Adams V, et al. Virulence Plasmids of Spore-Forming Bacteria. Microbiol. Spectr. 2014;2:1–24. doi: 10.1128/microbiolspec.PLAS-0024-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jansen, W., Grabowski, N., Gerulat, B. & Klein, G. Food Safety Hazards and Microbiological Zoonoses in European Meat Imports Detected in Border Inspection in the Period 2008–2013. Zoonoses and Public Health63, 53–61, 10.1111/zph.12204. Epub 2015 May 29 (2016). [DOI] [PubMed]
- 92.Al-Allaf MAA. Isolation of Bacillus spp. from some sources and study of its proteolytic activity. Tikrit J. Pure Sci. 2011;16:59–63. [Google Scholar]
- 93.FDA. Bacteriological Analytical Manual Chapter 14 Bacillus cereus U.S. Food and Drug Administration 10903 New Hampshire Avenue Silver Spring, MD 20993 1-888-INFO-FDA (1-888-463-6332), Page Last Updated: 08/06/2015.
- 94.Park YB, et al. Prevalence, genetic diversity, and antibiotic susceptibility of Bacillus cereus strains isolated from rice and cereals collected in Korea. J. Food Prot. 2009;72:612–617. doi: 10.4315/0362-028X-72.3.612. [DOI] [PubMed] [Google Scholar]
- 95.CLSI. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twentieth informational supplement. Document M100S20, CLSI. Wayne, PA (2010).
- 96.Gómez-Zorrilla S, et al. Antibiotic Pressure Is a Major Risk Factor for Rectal Colonization by Multidrug-Resistant Pseudomonas aeruginosa in Critically Ill Patients. Antimicrob. Agents Chemother. 2014;58:5863–5870. doi: 10.1128/AAC.03419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Magiorakos AP, et al. Multidrug resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 98.Mateo M, et al. Strong slime production is a marker of clinical significance in Staphylococcus epidermidis isolated from intravascular catheters. Eur. J. Clin. Microbiol. Infect. Dis. 2008;27:311–314. doi: 10.1007/s10096-007-0433-y. [DOI] [PubMed] [Google Scholar]
- 99.Stepanovic S, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 100.Ghelardi E, et al. Contribution of surfactin and SwrA to flagellin expression, swimming, and surface motility in Bacillus subtilis. Appl. Environ. Microbiol. 2012;78:6540–6544. doi: 10.1128/AEM.01341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghelardi E, et al. Swarming behavior of and hemolysin BL secretion by Bacillus cereus. Appl. Environ. Microbiol. 2007;73:4089–4093. doi: 10.1128/AEM.02345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wiwat C, Thiramanas R. Detection of Hemolysin BL Gene of Bacillus cereus Isolates. Mahidol Univ. J. Pharm. Sci. 2014;41:22–30. [Google Scholar]
- 103.Buchanan RL, Schultz FJ. Comparison of the Tecra VIA kit, Oxoid BCET-RPLA kit and CHO cell culture assay for the detection of Bacillus cereus diarrhoeal enterotoxin. Lett. Appl. Microbiol. 1994;19:353–356. doi: 10.1111/j.1472-765X.1994.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 104.Sergeev N, et al. Microarray analysis of Bacillus cereus group virulence factors. J. Microbiol. Methods. 2006;65:488–502. doi: 10.1016/j.mimet.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 105.Forghani F, Kim JB, Oh DH. Enterotoxigenic Profiling of Emetic Toxin- and Enterotoxin-Producing Bacillus cereus, Isolated from Food, Environmental, and Clinical Samples by Multiplex PCR. J. Food Sci. 2014;79:M2288–M2293. doi: 10.1111/1750-3841.12666. [DOI] [PubMed] [Google Scholar]
- 106.R Core Team. R: A Language and Environment for Statistical Computing. R Found. Stat (2017).
- 107.Oksanen J, Blanchet F, Kindt R, Legendre P, O’Hara R. Vegan: community ecology package. R Packag. 2017;2:4–3. [Google Scholar]
- 108.Hervé, M. RVAideMemoire: Diverse Basic Statistical and Graphical Functions. R Packag. 0.9–57 Available at, https://cran.r-project.org/web/packages/RVAideMemoire/ (2016).
- 109.Lê S, Josse J, Husson F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008;25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 110.Kassambara, A. & Mundt, F. Factoextra: extract and visualize the results of multivariate data analyses. R Packag. version 1 (2016).
- 111.Wei, T. & Simko, V. The corrplot package. CRAN Repos. Available at, http://www.sthda.com/french/wiki/matrice-de-correlation-la-fonction-r-qui-fait-tout (2016).
- 112.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 113.Zhao X, Sandelin A. GMD: Measuring the distance between histograms with applications on high-throughput sequencing reads. Bioinformatics. 2012;28:1164–1165. doi: 10.1093/bioinformatics/bts087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.